Abstract
Background
This study aimed to investigate the molecular mechanism of Tongfengding capsule (TFDC) in treating immune-inflammatory diseases of gouty arthritis (GA) and interleukin-1-beta (IL-1β) inhibitors by using network pharmacology, molecular docking, and cell experiments.
Methods
In this study, the compounds of TFDC and the potential inflammatory targets of GA were obtained from Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP), Online Mendelian Inheritance in Man (OMIM), and GeneCards databases. The TFDC-GA-potential targets interaction network was accomplished by the STRING database. The TFDC-active compound-potential target-GA network was constructed using Cytoscape software. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were used to further explore the GA mechanism and therapeutic effects of TFDC. Quantitative real-time PCR (qPCR) was used to verify whether the TFDC inhibited IL-1β in GA. Molecular docking technology was used to analyze the optimal effective compounds from the TFDC for docking with IL-1β.
Result
133 active compounds and 242 targets were screened from the TFDC, and 25 of the targets intersected with GA inflammatory targets, which were considered as potential therapeutic targets. Network pharmacological analysis showed that the TFDC active compounds such as quercetin, stigmasterol, betavulgarin, rutaecarpine, naringenin, dihydrochelerythrine, and dihydrosanguinarine had better correlation with GA inflammatory targets such as PTGS2, PTGS1, NOS2, SLC6A3, HTR3A, PPARG, MAPK14, RELA, MMP9, and MMP2. The immune-inflammatory signaling pathways of the active compounds for treating GA are IL-17 signaling pathway, TNF signaling pathway, NOD-like receptor signaling pathway, NF-kappa B signaling pathway, Toll-like receptor signaling pathway, HIF-1 signaling pathway, etc. The TFDC reduced IL-1β mRNA expression in GA by qPCR. Molecular docking results suggested that rutaecarpine was the most appropriate natural IL-1β inhibitor.
Conclusion
Our findings provide an essential role and bases for further immune-inflammatory studies on the molecular mechanisms of TFDC and IL-1β inhibitors development in GA.
1. Introduction
Gouty arthritis (GA), one of the most serious and common arthritis usually starting suddenly at night, is a lesion and inflammatory reaction caused by urate deposition in the joint capsule, synovial bursa, cartilage, bone, tendon, and other tissues due to hyperuricemia, which is mainly manifested in swelling, pain, and fever of joints and/or tissues around joints [1–3]. Repeated attacks may form tophi, leading to joint deformities, joint damages, and dyskinesia [3]. In recent years, the incidence of GA has been increasing worldwide, seriously affecting the quality of life and work efficiency of patients and causing a heavy medical burden to the society [4, 5]. At present, colchicine, nonsteroidal anti-inflammatory drugs, allopurinol, benzbromarone, febuxostat, etc., are common Western medicines used for treating GA, but they can cause serious adverse reactions and side effects such as renal function damage, abnormal liver function, gastrointestinal diseases, and cardiovascular and cerebrovascular diseases [6]. Interleukin-1 (IL-1) inhibitors are second-line drugs for GA, including direct inhibitors of interleukin-1 beta (IL-1β) (canakinumab and gevokizumab), selective inhibitors of IL-1 receptors (anakinra), and IL-1β receptor antagonist (rilonacept), but treatment-induced adverse reactions, especially those related to upper respiratory tract infections, abscesses, and gastrointestinal diseases, are still unavoidable [7]. Therefore, it is urgent to further develop new drugs that not only have good curative effects on GA but also have fewer side effects.
Tongfengding capsule (TFDC), which contains 8 kinds of traditional Chinese medicine, including Gentiana Macrophylla Pall (Qinjia), Phellodendri Chinrnsis Cortex (Huangbo), Corydalis Rhizoma (Yanhuosuo), Radix Paeoniae Rubra (Chishao), Cyathulae Radix (Chuanniuxi), Smilacis Glabrae Rhixoma (Tufuling), Plantaginis Semen (Cheqianzi), and Alisma Orientale (Sam.) Juz. (Zexie), is one of the Chinese patent medicines approved by the National Medical Products Administration (NMPA) of the People's Republic of China and listed in the Chinese Pharmacopoeia (2020) [8]. It can clear away heat and dampness, promote blood circulation, dredge collaterals, and calm pain, which is mainly used for treating gout. The study found that TFDC might inhibit the inflammatory response in treating gout by regulating the arachidonic acid metabolic pathway, especially its active compound apigenin could significantly inhibit the related targets of prostaglandin G/H synthase 2 (PTGS2), tumor necrosis factor-α (TNF-α), and interleukin-6 (IL-6) in this pathway [9]. Clinically, TFDC could improve blood uric acid levels in patients and reduce adverse reactions such as stomach discomfort and mild diarrhea, so as to better treat GA [10].
IL-1β, a key bone marrow-derived proinflammatory cytokine, involves in a variety of autoimmune inflammatory responses and cellular activities including cell proliferation, differentiation, and apoptosis [11]. When IL-1β is excessively produced, it can cause atherosclerosis, rheumatoid arthritis, gout, Parkinson's disease, Alzheimer's disease, tumors, and other diseases [12, 13]. In particular, IL-1β plays a key role in the occurrence and development of GA [14]. Urate deposition in GA promotes the release of IL-1β, which in turn promotes the influx of neutrophils into the joint to trigger joint inflammation [15–18]. Moreover, tophi themselves also have a large number of cells expressing IL-1β [15]. IL-1β also has been implicated in bone destruction and bone erosion in GA [15]. In view of this, some in-depth studies have revealed that the herbal medicine simiaosan can alleviate the symptoms of GA by regulating the NALP3/IL-1β signaling pathway, and that curcumin can suppress the expression of Nod-like receptor 3 (NLRP3) inflammasome to downregulate the level of IL-1β via inhibiting the activation of nuclear factor-kappa B (NF-κB) signaling pathway in monosodium urate (MSU)-induced GA [19, 20]. Anyway, there is an urgent need and important clinical value to develop inhibitors targeting IL-1β in GA with better efficacy and fewer side effects.
Network pharmacology, originally proposed by Andrew L. Hopkins in 2007, mainly studies the complex and diverse relationships among drugs, targets, diseases, and pathways, providing new methods for the development of new drugs and new ideas for the treatment of some diseases [21–23]. Molecular docking, developed with the continuous update of computer technology and the rapid increase of structural, chemical, and biological data related to available therapeutic targets, can be used to identify new compounds with therapeutic significance and contribute to drug discovery and development [24, 25]. This study firstly screened the active compounds of TFDC and then analyzed and summarized the possible targets of active compounds in treating GA. Next, the targets of active compounds and the pathways of targets that might play a therapeutic role were investigated by using network pharmacology, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. And then, the therapeutic effect of TFDC on IL-1β, an important target in the pathological mechanism of GA, was verified by the experiment. Finally, the optimal active compounds of TFDC docked with IL-1β were analyzed by molecular docking technology in order to explore and develop novel optimum compounds that could inhibit IL-1β in GA. Taken together, this study aimed to provide theoretical basis and more research ideas for the molecular mechanism of TFDC against GA and discover more natural compounds as IL-1β inhibitors that can be used to better treat GA with fewer side effects. The workflow is shown in Figure 1.
Figure 1.
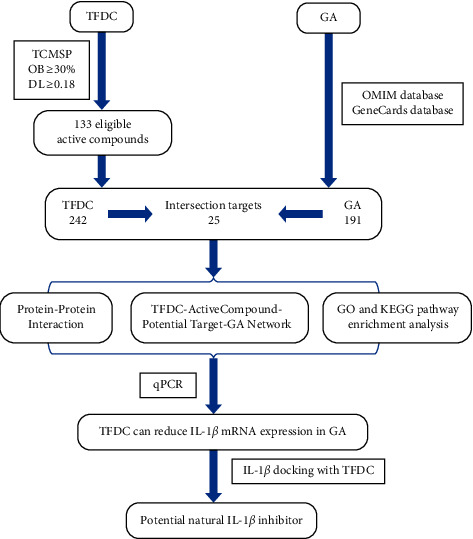
The workflow of IL-1β inhibitor prediction in GA.
2. Method and Materials
2.1. Identification of Active Compounds and Related Targets in TFDC
The Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, https://tcmspw.com/index.php) [26] was used to screen the active compounds and related targets of 8 Chinese herbal medicines in TFDC with the criteria of oral bioavailability (OB) ≥ 30% and drug-likeness (DL) ≥ 0.18. Then, the UniProt database (https://www.uniprot.org/) [27] was used to obtain the gene symbol and UniProt ID of the targets screened by TCMSP with the species selected as “Homo sapiens”.
2.2. Identification of TFDC Targets Related to GA
The GA-related targets were collected from GeneCards (https://www.genecards.org/) [28] and OMIM (Online Mendelian Inheritance in Man, https://omim.org/search/advanced/geneMap) [29] with the keywords “gouty arthritis”. Then, the gene symbol and UniProt ID of the GA-related targets were obtained from UniProt as well. Potential targets of TFDC for GA were acquired through the Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/) intersection.
2.3. Construction of Protein-Protein Interaction (PPI) Network
The TFDC-GA-potential targets were processed by the STRING database (https://string-db.org/) [30] with the species limited to “Homo sapiens” and a confidence score of >0.9. Then, the PPI network of TFDC-GA-potential targets was imported into Cytoscape software (version 3.8.0) [31] for network analysis. Nodes represent targets in the PPI network map, and edges represent functional associations between potential targets.
2.4. Construction of TFDC-Active Compound-Potential Target-GA Network
To accurately elicit interactions between active compounds and its corresponding GA-related targets of TFDC, a visual network was established through Cytoscape software. Furthermore, the network topology parameters were applied to select the key compounds and targets.
2.5. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis
The packages “org.Hs.eg.db”, “DOSE”, “enrichplot”, “clusterProfiler” [32], and “ggplot2” in R software (version 4.1.1 for Windows) were used to carry out GO and KEGG pathway enrichment analyses and visualization for TFDC-GA-potential targets with P < 0.05.
