Abstract
Polynucleotide phosphorylase (PNPase) synthesis is translationally autocontrolled via an RNase III-dependent mechanism, which results in a tight correlation between protein level and messenger stability. In cells grown at 18°C, the amount of PNPase is twice that found in cells grown at 30°C. To investigate whether this effect was transcriptional or posttranscriptional, the expression of a set of pnp-lacZ transcriptional and translational fusions was analyzed in cells grown at different temperatures. In the absence of PNPase, there was no increase in pnp-lacZ expression, indicating that the increase in pnp expression occurs at a posttranscriptional level. Other experiments clearly show that increased pnp expression at low temperature is only observed under conditions in which the autocontrol mechanism of PNPase is functional. At low temperature, the destabilizing effect of PNPase on its own mRNA is less efficient, leading to a decrease in repression and an increase in the expression level.
Polynucleotide phosphorylase (polyribonucleotide:orthophosphate nucleotidyl transferase, EC 2.7.7.5) (PNPase) is a 3′-5′ exoribonuclease involved in mRNA degradation. It is expressed from two kinds of mRNAs: one originating from the promoter of the rpsO gene (immediately upstream) (13) and the other originating from its own promoter (16) (Fig. 1A). Like RNase III and RNase E, two Escherichia coli endoribonucleases, expression of PNPase is autoregulated at the posttranscriptional level. However, in this case, regulation occurs only when the pnp mRNAs are specifically processed by RNase III, 81 nucleotides upstream of the initiation codon (17, 18). This processing occurs during transcription or immediately after it and affects both transcripts equally. As a consequence, the two different mRNAs encoding PNPase become identical. The presence of PNPase triggers destabilization of its own mRNA, and a tight correlation has been observed between the degree of mRNA instability and the level of repression caused by the amount of PNPase in the cell (17–19). The mechanism of autoregulation implies an interaction between PNPase and the leader region of its own mRNA (5, 19). It has been shown previously that steady-state PNPase levels increase about threefold between 37 and 15°C and twofold between 37 and 23°C (8). Here, transcriptional and translational fusions were used to determine whether the increase in steady-state PNPase levels at low temperature involves a transcriptional and/or posttranscriptional control mechanism.
FIG. 1.
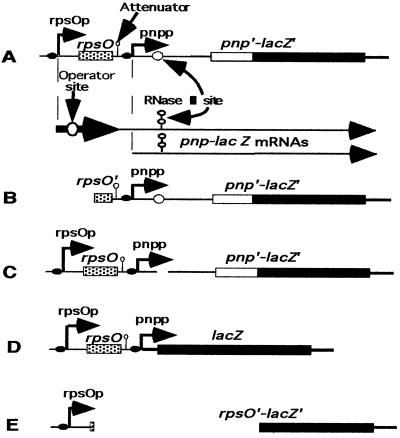
pnp-lacZ fusions and lac derivatives. Translational and transcriptional fusions used for pnp expression. (A) Expression of pnp is under the control of rpsO (rpsOp) and pnp (pnpp) promoters. The junction point between pnp and lacZ is located 183 nucleotides after the translation initiation starting point. (B) Derivatives of this construction. A PstI fragment of the fusion described for panel A was fused in frame to lacZ. In this fusion, the rpsO promoter and half of the rpsO structural gene were removed. (C) A deletion of 100 nucleotides in the pnp leader of the fusion described for panel A removes the RNase III cleavage site. (D) In this transcriptional fusion, lacZ is expressed from the rpsO and pnp promoters. (E) This translational fusion carries a ScaI-SmaI deletion removing all of the pnp DNA and creating an in-frame rpsO-lacZ translational fusion.
MATERIALS AND METHODS
Strains and plasmids.
