Abstract
Background
Although the changed lipid environment of the pilosebaceous unit and the growth of lipophilic Cutibacterium acnes (C. acnes) during puberty has long been considered as the trigger of acne vulgaris, the involvement of the interaction between the epidermal barrier integrity and the skin microbiome in this disease has not been fully elucidated.
Objective
The aim of this study was to analyze the differences in the epidermal barrier and skin microbiota in patients with acne vulgaris and their correlation.
Methods
The skin microbial samples and epidermal barrier data from 74 acne patients and 19 healthy subjects were collected in this cross-sectional study. The microbial diversity was analyzed based on a high-throughput sequencing approach that targets the V3-V4 region of the bacteria 16S ribosomal RNA genes.
Results
Compared with healthy controls, acne patients had significantly increased transepidermal water loss (TEWL), pH levels, sebum, porphyrins, and red areas, and reduced skin microbiome diversity according to the goods coverage diversity index (p = 0.021), Shannon diversity index (p = 0.037), and Simpson diversity index (p = 0.023). Moreover, the diversity gradually decreased with the increase in acne grading. Based on the principal coordinate analysis (PCoA) analysis plot, the skin microbiota of acne patients and healthy controls could be divided into two different sets, which could not be used to separate acne patients with different disease severity. Finally, this study found that both TEWL and sebum were negatively associated with the Shannon and Simpson diversity index. Meanwhile, the taxa Enhydrobacter and Stenotrophomonas were positively associated with TEWL, stratum corneum hydration, respectively.
Conclusion
This study demonstrated that acne vulgaris exists in patients with both damaged epithelial barriers and associated microbiota dysbiosis; the findings will help improve the understanding of the disease and may contribute to the development of better treatment options.
Keywords: epidermal barrier, skin microbiome
Introduction
It is known that there is a mutualistic relationship between physical, chemical, and immunological features of the skin barrier, and microbial populations may create pathogenicity under certain pathological conditions. In fact, dysbiosis of the skin microbiome and skin barrier has been linked to psoriasis, eczema, atopic dermatitis, and other similar disorders of the skin.1
Acne vulgaris is a chronic inflammatory skin disorder affecting 44–95% of adolescents and young adults, which is characterized by specific localization on sebaceous-gland-rich (SGR) skin regions, substantial socioeconomic burden, incidence of anxiety and depression, and negative impact on quality of life.2,3 Recently, dermatologists proposed that the development of acne vulgaris is driven by dynamic changes in the interactions between adolescent SGR skin and the microbial/chemical milieu relative to childhood development.4 During puberty, sebaceous gland proliferation in SGR skin and excessive sebum production, enhanced by increasing concentrations of androgen hormone, promote a lipid-rich environment and consequently, a microbial shift in this area.5 The skin immune system cannot tolerate the change in the microbiota populations, specifically the increasing relative abundance of lipophilic Cutibacterium acnes (C. acnes), leading to a state of inflammation, rather than homeostasis.4,5 In fact, the C. acnes that is predominant in sebaceous sites was previously not considered to be the trigger of acne, as patients with acne do not harbor more C. acnes than individuals without acne.6 The development of sequencing technology revealed the critical role of a tight equilibrium between members of the skin flora and among C. acnes phylotypes in acne onset.7,8
In addition to the lipid environment and lipophilic C. acnes, the interactions among epidermal barrier, disease severity, and skin microbiome as they relate to acne vulgaris have not yet been fully clarified. This study was designed to perform a comprehensive analysis of the correlations among epidermal barrier, skin microbiota, and disease severity in patients with acne vulgaris.