2.6. Cell Culture and Treatment
Human monocytic leukemia THP-1 cell was purchased from Procell Life Science & Technology Co., Ltd (Wuhan, Hubei Province, China). Cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (Gibco, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco, USA). They were maintained in an incubator at 37°C with 5% CO2 and saturated humidity, and the culture medium was replaced with complete culture medium every 2 or 3 days. The cells were treated with phorbol-12-myristate-13-acetate (PMA) (100 nM) for 3 h to induce their differentiation into resting M0 macrophage. A fresh complete culture medium was added and cultured for 24 h after washing the cells with phosphate-buffered saline (PBS). Differentiated THP-1 cells were then stimulated with MSU (50, 100, and 200 μg/mL) with optimal concentration and treated with TFDC at final concentrations of 25, 50, 75, 100, and 125 μg/mL as well as colchicine (positive drug, 2 μg/mL) simultaneously at 37°C for 24 h [33].
2.7. Cell Viability Analysis
THP-1 cells were harvested during the logarithmic growth phase and seeded into a 96-well plate at a density of 1 × 106 cells/well with a final volume of 100 μL. After treatment, 10 μL of CCK-8 was added to each well, and the plates were then incubated for additional 2 h. The absorbance at 450 nm was measured using a microplate reader. The experiments were performed in quadruplicate and repeated at least three times.
2.8. Quantitative Real-Time PCR (qPCR) Analysis
Total RNA was extracted from cells in 12-well plates, and cDNA was prepared according to the manufacturer's protocol. qPCR was amplified and measured on the BIO-RAD CFX96TM (BIO-RAD, USA) in the presence of SYBR Green. Then, the fluorescence values were collected, and a melting curve analysis was performed. The experiments were conducted at least three times. The sequences of IL-1β and β-actin primers are, respectively, as follow: Forward: CTCGCCAGTGAAATGATGGCT, Reverse: GTGGTGGTCGGAGATTCGTAG; and Forward: TGGCACCCAGCACAATGAA, Reverse: CTAAGTCATAGTCCGCCTAGAAGCA. The relative quantifications of genes expression were calculated using the 2−ΔΔCq method with β-actin served as a normalization control.
2.9. Verification between Active Compounds and Key Targets by Molecular Docking
Active compounds selected from the TFDC-active compound-potential target-GA network were molecularly docked with IL-1β receptor. Before performing the docking progress, the 2D structures of active compounds were acquired from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) [34] and optimized to save as 3D structures by Chem3D software. The 3D protein structure of IL-1β receptor was acquired from the RCSB PDB database (https://www.rcsb.org/) [35]. Then, AutoDockTools software was used to process molecular structures followed by AutoDockVina software used for molecular docking. Last, visualization of the docking results was carried out by PyMOL and Discovery Studio software.
2.10. Statistical Analysis
Data were expressed as the mean ± standard deviation (SD). Intergroup differences were determined using one-way ANOVA followed by Tukey's comparison tests. P < 0.05 was considered statistically significant. All data were processed using GraphPad Prism 8.3.0 software.
3. Results
3.1. Acquirement of Active Compounds in TFDC
Based on the criteria of OB ≥ 30% and DL ≥ 0.18, a total of 155 (after removing duplication: 133) eligible active compounds of TFDC were screened, of which 9 were from Cheqianzi (CQZ), 29 were from Chishao (CS), 4 were from Chuanniuxi (CNX), 37 were from Huangbo (HB), 2 were from Qinjia (QJ), 15 were from Tufuling (TFL), 49 were from Yanhuosuo (YHS), and 10 were from Zexie (ZX). The characteristics of all the screened out eligible active compounds in the TFDC are shown in Table 1.
Table 1.
Characteristics of eligible active compounds in TFDC.
| No. | Molecule ID | Molecule name | Molecular weight | OB (%) | DL | Herbs |
|---|---|---|---|---|---|---|
| 1 | MOL000791 | Bicuculline | 367.38 | 69.67 | 0.88 | YHS |
| 2 | MOL002668 | Worenine | 334.37 | 45.83 | 0.87 | HB |
| 3 | MOL001463 | Dihydrosanguinarine | 333.36 | 59.31 | 0.86 | YHS |
| 4 | MOL004225 | Pseudocoptisine | 320.34 | 38.97 | 0.86 | YHS |
| 5 | MOL001474 | Sanguinarine | 332.35 | 37.81 | 0.86 | YHS |
| 6 | MOL004231 | Tetrahydrocorysamine | 337.40 | 34.17 | 0.86 | YHS |
| 7 | MOL001458 | Coptisine | 320.34 | 30.67 | 0.86 | HB, YHS |
| 8 | MOL004230 | Stylopine | 323.37 | 48.25 | 0.85 | YHS |
| 9 | MOL004198 | 18797-79-0 | 367.43 | 46.06 | 0.85 | YHS |
| 10 | MOL000787 | Fumarine | 353.40 | 59.26 | 0.83 | HB, YHS |
| 11 | MOL004226 | 24240-05-9 | 353.40 | 53.75 | 0.83 | YHS |
| 12 | MOL002673 | Hispidone | 472.78 | 36.18 | 0.83 | HB |
| 13 | MOL000856 | Alisol C monoacetate | 514.77 | 33.06 | 0.83 | ZX |
| 14 | MOL007835 | Orobanchoside_qt | 476.47 | 55.99 | 0.82 | CQZ |
| 15 | MOL002660 | Niloticin | 456.78 | 41.41 | 0.82 | HB |
| 16 | MOL000853 | Alisol B | 444.72 | 36.76 | 0.82 | ZX |
| 17 | MOL006392 | Dihydroniloticin | 458.80 | 36.43 | 0.82 | HB |
| 18 | MOL006413 | Phellochin | 488.83 | 35.41 | 0.82 | HB |
| 19 | MOL000830 | Alisol B | 472.78 | 34.47 | 0.82 | ZX |
| 20 | MOL002636 | Kihadalactone A | 512.70 | 34.21 | 0.82 | HB |
| 21 | MOL000854 | Alisol C | 486.76 | 32.70 | 0.82 | ZX |
| 22 | MOL000832 | Alisol B 23-acetate | 446.74 | 32.52 | 0.82 | ZX |
| 23 | MOL000546 | Diosgenin | 414.69 | 80.88 | 0.81 | TFL |
| 24 | MOL004202 | Dehydrocavidine | 351.43 | 38.99 | 0.81 | YHS |
| 25 | MOL002656 | Dihydroniloticin | 458.80 | 36.43 | 0.81 | HB |
| 26 | MOL002670 | Cavidine | 353.45 | 35.64 | 0.81 | HB, YHS |
| 27 | MOL000831 | Alisol B monoacetate | 514.82 | 35.58 | 0.81 | ZX |
| 28 | MOL000862 | [(1S,3R)-1-[(2R)-3,3-dimethyloxiran-2-yl]-3-[(5R,8S,9S,10S,11S,14R)-11-hydroxy-4,4,8,10,14-pentamethyl-3-oxo-1,2,5,6,7,9,11,12,15,16-decahydrocyclopenta[a]phenanthren-17-yl]butyl] acetate | 514.82 | 35.58 | 0.81 | ZX |
| 29 | MOL001461 | Dihydrochelerythrine | 349.41 | 32.73 | 0.81 | YHS |
| 30 | MOL001921 | Lactiflorin | 462.49 | 49.12 | 0.80 | CS |
| 31 | MOL004190 | (−)-Alpha-N-methylcanadine | 354.46 | 45.06 | 0.80 | YHS |
| 32 | MOL001924 | Paeoniflorin | 480.51 | 53.87 | 0.79 | CS |
| 33 | MOL004228 | Saulatine | 396.47 | 42.74 | 0.79 | YHS |
| 34 | MOL006401 | Melianone | 470.76 | 40.53 | 0.78 | HB |
| 35 | MOL001454 | Berberine | 336.39 | 36.86 | 0.78 | HB, YHS |
| 36 | MOL002666 | Chelerythrine | 332.37 | 34.18 | 0.78 | HB |
| 37 | MOL002903 | (R)-canadine | 339.42 | 55.37 | 0.77 | YHS |
| 38 | MOL001455 | (S)-canadine | 339.42 | 53.83 | 0.77 | HB |
| 39 | MOL013352 | Obacunone | 454.56 | 43.29 | 0.77 | HB |
| 40 | MOL000849 | 16β-methoxyalisol B monoacetate | 544.85 | 32.43 | 0.77 | ZX |
| 41 | MOL007004 | Albiflorin | 480.51 | 30.25 | 0.77 | CS |
| 42 | MOL000449 | Stigmasterol | 412.77 | 43.83 | 0.76 | CS, HB, TFL, YHS |