Strains AB5311 (argE3 rpsL recA1 ΔlacX74) and AB5321 (argG6 arg3E his4 rpsL ΔlacX74) and their lysogenic derivatives GF5311, PF5311, GF5321, and GFΔRN5321 have been described previously (18). Briefly, GF5311 [AB5311 λ Φ(pnp′-lacZ′)1(Hyb)] and GF5321 [AB5321λ Φ(pnp′-lacZ′)1(Hyb)] are λ lysogens of AB5311 and AB5321, respectively, and harbor the same λGF1 phage, which carries a pnp-lacZ translational fusion between the proximal part of pnp and the distal part of lacZ (Fig. 1, fusion A). In this fusion, the chimeric messenger is transcribed from two promoters: one, pnpp, corresponding to that of the pnp gene, and the other, rpsOp, corresponding to that of rpsO, the gene just upstream of pnp. In strain PF5311 [AB5311 λ Φ(pnp′-lacZ′)2(Hyb)], a truncated derivative of this fusion expresses β-galactosidase from the pnp promoter only (Fig. 1, fusion B). GFΔRN5321 [AB5321 λ Φ(ΔRIII-pnp′-lacZ′)3(Hyb)] carries the same fusion as GF5311 and GF5321, but with a complete deletion of the RNase III cleavage site in the pnp leader (Fig. 1, fusion C) (18). CP5321, which will be described elsewhere, is a derivative of AB5321, which carries a deletion of 2,557 bp (C. Portier, unpublished data), removing the complete pnp gene and its own promoter, as well as the proximal part of the downstream gene nlpI (11). CP5321F [CP5321 λ Φ(pnp′-lacZ′)1(Hyb)] corresponds to CP5321 lysogenized by the λGF1 phage. A transcriptional fusion was constructed by cleaving the M13 phage derivative M13GF18 (18) with EcoRI and BglII. (A BglII site was created by changing 1 nucleotide 6 nucleotides downstream of the transcription start point of pnp.) The liberated fragment, which carries rpsOp and pnpp, was then inserted into plasmid pRS415 (22), previously cut by EcoRI and BamHI. An EcoRI-SacII fragment of this plasmid, derived from pRS415, was inserted into a λ phage as described previously (18). The resulting phage, which bears a transcriptional fusion expressing lacZ from both the rpsOp and pnpp promoters, was used to lysogenize strain CP5321. A monolysogenic strain was isolated, which was called FT5321[CP5321 λ Φ(rpsOp-pnpp-lacZ)4] (Fig. 1, fusion D). AB5312 [AB5311 λ Φ(rpsO′-lacZ′)1(Hyb)] is a lysogenic derivative of AB5311 that carries an rpsO-lacZ translational fusion (Fig. 1, fusion E) (14). Plasmid pBP111 harbors the complete rpsO-pnp operon (16), whereas pBPΔ10 carries only pnp (18).
Cultures.
Cultures were grown in Luria-Bertani medium according to the method of Miller (10). For β-galactosidase measurements, lysogenic cells carrying a translational fusion were grown overnight at 30°C in MOPS (morpholinepropanesulfonic acid) medium (15), diluted to an optical density at 600 nm (OD600) of 0.05, and then grown to OD600s of 0.2 and 0.4 at 30, 20, or 15°C before aliquots were removed for assay. For each strain, the effect of PNPase (or protein S15) overproduction in trans was measured by introduction of plasmid pBP111, with plasmid pBR322 used as a control. Ampicillin was used at 100 μg/ml of culture to maintain pBP111 and at 25 μg/ml to maintain pBPΔ10.
RNA extraction and primer extension.
Total RNA from aliquots of cultures grown in MOPS medium was extracted with hot phenol as previously described (15). Samples of total RNA (10 μg) were hybridized at 60°C with 20 pmol of an oligonucleotide complementary to the lacZ gene (M13/pUC sequencing primers [−40] or [−47], from New England Biolabs, Inc.) and 5′ labeled with [γ-32P]ATP (3 MCi/mol; 111 PBq/mol) as described by Sanson and Uzan (21).
DNA sequencing and electrophoresis.
Sequencing was carried out according to the method of Sanger (20). Products were separated on 6% polyacrylamide gels containing 6 M urea after migration at 1,200 V. The lengths of primer extension products were determined by comparison with a sequence of the corresponding DNA with the same oligonucleotide primer.
β-Galactosidase assays.
β-gactosidase levels were measured as described by Miller and are expressed in Miller units (10).
Western blots.
Aliquots of cells grown at 30 and 18°C to an OD600 of 0.5 were harvested and broken by ultrasonic treatment in a buffer containing 100 mM Tris-HCl (pH 8.0). Protein concentration was measured by using the bicinchoninic acid protein assay from Pierce. Protein (0.5 and 1 μg) was mixed with denaturation buffer and electrophoresed on sodium dodecyl sulfate-polyacrylamide (12.5%) gels as described previously (16). The gel was then fixed in a transfer buffer containing 25 mM Tris-HCl (pH 8.3), 150 mM glycine, and 20% methanol for 10 min. Transfer to a Hybond-C Super membrane (Amersham) was carried out overnight at 120 mA in transfer buffer. The membrane was immersed for 2 h in a blocking buffer containing 80 mM Na2HPO4, 20 mM NaH2PO4, and 100 mM NaCl, to which was added 10% dried skimmed milk. After two washes for 5 min in blocking buffer containing 0.1% Tween, the membrane was incubated with diluted PNPase antibodies in blocking buffer for 2 h, washed two times for 5 min in the same buffer containing 0.1% Tween, and incubated for 2 h in the presence of 40.5 kBq of 131I-protein A (specific activity, >30 μCi/μg [ICN]) in blocking buffer. After two more washes, the membrane was exposed to a PhosphorImager screen, and band intensities were measured.