Patients and Methods
Patients
A total of 74 acne patients, mainly with lesions on their cheeks, and 19 healthy control subjects were prospectively enrolled in this study. All participants were students from colleges and universities in Luzhou City, Sichuan province and medical staff at the Southwest Medical University between November 2018 and September 2019. The severity of acne vulgaris was evaluated according to the Global Acne Grading System (GAGS).9 In this study, the severity of mild, moderate, severe acne was defined as S1 (score: 1–18), S2 (score: 19–30), and S3 (score: ≥31). The subjects responded to a written questionnaire, administered by a well-trained study coordinator who went over each question with the subjects. None of the participants had received any treatment, such as isotretinoin, antibiotics, or phototherapy, within the preceding 3 months. Those with other skin diseases, such as acne rosacea, contact dermatitis, eczema, psoriasis, or serious internal disease, were excluded. Meanwhile, the daily facial cleaning habits of each participant were recorded, and those that indicated long-term use of cleaning products with antibacterial activity or oil removal function were also excluded. All participants were asked to avoid bathing or applying any topical agent for 24 h prior to the examination visit. The study was approved by the ethical review board of the hospital affiliated to Southwest Medical University, China, and written informed consent was obtained from all study participants.
Evaluation of Epidermal Barrier Function and Disease Features
The epidermal barrier function calculated as transepidermal water loss (TEWL), skin pH, and the casual sebum level were detected on the cheeks of participants by noninvasive techniques. At sampling sites, the TEWL was measured using the Tewameter® TM 300, the hydration of the stratum corneum (SCH) was tested using the Corneometer® CM 825, and the skin pH and the casual sebum level were detected using the Skin-pH-Meter® pH 905 and the Sebumeter®(Courage+Khazaka electronic GmbH, Cologne, Germany), respectively. Additionally, the porphyrins and the red areas were measured by the VISIA-CR™ imaging system (Canfield Scientific Inc., USA). Notably, the higher the percentage of porphyrins and red areas, the less severity of the disease.10 All measurements were conducted according to the manufacturer’s instructions. The average values were calculated, and the mean of three measurements was used for analysis.
Skin Microbial Sample Collection
Samples were obtained from 4 cm2 areas from each subject on both sides of the cheek, using sterile cotton swabs. The skin area was rubbed 50 times for 30 seconds with three sterile cotton swabs: 25 times in one direction and 25 times perpendicular to that direction. The swabs were placed in a sterile cryopreservation tube that contained 0.9% sodium chloride solution and stored immediately at −80 °C until DNA extraction.
DNA Extraction and PCR Amplification
Microbial DNA was extracted from swab samples using the QIAamp Fast DNA Stool Mini Kit according to the manufacturer’s instruction. The V3-V4 region of the bacteria 16S ribosomal RNA genes was amplified by PCR (95 °C for 3 min; followed by 30 cycles at 98 °C for 20s, 58 °C for 15s, and 72 °C for 20s; and a final extension at 72 °C for 5 min) using primers 341F 5’-CCTACGGGRSGCAGCAG)-3’ and 806R 5’-GGACTACVVGGGTATCTAATC-3’. PCR reactions were performed in a 30 μL mixture containing 15 μL of 2×KAPA Library Amplification ReadyMix, 1 μL of each primer (10 μM), 50 ng of template DNA, and ddH2O.
Illumina HiSeq PE250 Sequencing and Sequence-Based Microbiota Analysis
Amplicons were extracted from 2% agarose gels and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to the manufacturer’s instructions and quantified using Qubit®2.0 (Invitrogen, USA). After the preparation of the library, these tags were sequenced on a HiSeq PE250 platform (Illumina, Inc., CA, USA) for paired-end reads of 250 bp, which were overlapped on their 3 ends for concatenation into original longer tags. 16S tags were restricted between 250 and 500 bp so that the average Phred score of bases was no worse than 30 (Q30), with no more than 1 ambiguous N. Only the tags with frequency more than 1, which tend to be more reliable, were clustered into OTUs, each of which had a representative tag. The OTUs were clustered with 97% similarity using UPARSE (http://drive5.com/uparse/), and chimeric sequences were identified and removed using Usearch (version 7.0). Each representative tag was assigned to a taxa by RDP Classifier (http://rdp.cme.msu.edu/) against the RDP database (http://rdp.cme.msu.edu/) using a confidence threshold of 0.8. Rarefaction curves were generated with QIIME to test the current sequencing depth. The α diversity, calculated by goods_coverage, Shannon, and Simpson diversity indices, which represents the species abundance in a single sample, was analyzed among groups using R3.1.0. The communities were compared based on phylogenetic distances using the weighted UniFrac metric to represent β diversity. Principal coordinate analysis (PCoA) was performed on the resulting matrix of distances between each pair of samples using R3.1.0. Multi-response permutation procedures (MRPP), which test for significant differences between sampling groups, have been described in detail previously.11 Based on the UniFrac phylogenetic distance, significance testing for the clustering of samples in the study was carried out by one-way analysis of similarities (ANOSIM).
