Abstract
Turnover and recycling of the cell wall murein represent a major metabolic pathway of Escherichia coli. It is known that E. coli efficiently reuses, i.e., recycles, its murein tripeptide, l-alanyl-γ-d-glutamyl-meso-diaminopimelate, to form new murein. However, the question of whether the cells also recycle the amino sugar moieties of cell wall murein has remained unanswered. It is demonstrated here that E. coli recycles the N-acetylglucosamine present in cell wall murein degradation products for de novo murein and lipopolysaccharide synthesis. Furthermore, E. coli also recycles the anhydro-N-acetylmuramic acid moiety by first converting it into N-acetylglucosamine. Based on the results obtained by studying mutants unable to recycle amino sugars, the pathway for recycling is revealed.
Escherichia coli breaks down about 40% of its cell wall murein (peptidoglycan) each generation (8, 15). This phenomenon has been termed turnover. However, the true turnover rate is masked and appears to be only 3 to 10% per generation because the murein tripeptide is efficiently recycled (10, 15, 24). The recycling pathway for the reuse of the murein tripeptide begins when anhydromuropeptides are formed upon breakdown of the murein sacculus by lytic transglycosylases and endopeptidases during active growth (30). The principal anhydromuropeptide is N-acetylglucosaminyl-β-1,4-anhydro-N- acetylmuramyl-l-alanyl-γ-d-glutamyl-meso-diaminopimelate-d-alanine. The anhydromuropeptides are imported into the cytoplasm via AmpG permease (15) and then are cleaved by LdcA, an l, d-carboxypeptidase (32), by AmpD, an anhydromuramyl-l-alanine amidase (14, 16), and by NagZ, a β-N-acetylglucosaminidase (4, 31, 33, 34), to release N-acetylglucosamine (GlcNAc), anhydro-N-acetylmuramic acid (anhMurNAc), d-alanine, and the murein tripeptide. The murein tripeptide is promptly ligated to UDP-N-acetylmuramic acid (UDP-MurNAc) by murein peptide ligase (Mpl) to return the tripeptide to the biosynthetic pathway for murein synthesis (23). In view of the fact that intact murein tripeptide is recycled, the question naturally arises as to whether GlcNAc and anhMurNAc are also recycled. The results presented here indicate that both amino sugar moieties are reused. AnhMurNac is converted to GlcNAc by an unknown enzymatic reaction(s) before reentering the biosynthetic pathway.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used are listed in Table 1. All are derivatives of E. coli K-12. Bacteria were grown at 37°C with aeration by shaking.
TABLE 1.
E. coli strains used in this work
| Strain | Genotype | Reference or source |
|---|---|---|
| TP71 | F−lysA opp araD139 rpsL150 relA1 thi-1 deoC1 ptsF25 flbB5301 rbsR Δ(argF-lac) | 15 |
| IBPC5321 | F−thi-1 argG6 argE3 his-4 xyl-5 mtl-1 tsx-29 rpsL ΔlacX74 | 27 |
| IBPC546 | IBPC5321 nagB::Kan | 27 |
| TP71B | TP71 nagB::Kan | This work |
| IBPC571 | IBPC5321 nagB2 zbf-507::Tn10 | 28 |
| TP72 | TP71 ampG::Kan | 15 |
| TP72B | TP72 nagB2 zbf-507::Tn10 | This work |
| TP77 | TP71 nagZ::Cm | 4 |
| TP77B | TP77 nagB::Kan | This work |
| IBPC590 | IBPC5321 ΔnagEBACD::Tet | 28 |
| TP80 | TP71 ΔnagEBACD::Tet | This work |
Construction of E. coli strains lacking GlcN deaminase (nagB) and GlcNAc deacetylase (nagA).