RESULTS
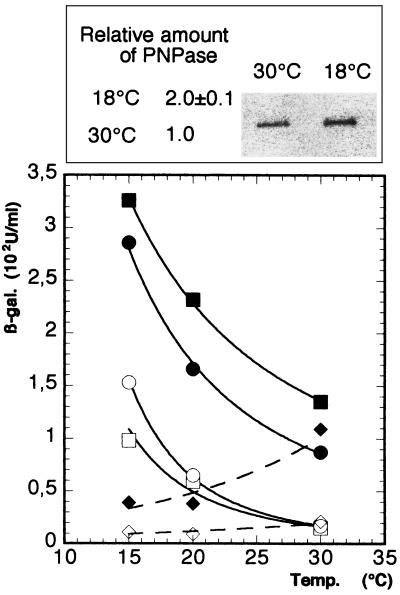
Enhanced expression of PNPase at low temperatures in a wild-type strain is also observed with a pnp-lacZ translational fusion. Strain GF5311[AB5311 λ Φ(pnp′-lacZ′)1(Hyb)] (18), which carries a pnp-lacZ translational fusion expressed from the rpsO and pnp promoters (Fig. 1, fusion A), was grown at 30°C overnight, diluted and divided in 2, with one-half incubated at 30°C and the other at 18°C. Cultures were harvested at an OD600 of 0.5 (about 8 h at 18°C). Aliquots were then taken to measure the amount of PNPase by Western blotting. A twofold increase in PNPase protein levels was observed at 18°C compared to those at 30°C (Fig. 2, insert). A very similar result was obtained by measuring the levels of β-galactosidase produced from the fusion at 20 and 30°C; expression was increased about twofold at the lower temperature (Table 1). Since the level of expression of the pnp-lacZ fusion correlated well with the intracellular level of PNPase, the subsequent experiments were performed with pnp-lacZ fusions.
FIG. 2.
Expression of translational fusions at different temperatures in the presence of different amounts of PNPase. The values indicated in Table 1 are plotted. In GF5311, the pnp-lacZ fusion (Fig. 1, fusion A) is expressed from the rpsO and pnp promoters, whereas in PF5311, it is expressed only from the pnp promoter (fusion B). AB5312 carries an rpsO-lacZ fusion and was used as a control (fusion E). Squares, GF5311; circles, PF5311; diamonds, AB5312. Solid symbols represent expression in the presence of pBR322 (plasmid control), and open symbols represent expression in the presence of pBP111 (overproducing PNPase and S15). An insert shows a Western blot of PNPase. Crude extracts (1 μg) from cells grown at 18 and 30°C were separated by gel electrophoresis and blotted onto a membrane, and the PNPase protein was detected by antisera as described in Materials and Methods. The relative amount of PNPase in strain GF5311 at 18°C is indicated, taking the value at 30°C as 100%.
TABLE 1.
Effect of low temperatures on the expression of two pnp-lacZ fusions under conditions of PNPase overexpression
| Straina | Plasmid | β-Galactosidase level (fold increase)b in Miller units
|
||
|---|---|---|---|---|
| 30°C | 20°C | 15°C | ||
| GF5311 | pBR322 | 135 ± 8 | 232 ± 2 (1.7) | 326 ± 17 (2.4) |
| pBP111(pnp+) | 15 ± 3 | 59 ± 12 (3.9) | 98 ± 1.7 (6.5) | |
| Repression level | 9 | 4 | 3.3 | |
| PF5311 | pBR322 | 87 ± 19 | 166 ± 13 (1.9) | 286 ± 40 (3.3) |
| pBP111 | 17 ± 0.7 | 65 ± 10 (3.8) | 153 ± 9 (9) | |
| Repression level | 5.1 | 2.5 | 1.9 | |
| AB5312 | pBR322 | 109 ± 4 | 38 ± 1.4 (0.3) | 39 ± 9 (0.3) |
| pBP111 (rpsO+) | 21 ± 2 | 9 ± 0.6 (0.4) | 11 ± 0.6 (0.5) | |
| Repression level | 5.2 | 4.2 | 3.5 | |
Autocontrol of pnp expression is decreased at low temperature.
To see whether the increase in expression at low temperature was related to autocontrol, the steady-state β-galactosidase levels of the translational fusion were measured at 15, 20, and 30°C in strains transformed either with a plasmid overexpressing PNPase (pBP111) or with the vector pBR322 as a control. The results are shown Table 1 and Fig. 2. At low growth temperatures, expression of the pnp-lacZ fusion was increased relative to that measured at 30°C in both strains, i.e., whether PNPase is expressed solely from its chromosomal gene (Fig. 2) or whether it was overexpressed from the high-copy-number plasmid pBP111. The lower the growth temperature was, the greater the increase in expression observed was. In strains overexpressing PNPase, expression was 3.9-fold higher at 20°C and 6.5-fold higher at 15°C than the levels measured at 30°C. In the control strain, these increases were 1.7- and 2.4-fold at 20 and 15°C, respectively. The level of repression by PNPase (i.e., the level of expression in cells containing pBR322 divided by the expression level in cells containing pBP111), on the other hand, decreased with temperature. At 30°C, overexpression of PNPase caused a ninefold decrease in pnp-lacZ expression, whereas at 15°C, repression was only 3.3-fold (Table 1).