| 43 | MOL004355 | Spinasterol | 412.77 | 42.98 | 0.76 | CS |
| 44 | MOL001663 | (4aS,6aR,6aS,6bR,8aR,10R,12aR,14bS)-10-hydroxy-2,2,6a,6b,9,9,12a-heptamethyl-1,3,4,5,6,6a,7,8,8a,10,11,12,13,14b-tetradecahydropicene-4a-carboxylic acid | 456.78 | 32.03 | 0.76 | CQZ |
| 45 | MOL002776 | Baicalin | 446.39 | 40.12 | 0.75 | CS |
| 46 | MOL006999 | Stigmast-7-en-3-ol | 414.79 | 37.42 | 0.75 | CS |
| 47 | MOL002643 | Delta 7-stigmastenol | 414.79 | 37.42 | 0.75 | HB |
| 48 | MOL000359 | Sitosterol | 414.79 | 36.91 | 0.75 | CQZ, CS, QJ, TFL, YHS, ZX |
| 49 | MOL005869 | Daucosterol_qt | 414.79 | 36.91 | 0.75 | CQZ |
| 50 | MOL000358 | Beta-sitosterol | 414.79 | 36.91 | 0.75 | CS, CNX, HB, QJ, TFL |
| 51 | MOL001771 | Poriferast-5-en-3beta-ol | 414.79 | 36.91 | 0.75 | HB |
| 52 | MOL013119 | Enhydrin | 464.51 | 40.56 | 0.74 | TFL |
| 53 | MOL013118 | Neoastilbin | 450.43 | 40.54 | 0.74 | TFL |
| 54 | MOL004575 | Astilbin | 450.43 | 36.46 | 0.74 | TFL |
| 55 | MOL004234 | 2,3,9,10-Tetramethoxy-13-methyl-5,6-dihydroisoquinolino[2,1-b]isoquinolin-8-one | 381.46 | 76.77 | 0.73 | YHS |
| 56 | MOL006422 | Thalifendine | 322.36 | 44.41 | 0.73 | HB |
| 57 | MOL002894 | Berberrubine | 322.36 | 35.74 | 0.73 | HB |
| 58 | MOL001460 | Cryptopin | 369.45 | 78.74 | 0.72 | YHS |
| 59 | MOL004210 | (1S,8′R)-6,7-dimethoxy-2-methylspiro[3,4-dihydroisoquinoline-1,7′-6,8-dihydrocyclopenta[g][1,3]benzodioxole]-8′-ol | 369.45 | 43.95 | 0.72 | YHS |
| 60 | MOL005043 | Campest-5-en-3beta-ol | 400.76 | 37.58 | 0.71 | CS |
| 61 | MOL005438 | Campesterol | 400.76 | 37.58 | 0.71 | HB |
| 62 | MOL004224 | Pontevedrine | 381.41 | 30.28 | 0.71 | YHS |
| 63 | MOL004567 | Isoengelitin | 434.43 | 34.65 | 0.70 | TFL |
| 64 | MOL002659 | Kihadanin A | 486.56 | 31.60 | 0.70 | HB |
| 65 | MOL004191 | Capaurine | 371.47 | 62.91 | 0.69 | YHS |
| 66 | MOL000793 | C09367 | 325.39 | 47.54 | 0.69 | YHS |
| 67 | MOL002671 | Candletoxin A | 608.79 | 31.81 | 0.69 | HB |
| 68 | MOL004195 | Corydaline | 369.50 | 65.84 | 0.68 | YHS |
| 69 | MOL004204 | Dehydrocorydaline | 366.47 | 41.98 | 0.68 | YHS |
| 70 | MOL004214 | Isocorybulbine | 368.51 | 40.18 | 0.66 | YHS |
| 71 | MOL000785 | Palmatine | 352.44 | 64.60 | 0.65 | HB, YHS |
| 72 | MOL000762 | Palmidin A | 510.52 | 35.36 | 0.65 | HB |
| 73 | MOL004071 | Hyndarin | 355.47 | 73.94 | 0.64 | YHS |
| 74 | MOL004203 | Dehydrocorybulbine | 352.44 | 46.97 | 0.63 | YHS |
| 75 | MOL004216 | 13-Methylpalmatrubine | 352.44 | 40.97 | 0.63 | YHS |
| 76 | MOL002672 | Hericenone H | 580.88 | 39.00 | 0.63 | HB |
| 77 | MOL004209 | 13-Methyldehydrocorydalmine | 352.44 | 35.94 | 0.63 | YHS |
| 78 | MOL002662 | Rutaecarpine | 287.34 | 40.30 | 0.60 | HB |
| 79 | MOL004199 | Corynoloxine | 365.41 | 38.12 | 0.60 | YHS |
| 80 | MOL004196 | Corydalmine | 340.45 | 52.50 | 0.59 | YHS |
| 81 | MOL004205 | Dehydrocorydalmine | 338.41 | 43.90 | 0.59 | YHS |
| 82 | MOL000790 | Isocorypalmine | 341.44 | 35.77 | 0.59 | HB, YHS |
| 83 | MOL004220 | N-methyllaurotetanine | 341.44 | 41.62 | 0.56 | YHS |
| 84 | MOL004233 | ST057701 | 341.44 | 31.87 | 0.56 | YHS |
| 85 | MOL004221 | Norglaucing | 341.44 | 30.35 | 0.56 | YHS |
| 86 | MOL004197 | Corydine | 341.44 | 37.16 | 0.55 | YHS |
| 87 | MOL004193 | Clarkeanidine | 327.41 | 86.65 | 0.54 | YHS |
| 88 | MOL004208 | Demethylcorydalmatine | 327.41 | 38.99 | 0.54 | YHS |
| 89 | MOL000217 | (S)-scoulerine | 327.41 | 32.28 | 0.54 | YHS |
| 90 | MOL007003 | Benzoylpaeoniflorin | 584.62 | 31.14 | 0.54 | CS |
| 91 | MOL007025 | Isobenzoylpaeoniflorin | 584.62 | 31.14 | 0.54 | CS |
| 92 | MOL004763 | Izoteolin | 327.41 | 39.53 | 0.51 | YHS |
| 93 | MOL012298 | Rubrosterone | 334.45 | 32.69 | 0.47 | CNX |
| 94 | MOL007014 | 8-Debenzoylpaeonidanin | 390.43 | 31.74 | 0.45 | CS |
| 95 | MOL004200 | Methyl-[2-(3,4,6,7-tetramethoxy-1-phenanthryl)ethyl]amine | 355.47 | 61.15 | 0.44 | YHS |
| 96 | MOL007008 | 4-Ethyl-paeoniflorin_qt | 332.38 | 56.87 | 0.44 | CS |
| 97 | MOL002641 | Phellavin_qt | 374.42 | 35.86 | 0.44 | HB |
| 98 | MOL007012 | 4-o-Methyl-paeoniflorin_qt | 332.38 | 56.70 | 0.43 | CS |
| 99 | MOL001002 | Ellagic acid | 302.20 | 43.06 | 0.43 | CS |
| 100 | MOL001925 | Paeoniflorin_qt | 318.35 | 68.18 | 0.40 | CS |
| 101 | MOL002651 | Dehydrotanshinone II A | 292.35 | 43.76 | 0.40 | HB |
| 102 | MOL012286 | Betavulgarin | 312.29 | 68.75 | 0.39 | CNX |
| 103 | MOL001131 | Phellamurin_qt | 356.40 | 56.60 | 0.39 | HB |
| 104 | MOL001918 | Paeoniflorgenone | 318.35 | 87.59 | 0.37 | CS |
| 105 | MOL007016 | Paeoniflorigenone | 318.35 | 65.33 | 0.37 | CS |
| 106 | MOL006996 | 1-o-Beta-d-glucopyranosylpaeonisuffrone_qt | 332.38 | 65.08 | 0.35 | CS |
| 107 | MOL004232 | Tetrahydroprotopapaverine | 329.43 | 57.28 | 0.33 | YHS |
| 108 | MOL007005 | Albiflorin_qt | 318.35 | 48.70 | 0.33 | CS |
| 109 | MOL007018 | 9-Ethyl-neo-paeoniaflorin A_qt | 334.40 | 64.42 | 0.30 | CS |
| 110 | MOL006992 | (2R,3R)-4-methoxyl-distylin | 318.30 | 59.98 | 0.30 | CS |
| 111 | MOL002464 | 1-Monolinolein | 354.59 | 37.18 | 0.30 | ZX |
| 112 | MOL006994 | 1-o-Beta-d-glucopyranosyl-8-o-benzoylpaeonisuffrone_qt | 302.35 | 36.01 | 0.30 | CS |
| 113 | MOL007813 | Dihydrotricetin | 304.27 | 58.12 | 0.28 | CQZ |
| 114 | MOL000098 | Quercetin | 302.25 | 46.43 | 0.28 | CQZ, CNX, HB, TLF, YHS |
| 115 | MOL002644 | Phellopterin | 300.33 | 40.19 | 0.28 | HB |
| 116 | MOL013117 | 4,7-Dihydroxy-5-methoxyl-6-methyl-8-formyl-flavan | 314.36 | 37.03 | 0.28 | TFL |
| 117 | MOL007819 | Hypolaetin | 302.25 | 33.24 | 0.28 | CQZ |
| 118 | MOL004580 | cis-Dihydroquercetin | 304.27 | 66.44 | 0.27 | TFL |
| 119 | MOL013129 | (2R,3R)-2-(3,5-dihydroxyphenyl)-3,5,7-trihydroxychroman-4-one | 304.27 | 63.17 | 0.27 | TFL |
| 120 | MOL001736 | (−)-Taxifolin | 304.27 | 60.51 | 0.27 | TFL |
| 121 | MOL004576 | Taxifolin | 304.27 | 57.84 | 0.27 | TFL |
| 122 | MOL001735 | Dinatin | 300.28 | 30.97 | 0.27 | CQZ |
| 123 | MOL006990 | (1S,2S,4R)-trans-2-hydroxy-1,8-cineole-B-D-glucopyranoside | 332.44 | 30.25 | 0.27 | CS |
| 124 | MOL004215 | Leonticine | 327.46 | 45.79 | 0.26 | YHS |
| 125 | MOL002652 | delta7-Dehydrosophoramine | 242.35 | 54.45 | 0.25 | HB |
| 126 | MOL000492 | (+)-Catechin | 290.29 | 54.83 | 0.24 | CS |
| 127 | MOL007836 | Plantaginin_qt | 288.27 | 54.04 | 0.24 | CQZ |
| 128 | MOL007022 | Evofolin B | 318.35 | 64.74 | 0.22 | CS |
| 129 | MOL004328 | Naringenin | 272.27 | 59.29 | 0.21 | TFL |
| 130 | MOL002714 | Baicalein | 270.25 | 33.52 | 0.21 | CS |
| 131 | MOL002663 | Skimmianin | 259.28 | 40.14 | 0.20 | HB |
| 132 | MOL000622 | Magnograndiolide | 266.37 | 63.71 | 0.19 | HB |
| 133 | MOL002883 | Ethyl oleate (NF) | 310.58 | 32.40 | 0.19 | CS |
OB : oral bioavailability; DL : drug-likeness; YHS : Yanhuosuo; HB : Huangbo; ZX : Zexie; CQZ : Cheqianzi; TFL : Tufuling; CS : Chishao; CNX : Chuanniuxi; QJ : Qinjia.