In order to study the turnover of amino sugars from murein, strains were constructed that lacked nagB, the structural gene for glucosamine (GlcN) deaminase, required for the deamination of GlcN-6-phosphate (GlcN-6-P). In the absence of the deaminase, GlcN cannot be degraded; hence, cells labeled with 3H-GlcN will have label only in products containing GlcN or muramic acid. E. coli strains TP71 and TP77 (nagZ::Cm) were transduced to kanamycin resistance with P1 grown on E. coli IBPC546 (nagB::Kan). Kanr strains TP71B and TP77B were isolated and tested for their ability to grow on M9 minimal medium agar (29) with GlcNAc as a carbon source. NagB mutants are unable to grow on minimal medium with GlcNAc as a carbon source. TP71B and TP77B failed to grow on GlcNAc minimal medium, confirming that in both strains nagB::Kan had replaced nagB. TP72 (ampG::Kan) (15) was transduced to tetracycline resistance with P1 grown on E. coli IBPC571, a strain carrying zbf507::Tn10 and mutation nagB2, which is 25% cotransducible with zbf507::Tn10. TP72B is a Tetr transductant that is unable to grow on M9 agar with GlcNAc as a carbon source and hence lacks nagB. TP80 is a derivative of TP71 transduced to Tetr with P1 grown on IPBC590 (ΔnagEBACD::Tet). It is unable to grow on GlcNAc minimal medium. The relevant deletions in TP80 are nagA, the structural gene for GlcNAc-6-P deacetylase, and nagB, the structural gene for GlcN deaminase. The other genes in the deleted operon are nagC, which encodes a repressor for the nag regulon, nagE, which encodes a specific transporter for GlcNAc, and nagD, whose function is unknown.
Design of chase experiments.
Chase experiments with L broth to measure the rate of loss of label from sacculi were done as follows. Eight milliliters of L broth (10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter) supplemented with 2 mM MgCl2, 0.25 × M9 salts, and 5 or 8 μCi of d-6-3H-GlcN (NEN Life Sciences Products, Boston, Mass.) was inoculated with 40 μl of an overnight culture of the test strain and incubated at 37°C with aeration. When the optical density at 600 nm reached approximately 0.25, the cells were collected on a 0.45-μm-pore-size membrane filter, washed extensively, and resuspended in 34 ml of nonradioactive medium to start the chase. Five-milliliter aliquots were collected after zero, one, two, and three generations of growth and added directly to 3 ml of 10% sodium dodecyl sulfate (SDS). After incubation at 95°C for 30 min, the samples of sacculi were filtered onto 0.22-μm-pore-size 47-mm filters and washed six times with 3-ml portions of water. The filters containing the sacculi were dried and immersed in scintillation fluid, and counts were determined.
Chase experiments with minimal medium were designed so that the radioactive components present in the spent medium, the intracellular pool, the cell wall sacculi, and the SDS-soluble material could be analyzed. Eight milliliters of M9 minimal medium containing 0.6% glycerol, 0.1% Casamino Acids, 2 mM MgCl2, 2 μg of thiamine per ml, 20 μg of lysine per ml, and 8 μCi of 3H-GlcN was inoculated with 80 μl of an overnight culture and incubated at 37°C with aeration. When the optical density at 600 nm reached 0.25 (about four generations of growth), the cells were filtered onto 0.45-μm-pore-size 24-mm membrane filters and washed five times with 2 ml of 0.85% saline. The filters were suspended in 34 ml of M9 minimal medium lacking 3H-GlcN to begin the chase. Eight milliliters of culture was harvested by centrifugation after zero, one, two, and three generations of growth. The cells were suspended in 2 ml of cold water, heated at 95°C for 5 min, and centrifuged to obtain the hot water extract. The cell residue was suspended in 2.5 ml of 4% SDS and heated at 95°C for 30 min to free the sacculi, which were recovered by centrifugation for 30 min at 30,000 rpm in a Beckman TL-100 centrifuge. The spent medium, the hot water extract containing the intracellular material, the sacculi, and the SDS-soluble material, in which 3H-GlcN is predominantly present in lipopolysaccharides (LPS), were analyzed.
HPLC analysis.
High-pressure liquid chromatography (HPLC) was performed as previously described (4). This procedure involved a C18 reverse-phase column and a gradient to 10% acetonitrile over 50 min. Fractions were collected every 0.5 min at a flow rate of 0.5 ml/min. Under these conditions, a mixture containing GlcN, GlcNAc, GlcN-P, GlcNAc-P, and UDP-GlcNAc is recovered in fractions 15 to 26; the anhMurNAc peak is at fraction 31, the GlcNAc-anhMurNAc peak is at fraction 41, and the UDP-MurNAc pentapeptide peak is at fraction 83.
Analysis of the mixture of GlcN-containing derivatives by TLC.