Similar results were obtained with PF5311 [AB5311 λ Φ(pnp′-lacZ′)2(Hyb)], a strain expressing the pnp-lacZ fusion from the pnp promoter only (Fig. 1, fusion B), after transformation with pBP111 and pBR322. The absolute levels of expression were lower, due to the lack of the rpsO promoter (Table 1, compare GF5311/pBR322 to PF5311/pBR322). In the control strain, PF5311/pBR322, expression was 1.9-fold higher at 20°C and 3.3-fold higher at 15°C, compared to the levels of β-galactosidase measured at 30°C (Table 1 and Fig. 2). In strain PF5311/pBP111, which overexpresses PNPase, expression was 3.8-fold higher at 20°C and 9-fold higher at 15°C. As with the fusion driven by both the rpsO and pnp promoters, as expression of the fusion increased, the repression level decreased with temperature. At 30°C, overexpression of PNPase caused a 5.1-fold decrease in pnp-lacZ expression, whereas at 20 and 30°C, the levels of repression were only 2.5- and 1.9-fold, respectively (Table 1 and Fig. 2). The fact that the patterns of expression and repression were the same, whether the fusion was expressed from one (PF5311) or two promoters (GF5311), suggests that increased expression at low temperature was linked either to a cold inducibility of the pnp promoter or to a decrease in the efficiency of the posttranscriptional autoregulatory mechanism.
The plasmid pBP111, used to overexpress PNPase, also overexpresses ribosomal protein S15, the translational repressor of the expression of its own gene, rpsO (14). To demonstrate that the effect described above was specific to the expression of the pnp gene, expression of the rpsO gene was measured under the same conditions. Hence, expression of an rpsO-lacZ translational fusion, carried by strain AB5312 (Fig. 1, fusion E), was measured in cells containing pBP111 and the control vector, pBR322, at 30, 20, and 15°C (Fig. 2). In contrast to pnp, expression of rpsO decreased at temperatures below 30°C. β-Galactosidase expression of the rpsO-lacZ fusion decreased from 21 Miller units at 30°C to 11 Miller units at 15°C (twofold) in cells containing pBP111 (Table 1 and Fig. 2), whereas expression decreased from 109 Miller units to 39 Miller units (around threefold) in cells containing pBR322 (Table 1 and Fig. 2). Although the repression of rpsO-lacZ expression by the S15 protein was also decreased at lower temperatures, this decrease was limited (5.2-fold at 30°C versus 3.5-fold at 15°C) and was not as strong as that measured with the pnp-lacZ fusions. Together, these results show that expression of the ribosomal protein S15 at low temperature is different from that of PNPase and that neither the S15 autoregulatory mechanism nor the rpsO promoter was involved in cold-stimulated expression of pnp.
Increased expression of a pnp-lacZ translational fusion at low temperature is not observed in the absence of PNPase.
To obtain further evidence that expression observed at low temperature was linked to the autocontrol mechanism, expression of the pnp-lacZ fusion was examined in a strain lacking PNPase and was thus fully derepressed. Strains AB5321(pnp+) and CP5321(Δpnp), were lysogenized by the λ phage carrying the pnp-lacZ fusion (Fig. 1, fusion A), giving GF5321 [AB5321 λ Φ(pnp′-lacZ′)1(Hyb)] and CP5321F [CP5321 λ Φ(pnp′-lacZ′)1(Hyb)], respectively. Their β-galactosidase levels were measured in cultures grown at 18 and 30°C. At 30°C, the wild-type strain, GF5321, produced low levels (175 Miller units of β-galactosidase) (Table 2), whereas in the Δpnp strain, CP5321F, this expression level was increased 10-fold (Table 2). At 18°C, expression was increased twofold in strain GF5321 compared to that at 30°C, whereas expression remained unchanged in strain CP5321F. This result indicates that an intact pnp gene is necessary for increased expression at low temperature, in support of the idea that the autocontrol mechanism is involved in this phenomenon.
TABLE 2.
Effect of temperature on the expression of different lacZ fusions
| Fusiona | PNPase expression | β-Galactosidase level (fold increase)b in Miller units
|
|
|---|---|---|---|
| 30°C | 18°C | ||
| A | pnp+ | 175 ± 45 | 309 ± 97 (1.8) |
| A | Δpnp | 1,762 ± 481 | 1,787 ± 301 (1.0) |
| D | Δpnp | 974 ± 4 | 1,259 ± 3 (1.3) |
| C (ΔRIII) | pnp+ | 694 ± 134 | 697 ± 25 (1.0) |
| C (ΔRIII) | pnp+++c | 634 ± 20 | 677 ± 21 (1.1) |
Fusions are represented in Fig. 1.
Fold increase in fusion expression is given relative to the value at 30°C.
pnp +++, overproduction of PNPase (presence of plasmid pBPΔ10).
In the absence of RNase III processing of the pnp leader, no increased expression can be observed at low temperature.
