Abstract
Agrobacterium tumefaciens uses a type IV secretion system to deliver oncogenic nucleoprotein particles and effector proteins, such as the multifunctional VirE2 protein, to plant cells. In this study, we examined the function of virE1 and its product, the VirE1 secretion chaperone, in mediating VirE2 export. A nonpolar virE1 null mutant accumulated low levels of VirE2, and trans expression of virE1 in this mutant only partially restored VirE2 abundance. Deletion of virE1 did not affect transcription but decreased translation of virE2, as shown by analysis of lacZ transcriptional and translational fusions. VirE2 was stable for a prolonged period, more than 6 h, when it was expressed in cis with virE1, and it exhibited half-lives of about 2 h when it was expressed in trans with virE1 and less than 10 min when it was expressed in the absence of virE1, as shown by pulse-chase experiments. VirE1 stabilized VirE2 via an interaction with a domain near the N terminus of VirE2, as shown by analyses of VirE2 truncation and insertion mutants synthesized in A. tumefaciens. VirE1 self-association was demonstrated by using bacteriophage λ cI repressor fusion and pull-down assays, and evidence of VirE1 homomultimerization in vivo was obtained by native polyacrylamide gel electrophoresis and gel filtration chromatography. A putative VirE1-VirE2 complex with a molecular mass of about 70 to 80 kDa was detected by gel filtration chromatography of extracts from wild-type cells, whereas higher-order VirE2 complexes or aggregates were detected in extracts from a virE1 mutant. Taken together, our findings show that virE1 contributes in several ways to VirE2 export:(i) virE1 regulates efficient virE2 translation in the context of expression from the native PvirE promoter; (ii) the VirE1 secretion chaperone stabilizes VirE2, most probably via an interaction with an N-terminal domain; and (iii) VirE1 forms a VirE1-VirE2 complex with a predicted 2:1 stoichiometry that inhibits assembly of higher-order VirE2 complexes or aggregates.
Agrobacterium tumefaciens transfers at least three macromolecular substrates, oncogenic T-DNA, VirE2 single-stranded DNA-binding protein (SSB), and VirF protein, to plant cells during the course of infection (8). Substrate transfer is mediated by a type IV secretion system assembled from the products of the ∼9.5-kbp virB operon and the virD4 gene (29). This transfer system is a bona fide conjugation apparatus, as suggested by its ancestral relatedness to transfer systems (Tra) of several broad-host-range plasmids and as demonstrated by its ability to transfer the mobilizable IncQ plasmid RSF1010 to bacterial and plant recipient cells. In recent years, other type IV systems have been shown to contribute to the virulence of several mammalian pathogens. The pathogens utilizing type IV systems during infection include Helicobacter pylori (12), Bordetella pertussis (3), Brucella spp. (31, 42), Bartonella henselae (35, 40), and Legionella pneumophila (47). A type IV system of H. pylori exports CagA to the cytosol of mammalian cells (32, 41, 44), whereas a related system exports pertussis toxin across the outer membrane of B. pertussis (3). Other type IV systems are thought to export effector molecules, whose identities are currently unknown, to the mammalian cell cytosol.
Mechanistic studies of the A. tumefaciens T-DNA transfer system and the related conjugal transfer systems of the IncP, IncN, and IncW broad-host-range plasmids have contributed important structure and function information about the type IV systems (8, 29). An area of special interest concerns the mechanism of substrate processing and presentation to the mating channel. Recent work has shown that these systems translocate two general classes of substrates: (i) conjugative plasmids and T-DNA that are translocated as transfer intermediates composed of a single-stranded molecule of DNA covalently bound at the 5′ end by a nicking enzyme (4, 7) and (ii) proteins that are translocated independent of DNA (25, 46, 50). Interestingly, export of DNA and protein substrates via type IV machines appears to require common as well as distinct factors at the early stages of substrate processing. In A. tumefaciens, T-DNA translocation requires the VirD2 nicking enzyme and the VirD1 auxiliary factor for cleavage at the T-DNA borders, the VirD4 protein, which is thought to physically link the T-DNA processing system and the mating channel, and two membrane-associated proteins, VirC1 and VirC2, whose contributions to T-DNA transfer are largely undetermined (23, 53). By contrast, translocation of protein substrates, such as VirE2 SSB and VirF, does not require VirD1, VirD2, or the VirC proteins but is still dependent on VirD4 (7, 9, 46, 53). In addition, translocation of VirE2 requires a small upstream gene encoding the putative VirE1 secretion chaperone.
Definition of the role of VirE1 for VirE2 export has been the subject of recent investigations. VirE1 interacts with VirE2, as shown by the yeast two-hybrid screen (15, 45, 52) and pull-down (15) and immunoprecipitation (52) assays. An interaction domain was localized to the C-terminal half of VirE2, possibly overlapping with regions required for single-stranded DNA binding and for VirE2 self-association (15, 45, 52). An early study suggested that VirE1 stabilizes VirE2 when it is synthesized in Escherichia coli (30), although recent work has led to conflicting results regarding this activity in A. tumefaciens (15, 16). There is also some evidence that VirE1 prevents VirE2 from aggregating in solution (15). VirE1 is a small, acidic protein with an amphipathic α-helix at its C terminus, the proposed site for interaction with VirE2 (45). Of considerable interest, these physical and functional properties of VirE1 are also features of the Syc chaperone or bodyguard proteins required for export of effector proteins via the type III secretion systems of plant and mammalian pathogens (6, 36).
In this study, we further defined the role of virE1 for VirE2 translocation. Our findings support a model in which virE1 acts at the level of translation to ensure efficient virE2 expression and VirE1 protein acts posttranslationally to stabilize VirE2 and prevent the formation of higher-order nonproductive complexes or aggregates in A. tumefaciens.
MATERIALS AND METHODS
Enzymes, chemicals, and reagents.
Restriction endonucleases were purchased from Promega (Madison, Wis.), New England Biolabs (Beverly, Mass.), or GIBCO-BRL (Grand Island, N.Y.). The Klenow fragment of E. coli DNA polymerase I and T4 DNA ligase were obtained from Promega. Isopropyl-β-d-thiogalactopyranoside (IPTG), phenylmethylsulfonyl fluoride (PMSF), carbenicillin, kanamycin, Triton X-100, Coomassie brilliant blue R250, p-nitroblue tetrazolium, and 5-bromo-4-chloro-3-indolylphosphate (BCIP) were purchased from Sigma Chemical Co. (St. Louis, Mo.). Acetosyringone (3′,5′-dimethoxy-4′-hydoxyacetophenone) (AS) was obtained from Aldrich (Milwaukee, Wis.). Alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G and a Bradford protein assay kit were obtained from Bio-Rad Laboratories (Hercules, Calif.).