Fractions 15 to 26 from the HPLC analyses were pooled, lyophilized, and dissolved in sufficient water to yield a concentration of ∼5,000 cpm per μl. Five microliters was digested with 5 U of calf intestinal alkaline phosphatase at 37°C for 30 min. The digest and a 5-μl untreated sample were fractionated by thin-layer chromatography (TLC) on plastic sheets (20 by 20 cm) covered with 0.1—mm-thick cellulose (EM Science, Cherry Hill, N.J.). The chromatograph was run for approximately 4.5 h in 80% ethanol–20% 2 M NH4OH with the solvent front ascending 14 to 15 cm. Under these conditions, a GlcN standard has an Rf of 0.50 and a GlcNAc standard has an Rf of 0.62, while the phosphorylated derivatives remained within 2 cm of the origin. Thus, the radioactivity from the untreated sample at an Rf of 0.5 was considered to be GlcN, that at an Rf of 0.62 was considered to be GlcNAc, and the increase in radioactivity at these positions following phosphatase treatment was taken as a measure of the respective phosphorylated derivatives.
RESULTS
Turnover of amino sugars from E. coli strains labeled with 3H-GlcN in L broth.
Table 2 shows the rate of loss of radioactive label from the sacculi of E. coli cells growing in L broth after being labeled with either 3H-GlcN or 3H-Dap (diaminopimelic acid). The rate of loss from the ampG strains, which are unable to import anhydromuropeptides (15), is 30 to 40% per generation, regardless of whether the cells are labeled with 3H-GlcN or 3H-Dap. (The rate of loss is smaller initially because the cytoplasmic pool of radioactive intermediates is being converted to murein.) AmpG+ TP71B (nagB) cells whose murein is labeled with GlcN also lose amino sugars at a rate (∼30% per generation) approaching that which occurs when recycling is prevented by the absence of AmpG permease (Table 2). In contrast, 3H-Dap-labeled wild-type cells (TP71) lose less than 10% of their murein per generation. These results seem to clearly indicate that the amino sugars are not recycled. However, as reported here, if one analyzes the biosynthetic intermediates and their conversion to LPS as well as murein by cells growing in minimal medium, it becomes clear that the amino sugars are, in fact, recycled. The reason for the misleadingly high turnover of murein amino sugars by cells growing in L broth is presented in the Discussion.
TABLE 2.
Turnover of 3H-GlcN-labeled murein and 3H-Dap-labeled murein
Recycling of amino sugars.
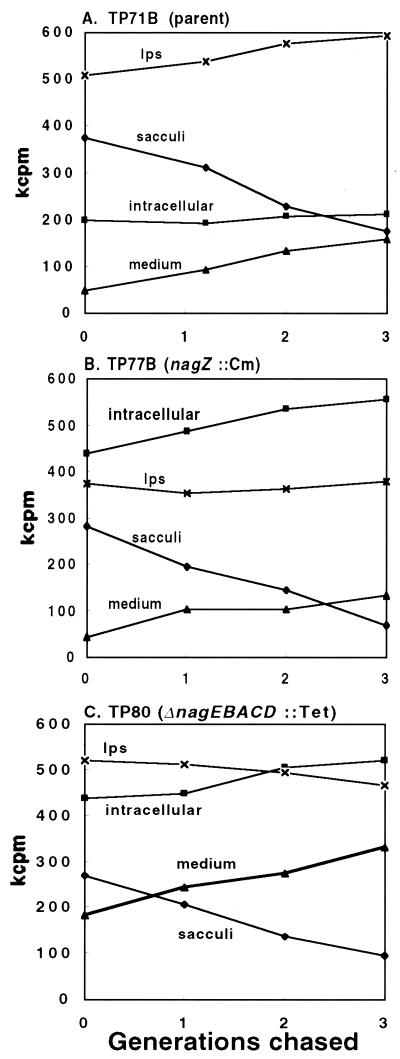
Recently, it was found that E. coli strain TP77, which lacks nagZ, the β-N-acetylglucosaminidase gene, contains large amounts of the disaccharide GlcNAc-anhMurNAc in its cytoplasm (4). Since anhMurNAc can arise from the cell wall murein only through the action of lytic transglycosylases (30), all of this disaccharide must have been derived from the sacculus. As noted earlier, the anhydrodisaccharide-peptides released from the sacculus by lytic transglycosylases are transported into the cell via AmpG permease. In the cytoplasm, AmpD amidase cleaves the bond between the disaccharide and the l-alanine of the peptides. If NagZ is absent, the amino sugars are trapped as GlcNAc-anhMurNAc; thus, obviously, a strain with such characteristics would be unable to recycle amino sugars. The nagZ null strain and the parent strain were therefore compared for their ability to recycle amino sugars in a chase experiment with cells labeled with 3H-GlcN. In these experiments, M9 minimal medium was used with glycerol as the carbon source. Note that in all experiments reported here, chase simply means continued growth in the absence of added radioactive GlcN. Figure 1 compares the amount of label in the sacculi, the LPS fraction, the intracellular pool, and the spent medium from parent TP71B (nagB) cells during three generations of chase with that in the same fractions from the nagZ::Cm mutant and from the ΔnagEBACD::Tet mutant. These experiments demonstrated in three different ways that recycling occurred in the parent strain.