3.2. Acquirement of the Potential Therapeutic Targets and Analysis of the PPI Network
A total of 207 GA-related targets were obtained from GeneCards and OMIM databases. After removing duplicate targets, 191 GA-related targets in total were collected at last. Similarly, 242 unrepeated targets of 133 active compounds in the TFDC were obtained from TCMSP database. The Venny 2.1 platform was used to intersect the GA-related and TFDC-related targets. 25 potential therapeutic targets were obtained (Figure 2(a)) (Table 2). Sequentially, the above targets were imported into the STRING database to obtain the PPI network (Figure 2(b)). The above targets of the PPI network results were identified and listed by the R package “Venn Diagram” (Figure 2(c)). These genes include mitogen-activated protein kinase 14 (MAPK14), interleukin-1 beta (IL1B, IL-1β), prostaglandin G/H synthase 1 (PTGS1), peroxisome proliferator activated receptor gamma (PPARG), transcription factor p65 (RELA), and matrix metalloproteinase-9 (MMP9). Among them, IL-1β is at the core because it is most associated with other targets in the PPI network, which indicates that IL-1β plays a significant role in the occurrence and development of GA.
Figure 2.
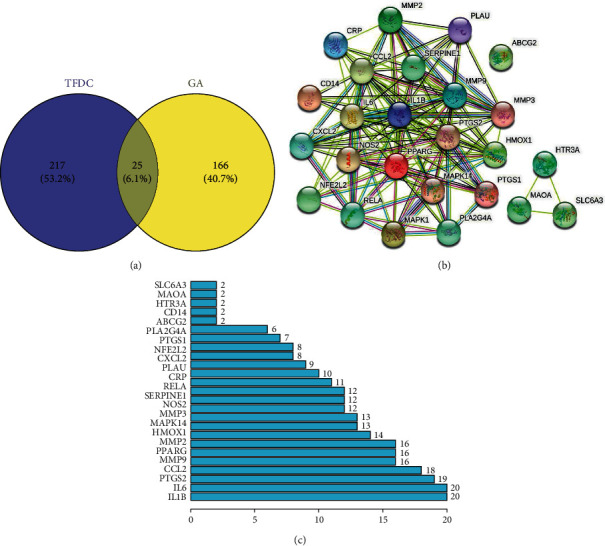
Potential therapeutic targets and PPI network map of TFDC for GA. (a) The Venny results of potential therapeutic targets of TFDC for GA. (b) The PPI network map of 25 targets. (c) Count and list of the above targets of PPI network map.
Table 2.
25 potential therapeutic targets of TFDC for GA.
| No. | Target | Symbol | Entrez ID |
|---|---|---|---|
| 1 | 72 kDa type IV collagenase | MMP2 | 4,313 |
| 2 | Plasminogen activator inhibitor 1 | SERPINE1 | 5,054 |
| 3 | Mitogen-activated protein kinase 14 | MAPK14 | 1,432 |
| 4 | Urokinase-type plasminogen activator | PLAU | 5,328 |
| 5 | ATP-binding cassette sub-family G member 2 | ABCG2 | 9,429 |
| 6 | Heme oxygenase 1 | HMOX1 | 3,162 |
| 7 | Nitric oxide synthase, inducible | NOS2 | 4,843 |
| 8 | Stromelysin-1 | MMP3 | 4,314 |
| 9 | Cytosolic phospholipase A2 | PLA2G4A | 5,321 |
| 10 | Mitogen-activated protein kinase 1 | MAPK1 | 5,594 |
| 11 | Prostaglandin G/H synthase 2 | PTGS2 | 5,743 |
| 12 | C-X-C motif chemokine 2 | CXCL2 | 2,920 |
| 13 | Interleukin-1 beta | IL1B | 3,553 |
| 14 | Amine oxidase [flavin-containing] A | MAOA | 4,128 |
| 15 | Prostaglandin G/H synthase 1 | PTGS1 | 5,742 |
| 16 | 5-Hydroxytryptamine receptor 3A | HTR3A | 3,359 |
| 17 | Peroxisome proliferator activated receptor gamma | PPARG | 5,468 |
| 18 | C-reactive protein | CRP | 1,401 |
| 19 | C–C motif chemokine 2 | CCL2 | 6,347 |
| 20 | Transcription factor p65 | RELA | 5,970 |
| 21 | Interleukin-6 | IL6 | 3,569 |
| 22 | Sodium-dependent dopamine transporter | SLC6A3 | 6,531 |
| 23 | Nuclear factor erythroid 2-related factor 2 | NFE2L2 | 4,780 |
| 24 | Matrix metalloproteinase-9 | MMP9 | 4,318 |
| 25 | Monocyte differentiation antigen CD14 | CD14 | 929 |
3.3. Construction and Analysis of the TFDC-Active Compound-Potential Target-GA Network
The potential targets and their corresponding eligible active compounds were entered into the Cytoscape software to obtain a TFDC-active compound-potential target-GA network (Figure 3). A total of 104 nodes and 332 edges were obtained from the TFDC-active compound-potential target-GA network. The results of network topology analysis showed that the top 10 targets, namely, prostaglandin G/H synthase 2 (PTGS2), PTGS1, nitric oxide synthase, inducible (NOS2), sodium-dependent dopamine transporter (SLC6A3), 5-hydroxytryptamine receptor 3A (HTR3A), PPARG, MAPK14, RELA, MMP9, and 72 kDa type IV collagenase (MMP2), have a higher degree, which indicates that they play important roles in this interaction network (degree ≥ 4) (Table 3). Besides, the top 7 active compounds, namely, quercetin, stigmasterol, betavulgarin, rutaecarpine, naringenin, dihydrochelerythrine, and dihydrosanguinarine, also have a higher degree, which have remarkable significance in the network (degree ≥ 6) (Table 4).
Figure 3.
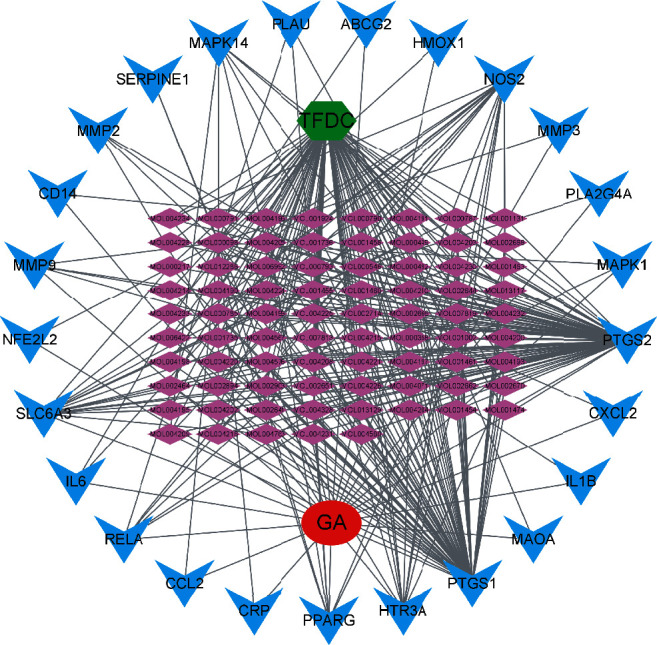
The TFDC-active compound-potential target-GA network. The green node represents TFDC. The red node represents GA. Purple nodes represent active compounds. Blue nodes represent targets. Gray lines represent interconnections between nodes and nodes.
Table 3.
Top 10 high-degree targets in the network.
| No. | Target | Symbol | Degree |
|---|---|---|---|
| 1 | Prostaglandin G/H synthase 2 | PTGS2 | 75 |
| 2 | Prostaglandin G/H synthase 1 | PTGS1 | 66 |
| 3 | Nitric oxide synthase, inducible | NOS2 | 20 |
| 4 | Sodium-dependent dopamine transporter | SLC6A3 | 17 |
| 5 | 5-Hydroxytryptamine receptor 3A | HTR3A | 11 |
| 6 | Peroxisome proliferator activated receptor gamma | PPARG | 9 |
| 7 | Mitogen-activated protein kinase 14 | MAPK14 | 8 |
| 8 | Transcription factor p65 | RELA | 7 |
| 9 | Matrix metalloproteinase-9 | MMP9 | 5 |
| 10 | 72 kDa type IV collagenase | MMP2 | 4 |
Table 4.
Top 7 high-degree active compounds in the network.
| No. | Molecule ID | Molecule name | Molecular weight | OB (%) | DL | Herbs | Degree |
|---|---|---|---|---|---|---|---|
| 1 | MOL000098 | Quercetin | 302.25 | 46.43 | 0.28 | CQZ, CNX, HB,TFL, YHS | 19 |
| 2 | MOL000449 | Stigmasterol | 412.77 | 43.83 | 0.76 | CS, HB, TFL, YHS | 6 |
| 3 | MOL012286 | Betavulgarin | 312.29 | 68.75 | 0.39 | CNX | 6 |
| 4 | MOL002662 | Rutaecarpine | 287.34 | 40.30 | 0.60 | HB | 6 |
| 5 | MOL004328 | Naringenin | 272.27 | 59.29 | 0.21 | TFL | 6 |
| 6 | MOL001461 | Dihydrochelerythrine | 349.41 | 32.73 | 0.81 | YHS | 6 |
| 7 | MOL001463 | Dihydrosanguinarine | 333.36 | 59.31 | 0.86 | YHS | 6 |
OB : oral bioavailability; DL : drug-likeness; CQZ : Cheqianzi; CNX : Chuanniuxi; HB : Huangbo; TFL : Tufuling; YHS : Yanhuosuo; CS : Chishao.