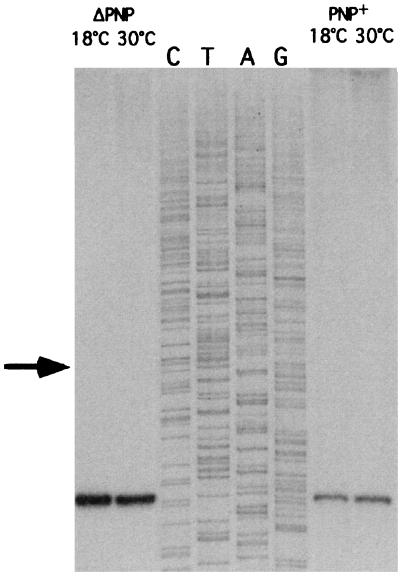
If autocontrol, not PNPase per se, were essential for increased pnp expression at low temperature, mutations in cis in the pnp leader, which abolish autocontrol, should not result in increased expression in cells grown at 18°C. It is known that autocontrol of pnp expression is abolished in the absence of RNase III cleavage in the pnp leader. β-Galactosidase activity was thus measured in a strain that carries a deletion of the RNase III cleavage site, GFΔRN5321 [IBPC5321 λ Φ(ΔRIII-pnp′-lacZ′)3(Hyb)] (18) (Fig. 1, fusion C), at 30 and 18°C, with or without overproduction of PNPase in trans. Table 2 shows that the β-galactosidase levels in these strains were identical at 30°C (697 Miller units) and at 18°C (694 Miller units), whether or not PNPase was overexpressed from pBPΔ10, a plasmid that carries pnp under its own promoter and known to overproduce PNPase (18). These results clearly show that increased expression of the pnp-lacZ fusion does not occur at low temperature in the presence of a cis mutation abolishing autocontrol, even in the presence of PNPase. The experiment described above did not exclude the possibility that the cleavage efficiency of the pnp leader by RNase III was decreased at low temperature, leading to incomplete processing and, hence, increased expression. To test this hypothesis, the pnp leader expressed from the pnp-lacZ fusion at 18 and 30°C was analyzed by primer extension in the presence (GF5321) and absence (CP5321F) of PNPase. Figure 3 shows that only fully processed 5′ ends were present in all cases, regardless of the temperature or the concentration of PNPase. Thus, increased expression at low temperature is not linked to a defect in processing of the pnp leader by RNase III, but requires a functional autocontrol mechanism.
FIG. 3.
Processing efficiency of the pnp-lacZ mRNA by RNase III at 30 and 18°C. Cells were grown at 30 and 18°C to an OD600 of 0.5, and total RNA was extracted. Primer extensions were done with an oligonucleotide hybridizing to the lac part of the message, downstream of the polylinker position. Bands indicate the positions of fragments processed by RNase III. No bands upstream of the RNase III cleavage site corresponding to the position of fragments initiated at the transcription start point are present (arrow), regardless of the growth temperature. In CP5321F, the pnp gene was deleted. Strain GF5321 carries a chromosomal copy of the pnp gene.
The amount of pnp-lacZ mRNA correlates with the expression level of the fusion.
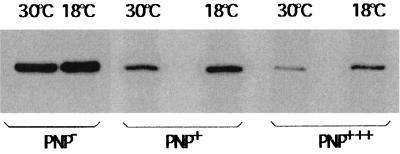
To see whether the pnp-lacZ mRNA level correlated with expression of the fusion at low temperature, the amount of pnp-lacZ mRNA was measured at 30 and 18°C in the presence and absence of PNPase. Cultures of GF5321[IBPC λ Φ(pnp′-lacZ′)1(Hyb)] and CP5321F [CP5321 λ Φ(pnp′-lacZ′)1(Hyb)] (Fig. 1, fusion A) were grown at 18 and 30°C. Total RNA was extracted, and the amounts of pnp-lacZ mRNA were estimated by primer extension. In the absence of PNPase (strain CP5321F), a large amount of pnp-lacZ mRNA was detected, which did not vary significantly between 30 and 18°C (Fig. 4). A quite different result was observed in the presence of PNPase. In this case, the amount of pnp-lacZ mRNA observed was smaller than that in the absence of PNPase, and more mRNA was observed at 18°C than at 30°C. The amount of pnp-lacZ mRNA decreased further upon transformation of strain GF5321 with a multicopy plasmid (pBP111) containing the pnp gene. However, once again, more mRNA was observed at 18°C than at 30°C. Thus, the amount of pnp-lacZ mRNA correlates well with the amount of β-galactosidase measured from the fusions in CP5321F and GF5321.
FIG. 4.
Amount of pnp-lacZ mRNA at 18 and 30°C in the presence of different amounts of PNPase. Amounts of mRNA were measured after separation on a 6% polyacrylamide sequencing gel. PNP−, strain CP5321F (lacking PNPase); PNP+, strain GF5321 with a chromosomal copy of pnp and transformed by pBR322 (control); PNP+++, strain GF5321 strain transformed with plasmid pBP111 carrying a pnp gene.