FIG. 1.
Chase of parent and mutant strains prelabeled with 3H-GlcN. The results for each strain are the average of two experiments.
Three lines of evidence that amino sugars are recycled. (i) Amount of radiolabel accumulating in mutant cells.
A striking result in these experiments was the large amount of radiolabel that accumulated intracellularly in both mutant strains compared to the parent strain (Fig. 1). There was about 2.5-fold more radiolabeled amino sugars in the cytoplasm of the mutants after three generations of chase than in that of the parent. Considered another way, during the labeling and chase periods, about 90% of the labeled murein was degraded by the mutants without the amino sugars being reused. This finding indicates that the ΔnagEBACD::Tet strain as well as the nagZ::Cm strain cannot recycle amino sugars. Conversely, the parent strain must have recycled some of its amino sugars, since the products of turnover did not accumulate.
(ii) The parent synthesizes radioactive LPS during the chase.
As shown in Fig. 1, the parent strain gained radioactive LPS during the chase, whereas the nagZ::Cm mutant and the ΔnagEBACD::Tet mutant did not. These results clearly demonstrate that amino sugars can be recycled by the parent and confirm that the ΔnagEBACD::Tet strain and the nagZ::Cm strain, which do not incorporate more label in LPS during the chase, do not recycle amino sugars. Incidentally, in addition to the LPS component, the SDS-soluble fraction may contain a small amount of lipoprotein-bound GlcNAc-anhMurNAc-tripeptide, equivalent to 10% of the murein, as judged by the 3H-Dap content of the SDS-soluble fraction from 3H-Dap-labeled cells.
(iii) The apparent turnover of amino sugars from the sacculi of the parent is low compared to that of the mutants, which cannot recycle amino sugars.
Careful inspection of the data in Fig. 1 reveals that the nagZ::Cm mutant lost 75% of the label from its sacculi after three generations and that nearly all of this label, which was trapped as dissacharide, remained in the cytoplasm, although some escaped to the medium. In fact, the material that accumulated in the spent medium of the nagZ::Cm mutant was essentially all GlcNAc-anhMurNAc (data not shown). The ΔnagEBACD::Tet mutant lost 65% of the label from its murein during three generations of chase, but the parent strain lost only 50%. These results suggest that the parent must have used some of the amino sugars released by turnover of the sacculi to form new murein.
Absence of anhMurNAc in the intracellular pool.
The amounts of the various amino sugar-containing compounds in the intracellular pools from the parent and mutant strains are summarized in Table 3. There are several notable results. Almost all of the amino sugars released from the murein of the nagZ::Cm strain were trapped in the form of GlcNAc-anhMurNAc, and the intermediates present at the start of the chase were used, leaving the pool of intermediates largely depleted after two generations of chase. However, the other two strains, which possess NagZ β-N-acetylglucosaminidase, cleaved the disaccharide to release anhMurNAc. Nevertheless, little if any free anhMurNAc was present in any of the cells. The conclusion to be drawn from these results is that E. coli efficiently converts anhMurNAc to GlcNAc (or a compound that behaves like GlcNAc and/or GlcNAc-P in our tests). Because TP80 lacks NagA deacetylase, it totally lacked GlcN and GlcN-P in its cytoplasm (Table 3). Instead, TP80 accumulated large amounts of GlcNAc and GlcNAc-P. These results again clearly indicate that GlcNAc cannot be recycled in the absence of NagA.
TABLE 3.
Amino sugar-containing compounds present in the cytoplasm of nagB cells before and after a chase
| Compound | Amt (Kcpm) of compound in the following strain after the indicated no. of generations:
|
|||||
|---|---|---|---|---|---|---|
| TP71B (parent)
|
TP77B (nagZ::Cm)
|
TP80 (ΔnagEBACD::Tet)
|
||||
| 0 | 3 | 0 | 2 | 0 | 3 | |
| GlcN | 8 | 17 | 1 | <1 | <1 | <1 |
| GlcN-P | 23 | 10 | 7 | <1 | <1 | <1 |
| GlcNAc | 73 | 55 | 27 | 4 | 176 | 275 |
| GlcNAc-P | 126 | 149 | 40 | 9 | 95 | 207 |
| anhMurNAc | <5 | <4 | <5 | <10 | <1 | <1 |
| GlcNAc-anhMurNAc | 3 | 2 | 473a | 914a | <2 | <2 |
| UDP-MurNAc pentapeptide | 16 | 12 | 8 | 2 | <1 | <1 |
Includes GlcNAc-anhMurNAc from spent medium. The values are not comparable to those for the other genotypes, since five times more 3H-GlcN was used for labeling.