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study and their relevant characteristics are listed in Table 1. E. coli strains were maintained on Luria-Bertani medium, and A. tumefaciens strains were maintained on MG/L medium or on AB minimal salts medium (52). Media were supplemented with antibiotics as follows: for E. coli, 50 μg of chloramphenicol per ml; and for E. coli and A. tumefaciens, 100 μg of carbenicillin per ml, 100 μg of kanamycin per ml, and 5 μg of tetracycline per ml. For induction of vir genes, A. tumefaciens cells were grown in MG/L medium to an optical density at 600 nm (OD600) of 0.6, harvested by centrifugation, and inoculated at an initial OD600 of 0.25 into induction medium composed of AB minimal salts medium (pH 5.5), 1 mM phosphate, and 200 μM AS (52). Cultures were incubated with shaking at 22°C for 18 h and then harvested for protein analysis.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA relA1 | GIBCO-BRL |
| BL21(DE3) | F−ompT hsdsB(rB− mB−) dcm gal λ (DE3) | Novagen |
| AG1688 | MC1061 F′128 lac1qlacZ::Tn5 | J. Hu |
| A. tumefaciens strains | ||
| A136 | Strain C58 cured of pTiC58 | 49 |
| A348 | A136 containing octopine Ti plasmid pTiA6NC | 21 |
| PC1000 | A348 derivative with the virB operon deleted from pTiA6NC | 19 |
| KE1 | A348 with a Kanr cassette in place of the virE operon | 30 |
| At12516 | A348 with a Spcr cassette in place of the virE2 gene | 20 |
| PC3001 | A348 with a virE1 gene deletion | This study |
| LBA4404 | pTiA6NC lacking T-DNA | 33 |
| Bacteriophage λKH54 | cI− lambda phage | 27 |
| Plasmid vectors | ||
| pUC118 | Crbr, cloning vector | 39 |
| pUC119 | Crbr, cloning vector | 39 |
| pBSIISK+ | Crbr, cloning vector | Stratagene |
| pBSIISK+.NcoI | Crbr, pBSIISK+ with an NcoI site at the translational start site of lacZα | 2 |
| pBSIISK+.NdeI | Crbr, pBSIISK+ containing an NdeI site at the translational start site of lacZα | 2 |
| pET15b | Crbr, cloning vector for construction of N-terminal His-tagged proteins | Novagen |
| pGEX-2T | Crbr, cloning vector for construction of N-terminal GSTa-tagged proteins | Pharmacia |
| pSW172 | Tetr, broad-host-range IncP cloning vector | 5 |
| pXZ151 | Kanr, pSW172 with an nptII gene | 52 |
| pPC914KS+.NdeI | Crbr, pBSIIKS+ with PvirB::virB1, NdeI site at virB1 start site | 2 |
| pPC914KS+.NcoI | Crbr, pBSIIKS+ with PvirB::virB1, NcoI site at virB1 start site | This study |
| pMC1871 | Tetr, lacZ translational fusion vector, promoterless lacZ gene lacking a ribosome-binding site and first eight nonessential codons | Pharmacia Biotech |
| pRS551 | Crbr, lacZYA transcriptional fusion vector | N. Kleckner |
| pBB50 | Kanr Chlr, pBCSK+ with sacBR and nptII genes in polylinker | 2 |
| pZ150 | Crbr, pBR322-based vector | 26 |
| pTAD180 | Crbr, pBR322-based vector expressing PlacUV5::cI′-virB11 | 13 |
| pZ157 | Crbr, pBR322-based vector expressing PlacUV5::cI | 26 |
| pKH101 | Crbr, pBR322-based vector expressing PlacUV5::cI[1–102] | 26 |
| virE expression plasmids | ||
| pXZaE1 | Crbr, pACT2.2 yeast vector with virE1 gene | 52 |
| pXZ27 | Crbr, pBSIIKS+ expressing PvirB::virE2 | 52 |
| pXZ46 | Crbr, pBSIISK+ expressing Plac::virE2 | 52 |
| pXZ761-769 series | Crbr, pBSIISK+.NdeI with PvirB::virE2.i31 insertions | 52 |
| pXZ201-212 series | Crbr, pBSIIKS+ with virE2 truncations expressed from PvirB | 52 |
| pXZ420 | Tetr, pSW172 with PvirE::virE2 | X.-R. Zhou |
| pXZ425 | Tetr, pSW172 with PvirB::virE2 | 52 |
| pXZ426 | Tetr, pSW172 with PvirE::virE1 | X.-R. Zhou |
| pPC725 | Chlr, pBCSK+.NdeI with Plac::virE2, NdeI site at 5′ end of virE2 | 52 |
| pPC730 | Crbr, pET-15b expressing PT7::virE2 by insertion of a 1.6-kb NdeI-XhoI fragment containing virE2 gene from pPC725 | This study |
| pPC731 | Crbr, pBSIIKS+.NdeI with virE1 and virE2 coexpressed from PvirE | 52 |
| pPC732 | Crbr, pBSIIKS+.NdeI with virE2 expressed from PvirE | 52 |
| pSM358 | Kanr Crbr, virE2′-lacZ translational fusion as a result of Tn3HoHo1 insertion, IncP replicon | 43 |
| pSF1 | Kanr, pBB50 with PvirE::virE2 on a 2.5-kb XbaI-XhoI fragment (blunt ended) in SacI site | This study |
| pZD1 | Crbr, pBSIIKS+ with virE2Δ39-84 expressed from PvirB | This study |
| pZZ1 | Crbr, pBR322 plasmid expressing PlacUV5::cI′-virE2, 1.6-kb NdeI-XhoI fragment containing virE2 from pPC725 substituting for virB11 fragment of pTAD180 | This study |
| pZZ2 | Crbr, pBR322 plasmid expressing PlacUV5::cI′-virE1, 0.75-kb NdeI-XhoI fragment containing virE1 from pXZaE1 substituting for virB11 fragment of pTAD180 | This study |
| pZZ3 | Crbr, pET-15b expressing PT7::virE1 by insertion of a 1-kb NdeI-XhoI fragment containing virE1 gene from pXZ426 | This study |
| pZZ4 | Crbr, pBSIIKS+.NcoI expressing Plac::his-virE1 by insertion of a 1.1-kb NcoI-XhoI fragment from pZZ3 | This study |
| pZZ6 | Crbr, pGEX-2T expressing Ptac::gst-virE1 by insertion of 1.0-kb NdeI (blunt-ended)-EcoRI fragment carrying virE1 gene from pXZ426 into SmaI-EcoRI sites of pGEX-2T | This study |
| pZZ9 | Crbr, pBSIIKS+ expressing PvirB::virE1 by substitution of 1.0-kb NdeI-XhoI fragment from pZZ3 for virB1 fragment of pPC914KS+ | This study |
| pZZ12 | Crb+, pBSIIKS+ expressing PvirB::his-virEI by substitution of 1.2-kb NcoI-EcoRI fragment from pZZ3 for virB1 fragment of pPC914KS+.NcoI | This study |
| pZZB13 | Crbr Tetr, pZZ12/pXZ426 cointegrate plasmid expressing PvirB::his-virE1 and PvirE::virE1 | This study |
| pZZ14 | Crbr, pBSIIKS+.NdeI expressing PvirE::virE1 virE2′lacZYA transcriptional fusion | This study |
| pZZ15 | Crbr, pBSIIKS+.NdeI expressing PvirE::virE2′ lacZYA transcriptional fusion | This study |
| pZZ16 | Crbr, pBSIIKS+.NdeI expressing PvirE::virE1 virE2′-′lacZ translationalfusion | This study |
| pZZ17 | Crbr, pBSIIKS+.NdeI expressing PvirE::virE2′-′lacZ translational fusion | This study |
| pZZB45 | Crbr Tetr, pZZ4/pXZ425 cointegrate plasmid expressing Plac::his-virE1 and PvirB::virE2 | This study |
| pZZB46 | Crbr Tetr, pZZ12/pXZ425 cointegrate plasmid expressing PvirB::his-virE1 and PvirB::virE2 | This study |
| pZZB65 | Crbr Tetr, pZZ12/pXZ420 cointegrate plasmid expressing PvirB::his-virE1 and PvirE::virE2 | This study |
| pZZB66 | Crbr Tetr, pZZ9/pXZ420 cointegrate plasmid expressing PvirB::virE1 and PvirE::virE2 | This study |
| pZZB67 | Crbr Tetr, pXZ426/pXZ27 cointegrate plasmid expressing PvirE::virE1 and PvirB::virE2 | This study |
| pZZB68 | Crbr Tetr, pXZ426/pPC732 cointegrate plasmid expressing PvirE::virE1 and PvirE::virE2 | This study |
GST, glutathione-S-transferase.
Recombinant DNA techniques.
DNA manipulations and DNA electrophoresis were performed as described by Sambrook et al. (39). A. tumefaciens cells were transformed by electroporation, and plasmids were recovered for physical characterization as previously described (51). DNA sequencing was carried out at the DNA Core Facility of the Department of Microbiology and Molecular Genetics, University of Texas-Houston Medical School, Houston, Tex., with an ABI 373A DNA sequencer (Perkin-Elmer, Applied Biosystems Division) by using Taq polymerase in a thermal cycling reaction. PCR was performed with a Perkin-Elmer Cetus DNA thermocycler by using Pwo DNA polymerase from Boehringer (Mannheim, Germany). Oligonucleotides were purchased from Sigma-Genosys (Woodlands, Tex.).
Construction of virE1 deletion strain PC3001.
To delete virE1 from pTiA6NC, a 2.5-kb XbaI-XhoI fragment (made blunt ended with the Klenow fragment) from pPC732 (carries PvirE::virE2) was introduced at the ScaI site of sacBR suicide plasmid pBB50. The new plasmid, pSF1, was recombined onto pTiA6NC by a single crossover event, and sucrose counterselection was used to enrich for double-crossover recombinants lacking the pSF1 plasmid. The virE1 deletion in strain PC3001 was confirmed by sequencing a PCR product resulting from amplification across the putative ΔvirE1 junction site with primers complementary to the sequences in the virE promoter (5′-TCAAGACCCGAGTATGGATG-3′) and the virE2 gene (5′-TGCCAGAAAGATCCATCGTC-3′).
Construction of virE expression plasmids.