The decay rate of the pnp-lacZ mRNA is decreased at low temperature.
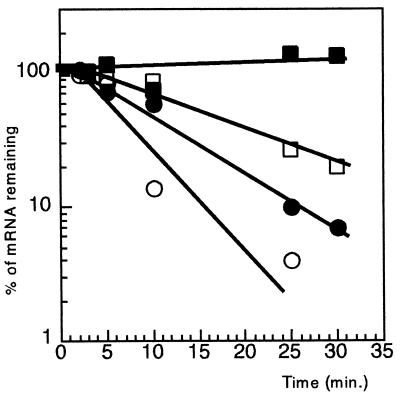
To see whether the variations in the amount of pnp-lacZ mRNA were the result of changes in decay rate or transcription rate, the stability of the pnp-lacZ mRNA was measured in strains GF5321 and CP5321F grown at 18 and 30°C. The decay rate was measured by plotting the percentage of pnp-lacZ mRNA remaining versus time. Figure 5 shows that pnp-lacZ mRNA was more stable at 18°C than at 30°C. In addition, in the absence of PNPase, the half-life of the pnp-lacZ mRNA increased from 4 min to 11 min at 30°C and from 7 min to more than 60 min at 18°C. Thus, at a given temperature, pnp-lacZ mRNA was more stable in the Δpnp background than pnp+, suggesting that pnp-lacZ mRNA is destabilized by PNPase and that this destabilization is decreased in cultures grown at low temperature.
FIG. 5.
Decay rates of pnp-lacZ messengers at 18 and 30°C with and without PNPase. Cells were grown overnight at 30°C in MOPS medium, diluted to an OD600 of 0.05 to 0.08, and incubated at either 30 or 18°C. At an OD600 of 0.5, rifampin was added to the cultures to 0.5 mg/ml (time zero). Next, 2-ml aliquots were taken at different times, total RNA was extracted, and pnp-lacZ mRNA was estimated by primer extension with a lacZ probe. The results were analyzed on a polyacrylamide sequencing gel. A graphical representation of the results is shown. The intensity of the bands was quantified with a PhosphorImager, and the values were plotted versus time. Circles, cultures grown at 30°C; squares, cultures grown at 18°C; open symbols, GF5321 (pnp+); solid symbols, CP5321F (pnp).
To see whether this stabilization was specific to pnp-lacZ mRNA, the decay rate of the rpsO-lacZ mRNA was also analyzed at 30 and 18°C with strain AB5312 (14). A strong stabilization of this mRNA was also observed at low temperature (data not shown). This result was not consistent with the low level of expression of the rpsO-lacZ fusion at 18°C and suggests that the decay rates of different mRNAs can be decreased at low temperature, without leading to increased expression of the encoded protein.
The pnp promoter is not low-temperature inducible.
The results described above did not exclude the possibility of an effect of low temperature on the pnp promoter. To see whether the pnp promoter was low-temperature inducible, β-galactosidase levels were measured in the transcriptional fusion FT5321 [CP5321 λ Φ(rpsOp-pnpp-lacZ)4]. In this strain, β-galactosidase was expressed from rpsOp and pnpp by using the Shine-Dalgarno sequence of lacZ (Fig. 1, fusion D). Table 2 shows that the level at 30°C was nearly identical to that observed at 18°C, suggesting that expression of the pnp-lacZ fusion at low temperature was not dependent upon a temperature inducible pnp promoter.
DISCUSSION
In this work, expression of PNPase at low temperature was studied by comparing levels of expression of pnp-lacZ translational fusions in cells growing at different temperatures below 30°C. By analyzing cells growing at (or near) steady state and at 18°C, complications linked either to the transient metabolic changes that occur during the first 2 h after cold shock or to any changes which might be associated with survival adaptation when strains lacking PNPase are incubated at 16°C (very close to the minimal growth temperature) or lower (9 [see below]) should be avoided.
The increase in pnp-lacZ expression at low temperature is linked to a change in the efficiency of PNPase autocontrol.