DISCUSSION
Fate of murein amino sugars that are turned over.
The data in Fig. 1A show that the parent strain lost amino sugars from its murein at an average rate of 19% per generation. This rate is considerably lower than the 37% rate of loss per generation observed when recycling of the amino sugars is completely blocked (Fig. 1B: nagZ mutant) and indicates that recycling of amino sugars occurs. However, 19% is a relatively high rate of loss and appears inconsistent with the apparent efficient reutilization indicated by maintenance of the intracellular pool of intermediates. The explanation lies in the fact that about 60% of the amino sugars are used to synthesize LPS and hence are not available for de novo murein synthesis. The higher rate of loss of amino sugars (19% per generation) than of Dap (<10%) is also due to the fact that many of the available amino sugars are channeled into LPS, whereas all of Dap can only be recycled into murein.
The higher rate of loss of amino sugars from the sacculi of TP71B cells growing in L broth compared to those growing in M9 minimal medium (>30% versus 19%) can be explained by the presence of GlcN in L broth (derived from glycoproteins, hyaluronic acid, and mucopolysaccharides present in the tryptone used to make L broth). The uptake of GlcN rapidly dilutes the specific activity of the GlcN-6-P pool and thus greatly reduces or prevents the recycling of 3H-GlcNAc.
Fate of anhMurNAc.
It is clear from the results in Table 3 that little or no free anhMurNAc is present in the cells at any time. The ΔnagEBACD::Tet strain, which cannot recycle amino sugars because it cannot convert GlcNAc-6-P to GlcN-6-P, might be expected to accumulate equal amounts of anhMurNAc and GlcNAc. However, the ΔnagEBACD::Tet strain has no detectable anhMurNAc. Instead, the pool contains large amounts of GlcNAc and GlcNAc-P. Analysis of the spent medium indicated that it also totally lacks anhMurNAc. The conclusion to be drawn from these data is that anhMurNAc is rapidly converted to GlcNAc. There is one caveat. The identification of GlcNAc and GlcNAc-P is based solely on their behavior in HPLC and TLC. Supporting evidence that anhMurNAc was converted to GlcNAc lies in the fact that significantly more label was incorporated into LPS and murein than was released from murein in the form of GlcNAc.
In a preliminary test to examine whether E. coli could enzymatically convert anhMurNAc to GlcNAc, 3H-anhMurNAc was incubated with the soluble fraction from E. coli cells. Upon incubation for 2 h at 37°C, 7% of anhMurNAc was converted to material that eluted with GlcNAc in HPLC. Further work is in progress to identify the enzyme(s) involved in converting anhMurNAc to GlcNAc. Conversion of anhMurNAc to GlcNAc requires cleavage of an ether bond. The only β-etherases of microbial origin described in the literature are ones produced by Pseudomonas paucimobilis (18, 19). These attack a model compound from lignin, and for cleavage to occur, a carbonyl group must be adjacent to the target ether bond. Tantalizingly, that is exactly the situation in anhMurNAc. One might imagine that gram-negative bacteria, since they appear to have the genes for the recycling pathway (4), are able to degrade anhMurNAc and that P. paucimobilis has evolved modified forms of the enzyme that can metabolize lignin compounds.
Role of NagA.
All three lines of evidence demonstrate that TP80 (ΔnagEBACD::Tet) cannot recycle the amino sugars derived from the sacculi. The relevant gene in this strain is nagA, the structural gene for GlcNAc-6-P deacetylase. Without this enzyme the cell cannot reutilize GlcNAc, because E. coli can isomerize only GlcN-6-P. As demonstrated in some careful studies by Mengin-Lecreulx and van Heijenoort, E. coli is unable to convert GlcNAc-6-P to GlcNAc-1-P (22). Instead, E. coli uses GlmM, a phosphoglucosamine mutase, to convert GlcN-6-P to GlcN-1-P (22). Thereafter, in the normal pathway for UDP-GlcNAc synthesis, GlmU acetylates GlcN-1-P and also catalyzes the reaction of GlcNAc-1-P with UTP to form the UDP-GlcNAc intermediate necessary for the synthesis of murein and LPS (20, 21).