3.4. GO and KEGG Pathway Enrichment Analysis
In order to elucidate the biological mechanisms of TFDC against GA, GO and KEGG pathway enrichment analyses were performed by using clusterProfiler in R. The 25 potential targets of TFDC for treating GA were input into the R, and a total of 1209 GO terms with remarkable significance were obtained, including 1100 biological process (BP) terms, 95 molecular function (MF) terms, and 14 cellular component (CC) terms. The results of GO enrichment analysis mainly include cellular response to lipopolysaccharide, cellular response to molecule of bacterial origin, membrane raft, membrane microdomain, heme binding, tetrapyrrole binding, etc. (Figure 4). Additionally, a total of 93 enriched KEGG pathways were obtained, mainly including interleukin-17 (IL-17) signaling pathway, tumor necrosis factor (TNF) signaling pathway, NOD-like receptor signaling pathway, NF-κB signaling pathway, Toll-like receptor signaling pathway, etc. (Figure 5). What's more, the vital IL-17 signaling pathway and TNF signaling pathway are shown in Figure 6 and Figure 7, respectively.
Figure 4.
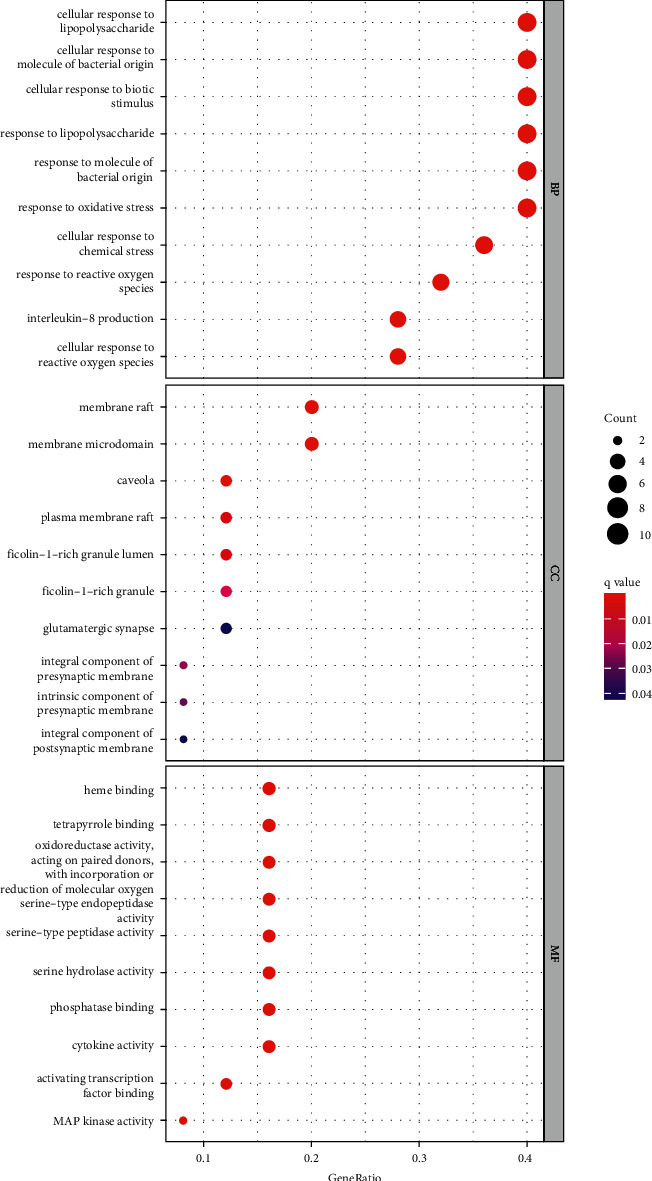
GO enrichment analysis of potential targets of TFDC in GA.
Figure 5.
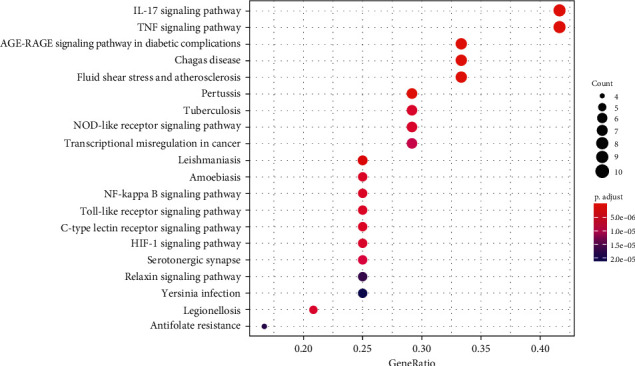
KEGG pathway enrichment analysis of potential targets of TFDC in GA.
Figure 6.
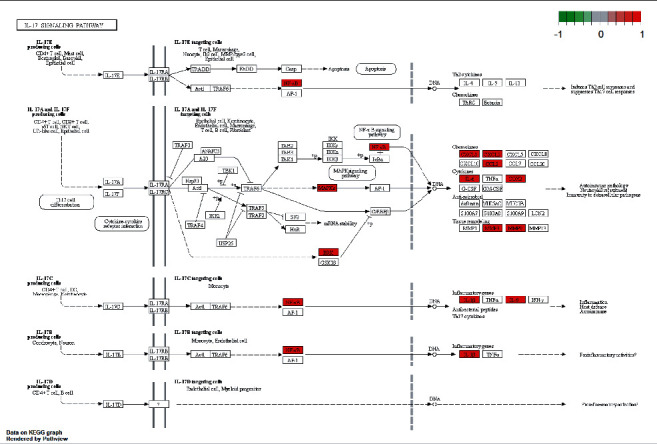
The IL-17 signaling pathway of potential targets of TFDC in GA. Arrows indicate upstream and downstream relationships between targets. The red color represents TFDC-related targets in the network.
Figure 7.
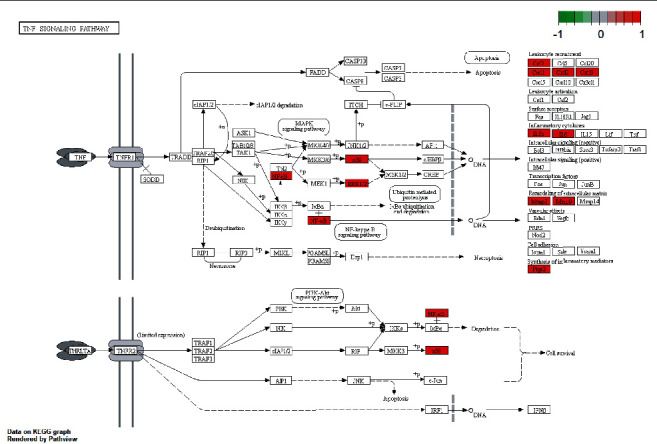
The TNF signaling pathway of potential targets of TFDC in GA. Arrows indicate upstream and downstream relationships between targets. The red color represents TFDC-related targets in the network.
3.5. Effect of TFDC on Cell Viability
The effects of TFDC and MSU on THP-1 cells were respectively evaluated with a CCK-8 assay and are shown in Figure 8(a) and 8(b). According to the result of OD value, TFDC at final concentrations of 25, 50, 75, 100, and 125 μg/mL had little effect on the viabilities of THP-1 cells (P > 0.05). THP-1 cells stimulated with MSU (50 and 100 μg/mL) showed insignificant differences compared with the control group (P > 0.05), but that with 200 μg/mL MSU obviously decreased the viabilities of THP-1 cell (P < 0.01). Based on the above results, 200 μg/mL was the optimum induction dosage in further experiments. For anti-inflammatory activity, MSU-induced THP-1 cells treated with TFDC among 25, 50, 75, 100, and 125 μg/mL exhibited different degrees of protection (Figure 8(c)). 50 μg/mL TFDC had the best protective effect on the viabilities of MSU-induced THP-1 cells, and the concentration of TFDC was defined to 50 μg/mL for qPCR verification.
Figure 8.
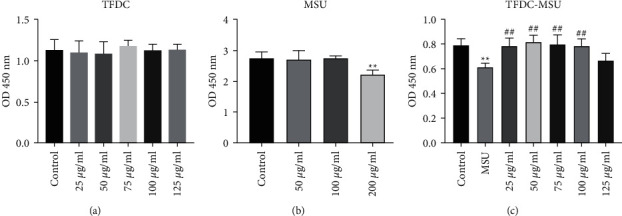
Effects of TFDC on MSU-induced THP-1 cell viability. (a) THP-1 cells were exposed to TFDC at various concentrations for 24 h. (b) THP-1 cells were exposed to MSU at various concentrations for 24 h. (c) Protective effects of TFDC on the viabilities of MSU-induced THP-1 cells. Cell viability was assessed by CCK-8 assay and expressed relative to untreated control cells. ∗∗P < 0.01 versus control group. ##P < 0.01 versus MSU group.
3.6. Effect of TFDC on IL-1β
By observation under microscope, the cell density of the MSU group clearly reduced compared with that of the control group, while the cell densities of the colchicine group and the TFDC group both significantly exceeded that of the MSU group (Figure 9(a)). In order to further validate whether TFDC could inhibit IL-1β in GA against inflammation, mRNA expression of IL-1β was examined by qPCR analysis. The IL-1β mRNA expression in the MSU group was remarkably higher than that in the control group (P < 0.001) (Figure 9(b)). Compared with the MSU group, the IL-1β mRNA expressions in the colchicine group and the TFDC group were separately apparently decreased (P < 0.001) (Figure 9(b)). The results suggested that the anti-inflammation of TFDC on GA was associated with inhibition of IL-1β mRNA expression.
Figure 9.