In Δpnp strains, in which pnp-lacZ translational fusions are derepressed, pnp expression is the same at 18°C as at 30°C. In contrast, in wild-type strains, where pnp expression is autocontrolled, expression of pnp-lacZ fusions is increased at 18°C. In cells overexpressing PNPase, the repression level drops as expression of the fusion increases with decreasing temperature. This phenomenon is dependent on the presence of a functional RNase III cleavage site. These observations suggest that increased pnp expression at low temperature is linked to autocontrol. An identical conclusion was reached very recently by Beran and Simons (1) while studying PNPase expression after cold shock. Since the efficiency of the RNase III processing of the pnp messengers, which is a prerequisite for repression by PNPase (15, 17), is not affected, the increased expression most likely results from changes in the second step of autocontrol (19). The simplest explanation to account for this effect is to suppose that PNPase is less efficient in its ability to repress expression of the fusion when cells are grown at low temperature. PNPase is thought to interact with its own mRNA to trigger its degradation, since a tight inverse relationship between the amount of PNPase in the cell and pnp-lacZ mRNA stability has been observed (15, 18, 19). Consistent with this hypothesis, a mutation in the KH RNA binding domain of PNPase derepresses pnp expression and increases the level of pnp mRNA (5). If increased expression were the result of a decreased interaction between PNPase and the processed pnp leader at low temperature, it would account for the inverse correlation between temperature and expression as well as the differential increase in expression of the fusion with increasing amount of repressor (PNPase). As a result, the degradation rate of the pnp-lacZ mRNA would be progressively decreased with temperature, increasing the amount of pnp-lacZ mRNA and, hence, expression of the fusion. Consistent with this idea, the amount of pnp-lacZ mRNA is higher at 18°C than at 30°C, and this variation is linked to the amount of PNPase, suggesting that pnp-lacZ mRNA stability is under PNPase control, even at low temperature. Accordingly, the decay rate of pnp-lacZ mRNA is increased in the presence of PNPase, but less so at low temperature. In the absence of PNPase, full stabilization of the pnp-lacZ mRNA is observed at 18°C, but not at 30°C, suggesting that pnp mRNA decay can occur mainly via a PNPase-dependent mechanism at low temperature. However, this stabilization did not lead to an increase in pnp-lacZ mRNA levels compared to those observed at 30°C. It is possible that the increase in mRNA stability was compensated for by a decrease in the transcription rate.
Other factors that might affect expression at low temperature were tested by exchanging the rpsO and pnp promoters for the cold-insensitive (6) lacUV5 promoter (data not shown) or by substituting the pnp Shine-Dalgarno region with the corresponding lac sequence. In neither case was expression of a pnp-lacZ fusion increased at low temperature in a Δpnp background, showing that the increase in expression observed was not the result of an increase in the transcription or translation rate.
Low temperature stabilizes mRNAs.
Stabilization of pnp mRNA at low temperature was also observed in E. coli (1, 23) and in strain K122 of Photorhabdus sp. (3). Indeed, stabilization of other mRNAs, including rpsO mRNA (3; data not shown), cspA mRNA, and cspA-lacZ mRNA (6) in cells grown at low temperature or following cold shock, has previously been observed, suggesting a general slowdown in mRNA degradation at low temperature. Interestingly, mRNA stabilization was not always followed by an increase in the encoded protein. This was the case for protein S15, suggesting a specific difference in the translation efficiency of mRNAs from cold shock and non-cold shock proteins.
The transient increase in pnp mRNA stability observed after cold shock (1, 23) may be a specific property of pnp mRNA under these conditions. When temperature drops, pnp mRNA is instantaneously, but transiently stabilized (1). However, as shown previously, disruption of the pnp gene either in Bacillus subtilis (pnpA::mini-Tn10) or in E. coli (pnp::Tn5) (12) prevents cell growth at or below 15.8°C (9) and at 5°C in the psychrotrophic bacterium Yersinia enterocolitica (7). Nevertheless, some residual growth appears to occur after a cold shock at 15°C in an E. coli strain expressing inactivated PNPase (1). Thus, besides cold adaptation, some survival metabolism might be induced in cells growing at 16°C, which could affect the synthesis rate of PNPase mRNA at cold temperatures. Under these conditions, it is difficult to compare the results observed with pnp mutant cells growing at steady state at 18°C, but with the same cells incubated at or below 16°C unable to grow or growing very slowly. These particular conditions, added to the use of a different pnp mutant, might be the explanation of why Zangrossi and coworkers (23) find that autogenous control of PNPase synthesis during cold shock is regulated at the level of transcription elongation.
The essential role of PNPase at low temperature remains to be clearly established. One can imagine that secondary structures of mRNAs are stabilized at low temperatures, which might impede RNA degradation in the absence of this enzyme. In the absence of mRNA degradation, depletion of nucleoside diphosphate pools would be expected, which, in turn, would slow down DNA replication (4). The importance of these functions might account for the necessity to increase synthesis of PNPase at low temperature. Whether PNPase acts alone or in the context of the degradosome (2) also remains an open question. The large excess of PNPase relative to other components of the degradosome might argue in favor of the former hypothesis.
ACKNOWLEDGMENTS
We thank Ciaran Condon for careful reading and suggestions to improve the manuscript.
This research was supported by the CNRS (UPR9073).