Pathway for recycling of murein amino sugars.
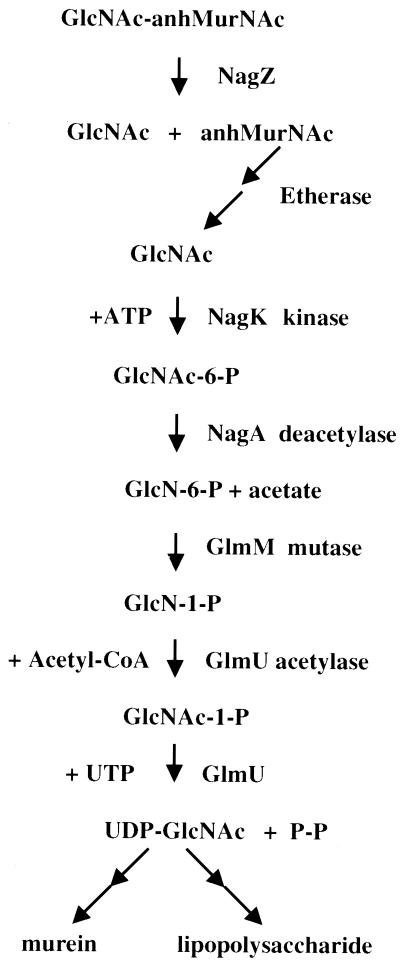
Based on the results reported here, the pathway for recycling amino sugars from cell wall murein is as shown in Fig. 2. After release in the cytoplasm, anhMurNAc is converted to GlcNAc by an unknown series of reactions. GlcNAc is phosphorylated by GlcNAc kinase to form GlcNAc-6-P (1). Incidentally, the kinase was purified and its properties were studied over 35 years ago (1); the gene, referred to here as nagK, has not yet been identified. An alternative to direct phosphorylation of GlcNAc by NagK would be to export GlcNAc and immediately reimport GlcNAc via NagE and the phosphotransferase system to produce GlcNAc-6-P. The possible contribution of this alternative pathway to the formation of GlcNAc-6-P has not been evaluated. After phosphorylation, GlcNAc-6-P is deacetylated by NagA to form GlcN-6-P. GlcN-6-P is a normal intermediate in the pathway for the synthesis of UDP-GlcNAc. GlmM isomerizes it to GlcN-1-P. GlmU acetylates GlcN-1-P and, in the presence of UTP, catalyzes the formation of UDP-GlcNAc. UDP-GlcNAc is then used for the synthesis of UDP-MurNAc, murein, and LPS. Note that E. coli and, presumably, other gram-negative bacteria appear to have evolved at least five genes essential for the recycling of GlcNAc and anhMurNAc (nagZ, nagA, nagK, a putative β-etherase gene, and a putative anhydrase gene). One must conclude that such a pathway has a useful purpose in preserving the species.
FIG. 2.
Pathway for recycling of GlcNAc and anhMurNAc. Etherase represents unidentified enzymes that hydrolyze the anhydro bond and cleave the ether linkage in anhMurNAc to yield GlcNAc. NagK, N-Acetyl-d-glucosamine kinase (1). CoA, coenzyme A.
The unanswered question: Why does E. coli turn over its cell wall?
Judging from the rate of loss of amino sugars and Dap from mutants blocked in the recycling pathway, 30 to 45% of the murein is turned over each generation. If one takes into consideration that the poles of the cell are relatively stable (3, 7, 17), then well over half of the murein of the side wall is turned over each generation.
There is no obvious explanation for this seemingly unnecessary degradation process. Some years ago, in an attempt to account for the turnover, a novel “3-for-1” model for the elongation of the sacculus was suggested (11). According to this hypothesis, three new strands would be assembled and cross-linked to each other underneath the existing murein strands. The outer two strands would simultaneously be cross-linked to existing strands directly above themselves in the murein sacculus. However, the middle new strand would not be cross-linked to the old strand above it. Instead, the old strand would be digested by an endopeptidase and lytic transglycosylase (thereby accounting for the high turnover), and the three new strands would be drawn into the elongating sacculus. The problem with this proposal is that it is well established that new strands attach only to preexisting strands during elongation (5, 6, 25). Hence, the three new stands cannot be cross-linked to each other. To take this fact into consideration, the “3-for-1” model was modified by assuming that the middle strand, termed the primer strand, already existed as a precursor (12, 13). However, this situation would require an amount of unattached primer strands equivalent to half of the murein sacculus side walls, and these strands somehow would have to avoid degradation in the periplasm while waiting to be utilized. In fact, there is strong evidence that no pool of free-floating strands exists (2, 9). Thus, the modified 3-for-1 model is highly unlikely.