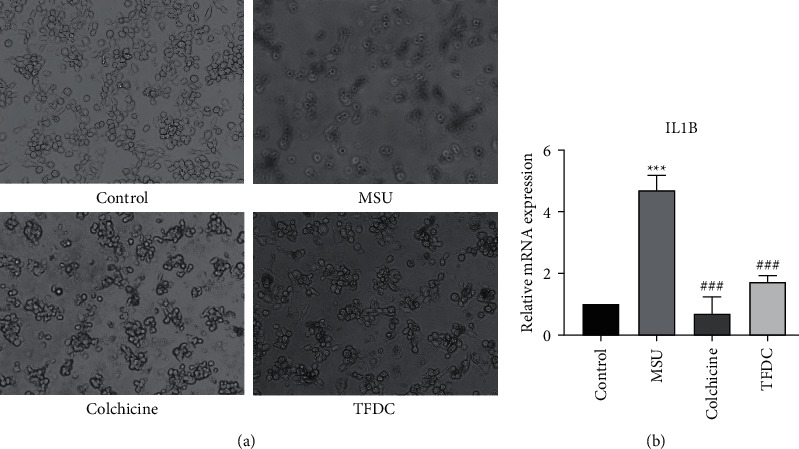
TFDC protects THP-1 cells against MSU-induced inflammation by affecting the expression of IL-1β. (a) Effects of TFDC on MSU-induced THP-1 cells. (b) Statistical analysis of the effect of TFDC on the mRNA expression level of IL-1β. Data are presented as the mean ± SD (n = 3). ∗∗∗P < 0.001 versus control group. ###P < 0.001 versus MSU group.
3.7. Molecular Docking Results of Active Compounds and IL-1β
The top 7 high-degree active compounds from the TFDC-active compound-potential target-GA network bind IL-1β to varying degrees (Table 5). The lower the binding energy, the stronger and more stable the interaction between the active compounds and receptor. The binding energy of rutaecarpine, dihydrosanguinarine, stigmasterol, naringenin, quercetin, dihydrochelerythrine, and betavulgarin increased sequentially, indicating that rutaecarpine had the strongest and most stable binding affinity toward IL-1β. The diagrams of the binding of rutaecarpine, dihydrosanguinarine, stigmasterol, naringenin, quercetin, dihydrochelerythrine, and betavulgarin to IL-1β are shown in Figure 10. As shown in the figure, for example, rutaecarpine formed van der Waals with IL-1β protein structure 5R8Q amino acid residue A chain LEU82, formed π-σ interaction with amino acid residue A chain THR79, formed π-lone pair interaction with amino acid residue A chain VAL132, formed amide-π stacked interaction with amino acid residue A chain GLN81, and formed alkyl and π-alkyl interactions with amino acid residue A chain PRO131 and LEU80. These interactions reduced the energy required for binding, which increased the affinities between the active compounds and IL-1β protein structure 5R8Q to make them easier to bind.
Table 5.
Molecular docking results of 7 active compounds in TFDC binding with IL-1β.
| Targets | PDB ID | Molecule ID | Molecule name | Binding energy (kcal/mol) |
|---|---|---|---|---|
| IL-1β (IL1B) | 5R8Q | MOL002662 | Rutaecarpine | −8.5 |
| MOL001463 | Dihydrosanguinarine | −8.2 | ||
| MOL000449 | Stigmasterol | −7.6 | ||
| MOL004328 | Naringenin | −7.4 | ||
| MOL000098 | Quercetin | −7.2 | ||
| MOL001461 | Dihydrochelerythrine | −7.0 | ||
| MOL012286 | Betavulgarin | −6.8 |
Figure 10.
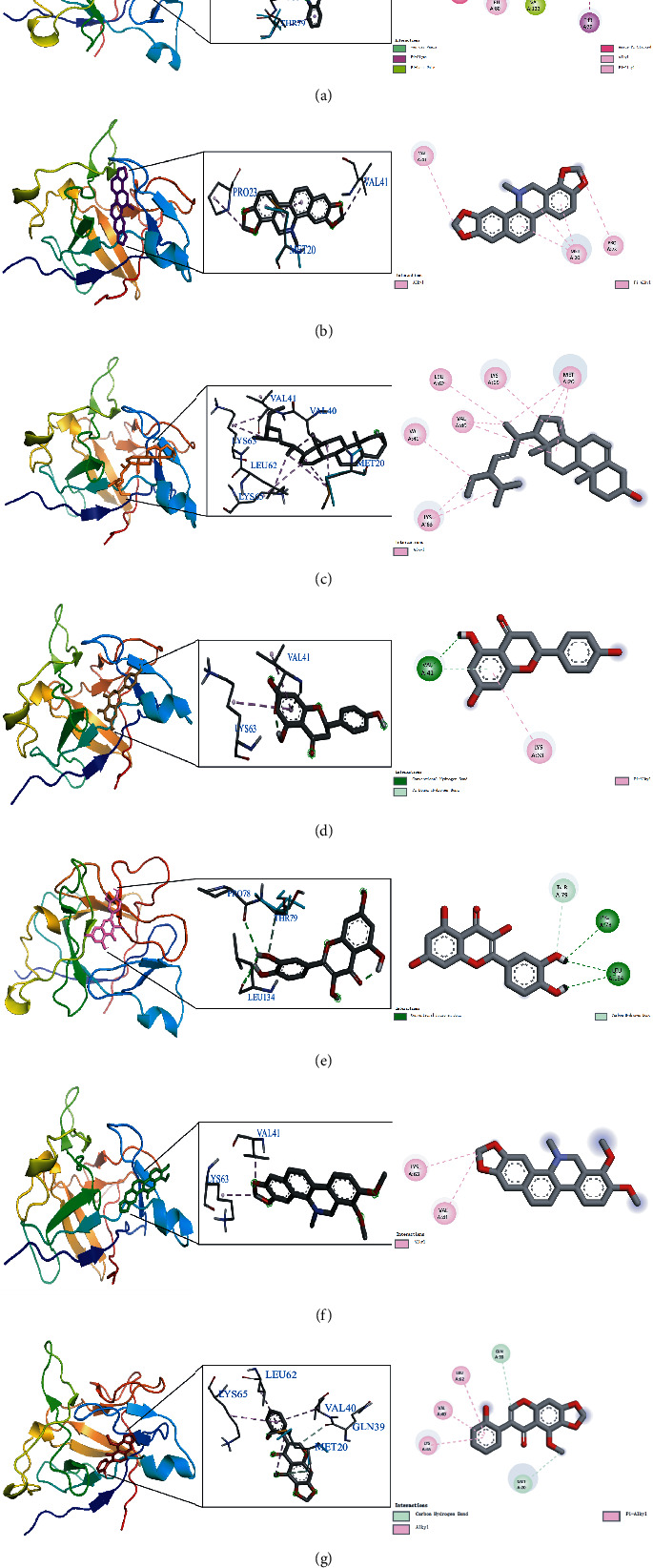
The diagram of the binding of rutaecarpine (a), dihydrosanguinarine (b), stigmasterol (c), naringenin (d), quercetin (e), dihydrochelerythrine (f), and betavulgarin (g) with IL-1β.
4. Discussion
GA, as a chronic inflammatory disease caused by the deposition of urate in the joints, is mainly characterized by redness, swelling, heat, and pain in the affected joints [36]. Repeated attacks can form tophi, leading to bone erosion, destroying the joints, and causing movement obstacles or severe disability, which have a serious negative impact on the quality of life of patients and bring huge psychological and economic burden to patients [37, 38]. It has been clear that the pathology of GA involves the production and release of the inflammatory factor IL-1β, and the progression of GA can be mediated by IL-1β [39, 40]. In this study, the PPI network revealed that the potential targets of TFDC for GA are interacting rather than isolated, resulting in a synergistic therapeutic effect between the targets. Moreover, the network also showed that IL-1β was the most associated target with other targets, indicating that IL-1β was the most critical target in TFDC treating GA. Based on this, the qPCR experiment also confirmed that TFDC could obviously inhibit IL-1β, playing an important role in the occurrence and development of GA, which provided strong experimental evidence support for confirming that TFDC could indeed treat GA.
In addition, in this study, the analysis results of the TFDC-active compound-potential target-GA network also found that these seven active compounds quercetin, stigmasterol, betavulgarin, rutaecarpine, naringenin, dihydrochelerythrine, and dihydrosanguinarine and these 10 targets PTGS2, PTGS1, NOS2, SLC6A3, HTR3A, PPARG, MAPK14, RELA, MMP9, and MMP2 played important roles in treating GA, which are worthy of further research in the future. Studies found that quercetin had a significant joint protective effect in gouty arthritis and, in the mouse model of gouty arthritis induced by monosodium urate crystals (MSU), exerted analgesic and anti-inflammatory effects in a naloxone-sensitive manner [41, 42]. In MSU-induced rat models with GA, quercetin alleviated edema, reduced the histological signs of acute inflammation, inhibited leukocyte recruitment, and reduced the level of chemokines in a dose-dependent manner, which exhibited a strong anti-inflammatory effect [43]. Previous studies pointed out that stigmasterol could reduce the serum uric acid level of hyperuricemia mice by inhibiting the activity of hepatic xanthine oxidase and significantly reduce the claw edema caused by MSU, which could be a promising drug for treating gouty arthritis, hyperuricemia, and inflammation [44]. At present, there are no research reports on treating gouty arthritis with betavulgarin, rutaecarpine, naringenin, dihydrochelerythrine, and dihydrosanguinarine, suggesting that these five active compounds can be used as promising candidate drugs for follow-up experimental research to verify the efficacy of GA. Once experimental studies confirm that these active compounds can be effective in the GA model, they will be essential discoveries and provide more possibilities for drug development of IL-1β inhibitors. Not only that, the verification results of molecular docking showed that rutaecarpine was confirmed to be the active compound that most easily bound to the IL-1β receptor, which is a natural IL-1β inhibitor with great therapeutic potential and potential research value. Moreover, the remaining 6 active compounds were also relatively easy to bind to the IL-1β receptor, and all had good affinities. Many studies showed that inhibition of PTGS2 (i.e., COX-2), NOS2, MAPK14, MMP9, MMP2, and activation of PPARG could improve inflammation in GA models, suggesting that these targets had potential importance in the pathogenesis of GA [45–51].