Statistical Analysis
Statistical analysis of skin microbial differences among different groups was performed by Wilcoxon rank sum test t and the Kruskal–Wallis rank sum test using R3.1.0. The discrepancy of skin barrier parameters and GAGS scores among different groups were determined by the Kruskal–Wallis test in IBM SPSS Statistics 21. All p-values reported were two-sided, and P < 0.05 was considered statistically significant. We also applied the calculated 95% confidence interval (CI) to control the false discovery rate. Pearson’s correlation was used to calculate the correlation coefficient, which was displayed by heatmap.
Results
Clinical Characteristics of Patients and Healthy Controls
This study included 74 patients with mild, moderate, and severe acne vulgaris and 19 healthy control subjects. The background information about the patients and controls is listed in Table 1. Compared with healthy controls, all acne patients had significantly increased TEWL, pH, sebum, porphyrins, and red areas (P < 0.05).
Table 1.
Background of Patients with Acne Vulgaris and Healthy Controls
| Factors | HC (n =19) | S1 (n =42) | S2 (n = 14) | S3 (n = 18) | P‐value |
|---|---|---|---|---|---|
| BMI(kg/m 2), mean ± SD | 20.02±1.63 | 23.94±16.63 | 20.27±1.75 | 20.87±1.75 | 0.586 |
| Course of the disease, months, mean ± SD | – | 42.02±31.23 | 51.14±25.12 | 46.72±23.36 | 0.370 |
| GAGS scores, mean ± SD | – | 15.36±2.37 | 21.5±4.43 | 25.83±5.03 | 0.000 |
| SCH, mean ± SD | 63.06±16.04 | 53.73±17.25 | 49.05±15.85 | 57.67±15.94 | 0.134 |
| TEWL(g/hm 2), mean ± SD | 15.85±4.13 | 18.16±8.15 | 17.93±6.78 | 22.12±6.65 | 0.025 |
| pH, mean ± SD | 5.40±0.49 | 5.53±0.38 | 5.47±0.26 | 5.83±0.43 | 0.01 |
| Sebum, mean ± SD | 46.26±28.37 | 83.86±52.15 | 84.36±44.40 | 75.15±17.29 | 0.005 |
| Porphyrin (%), mean ± SD | 61.82±24.62 | 40.38±22.68 | 39.74±20.72 | 40.89±18.16 | 0.011 |
| Red area (%), mean ± SD | 67.81±17.76 | 27.51±16.66 | 20.32±18.26 | 9.25±1.36% | 0.000 |
Abbreviations: HC, healthy controls; S1, mild acne; S2, moderate acne; S3, severe acne; BMI, Body Mass Index; SCH, Stratum Corneum Hydration; TEWL, Transepidermal Water Loss; GAGS, Global Acne Grading System.