Plasmids expressing virE derivatives were constructed as follows. Both lacZ transcriptional and translational fusions to virE2 were made by inserting cassettes at the StuI site at bp 36 of virE2. For the lacZ transcriptional fusions, StuI-XhoI fragments of pPC731 and pPC732 were replaced by a 6.3-kbp SmaI-SalI fragment containing the lacZYA cassette from pRS551 to generate pZZ14 (PvirE::virE1 virE2′ lacZYA) and pZZ15 (PvirE::virE2′ lacZYA). For lacZ translational fusions, StuI-HindIII fragments of pPC731 and pPC732 were replaced by the SmaI-HindIII fragments containing the ′lacZ cassette from pMC1871 to generate pZZ16 (PvirE::virE1 virE2′-′lacZ) and pZZ17 (PvirE::virE2′-′lacZ). In these constructs, 12 N-terminal residues of VirE2 were fused to β-galactosidase. For production of His-VirE1, the virE1 gene carried on a 1.0-kbp NdeI-XhoI fragment from pXZ426 was introduced into pET15b to make pZZ3. Plasmid pPC914KS+. NcoI was constructed as previously described for pPC914KS+. NdeI (2), except that an NcoI site was introduced at the virB1 start site by oligonucleotide-directed mutagenesis. For expression of his-virE1 from PvirB, a ∼1.1-kbp NcoI-XhoI fragment from pZZ3 was introduced into pPC914KS+. NcoI, replacing virB1. For production of GST-VirE1, the virE1 gene carried on a ∼1.0-kbp NdeI (blunt-ended)-EcoRI restriction fragment from pXZ426 was introduced into SmaI-EcoRI-digested pGEX-2T, resulting in pZZ6. For production of His-VirE2, the virE2 gene carried on a ∼1.6-kbp NdeI-XhoI restriction fragment from pPC725 was introduced into pET15b to make pPC730. Plasmids with ColE1 replication origins that were ligated to IncP broad-host-range plasmid pSW172 or pXZ151 for introduction into A. tumefaciens were designated by using the ColE1 plasmid name plus a B to indicate ligation to the broad-host-range plasmid.
Protein analysis, immunoblotting, and cell fractionation.
Proteins were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) or with a Tricine-SDS-PAGE system as previously described (37). Vir proteins were visualized by SDS-PAGE, transfer of the proteins to nitrocellulose membranes, and development of immunoblots with goat anti-rabbit antibodies conjugated to alkaline phosphatase and histochemical substrates. For enhanced sensitivity, blots were developed with anti-rabbit antibodies conjugated to horseradish peroxidase, and antibody-antigen interactions were visualized by chemiluminescence (Amersham, Arlington Heights, Ill.). Proteins were loaded on a per-cell-equivalent basis to compare VirE protein abundance in different strains. Molecular size markers were obtained from GIBCO-BRL. Fractionation of A. tumefaciens into soluble material (cytoplasm and periplasm) and insoluble material (cytoplasmic and outer membranes) was carried out as previously described (19). Subcellular fractions were applied to SDS-polyacrylamide gels on a per-cell-equivalent basis.
Purification of tagged VirE proteins.
Anti-VirE1 and anti-VirE2 antibodies were generated as follows. IPTG-induced BL21(DE3) cells transformed with pPC730 and pZZ6 were used as the starting materials for purification of His-VirE2 and GST-VirE1, respectively; this was done by performing immobilized Ni2+ metal affinity chromatography (IMAC) and glutathione affinity chromatography, respectively, as recommended by the manufacturers (Ni2+ resin from Novagen and glutathione resin from Pharmacia). Purified His-VirE2 and GST-VirE1 were sent to Cocalico Biologicals, Inc. (Reamstown, Pa.) to produce antibodies in New Zealand White rabbits. His-VirE1 was purified by IMAC with a Co2+ resin (Talon resin; Clontech) for analyses of His-VirE1 complexes.
Measurement of VirE2 turnover in AS-induced A. tumefaciens cells.
A. tumefaciens strains were grown to an OD600 of 0.5 in AB minimal medium (pH 7.0), washed, pelleted, and resuspended at a 1:5 dilution in induction medium. Cells were induced for 6 h (required because there was a long lag in induction of vir gene expression [5]), pelleted, and resuspended in 5 ml of induction medium supplemented with Pro-Mix ([35S]methionine-[35S]cysteine; Pharmacia) at a final concentration of 5 μCi/ml. Cells were pulse-labeled for 30 min at 22°C with shaking, and then excess cold methionine and cysteine were added at final concentrations of 25 mM. A 1-ml culture aliquot was removed (zero time), and aliquots were removed at various times during continued incubation of the cell cultures at 22°C with shaking. Washed cell pellets were resuspended in 600 μl of a solution containing 50 mM Tris (pH 8.0), 50 mM MgCl2, 20% sucrose, 0.1 mg of DNase per ml, 0.1 mg of RNase per ml, and 1 mM PMSF. Cells were broken by sonication and centrifuged at 14,000 × g for 15 min to remove the cell debris. Anti-VirE2 antisera and protein A-Sepharose were used to precipitate VirE2 protein from the clarified culture supernatants (24). Briefly, 5 μl of VirE2 antisera was incubated with culture supernatants at 4°C for 3 h, and then 50 μl of 10% (wt/vol) protein A–Sepharose in Tris buffer (50 mM Tris [pH 8.0], 1 mM PMSF) was added and the slurry was incubated at 4°C for 2 h. Precipitable complexes were recovered by centrifugation at 10,000 × g for 1 min. The pellet was washed with 1 ml of Tris-EDTA buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 5 mM EDTA, 1 mM PMSF) and resuspended in 25 μl of 50 mM Tris (pH 8.0) and 25 μl of SDS loading buffer. The resuspended material was boiled and analyzed for the presence of VirE2 by SDS-PAGE and immunoblotting. VirE2 abundance was estimated by scanning densitometry of autoradiographs.
Gel filtration chromatography and native PAGE.
Total soluble proteins from A. tumefaciens extracts or soluble His-VirE1 purified by Co2+ affinity chromatography was applied at a concentration of 5 mg of protein/ml of 50 mM Tris (pH 8)–50 mM MgCl2 to a gel filtration column (30 by 1.5 cm) prepared with Toyopearl MW55 resin (TosoHaas, Montgomeryville, Pa.) according to the manufacturer's instructions. Material was fractionated with a Pharmacia Gradi-frac chromatography system at a flow rate of 0.6 ml/min, and 1.2-ml fractions were collected. The fractions were analyzed for the presence of VirE1 or VirE2 by SDS-PAGE and immunostaining as described above. The protein molecular size markers used included apoferritin (443 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (BSA) (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12 kDa). Size markers were electrophoresed both together with and independent of the test sample.
For native gel electrophoresis, purified His-VirE1 was electrophoresed through a nondenaturing 15% polyacrylamide gel, and the gel lane containing His-VirE1 was excised and placed horizontally on a Tricine–SDS–16.5% polyacrylamide gel for resolution in a second dimension under denaturing conditions. The molecular sizes of the His-VirE1 complexes were estimated by comparison of His-VirE1 migration with migration of the native protein markers BSA (66 kDa), ovalbumin (45 kDa), trypsin inhibitor (20 kDa), and cytochrome c (12 kDa); these markers were subjected to the same two-dimensional electrophoresis procedure used for resolution of His-VirE1 complexes. VirE1 was identified by Coomassie blue staining and development of immunoblots with anti-VirE1 antisera, and the molecular size markers were identified by Coomassie blue staining.
Enzyme assays.
A. tumefaciens KE1 and At12516 cells expressing the lacZ gene fusions were grown to the mid-log phase in MG/L broth and induced for expression of the vir genes as described above. β-Galactosidase activities (in Miller units) were determined as previously described (13). At least three independent assays were performed in triplicate for each strain.
Phage immunity assay.
Transformants of E. coli AG1688 expressing wild-type or chimeric repressors were each cross-streaked against >106 PFU of lytic phage λKH54 spread on one-half of the surface of a Luria-Bertani agar plate. After incubation overnight at 37°C, the streaks were examined visually for bacterial growth in the presence of the phage. Immunity was defined by the ability of strains to grow in the presence of the phage (26, 37).
Virulence assays.
Strains of A. tumefaciens were tested for virulence on uniformly wounded Kalanchoe daigremontiana leaves (51). The controls for the tumorigenesis assays included coinoculation of the same leaf with wild-type A348 and strain A136 lacking plasmid Ti. Virulence was scored in terms of tumor size and time course of tumor appearance. Assays were repeated at least four times for each strain on separate leaves. Tumors were photographed 4 to 5 weeks after inoculation.
RESULTS
Effects of cis and trans expression of virE1 on virE2 expression.
In a previous study, we determined that expression of virE2 from a PvirE promoter without cis coexpression of the upstream virE1 gene resulted in accumulation of low levels of VirE2 protein (52). To further define the specific contributions of virE1 to virE2 expression, we first compared steady-state levels of VirE2 in an A. tumefaciens ΔvirE1 mutant expressing various combinations of virE1 and virE2 from Ti or IncP plasmids (Fig. 1A).
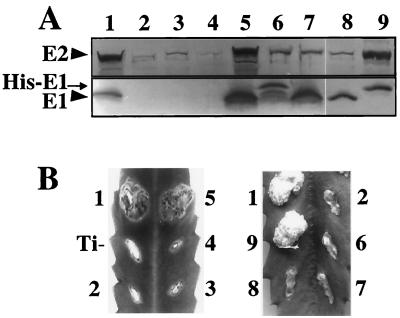
FIG. 1.