Another proposed rationale for recycling is that fluctuations in the levels of recycling intermediates might provide a sensing system for monitoring the condition of the murein sacculus (15, 26). This would allow the cell to respond should the sensor report accelerated breakdown of the sacculus. However, it is not obvious why a sensor should require a high rate of turnover. Thus, there is still no clear understanding of why turnover occurs.
Likewise, the reason for recycling is unknown. However, the fact that the cell has devoted four or more genes exclusively to the function of amino sugar recycling indicates that recycling has survival value.
ACKNOWLEDGMENTS
I am very grateful to Jacqueline Plumbridge for the nag mutants, which greatly facilitated this work, and also for helpful advice. I thank Debabrata RayChaudhuri for helpful discussions and critical reading of the manuscript.
This work was supported in part by Public Health Service grant GM51610 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Asensio C, Ruiz-Amil M. N-acetyl-d-glucosamine kinase. II. Escherichia coli. Methods Enzymol. 1966;IX:421–425. [Google Scholar]
- 2.Burman L G, Park J T. Changes in the composition of Escherichia coli murein as it ages during exponential growth. J Bacteriol. 1983;155:447–453. doi: 10.1128/jb.155.2.447-453.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burman L G, Raichler J, Park J T. Evidence for diffuse growth of the cylindrical portion of the Escherichia coli murein sacculus. J Bacteriol. 1983;155:983–988. doi: 10.1128/jb.155.3.983-988.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng Q, Li H, Merdek K, Park J T. Molecular characterization of the β-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J Bacteriol. 2000;182:4836–4840. doi: 10.1128/jb.182.17.4836-4840.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper S, Hseih M-L, Gunther B. Mode of peptidoglycan synthesis in Salmonella typhimurium: single-strand insertion. J Bacteriol. 1988;170:3509–3512. doi: 10.1128/jb.170.8.3509-3512.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.deJonge B L M, Wientjes F B, Jurida I, Driehuis F, Wouters J T M, Nanninga N. Peptidoglycan synthesis during the cell cycle of Escherichia coli: composition and mode of insertion. J Bacteriol. 1989;171:5783–5794. doi: 10.1128/jb.171.11.5783-5794.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Pedro M A, Quintela J C, Höltje J-V, Schwartz H. Murein segregation in Escherichia coli. J Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodell E W. Recycling of murein by Escherichia coli. J Bacteriol. 1985;163:305–310. doi: 10.1128/jb.163.1.305-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodell E W, Markiewicz Z, Schwarz U. Absence of oligomeric murein intermediates in Escherichia coli. J Bacteriol. 1983;156:130–135. doi: 10.1128/jb.156.1.130-135.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodell E W, Schwarz U. Release of cell wall peptides into the culture medium of exponentially growing Escherichia coli. J Bacteriol. 1985;162:391–397. doi: 10.1128/jb.162.1.391-397.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Höltje J-V. “Three for one”—a simple growth mechanism that guarantees a precise copy of the thin, rod-shaped murein sacculus of Escherichia coli. In: De Pedro M A, Holtje J-V, Loffelhardt W, editors. Bacterial growth and lysis. New York, N.Y: Plenum Press; 1993. pp. 419–426. [Google Scholar]
- 12.Höltje J-V. A hypothetical holoenzyme involved in the replication of the murein sacculus of Escherichia coli. Microbiology. 1996;142:1911–1918. doi: 10.1099/13500872-142-8-1911. [DOI] [PubMed] [Google Scholar]
- 13.Höltje J-V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Höltje J-V, Kopp U, Ursinus A, Wiedemann B. The negative regulator of β-lactamase induction AmpD is a N-acetyl-anhydromuramyl-l-alanine amidase. FEMS Microbiol Lett. 1994;122:159–164. doi: 10.1111/j.1574-6968.1994.tb07159.x. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs C, Huang L-J, Bartowsky E, Normark S, Park J T. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 1994;13:4684–4694. doi: 10.1002/j.1460-2075.1994.tb06792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs C, Joris B, Jamin M, Klarsov K, van Beemen J, Mengin-Lecreulx D, van Heijenoort J, Park J T, Normark S, Frere J-M. AmpD, essential for both β-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol Microbiol. 1995;15:553–559. doi: 10.1111/j.1365-2958.1995.tb02268.x. [DOI] [PubMed] [Google Scholar]