Decreased Diversity of Skin Microbiome in Patients with Acne Vulgaris
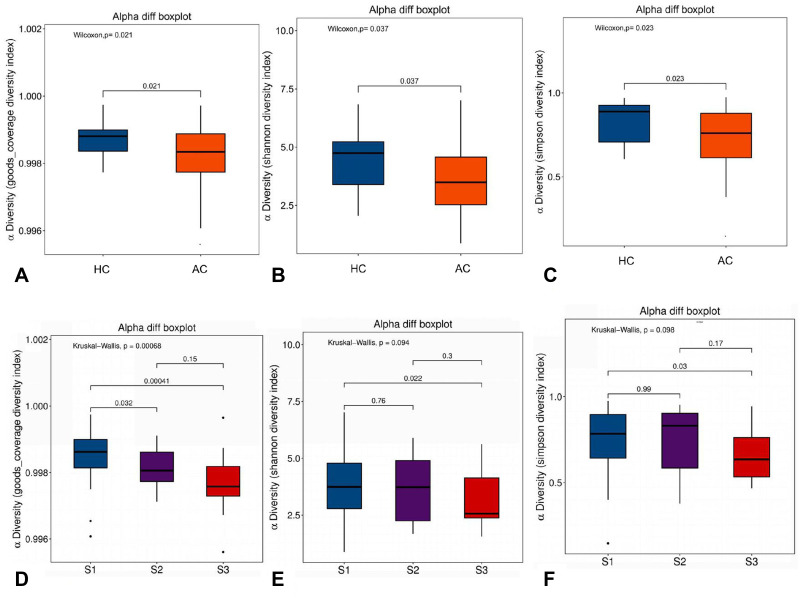
A total of 93 swab samples were obtained from the study population for sequencing. Compared with healthy controls, microbial diversity was significantly reduced in acne patients, as calculated by the goods coverage diversity index (p = 0.021), Shannon diversity index (p = 0.037), and Simpson diversity index (p = 0.023) (Figure 1A–C). When acne patients were divided into subgroups according to disease severity, the diversity of the skin microbiome gradually decreased with the increase in acne grading (Figure 1D–F).
Figure 1.
Comparison of different diversity indices between acne patient samples and healthy control samples, and between samples from patients with different severity of acne. Goods coverage, Shannon, and Simpson diversity index, representing community richness, were calculated for acne (AC), healthy control (HC), and the severity of acne, defined as S1 (score: 1–18), S2 (score: 19–30), and S3 (score: ≥31). (A) Goods coverage diversity index (p=0.021) between AC and HC. (B) Shannon diversity index (p=0.037) between AC and HC. (C) Simpson diversity index (p=0.023) between AC and HC. (D) Goods coverage diversity index (p=0.00068) among acne patients with different severity, Goods coverage diversity index (p=0.032) between S1 and S2, goods coverage diversity index (p=0.00041) between S1 and S3. (E) Shannon diversity index (p=0.094) among acne patients with different severity, Shannon diversity index (p=0.022) between S1 and S3. (F) Simpson diversity index (p=0.098) among acne patients with different severity, Simpson diversity index (p=0.03) between S1 and S3.
Analysis of Bacterial Differences Between Acne Patients and Healthy Control Subjects
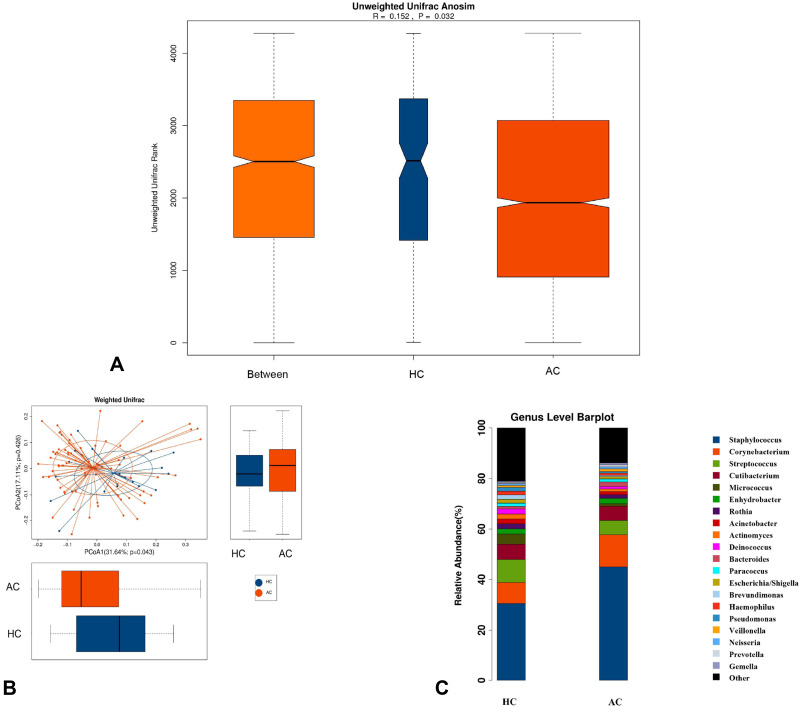
To investigate the different structures of skin microbes between the acne patients and healthy control subjects, we calculated the UniFrac phylogenetic distances of the microbe compositions among participants. ANOSIM analysis indicated the significance of clustering samples into acne patients and healthy controls (R = 0.152, p = 0.032) (Figure 2A). Based on the PCoA plot, it was shown that acne patients and healthy controls could be divided into two different sets by skin microbiota (Figure 2B). MRPP analysis indicated a significant difference between acne and control samples (p = 0.036).