Effect of virE1 on VirE2 protein accumulation. (A) VirE protein abundance as determined by immunoblot analysis. Lane 1, wild-type strain A348; lane 2, ΔvirE1 mutant PC3001; lane 3, PC3001(pPCB732) merodiploid with PvirE::virE2 carried on Ti and IncP replicons; lane 4, PC3001(pXZB46) merodiploid with PvirE::virE2 (Ti) and Plac::virE2 (IncP); lane 5, PC3001(pPCB731) merodiploid with PvirE::virE2 (Ti) and PvirE::virE1 virE2 (IncP); lane 6, PC3001(pZZB12) with PvirE::virE2 (Ti) and PvirB::his-virE1 (IncP); lane 7, PC3001 (pXZ426) with PvirE::virE2 (Ti) and PvirE::virE1 (IncP); lane 8, PC3001(pZZB68) with PvirE::virE2 (Ti) and both PvirE::virE1 and PvirE::virE2 (IncP); lane 9, PC3001(pZZB46) with PvirE::virE2 (Ti) and both PvirB::his-virE1 and PvirB::virE2 (IncP). (B) Virulence of virE mutants as determined by inoculation of wounded K. daigremontiana leaves. The numbers refer to the strains in the corresponding lanes in panel A. Ti, leaf wound site inoculated with negative control strain A136.
ΔvirE1 strain PC3001 did not synthesize VirE1 and accumulated very low levels of VirE2 (Fig. 1A, lane 2). PC3001 also was avirulent when it was inoculated onto wounded K. daigremontiana leaves and failed to export VirE2, as determined by a coinfection (34) assay (Fig. 1B, #2, and data not shown). We were not able to significantly increase the level of VirE2 or restore virulence of PC3001 cells by (i) expression of virE2 from an IncP replicon in the absence of virE1 (pPCB732 [PvirE::virE2] or pXZB46 [Plac::virE2]) (Fig. 1A; Fig. 1B, #3 and #4), (ii) expression of virE1 from an IncP plasmid (pZZB12 [PvirB::his-virE1] or pXZ426 [PvirE::virE1]) (Fig. 1B, #6 and #7), or (iii) expression of both virE1 and virE2 from separate promoters on an IncP plasmid (pZZB68 [PvirE::virE1 and PvirE::virE2]) (Fig. 1B, #8). By contrast, VirE2 abundance and virulence of PC3001 were restored to wild-type levels by coexpression of both virE1 and virE2 from the same promoter on an IncP replicon (pPCB731 [PvirE::virE1 virE2]) (Fig. 1A; Fig. 1B, #5). Together, these results indicate that virE1 must be coexpressed in cis with virE2 for synthesis of functional VirE2 when gene expression is controlled by the native PvirE promoter.
In addition, as demonstrated previously (52), we identified one case in which virE2 expression from a heterologous promoter in the absence of virE1 cis coexpression yielded functional VirE2. As shown for PC3001(pZZB46 [PvirB::his-virE1 PvirB::virE2]), expression of virE2 and his-virE1 from independent PvirB promoters resulted in high levels of VirE2 and restoration of virulence (Fig. 1A; Fig. 1B, #9). Note that PC3001 expressing PvirB::virE2 without cis or trans expression of virE1 (or his-virE1) was avirulent (data not shown). Therefore, the finding that virulence of PC3001 was restored by introduction of plasmid pZZB46 established that His-VirE1 was fully functional. The basis for our finding that virE2 expression from PvirB yields functional VirE2 independent of virE1 cis coexpression is explored further below.
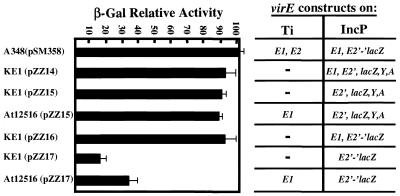
Next, we examined the effects of cis and trans expression of virE1 on transcription and translation of virE2 with the lacZ fusions shown in Fig. 2. The β-galactosidase activities of strains were compared to that of A348(pSM358), which expresses a virE2′-′lacZ translational fusion from an IncP plasmid. ΔvirE operon strain KE1 carrying pZZB14 or pZZB15 had β-galactosidase activity comparable to that of A348(pSM358). Plasmids pZZB14 and pZZB15 express virE2′-′lacZ transcriptional fusions with and without the upstream virE1 gene, respectively. Similarly, ΔvirE2 mutant At12516 carrying pZZB15 exhibited a high level of β-galactosidase activity (Fig. 2). These data show that cis expression of virE1 is not required for efficient virE2 transcription. Furthermore, production of VirE1 does not influence transcription from the native PvirE promoter positively or negatively.
FIG. 2.
β-Galactosidase (β-Gal) activities of the lacZ fusion strains expressed as percentages of the activity of strain A348(pSM358). Experiments were done in triplicate, and standard deviations are indicated by error bars. The virE genes expressed from the PvirE promoter on Ti and IncP plasmids are shown on the right.
Plasmids pZZB16 and pZZB17 express virE2′-′lacZ translational fusions with and without the upstream virE1 gene, respectively. These constructs encode the N-terminal 12 residues of VirE2 fused to β-galactosidase. Strain KE1 carrying pZZB16 exhibited β-galactosidase levels similar to those of the isogenic strain expressing the virE2′ lacZ transcriptional fusion. However, both KE1 and At12516 carrying pZZB17 exhibited appreciably lower β-galactosidase activities; these activities were approximately six- and threefold less, respectively, than the KE1(pZZB16) activity (Fig. 2). Expression of virE1 and virE2′-′lacZ from independent PvirE promoters on an IncP plasmid in KE1 cells yielded β-galactosidase activities comparable to that of strain At12516(pZZB17) (data not shown). Together, these findings demonstrate that the presence of the upstream virE1 coding sequence strongly influences the efficiency of virE2 translation. The virE2 start codon is separated by only 4 bp from the virE1 stop codon; therefore, one explanation for these findings is that translation through virE1 facilitates ribosomal loading at the virE2 Shine-Dalgarno sequence. The slightly elevated (about twofold-higher) β-galactosidase activity of At12516(pZZB17) cells compared to KE1(pZZB17) cells could reflect a possible stabilizing activity of VirE1 protein for the VirE2::β-galactosidase fusion, although this seems unlikely in view of findings described below which indicated that VirE1 stabilizes VirE2 via an interaction with a domain located between residues 39 and 84. Alternatively, VirE1 might slightly enhance VirE2 translation through a direct interaction with mRNA or the ribosome.
VirE1 chaperone stabilizes VirE2 in vivo.
Next, we examined the contribution of the VirE1 chaperone to the stability of VirE2. As shown above, PC3001 expressing virE1 or his-virE1 from an IncP replicon accumulated higher levels of VirE2 than parental PC3001 cells. Similar results were obtained when virE1 and virE2 were expressed from separate Ptac promoters (15). Furthermore, even though cells lacking virE1 accumulated high levels of VirE2 when virE2 was expressed from the PvirB promoter, the VirE2 levels were still appreciably higher in isogenic cells expressing virE1 (see below). A previous study showed that upon expression of virE2 from Ptac, detection of VirE2 in gels was enhanced by treatment of cell extracts with urea prior to electrophoresis (15). In our studies, urea treatment did not appreciably alter the VirE2 protein content upon electrophoresis of extracts from cells expressing virE2 from the PvirE or PvirB promoters (data not shown).
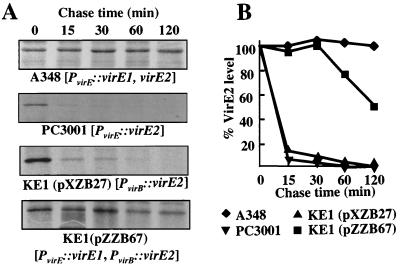
To test directly whether VirE1 influences VirE2 turnover, we monitored the stability of radiolabeled VirE2 in wild-type A348 and virE mutant strains. As shown in Fig. 3, labeled VirE2 was stable in induced wild-type cells. We were able to detect some protein degradation only after 6 h following the pulse-labeling period (data not shown). VirE2 was less stable, with a half-life of about 2 h in KE1(pZZB67 [PvirE::virE1 PvirB::virE2]) cells expressing the two genes from separate promoters. In striking contrast, VirE2 was degraded with half-lives of less than 10 min in two strains lacking virE1, PC3001 and KE1(pXZB27 [PvirB::virE2]). These findings clearly show that the VirE1 chaperone contributes to stabilization of VirE2. Furthermore, the stabilizing effect appears to be optimal when virE1 and virE2 are coexpressed from the same promoter. Finally, the presence of low levels of VirE2 at zero time following pulse-labeling of PC3001 cells is further evidence that virE1 contributes to efficient virE2 translation.
FIG. 3.
Effect of virE1 expression on VirE2 protein turnover in A. tumefaciens. (A) Autoradiographs showing VirE2 precipitated from extracts of radioactively labeled A. tumefaciens cells incubated for the chase times indicated. The strains expressing virE genes are indicated below the autoradiographs. (B) Graphic representation of the results expressed as the percentage of VirE2 detected at zero time for each strain. See text for experimental details.