- 17.Koch A L, Woldringh C L. The metabolic inertness of the pole wall of a gram-negative rod. J Theor Biol. 1994;171:415–425. [Google Scholar]
- 18.Masai E, Katayama Y, Kawai S, Nishikawa S, Yamasaki M, Morohoshi N. Cloning and sequencing of the gene for a Pseudomonas paucimobilis enzyme that cleaves β-aryl ether. J Bacteriol. 1991;173:7950–7955. doi: 10.1128/jb.173.24.7950-7955.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masai E, Katayama Y, Kubota S, Kawai S, Yamasaki M, Morohoshi N. A bacterial enzyme degrading the model lignin compound β-etherase is a member of the glutathione-S-transferase superfamily. FEBS Lett. 1993;323:135–140. doi: 10.1016/0014-5793(93)81465-c. [DOI] [PubMed] [Google Scholar]
- 20.Mengin-Lecreulx D, van Heijenoort J. Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J Bacteriol. 1993;175:6150–6157. doi: 10.1128/jb.175.19.6150-6157.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mengin-Lecreulx D, van Heijenoort J. Copurification of glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities of Escherichia coli: characterization of the glmU product as a bifunctional enzyme catalyzing two subsequent steps in the pathway for UDP-N-acetylglucosamine synthesis. J Bacteriol. 1994;176:5788–5795. doi: 10.1128/jb.176.18.5788-5795.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mengin-Lecreulx D, van Heijenoort J. Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J Biol Chem. 1996;271:32–39. doi: 10.1074/jbc.271.1.32. [DOI] [PubMed] [Google Scholar]
- 23.Mengin-Lecreulx D, van Heijenoort J, Park J T. Identification of the mpl gene encoding UDP-N-acetylmuramate:l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J Bacteriol. 1996;178:5347–5352. doi: 10.1128/jb.178.18.5347-5352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J T. Turnover and recycling of the murein sacculus in oligopeptide permease-negative strains of Escherichia coli: indirect evidence for an alternative permease system and for a monolayered sacculus. J Bacteriol. 1993;175:7–11. doi: 10.1128/jb.175.1.7-11.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park J T. An overview of the assembly, turnover and recycling of the murein sacculus. In: De Pedro M A, Holtje J-V, Loffelhardt W, editors. Bacterial growth and lysis. New York, N.Y: Plenum Press; 1993. pp. 119–126. [Google Scholar]
- 26.Park J T. Why does Escherichia coli recycle its cell wall peptides? Mol Microbiol. 1995;17:421–426. doi: 10.1111/j.1365-2958.1995.mmi_17030421.x. [DOI] [PubMed] [Google Scholar]
- 27.Plumbridge J A. Repression and induction of the nag regulon of Escherichia coli K-12: the roles of nagC and nagA in maintenance of the uninduced state. Mol Microbiol. 1991;5:2053–2062. doi: 10.1111/j.1365-2958.1991.tb00828.x. [DOI] [PubMed] [Google Scholar]
- 28.Plumbridge J A. A dominant mutation in the gene for the Nag repressor of Escherichia coli that renders the nag regulon uninducible. J Gen Microbiol. 1992;138:1011–1017. doi: 10.1099/00221287-138-5-1011. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Shockman G D, Höltje J-V. Microbial peptidoglycan (murein) hydrolases. In: Ghuysen J M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science Publishers B.V.; 1994. pp. 131–166. [Google Scholar]
- 31.Templin M F. Characterization of a β-N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and β-lactamase induction. J Biol Chem. 2000;275:39032–39038. doi: 10.1074/jbc.M004797200. [DOI] [PubMed] [Google Scholar]
- 32.Templin M F, Ursinus A, Höltje J-V. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 1999;18:4108–4117. doi: 10.1093/emboj/18.15.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yem D W, Wu H C. Purification and properties of β-N-acetylglucosaminidase from Escherichia coli. J Bacteriol. 1976;125:324–331. doi: 10.1128/jb.125.1.324-331.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yem D W, Wu H C. Isolation of Escherichia coli K-12 mutants with altered levels of β-N-acetylglucosaminidase. J Bacteriol. 1976;125:372–373. doi: 10.1128/jb.125.1.372-373.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]