GO enrichment analysis of potential therapeutic targets revealed that they mainly involved biological functions such as cellular response to lipopolysaccharide, cellular response to molecule of bacterial origin, membrane raft, membrane microdomain, heme binding, and tetrapyrrole binding. Meanwhile, the main signaling pathways enriched for potential therapeutic targets were IL-17 signaling pathway, TNF signaling pathway, NOD-like receptor signaling pathway, NF-κB signaling pathway, Toll-like receptor signaling pathway, etc. These biological functions and signaling pathways were related to the occurrence and development of GA, which might be the mechanism of TFDC in the treatment of GA. In particular, IL-17 signaling pathway and TNF signaling pathway played an extremely vital role in exploring the therapeutic mechanisms obtained by the analysis. IL-17, an important proinflammatory cytokine associated with several autoimmune diseases, significantly increased in the serum of patients with GA in the early stage of acute exacerbations, positively correlated with disease activity, and also correlated with the IL-1β level in serum [52]. In the GA rat model, the ratio of Treg/Th17 in the spleen decreased with the occurrence of joint inflammation, suggesting that the imbalance of Treg/Th17 might be related to the pathogenesis of acute gouty arthritis [53]. The GA mouse model could produce Th17 cells and their related inflammatory cell chemokines such as IL-17, and the use of neutralizing antibodies against IL-17 could reduce joint swelling and leukocyte infiltration into inflammatory sites [54]. TNF-α is a member of the TNF family and participates in systemic inflammation. It was confirmed that IL-1β, TNF-α, IL-6, interleukin-8 (IL-8), and interleukin-4 (IL-4) concentrations in the serum of GA patients increased significantly, which might be related to the pathogenesis of GA [55]. It was found that high expression of Cyr61 protein could induce MSU-stimulated rat synovial cells to produce many inflammatory cytokines such as IL-1β, TNF-α, and IL-6 [56]. And TNF-α could promote the secretion of IL-1β in MSU-induced human neutrophils [57]. Besides, many studies showed that regulating NOD-like receptor signaling pathway, NF-κB signaling pathway, Toll-like receptor signaling pathway, and their representative targets such as NOD-, LRR-, and pyrin domain-containing protein 3 (NLRP3) inflammasome, Toll-like receptor-4 (TLR4), Toll-like receptor-2 (TLR2), IKKα, IκBα, and NF-κB, could effectively improve the joint inflammatory response of the GA model [20, 58–61]. These all indicate that the above signaling pathways are involved in the pathogenesis of GA and play a significant role in the development of the disease, by regulating which the disease progression of GA can be effectively controlled. At the same time, these all suggest that TFDC treating GA is accomplished through multifunction and multichannel synergy, and some functions and pathways are more important, which can be taken as a basis and reference for the direction of further research.
5. Conclusion
In conclusion, the TFDC has obvious advantages and significant efficacy in the treatment of GA, which is consistent with the published results of relevant clinical and experimental studies. In this study, the network pharmacology method was used to investigate the biological functions and signaling pathways of the targets of TFDC's active compounds in the treatment of GA. Simultaneously, the experiment clearly verified that the TFDC could significantly inhibit the key target IL-1β in GA. In addition, rutaecarpine, one of the active compounds in TFDC, had the best binding activity with IL-1β by the molecular docking method, which could be investigated as the most suitable natural IL-1β inhibitor. These findings have provided new clues and ideas for further research on the molecular biological mechanism of TFDC in the treatment of GA and offered a practical and reliable theoretical basis for the clinical treatment of GA by TFDC.
Acknowledgments
The work was financially supported by the National Natural Science Foundation of China (No.82004395), China Postdoctoral Science Foundation (No. 2018M633088), and Shenzhen Futian District Health and Public Welfare Scientific Research Project (Nos. FTWS2020058 and FTWS2020059).
Contributor Information
Ziyun Jiang, Email: jzy19870829@126.com.
Shaochuan Huo, Email: huoshaochuansz@163.com.
Data Availability
The data used and analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Authors' Contributions
Liying Zeng and Zekun Lin collected the data and wrote the original manuscript. Pan Kang performed experiments. Liying Zeng, Zekun Lin, Meng Zhang, Hongyu Tang, and Miao Li completed the literature research. Liying Zeng, Pan Kang, Kun Xu, and Yamei Liu contributed to drawing figures and designing tables. Shaochuan Huo and Ziyun Jiang reviewed the manuscript, contributed to the project design, and supervised the study.
References
- 1.Keller S. F., Mandell B. F. Management and cure of gouty arthritis. Medical Clinics of North America . 2021;105(2):297–310. doi: 10.1016/j.mcna.2020.09.013. [DOI] [PubMed] [Google Scholar]
- 2.Ragab G., Elshahaly M., Bardin T. Gout: an old disease in new perspective–a review. Journal of Advanced Research . 2017;8(5):495–511. doi: 10.1016/j.jare.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desai J., Steiger S., Anders H. J. Molecular pathophysiology of gout. Trends in Molecular Medicine . 2017;23(8):756–768. doi: 10.1016/j.molmed.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Yu K. H., Chen D. Y., Chen J. H., et al. Management of gout and hyperuricemia: multidisciplinary consensus in Taiwan. International Journal of Rheumatic Diseases . 2018;21(4):772–787. doi: 10.1111/1756-185x.13266. [DOI] [PubMed] [Google Scholar]
- 5.Wilson L., Saseen J. J. Gouty arthritis: a review of acute management and prevention. Pharmacotherapy: The Journal of Human Pharmacology and Drug Therapy . 2016;36(8):906–922. doi: 10.1002/phar.1788. [DOI] [PubMed] [Google Scholar]
- 6.Cronstein B. N., Sunkureddi P. Mechanistic aspects of inflammation and clinical management of inflammation in acute gouty arthritis, journal of clinical rheumatology: practical reports on rheumatic & musculoskeletal diseases. Journal of Clinical Rheumatology . 2013;19(1):19–29. doi: 10.1097/rhu.0b013e31827d8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galozzi P., Bindoli S., Doria A., Oliviero F., Sfriso P. Autoinflammatory features in gouty arthritis. Journal of Clinical Medicine . 2021;10(9) doi: 10.3390/jcm10091880.1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinese Pharmacopoeia Commission. Pharmacopoeia of the peoples Republic of China., Beijing: China medical science and technology press. 2020. pp. 1758–1759.
- 9.Yang W., Jiang X., Liu J., et al. Integrated strategy from in vitro, in situ, in vivo to in silico for predicting active constituents and exploring molecular mechanisms of tongfengding capsule for treating gout by inhibiting inflammatory responses. Frontiers in Pharmacology . 2021;12 doi: 10.3389/fphar.2021.759157.759157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hua D., Shaoqing D. Clinical effect analysis of benzbromarone combined with Tongfengding capsule in the treatment of gouty arthritis. China Medicine and Pharmacy . 2020;10(05):75–77. [Google Scholar]
- 11.Levescot A., Chang M. H., Schnell J., et al. IL-1β–driven osteoclastogenic Tregs accelerate bone erosion in arthritis. Journal of Clinical Investigation . 2021;131(18) doi: 10.1172/jci141008.141008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vijayaraj S. L., Feltham R., Rashidi M., et al. The ubiquitylation of IL-1β limits its cleavage by caspase-1 and targets it for proteasomal degradation. Nature Communications . 2021;12(1):p. 2713. doi: 10.1038/s41467-021-22979-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbate A., Toldo S., Marchetti C., Kron J., Van Tassell B. W., Dinarello C. A. Interleukin-1 and the inflammasome as therapeutic targets in cardiovascular disease. Circulation Research . 2020;126(9):260–1280. doi: 10.1161/circresaha.120.315937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L., Wang D., Liu M., et al. The development from hyperuricemia to gout: key mechanisms and natural products for treatment. Acupuncture and Herbal Medicine . 2022;2(1):25–32. doi: 10.1097/hm9.0000000000000016. [DOI] [Google Scholar]
- 15.Schlesinger N., Thiele R. G. The pathogenesis of bone erosions in gouty arthritis. Annals of the Rheumatic Diseases . 2010;69(11):1907–1912. doi: 10.1136/ard.2010.128454. [DOI] [PubMed] [Google Scholar]
- 16.Schlesinger N. Difficult-to-treat gouty arthritis: a disease warranting better management. Drugs . 2011;71(11):1413–1439. doi: 10.2165/11592290-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 17.Renaudin F., Orliaguet L., Castelli F., et al. Gout and pseudo-gout-related crystals promote GLUT1-mediated glycolysis that governs NLRP3 and interleukin-1β activation on macrophages. Annals of the Rheumatic Diseases . 2020;79(11):1506–1514. doi: 10.1136/annrheumdis-2020-217342. [DOI] [PubMed] [Google Scholar]
- 18.Ea H. K. Mechanisms of gout inflammation. Presse Medicale . 2011;40(9):36–843. doi: 10.1016/j.lpm.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X., Ding H., Hu G., Lu Y. Simiaosan alleviates the symptoms of gouty arthritis via the NALP3/IL-1β pathway. Molecular Medicine Reports . 2021;23(3):p. 223. doi: 10.3892/mmr.2021.11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X., Xu D., Sun D., Zhang T., He X., Xiao D. Curcumin ameliorates monosodium urate-induced gouty arthritis through Nod-like receptor 3 inflammasome mediation via inhibiting nuclear factor-kappa B signaling. Journal of Cellular Biochemistry . 2019;120(4):6718–6728. doi: 10.1002/jcb.27969. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Z., Chen B., Chen S., et al. Applications of network pharmacology in traditional Chinese medicine research. Evidence-based Complementary and Alternative Medicine . 2020;2020:1–7. doi: 10.1155/2020/1646905.1646905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo T. t., Lu Y., Yan S. k., Xiao X., Rong X. l., Guo J. Network pharmacology in research of Chinese medicine formula: methodology, application and prospective. Chinese Journal of Integrative Medicine . 2020;26(1):72–80. doi: 10.1007/s11655-019-3064-0. [DOI] [PubMed] [Google Scholar]