VirE1 stabilization is mediated by an interaction with the N terminus of VirE2.
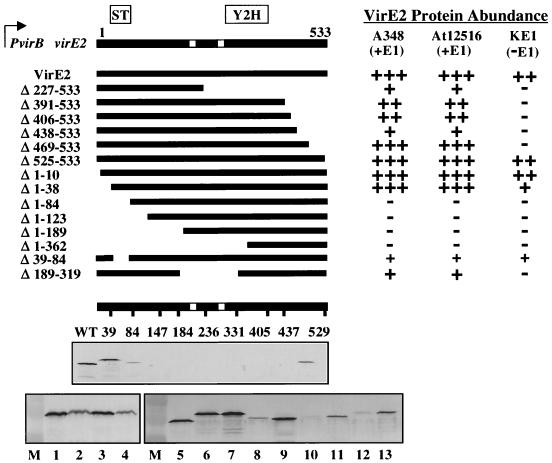
VirE1 interacts with a domain in the C-terminal half of VirE2, as shown by yeast two-hybrid screens (15, 45, 52). It has been proposed that a second interaction domain is localized in the N-terminal half of VirE2, but the boundaries of this domain could not be delineated because the N terminus of VirE2 self-activates transcription at the GAL4 promoter when it is fused to the GAL4 DNA-binding domain (45). To localize the region of VirE2 necessary for VirE1-mediated stabilization, we monitored steady-state accumulation of VirE2 truncation and insertion derivatives in isogenic virE1+ and virE1 strains as an indicator of intrinsic resistance to endogenous proteases. In the following experiments, we expressed the virE2 derivatives from the PvirB promoter. This is because initial studies showed that native VirE2 accumulates at appreciably higher levels when virE2 is expressed from PvirB than when virE2 is expressed from PvirE in the absence of virE1 cis coexpression (Fig. 1A and 4). Moreover, VirE2 protein levels approach wild-type levels in strains expressing virE2 from PvirB and virE1 from a separate promoter (Fig. 4, bottom). Use of the PvirB promoter therefore enabled us to uncouple translational effects of the virE1 gene from stabilizing effects of the VirE1 chaperone.
FIG. 4.
Abundance of VirE2 truncation and i31 peptide insertion derivatives upon introduction of expression constructs into wild-type strain A348 (virE1+ virE2+), At12516 (virE1+), and KE1 (virE). All derivatives were expressed from PvirB. The line at the top represents wild-type virE2 encoding the 533-residue SSB. The two nuclear localization sequences are indicated by white boxes; the VirE1 interaction domain was identified previously by yeast two-hybrid analyses (Y2H), and the second proposed interaction domain was identified by stabilization analyses (ST). Protein contents relative to wild-type VirE2 levels in A348(pXZB27 [PvirB::virE2]) cells are indicated by symbols ranging from +++ (wild-type levels) to − (no detectable protein); the content of the Δ39–84 mutant was detected by chemiluminescence and not by histochemical staining. At the bottom the numbers below the virE2 line representation correspond to the sites of the i31 peptide insertions. The level of each VirE2::i31 mutant in the KE1 host strain is shown in the lane below each number in the immunoblot. WT, pXZB27 [wild-type virE2 expressed from PvirB]. The immunoblots at the bottom show the protein levels in VirE2 and i31 derivatives. Lane 1, A348(pXZB27); lane 2, PC3001(pXZB27); lane 3, At12516(pXZB27); lane 4, KE1(pXZB27); lane 5, A348; lane 6, KE1(pXZB761 [VirE2.39::31]); lane 7, At12516(pXZB761); lane 8, KE1(pXZB762 [VirE2.84::i31]); lane 9, At12516(pXZB762); lane 10, KE1(pXZB768 [VirE2.147::i31]); lane 11, At12516(pXZB768); lane 12, KE1(pXZB763 [VirE2.184::i31]); lane 13, At12516(pXZB763). See reference 52 for levels of additional i31 mutant proteins in virE1+ strains.
Analyses of steady-state levels of VirE2 truncation and insertion derivatives synthesized in virE1 and virE1+ cells resulted in identification of several mutant classes (Fig. 4). One class resembled the native protein by showing virE1-enhanced accumulation. Mutants in this class included truncation derivatives composed of the first 227, 391, 406, 438, or 469 residues, mutants from which 9 N-terminal residues or 10 or 38 C-terminal residues had been deleted, internal deletion mutant Δ189–319, and all but one of a collection of mutants carrying an i31 peptide insertion (Fig. 4) (52). Class II mutants were undetectable even by chemiluminescence in virE1+ and virE1 strains; this class included truncation derivatives lacking 84 or more N-terminal residues. A class III mutant, VirE2Δ39–84, accumulated at comparable low levels detectable only by chemiluminescence in virE1 and virE1+ strains. A class IV mutant, VirE2.39::i31, accumulated at comparable high levels in both virE1+ and virE1 strains.
The phenotypes of the first two mutant classes together indicate that VirE1 exerts its stabilizing effect via an interaction with an N-terminal domain of VirE2. Conversely, the VirE1 interaction with the C-terminal domain identified by the yeast two-hybrid screening method is insufficient for VirE2 stabilization. Residues 11 to 38 appear to be important but are not essential for VirE1-mediated stabilization of VirE2. The phenotype of the class III mutation (virE1-independent accumulation of VirE2Δ39–84 at a low level) suggests that residues 39 to 84 are absolutely essential for VirE1-mediated stabilization of VirE2. The phenotype of the class IV mutation (virE1-independent accumulation of VirE2.39::i31 at a high level) is of considerable interest and is discussed further below.
VirE1 self-associates in vivo.
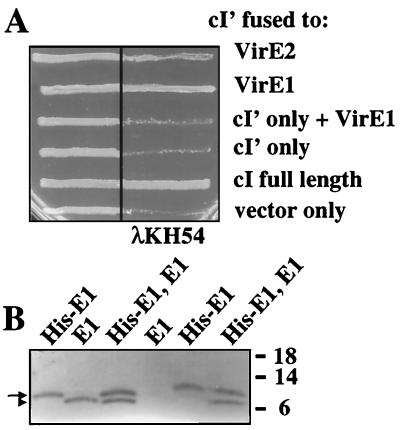
VirE1 self-activates transcription from a yeast GAL4 promoter when it is fused to the GAL4 DNA-binding domain, which prevents the use of the yeast two-hybrid screening method to assay for VirE1 self-association (45). To test for VirE1 self-association, we used λ cI repressor fusion and pull-down assays. cI repressor protein binds as a dimer to operator sites in λ early lytic gene promoters, and each monomer consists of an N-terminal DNA-binding domain and a C-terminal dimerization domain. The N-terminal domain binds DNA efficiently and represses transcription only when it is fused to the C-terminal dimerization domain or a heterologous dimerizing protein or peptide (26, 27).
The binding of full-length cI to target operator sites was demonstrated by growing AG1688(pZ157) cells in the presence of λ phage (Fig. 5A). By contrast, AG1688(pZ150) vector-only cells and AG1688(pKH101) cells expressing the N terminus of cI did not confer immunity to λ superinfection. AG1688(pZZ2) expressing cI′-virE1 grew in the presence of phage, and the level of immunity was approximately that of cells expressing full-length cI repressor protein (Fig. 5A). A fusion of VirE1 to cI′ was essential for operator binding because independent production of VirE1 and cI′ from separate promoters did not confer immunity (Fig. 5A). AG1688(pZZ1) cells expressing the corresponding fusion between the N-terminal DNA-binding domain of cI and full-length VirE2 also conferred phage immunity, which is consistent with previous reports of VirE2 self-association determined by yeast two-hybrid and immunoprecipitation assays (15, 45, 52). However, the cI′::VirE2 fusion protein seems to inefficiently dimerize or forms nonproductive aggregates in E. coli, as judged by the reduced growth of AG166(pZZ1) compared to that of unexposed cells or phage-exposed AG1688(pZZ2) cells (Fig. 5A).
FIG. 5.
VirE1 self-association. (A) cI repressor fusion assay. Cells were grown in the absence (left side) or presence (right side) of λKH54 phage. Dimerization of cI derivatives was assessed on the basis of immunity to phage infection. AG1688 cells expressing the proteins indicated carried the following plasmids: cI′::VirE2, pZZ1; cI′::VirE1, pZZ2; cI′ only and His-VirE1, pKH101 and pZZB4; cI′ only, pKH101; cI full length, pZ157; and vector only, pZ150. (B) His-tag pull-down assay. The first three lanes contained the total soluble proteins loaded onto the Co2+ columns; the other three lanes contained the proteins eluted from the columns. His-E1, KE1(pZZB12); E1, KE1(pXZ426); His-E1, E1, KE1(pZZB13). The arrow and the arrowhead indicate the positions of His-VirE1 and VirE1, respectively. The positions of molecular mass markers (in kilodaltons) are indicated on the right.