REFERENCES
- 1.Beran R, Simons R W. Cold-temperature induction of Escherichia coli polynucleotide phosphorylase occurs by reversal of its autoregulation. Mol Microbiol. 2001;39:112–125. doi: 10.1046/j.1365-2958.2001.02216.x. [DOI] [PubMed] [Google Scholar]
- 2.Carpousis A J, van Houwe G, Ehretsmann C, Krisch H M. Copurification of E. coli RNase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;11:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 3.Clarke D J, Dowds B C A. The gene coding for polynucleotide phosphorylase in Photorhabdus sp. strain K122 is induced at low temperature. J Bacteriol. 1994;176:3775–3784. doi: 10.1128/jb.176.12.3775-3784.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danchin A. Comparison between the Escherichia coli and Bacillus subtilis genomes suggests that a major function of polynucleotide phosphorylase is to synthesize CDP. DNA Res. 1997;4:9–18. doi: 10.1093/dnares/4.1.9. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Mena J, Das A, Sanchez-Trujillo A, Portier C, Montanez C. A novel mutation in the KH domain of polynucleotide phosphorylase affects autoregulation and mRNA decay in Escherichia coli. Mol Microbiol. 1999;33:235–248. doi: 10.1046/j.1365-2958.1999.01451.x. [DOI] [PubMed] [Google Scholar]
- 6.Goldenberg D, Azar I, Oppenheim A B. Differential mRNA stability of the cspA gene in the cold-shock response of Escherichia coli. Mol Microbiol. 1996;19:241–248. doi: 10.1046/j.1365-2958.1996.363898.x. [DOI] [PubMed] [Google Scholar]
- 7.Goverde R L J, Huis in't Veld J, Kusters J G, Mooi F R. The psychrotrophic bacterium Yersinia enterocolitica requires expression of pnp, the gene for polynucleotide phosphorylase, for growth at low temperature (5°C) Mol Microbiol. 1998;28:555–569. doi: 10.1046/j.1365-2958.1998.00816.x. [DOI] [PubMed] [Google Scholar]
- 8.Herendeen S L, VanBogelen R A, Neidhardt F C. Levels of major proteins of Escherichia coli during growth at different temperatures. J Bacteriol. 1979;139:185–194. doi: 10.1128/jb.139.1.185-194.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luttinger A, Hahn J, Dubnau D. Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol Microbiol. 1996;19:343–356. doi: 10.1046/j.1365-2958.1996.380907.x. [DOI] [PubMed] [Google Scholar]
- 10.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 11.Ohara M, Wu H C, Sankaran K, Rick P D. Identification and characterization of a new lipoprotein, NlpI, in Escherichia coli K-12. J Bacteriol. 1999;181:4318–4325. doi: 10.1128/jb.181.14.4318-4325.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Portier C. Isolation of a polynucleotide phosphorylase mutant using a kanamycin resistant determinant. Mol Gen Genet. 1980;178:343–349. doi: 10.1007/BF00270482. [DOI] [PubMed] [Google Scholar]
- 13.Portier C. Physical localisation and direction of transcription of the structural gene for Escherichia coli ribosomal protein S15. Gene. 1982;18:261–266. doi: 10.1016/0378-1119(82)90164-0. [DOI] [PubMed] [Google Scholar]
- 14.Portier C, Dondon L, Grunberg-Manago M. Translational autocontrol of the E. coli ribosomal protein S15. J Mol Biol. 1990;211:407–414. doi: 10.1016/0022-2836(90)90361-O. [DOI] [PubMed] [Google Scholar]
- 15.Portier C, Dondon L, Grunberg-Manago M, Régnier P. The first step in the functional inactivation of the E. coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5′ end. EMBO J. 1987;6:2165–2170. doi: 10.1002/j.1460-2075.1987.tb02484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Portier C, Migot C, Grunberg-Manago M. Cloning of E. coli pnp gene from an episome. Mol Gen Genet. 1981;183:298–305. doi: 10.1007/BF00270632. [DOI] [PubMed] [Google Scholar]
- 17.Régnier P, Portier C. Initiation, attenuation and RNase III processing of transcripts from the Escherichia coli operon encoding ribosomal protein S15 and polynucleotide phosphorylase. J Mol Biol. 1986;187:23–32. doi: 10.1016/0022-2836(86)90403-1. [DOI] [PubMed] [Google Scholar]
- 18.Robert-Le Meur M, Portier C. E. coli polynucleotide phosphorylase expression is autoregulated through an RNase III-dependent mechanism. EMBO J. 1992;11:2633–2641. doi: 10.1002/j.1460-2075.1992.tb05329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert-Le Meur M, Portier C. Polynucleotide phosphorylase of Escherichia coli induces the degradation of its RNase III processed messenger by preventing its translation. Nucleic Acids Res. 1994;22:397–403. doi: 10.1093/nar/22.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanson B, Uzan M. Dual role of the sequence-specific bacteriophage T4 endoribonuclease RegB. mRNA inactivation and mRNA destabilization. J Mol Biol. 1993;233:429–446. doi: 10.1006/jmbi.1993.1522. [DOI] [PubMed] [Google Scholar]
- 22.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–86. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 23.Zangrossi S, Briani F, Ghisotti D, Regonesi M-E, Totora P, Dehò G. Transcriptional and post-transcriptional control of polynucleotide phosphorylase during cold acclimation in Escherichia coli. Mol Microbiol. 2000;36:1470–1480. doi: 10.1046/j.1365-2958.2000.01971.x. [DOI] [PubMed] [Google Scholar]