- 23.Yuan H., Ma Q., et al. Liu G., Zhao X., Li W., Piao G. How can synergism of traditional medicines benefit from network pharmacology? Molecules . 2017;22(7) doi: 10.3390/molecules22071135.1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinzi L., Rastelli G. Molecular docking: shifting paradigms in drug discovery. International Journal of Molecular Sciences . 2019;20(18):p. 4331. doi: 10.3390/ijms20184331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Issa N. T., Badiavas E. V., Schürer S. Research techniques made simple: molecular docking in dermatology. The Journal of Investigative Dermatology . 2019;139(12):2400–2408. doi: 10.1016/j.jid.2019.06.129. [DOI] [PubMed] [Google Scholar]
- 26.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics . 2014;6(1):p. 13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium U. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Research . 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. D1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fishilevich S., Zimmerman S., Kohn A., et al. Genic insights from integrated human proteomics in gene cards. Database . 2016;2016 doi: 10.1093/database/baw030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amberger J. S., Bocchini C. A., Schiettecatte F., Scott A. F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Research . 2015;43(D1):D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szklarczyk D., Gable A. L., Nastou K. C., et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Research . 2021;49(D1):D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otasek D., Morris J. H., Bouças J., Pico A. R., Demchak B. Cytoscape Automation: empowering workflow-based network analysis. Genome Biology . 2019;20(1):p. 185. doi: 10.1186/s13059-019-1758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu G., Wang L. G., Han Y., He Q. Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology . 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao N., Qu J., He S. Exploring the therapeutic composition and mechanism of jiang-suan-chu-Bi recipe on gouty arthritis using an integrated approach based on chemical profile, network pharmacology and experimental support using molecular cell biology. Frontiers in Pharmacology . 2019;10 doi: 10.3389/fphar.2019.01626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S., Thiessen P. A., Bolton E. E., et al. PubChem substance and compound databases. Nucleic Acids Research . 2016;44(D1):D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose Y., Duarte J. M., Lowe R., et al. RCSB protein data bank: architectural advances towards integrated searching and efficient access to macromolecular structure data from the PDB archive. Journal of Molecular Biology . 2021;433(11) doi: 10.1016/j.jmb.2020.11.003.166704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cabău G., Crișan T. O., Kluck V., Popp R. A., Joosten L. A. B. Urate-induced immune programming: consequences for gouty arthritis and hyperuricemia. Immunological Reviews . 2020;294(1):92–105. doi: 10.1111/imr.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klooster P. M. T., Vonkeman H. E., van de Laar M. A. F. J. Disability due to gouty arthritis. Current Opinion in Rheumatology . 2012;24(2):139–144. doi: 10.1097/BOR.0b013e32834ff59d. [DOI] [PubMed] [Google Scholar]
- 38.Bardin T., Voshaar M. A. H. O., van de Laar M. A. F. J. The human and economic burden of difficult-to-treat gouty arthritis. Joint Bone Spine . 2015;82:S2–S8. doi: 10.1016/S1297-319X(15)30002-6. [DOI] [PubMed] [Google Scholar]
- 39.Steiger S., Harper J. L. Mechanisms of spontaneous resolution of acute gouty inflammation. Current Rheumatology Reports . 2014;16(1):p. 392. doi: 10.1007/s11926-013-0392-5. [DOI] [PubMed] [Google Scholar]
- 40.Fattori V., Staurengo-Ferrari L., Zaninelli T. H. IL-33 enhances macrophage release of IL-1β and promotes pain and inflammation in gouty arthritis. Inflammation Research . 2020;69(12):1271–1282. doi: 10.1007/s00011-020-01399-x. [DOI] [PubMed] [Google Scholar]
- 41.Goyal A., Agrawal N. Quercetin: a potential candidate for the treatment of arthritis. Current Molecular Medicine . 2021;10 doi: 10.2174/1566524021666210315125330. [DOI] [PubMed] [Google Scholar]
- 42.Ruiz-Miyazawa K. W., Staurengo-Ferrari L., Mizokami S. S. Quercetin inhibits gout arthritis in mice: induction of an opioid-dependent regulation of inflammasome. Inflammopharmacology . 2017;10 doi: 10.1007/s10787-017-0356-x. [DOI] [PubMed] [Google Scholar]
- 43.Huang J., Zhu M., Tao Y., et al. Therapeutic properties of quercetin on monosodium urate crystal-induced inflammation in rat. Journal of Pharmacy and Pharmacology . 2012;64(8):1119–1127. doi: 10.1111/j.2042-7158.2012.01504.x. [DOI] [PubMed] [Google Scholar]
- 44.Ferraz-Filha Z., Michel Araujo M., Ferrari F., Dutra I., Saude-Guimaraes D. Tabebuia roseoalba: in vivo hypouricemic and anti-inflammatory effects of its ethanolic extract and constituents. Planta Medica . 2016;82(16):1395–1402. doi: 10.1055/s-0042-105878. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y., Duan C., Chen H., et al. Inhibition of COX-2/mPGES-1 and 5-LOX in macrophages by leonurine ameliorates monosodium urate crystal-induced inflammation. Toxicology and Applied Pharmacology . 2018;351:1–11. doi: 10.1016/j.taap.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y., Tang H., Liu X., et al. Frontline Science: reprogramming COX-2, 5-LOX, and CYP4A-mediated arachidonic acid metabolism in macrophages by salidroside alleviates gouty arthritis. Journal of Leukocyte Biology . 2019;105(1):11–24. doi: 10.1002/jlb.3hi0518-193r. [DOI] [PubMed] [Google Scholar]
- 47.Lee H. P., Huang S. Y., Lin Y. Y., et al. Soft coral-derived lemnalol alleviates monosodium urate-induced gouty arthritis in rats by inhibiting leukocyte infiltration and iNOS, COX-2 and c-fos protein expression. Marine Drugs . 2013;11(12):99–113. doi: 10.3390/md11010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye X., Wu J., Zhang D., et al. How aconiti Radix cocta can treat gouty arthritis based on systematic pharmacology and UPLC-QTOF-MS/MS. Frontiers in Pharmacology . 2021;12 doi: 10.3389/fphar.2021.618844.618844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J., Chen G., Guo T. W., Qin W. Y., Jia P. Simiao Wan attenuates monosodium urate crystal-induced arthritis in rats through contributing to macrophage M2 polarization. Journal of Ethnopharmacology . 2021;275 doi: 10.1016/j.jep.2021.114123.114123 [DOI] [PubMed] [Google Scholar]
- 50.Chu S. C., Yang S. F., Lue K. H., Hsieh Y. S., Hsiao T. Y., Lu K. H. The clinical significance of gelatinase B in gouty arthritis of the knee. Clinica Chimica Acta . 2004;339(1-2):77–83. doi: 10.1016/j.cccn.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 51.Wang J., Chen G., Lu L., Zou H. Sirt1 inhibits gouty arthritis via activating PPARγ. Clinical Rheumatology . 2019;38(11):3235–3242. doi: 10.1007/s10067-019-04697-w. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y., Zhao Q., Yin Y., McNutt M. A., Zhang T., Cao Y. Serum levels of IL-17 are elevated in patients with acute gouty arthritis. Biochemical and Biophysical Research Communications . 2018;497(3):897–902. doi: 10.1016/j.bbrc.2018.02.166. [DOI] [PubMed] [Google Scholar]
- 53.Dai X. J., Tao J. H., Fang X., et al. Changes of Treg/Th17 ratio in spleen of acute gouty arthritis rat induced by MSU crystals. Inflammation . 2018;41(5):1955–1964. doi: 10.1007/s10753-018-0839-y. pp.1955-64. [DOI] [PubMed] [Google Scholar]
- 54.Raucci F., Iqbal A. J., Saviano A., et al. IL-17A neutralizing antibody regulates monosodium urate crystal-induced gouty inflammation. Pharmacological Research . 2019;147 doi: 10.1016/j.phrs.2019.104351.104351 [DOI] [PubMed] [Google Scholar]
- 55.Zhang T., Wang G., Zheng J., Li S., Xu J. Profile of serum cytokine concentrations in patients with gouty arthritis. Journal of International Medical Research . 2021;49(11) doi: 10.1177/03000605211055618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou M., Ze K., Hua L., et al. Cyr61 promotes inflammation of a gouty arthritis model in rats. Mediators of Inflammation . 2020;2020:1–13. doi: 10.1155/2020/8298615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yokose K., Sato S., Asano T., et al. TNF-α potentiates uric acid-induced interleukin-1β (IL-1β) secretion in human neutrophils. Modern Rheumatology . 2018;28(3):513–517. doi: 10.1080/14397595.2017.1369924. [DOI] [PubMed] [Google Scholar]
- 58.Lin Y., Luo T., Weng A., et al. Gallic acid alleviates gouty arthritis by inhibiting NLRP3 inflammasome activation and pyroptosis through enhancing Nrf2 signaling. Frontiers in Immunology . 2020;11 doi: 10.3389/fimmu.2020.580593.580593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen X., Gao Q., Zhou L., Wang Y., Sun R., Zhang Z. MiR-146a alleviates inflammation of acute gouty arthritis rats through TLR4/MyD88 signal transduction pathway. European Review for Medical and Pharmacological Sciences . 2019;23(21):9230–9237. doi: 10.26355/eurrev_201911_19415. [DOI] [PubMed] [Google Scholar]
- 60.Zhou Q., Lin F. F., Liu S. M., Sui X. F. Influence of the total saponin fraction from Dioscorea nipponica Makino on TLR2/4-IL1R receptor singnal pathway in rats of gouty arthritis. Journal of Ethnopharmacology . 2017;206:274–282. doi: 10.1016/j.jep.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 61.Liu Y., Zhu H., Zhou W., Ye Q. Anti-inflammatory and anti-gouty-arthritic effect of free Ginsenoside Rb1 and nano Ginsenoside Rb1 against MSU induced gouty arthritis in experimental animals. Chemico-Biological Interactions . 2020;332 doi: 10.1016/j.cbi.2020.109285.109285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used and analyzed during the current study are available from the corresponding author on reasonable request.