For the pull-down assay, VirE1 and His-VirE1 were cosynthesized in the ΔvirE operon mutant KE1. Initially studies established that His-VirE1 is a functional protein (Fig. 1) and that both native VirE1 and His-VirE1 partition with the soluble fraction of A. tumefaciens cells, which is consistent with the predicted cytoplasmic localization of this chaperone (Fig. 5B, lanes 1 to 3). His-VirE1 was retained by Co2+ columns upon fractionation of extracts from KE1(pZZB12) cells expressing his-virE1 or from KE1(pXZZB13) cells coexpressing his-virE1 and virE1 (lanes 5 and 6). In the latter case, native VirE1 also was retained by the column (lane 6). By contrast, VirE1 was not retained by the Co2+ column upon fractionation of extracts from KE1(pXZ426) cells expressing only virE1 (lane 4). These findings show that retention of VirE1 by the Co2+ column was dependent on the presence of His-VirE1, supporting the proposal that VirE1 self-associates in A. tumefaciens.
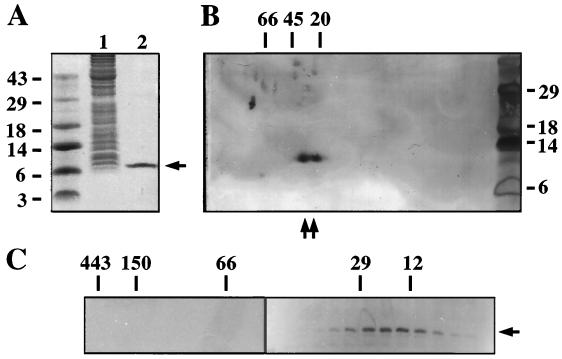
To determine the molecular size(s) of the VirE1 complex(es) in A. tumefaciens, we purified His-VirE1 from KE1(pZZB12) cells (Fig. 6A) for size fractionation by two-dimensional native denaturing gel electrophoresis and by gel filtration chromatography. Several native protein markers were similarly fractionated either independent of or together with His-VirE1. As shown in Fig. 6B, two VirE1-containing species were detected upon native denaturing gel electrophoresis, and the two species migrated close together between the 20-kDa trypsin inhibitor and 45-kDa ovalbumin size standards. The predicted molecular weight of His-VirE1 is 9,500, suggesting that at least one of the species is a homodimer. The larger complex might correspond to a form of the homodimer that migrates aberrantly or to a higher-order homomultimer.
FIG. 6.
Formation of purified His-VirE1. (A) His-VirE1 purification from A. tumefaciens KE1(pZZB12) by IMAC. Lane 1, total soluble proteins loaded onto the Co2+ columns; lane 2, His-VirE1 eluted from the column and visualized by Coomassie blue staining of the gel (arrow). The positions of molecular mass markers (in kilodaltons) are indicated on the left. (B) His-VirE1 complexes analyzed by native denaturing PAGE as described in the text. His-VirE1 complexes were detected by immunoblot analysis (arrows); the positions of molecular mass markers used for electrophoresis in the first dimension (in kilodaltons) are indicated at the top, and the positions of the molecular mass markers used for the second dimension (in kilodaltons) are indicated on the right. (C) His-VirE1 complexes analyzed by gel filtration chromatography. Purified His-VirE1 (1 mg) was fractionated as described in the text, and fractions were analyzed for the presence of His-VirE1 by immunoblot analysis (arrow). The positions of fractions corresponding to elution peaks for molecular mass markers (in kilodaltons) (see Materials and Methods) are indicated at the top.
His-VirE1 eluted from a gel filtration column with a predicted molecular mass of 20 to 30 kDa, as judged by comparisons with elution profiles for the 29-kDa BSA and 12-kDa cytochrome c size markers (Fig. 6C). Very small amounts of His-VirE1 were present in fractions whose sizes corresponded to the sizes expected for the monomer or complexes larger than a homodimer. These findings further suggest that His-VirE1 assembles as a homodimer in A. tumefaciens, at least when it is synthesized in the absence of VirE2 (see below).
VirE1-VirE2 complex formation in A. tumefaciens.
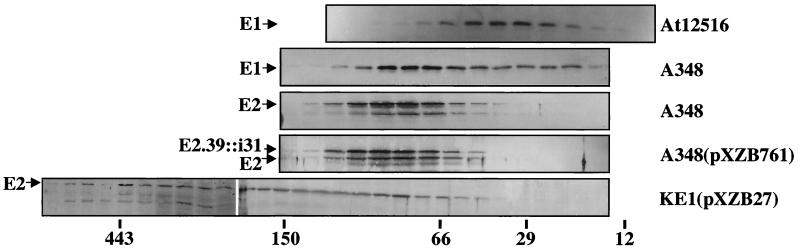
To further characterize VirE1 complex formation in A. tumefaciens, total soluble proteins from wild-type strain A348 and strains expressing either virE1 or virE2 were fractionated by gel filtration chromatography (Fig. 7). VirE1 from strain At12516 (ΔvirE2) eluted in fractions whose sizes corresponded to molecular masses of ∼30 to 40 kDa, which were slightly larger than the masses expected for a homodimer. This species might correspond to an aberrantly migrating homodimeric species or to a higher-order homo- or heteromultimer.
FIG. 7.
VirE1 and VirE2 complex formation in A. tumefaciens as analyzed by gel filtration chromatography. Total soluble proteins (5 mg) from the strains indicated on the right were applied to a gel filtration column and fractionated under identical conditions. The VirE proteins indicated on the left were detected by immunoblot analysis. The positions of elution peaks for molecular mass markers (in kilodaltons) are indicated at the bottom.
The distribution of VirE1 shifted upon cosynthesis with VirE2 in wild-type A348 cells (Fig. 7). VirE1 and VirE2 coeluted as a putative complex at about 70 to 80 kDa. Given the molecular masses of native VirE1 (7.5 kDa) and VirE2 (60 kDa), these findings suggest there is only one molecule of VirE2 in the presumed VirE1-VirE2 complex. The molecular mass of the complex and our evidence for VirE1 self-association further suggest that there are two molecules of VirE1 in the complex, although further studies are needed to confirm this proposed stoichiometry. VirE2 synthesized in the absence of VirE1 eluted in many fractions with sizes ranging from the size of the monomeric protein to more than 443 kDa. Therefore, VirE1-VirE2 complex formation probably is critical for preventing VirE2 aggregation or assembly as higher-order multimers. Finally, the functional VirE2.39::i31 mutant cofractionated with VirE2 upon chromatography of extracts from merodiploid strain A348(pXZB761) (Fig. 7) and from a ΔvirE2 strain expressing the i31 insertion mutant (data not shown). Thus, even though the Tyr.39::i31 mutation confers VirE1-independent stability, the mutant protein still apparently multimerizes with VirE1.
DISCUSSION
VirE2 contributes several functions to the A. tumefaciens infection process upon export from the bacterium (9, 46, 48). In a plant cell, VirE2 multimerizes with itself, the T-DNA–VirD2 transfer intermediate, and, presumably, plant proteins to yield a complex that is competent for intracellular trafficking to the site of T-DNA integration, the plant nuclear genome (9–11, 14, 22, 38). Of considerable further interest, purified VirE2 recently was shown to self-assemble as single-stranded DNA-specific channels in artificial membranes; it has been proposed that these channels facilitate translocation of substrates such as the T-DNA across the plant plasma membrane or through the plant cell (18). In a bacterium, VirE2 must be processed as a secretion-competent molecule. Minimally, this processing pathway must (i) configure VirE2 as a substrate recognizable by the type IV T-DNA transfer machine and (ii) prevent formation of VirE2 aggregates or pore-forming channels and nonproductive interactions with other exported effector molecules. In this study, we demonstrated that virE1 and its product play central roles in the second of these requisite processing activities.
Our findings indicate the virE1 and the VirE1 secretion chaperone contribute both translationally and posttranslationally to VirE2 export. A regulatory role for virE1 at the level of virE2 translation is supported by analyses of VirE2 abundance and stability in various A. tumefaciens mutants and by results of our lacZ fusion studies. The first line of investigation showed that in the context of the native PvirE promoter, virE1 and virE2 must be cis coexpressed for synthesis of abundant levels of VirE2 and for successful VirE2 export. In the absence of virE1 or when virE1 is expressed in trans to virE2, VirE2 accumulates at low levels and is very unstable. Our lacZ fusion studies further showed that virE1 mediates efficient translation of virE2 without affecting transcription. Based on our results, we propose that the virE1 gene plays an important role in production of export-competent VirE2 under native gene expression conditions. The fact that virE1 and virE2 are separated by only four nucleotides is compatible with a translational coupling model in which virE1 translation facilitates ribosomal loading at the virE2 Shine-Dalgarno sequence. Similar translational regulatory mechanisms have also been proposed for some of the chaperone genes associated with type III trafficking systems (28, 36).
A regulatory role for virE1 at the level of translation is appealing because a proposed biological consequence of translational coupling is that it facilitates formation of productive interactions between two or more subunits of a protein complex (17). In support of such an activity for production of export-competent VirE2, our pulse-chase studies showed that cis expression of virE1 and virE2 significantly favors formation of a stable VirE1-VirE2 complex, resulting in an appreciably longer half-life for VirE2 than when each gene is expressed from a separate promoter (Fig. 4). Furthermore, we present evidence that VirE1 exerts its stabilizing effect via an interaction with the N terminus of VirE2. Translational coupling might therefore serve as a mechanism to facilitate rapid association of newly synthesized VirE1 with the N terminus of VirE2, possibly as soon as this region of the protein emerges from the translational machinery. We propose that this association both stabilizes VirE2 and prevents N-terminally mediated protein misfolding and aggregation.
We identified one nonnative expression system in which trans expression of virE1 and virE2 can still result in synthesis of functional (i.e., export-competent) VirE2. In this case, virE2 must be expressed from the PvirB promoter, whereas virE1 can be expressed from any functional promoter. We suggested previously that virE2 expression from PvirB might simply yield a critical threshold level of VirE2 protein required for export (52). However, VirE2 abundance is not always correlated with virulence, and furthermore, we now have shown that VirE2 produced from PvirB is very unstable in the absence of VirE1. We therefore suggest an alternative possibility, that the PvirB promoter provides a cis-acting targeting function, perhaps an mRNA signal, that is normally provided by the 5′ untranslated region of PvirE and/or virE1. This proposed targeting function would serve to localize the virE2 translation machinery to the base of the T-DNA transfer channel to temporally and spatially coordinate virE2 translation and VirE2 translocation. Further comparative studies of the PvirE and PvirB promoter activities are needed to explore this and other possibilities.
In general, we found that the VirE1 chaperone stabilized VirE2 derivatives with an intact N terminus. If VirE1 exerts its stabilizing effect via a direct interaction with an N-terminal domain of VirE2, disruption of that domain should eliminate VirE1-mediated stability. This phenotype was observed with our class III mutation, VirE2Δ39–84. In contrast, the Ty39::i31 insertion mutation conferred VirE1-independent stability. We do not know the molecular basis for this finding, but one explanation is that an i31 insertion at this site interferes with formation of an N-terminal structure that targets the protein for degradation. If this is so, the Tyr39::i31 mutation might correspond to a functional mimic of chaperone binding to the N-terminal interaction domain, with the result that VirE2.39::i31 adopts a stable configuration independent of VirE1 binding. Interestingly, VirE1 is still required for VirE2.39::i31 export (52; data not shown). Furthermore, results of our gel filtration (Fig. 7) and yeast two-hybrid studies (52) suggest that a VirE1-VirE2.39::i31 complex is still assembled. Thus, while a VirE1 interaction is not needed for stabilization of this mutant protein, an interaction probably is required to prevent formation of nonproductive complexes or aggregates or to expose a substrate signal.
Intriguingly, VirE1 displays 21% identity and 35% similarity with the N terminus of VirE2 (Fig. 8). This level of sequence relatedness is also evident for the VirE1 and VirE2 proteins encoded by the pTiC58 plasmid of A. tumefaciens, as well as the pa megaplasmid of Rhizobium etli (data not shown). In addition, VirE1 and this region of VirE2 share a couple of physical features that may have biological importance. The VirE1 chaperones from different Ti or pa plasmid sources are highly acidic, with predicted pIs ranging from 4.7 to 5.2. Similarly, N termini of VirE2 from different Ti or pa plasmids are also highly acidic, with pIs in the range from 4.3 to 5.0, whereas full-length VirE2 proteins have predicted pIs of 6.1 to 6.6. VirE1 and the N terminus of VirE2 also possess hydrophilic N- and C-terminal regions and a hydrophobic central domain. Of further interest is the finding that residues 39 to 84 of VirE2, suggested by our studies to be important for a stabilizing interaction with VirE1, align with the C-terminal half of VirE1. Conversely, the C-terminal half of VirE1, which is predicted to adopt an amphipathic α-helix, was shown previously to mediate interactions with VirE2 (45). Taken together, these observations suggest that the VirE1 interaction with the N terminus of VirE2 might be mediated by conserved structural motifs present on both proteins.
FIG. 8.
Alignment of VirE1 with the N terminus of VirE2. The numbers correspond to positions in the VirE2 sequence. Identical residues are indicated by a black background; similar residues are indicated by a gray background. Alignment was performed with the ClustalX Multalign program.
Our studies showed that VirE1 forms at least two types of complexes. VirE1 can self-associate, at least in the absence of VirE2, and VirE1 also assembles as a VirE1-VirE2 complex with an apparent molecular mass of ∼70 to 80 kDa. The latter finding agrees with results of an early study which identified an ∼80-kDa complex with single-stranded DNA-binding activity in extracts of virE+ cells but not virE cells (22). Based on the predicted sizes of VirE1 (7.5 kDa) and VirE2 (60 kDa), the evidence for VirE1 self-association, and the estimated size of the putative VirE1-VirE2 complex, we suggest that the VirE1-VirE2 complex is composed of one molecule of VirE2 and two molecules of VirE1. Formation of such a VirE1-VirE2 complex clearly seems to be important for preventing assembly of higher-order VirE2 complexes or aggregates (Fig. 7) (15). Whether formation of this complex also is a prerequisite for delivery of the secretion substrate to the transfer channel awaits further study. It is intriguing to speculate that the VirE1 dimer interacts dynamically with VirE2, associating with SSB and then dissociating upon successful presentation of the substrate to the transfer channel. Whether a given VirE1 monomer or dimer can repetitively interact with newly synthesized VirE2 monomers destined for export is unknown; what minimally must occur is that VirE1 disengages from VirE2 prior to export.
Of final note, the multiple roles identified for virE1 and the VirE1 secretion chaperone for VirE2 secretion by this type IV transfer system are analogous to functions ascribed to the secretion chaperones of flagellar and type III secretion pathways (1, 6, 28, 36). Recent work on different members of the type III secretion family has demonstrated that there is considerable variation in the relative importance of mRNA and amino acid targeting signals, the contributions of chaperones for stabilization of effector proteins, and the number and locations of chaperone-binding domains on the effectors (36). Whether secretion chaperones are required for export of all type IV secretion substrates and whether these chaperones (or their cognate genes) provide targeting functions for delivery of the secretion substrates to the cognate type IV translocases are exciting areas for future study.
ACKNOWLEDGMENTS
We thank Xue-Rong Zhou for helpful discussions and construction of truncation and insertion mutations used in this study. We thank Brenda Graf, Simon Jakubowski, and Vitaliya Sagulenko for helpful discussions and Brenda Graf, Johnny Fernandez, and Sharon Fernandez for excellent technical assistance. We thank Jim Hu for providing advice, strains, plasmid constructs, and phage for the cI repressor fusion assays.
Work in our laboratory is supported by NIH grant GM48746.
REFERENCES
- 1.Anderson D M, Schneewind O. Type III machines of Gram-negative pathogens: injecting virulence factors into host cells and more. Curr Opin Microbiol. 1999;2:18–24. doi: 10.1016/s1369-5274(99)80003-4. [DOI] [PubMed] [Google Scholar]
- 2.Berger B R, Christie P J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns D L. Biochemistry of type IV secretion. Curr Opin Microbiol. 1999;2:25–29. doi: 10.1016/s1369-5274(99)80004-6. [DOI] [PubMed] [Google Scholar]
- 4.Byrd D R, Matson S W. Nicking by transesterification: the reaction catalysed by a relaxase. Mol Microbiol. 1997;25:1011–1022. doi: 10.1046/j.1365-2958.1997.5241885.x. [DOI] [PubMed] [Google Scholar]
- 5.Chen C-Y, Winans S C. Controlled expression of the transcriptional activator gene virG in Agrobacterium tumefaciens by using the Escherichia coli lac promoter. J Bacteriol. 1991;173:1139–1144. doi: 10.1128/jb.173.3.1139-1144.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng L W, Schneewind O. Type III machines of Gram-negative bacteria: delivering the goods. Trends Microbiol. 2000;8:214–220. doi: 10.1016/s0966-842x(99)01665-0. [DOI] [PubMed] [Google Scholar]
- 7.Christie P J. The Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie P J. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally-related to conjugation machines. Mol Microbiol. 2001;40:294–305. doi: 10.1046/j.1365-2958.2001.02302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christie P J, Ward J E, Winans S C, Nester E W. The Agrobacterium tumefaciens virE2 gene product is a single-stranded DNA-binding protein that associates with T-DNA. J Bacteriol. 1988;170:2659–2667. doi: 10.1128/jb.170.6.2659-2667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Citovsky V, DeVos G, Zambryski P. Single-stranded DNA binding protein encoded by the virE locus of Agrobacterium tumefaciens. Science. 1988;240:501–504. doi: 10.1126/science.240.4851.501. [DOI] [PubMed] [Google Scholar]
- 11.Citovsky V, Zupan J, Warnick D, Zambryski P. Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science. 1992;256:1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- 12.Covacci A, Telford J L, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 13.Dang T A T, Christie P J. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J Bacteriol. 1997;179:453–462. doi: 10.1128/jb.179.2.453-462.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das A. Agrobacterium tumefaciens virE operon encodes a single-stranded DNA-binding protein. Proc Natl Acad Sci USA. 1988;85:2909–2913. doi: 10.1073/pnas.85.9.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng W, Chen L, Peng W-T, Liang X, Sekiguchi S, Gordon M P, Nester E W. VirE1 is a specific molecular chaperone for the exported single-stranded-DNA-binding protein VirE2 in Agrobacterium. Mol Microbiol. 1999;31:1795–1807. doi: 10.1046/j.1365-2958.1999.01316.x. [DOI] [PubMed] [Google Scholar]
- 16.Dombek P, Ream W. Functional domains of Agrobacterium tumefaciens single-stranded DNA-binding protein VirE2. J Bacteriol. 1997;179:1165–1173. doi: 10.1128/jb.179.4.1165-1173.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Draper D E. Translation initiation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 902–908. [Google Scholar]
- 18.Dumas F, Duckely M, Pelczar P, van Gelder P, Hohn B. An Agrobacterium VirE2 channel for T-DNA transport into plant cells. Proc Natl Acad Sci USA. 2001;98:485–490. doi: 10.1073/pnas.011477898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez D, Dang T A T, Spudich G M, Zhou X-R, Berger B R, Christie P J. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J Bacteriol. 1996;178:3156–3167. doi: 10.1128/jb.178.11.3156-3167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fullner K J, Nester E W. Temperature affects the T-DNA transfer machinery of Agrobacterium tumefaciens. J Bacteriol. 1996;178:1498–1504. doi: 10.1128/jb.178.6.1498-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garfinkel D J, Simpson R B, Ream L W, White F F, Gordon M P, Nester E W. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell. 1981;27:143–153. doi: 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- 22.Gietl C, Koukolikova-Nicola Z, Hohn B. Mobilization of T-DNA from Agrobacterium to plant cells involves a protein that binds single-stranded DNA. Proc Natl Acad Sci USA. 1987;84:9006–9010. doi: 10.1073/pnas.84.24.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamilton C M, Lee H, Li P-L, Cook D M, Piper K R, Beck von Bodman S, Lanka E, Ream W, Farrand S K. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J Bacteriol. 2000;182:1541–1548. doi: 10.1128/jb.182.6.1541-1548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 25.Heinemann J. Genetic evidence of protein transfer during bacterial conjugation. Plasmid. 1999;41:240–247. doi: 10.1006/plas.1999.1392. [DOI] [PubMed] [Google Scholar]
- 26.Hu J, Kornacker M, Hochschild A. Escherichia coli one- and two-hybrid systems for the analysis and identification of protein-protein interactions. Methods. 2000;20:80–94. doi: 10.1006/meth.1999.0908. [DOI] [PubMed] [Google Scholar]
- 27.Hu J. Repressor fusions as a tool to study protein-protein interactions. Structure. 1995;3:431–433. doi: 10.1016/s0969-2126(01)00176-9. [DOI] [PubMed] [Google Scholar]
- 28.Karlinsey J E, Lonner J, Brown K L, Hughes K T. Translation/secretion coupling by type III secretion systems. Cell. 2000;102:487–497. doi: 10.1016/s0092-8674(00)00053-2. [DOI] [PubMed] [Google Scholar]
- 29.Lai E M, Kado C I. The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 2000;8:361–369. doi: 10.1016/s0966-842x(00)01802-3. [DOI] [PubMed] [Google Scholar]
- 30.McBride K E, Knauf V C. Genetic analysis of the virE operon of the Agrobacterium Ti plasmid pTiA6. J Bacteriol. 1988;170:1430–1437. doi: 10.1128/jb.170.4.1430-1437.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli M L, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 32.Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 33.Ooms G, Hooykaas P J J, Van Veen R J M, Van Beelen P, Regensburg-Tunik R, Schilperoort R A. Octopine Ti-plasmid deletion mutants of Agrobacterium tumefaciens with emphasis on the right side of the T-region. Plasmid. 1982;7:15–19. doi: 10.1016/0147-619x(82)90023-3. [DOI] [PubMed] [Google Scholar]
- 34.Otten L, De Greve H, Leemans J, Hain R, Hooykaas P, Schell J. Restoration of virulence of vir region mutants of Agrobacterium tumefaciens strain B6S3 by coinfection with normal and mutant Agrobacterium strains. Mol Gen Genet. 1984;195:159–163. [Google Scholar]
- 35.Padmalayam I, Karem K, Baumstark B, Massung R. The gene encoding the 17-kDa antigen of Bartonella henselae is located within a cluster of genes homologous to the virB virulence operon. DNA Cell Biol. 2000;19:377–382. doi: 10.1089/10445490050043344. [DOI] [PubMed] [Google Scholar]
- 36.Plano G, Day J, Ferracci F. Type III export: new uses for an old pathway. Mol Microbiol. 2001;40:284–293. doi: 10.1046/j.1365-2958.2001.02354.x. [DOI] [PubMed] [Google Scholar]
- 37.Rashkova S, Zhou X-R, Christie P J. Self-assembly of the Agrobacterium tumefaciens VirB11 traffic ATPase. J Bacteriol. 2000;182:4137–4145. doi: 10.1128/jb.182.15.4137-4145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossi L, Hohn B, Tinland B. Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 1996;93:126–130. doi: 10.1073/pnas.93.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Schmiederer M, Anderson B. Cloning, sequencing, and expression of three Bartonella henselae genes homologous to the Agrobacterium tumefaciens VirB region. DNA Cell Biol. 2000;19:141–147. doi: 10.1089/104454900314528. [DOI] [PubMed] [Google Scholar]
- 41.Segal E, Cha J, Lo J, Falkow S, Tompkins L. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc Natl Acad Sci USA. 1999;96:14559–14564. doi: 10.1073/pnas.96.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sieira R, Comerci D J, Sanchez D O, Ugalde R A. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J Bacteriol. 2000;182:4849–4855. doi: 10.1128/jb.182.17.4849-4855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stachel S E, Nester E W. The genetic and transcriptional organization of the vir region of the A6 Ti. EMBO J. 1986;5:1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stein M, Rappuoli R, Covacci A. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci USA. 2000;97:1263–1268. doi: 10.1073/pnas.97.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sundberg C D, Ream W. The Agrobacterium tumefaciens chaperone-like protein, VirE1, interacts with VirE2 at domains required for single-stranded DNA binding and cooperative interaction. J Bacteriol. 1999;181:6850–6855. doi: 10.1128/jb.181.21.6850-6855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vergunst A C, Schrammeijer B, den Dulk-Ras A, de Vlaam C M, Regensburg-Tuink T J, Hooykaas P J. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- 47.Vogel J P, Isberg R R. Cell biology of Legionella pneumophila. Curr Opin Microbiol. 1999;2:30–34. doi: 10.1016/s1369-5274(99)80005-8. [DOI] [PubMed] [Google Scholar]
- 48.Ward D, Zambryski P. The six functions of Agrobacterium VirE2. Proc Natl Acad Sci USA. 2001;98:385–386. doi: 10.1073/pnas.98.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watson B, Currier T C, Gordon M P, Chilton M D, Nester E W. Plasmid required for virulence of Agrobacterium tumefaciens. J Bacteriol. 1975;123:255–264. doi: 10.1128/jb.123.1.255-264.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkins B M, Thomas A T. DNA-independent transport of plasmid primase protein between bacteria by the I1 conjugation system. Mol Microbiol. 2000;38:650–657. doi: 10.1046/j.1365-2958.2000.02164.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhou X-R, Christie P J. Suppression of mutant phenotypes of the Agrobacterium tumefaciens VirB11 ATPase by overproduction of VirB proteins. J Bacteriol. 1997;179:5835–5842. doi: 10.1128/jb.179.18.5835-5842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou X-R, Christie P J. Mutagenesis of Agrobacterium VirE2 single-stranded DNA-binding protein identifies regions required for self-association and interaction with VirE1 and a permissive site for hybrid protein construction. J Bacteriol. 1999;181:4342–4352. doi: 10.1128/jb.181.14.4342-4352.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu J, Oger P M, Schrammeijer B, Hooykaas P J, Farrand S K, Winans S C. The bases of crown gall tumorigenesis. J Bacteriol. 2000;182:3885–3895. doi: 10.1128/jb.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]