Figure 2.
Comparison of the skin microbiota between acne patients and healthy controls. (A) Difference between and within groups of AC and HC were assessed by ANOSIM analysis. (B) Principal coordinate analysis (PCoA) plot with different relative abundances of OTUs between acne (AC) patients and healthy controls (HC). (C) The top 20 microbes with relative abundance in AC and HC at the genus level.
We conducted a comparative analysis of all the microbes at the genus level and found a total of 61 differential microbes between acne patients and healthy control subjects. The top 20 differential microbes are shown in Table 2. Microbes with a p-value <0.05 were considered to be significantly different. The 95% CIs of the mean abundances of those microbes among the two groups were also calculated to validate the result, which showed only 13 significantly different microbes. For the top 20 abundant genera of all participants, there were 7 different microbes between the patients and healthy control subjects. Compared with healthy controls, the relative abundance of Staphylococcus (P = 0.032) and Corynebacterium (P = 0.044) significantly increased, but the relative abundance of Acinetobacter (P = 0.005), Pseudomonas (P = 0.002), Brevundimonas (P = 0.01), Micrococcus (P = 0.021), and Escherichia/Shigella (P = 0.007) was significantly decreased in acne patients (Figure 2C).
Table 2.
Relative Abundance of the Top 20 Differential Microbes Between Acne Patients and Healthy controls. Only p-values <0.05 are Shown
| Taxonname | Mean(AC) | Mean(HC) | 95% CIs of (AC) | 95% CIs of (CN) | P-value | ||
|---|---|---|---|---|---|---|---|
| Staphylococcus | 4.15×10−1 | 2.89×10−1 | 3.62×10−1 | 4.68×10−1 | 1.96×10−1 | 3.82×10−1 | 0.032 |
| Corynebacterium | 1.18×10−1 | 7.71×10−2 | 8.92×10−2 | 1.47×10−1 | 1.60×10−2 | 1.38×10−1 | 0.044 |
| Micrococcus | 1.02×10−2 | 3.80×10−2 | 7.20×10−3 | 1.32×10−2 | 1.45×10−2 | 6.15×10−2 | 0.021 |
| Acinetobacter | 1.07×10−2 | 1.80×10−2 | 3.95×10−3 | 1.74×10−2 | 6.48×10−3 | 2.95×10−2 | 0.006 |
| Escherichia/Shigella | 8.05×10−3 | 1.42×10−2 | 5.77×10−3 | 1.03×10−2 | 8.31×10−3 | 2.00×10−2 | 0.004 |
| Pseudomonas | 5.91×10−3 | 1.40×10−2 | 4.62×10−3 | 7.21×10−3 | 7.10×10−3 | 2.10×10−2 | 0.002 |
| Brevundimonas | 4.94×10−3 | 1.59×10−2 | 1.76×10−3 | 8.12×10−3 | −3.44×10−4 | 3.21×10−2 | 0.011 |
| Faecalibacterium | 2.93×10−3 | 7.63×10−3 | 1.19×10−4 | 5.74×10−3 | −3.77×10−3 | 1.90×10−2 | 0.027 |
| Lautropia | 2.43×10−3 | 8.03×10−3 | 1.38×10−3 | 3.48×10−3 | 3.96×10−4 | 1.57×10−2 | 0.015 |
| Chryseobacterium | 2.51×10−3 | 4.86×10−3 | 1.09×10−3 | 3.93×10−3 | −9.42×10−4 | 1.07×10−2 | 0.019 |
| Streptophyta | 2.04×10−3 | 5.01×10−3 | 8.42×10−4 | 3.23×10−4 | 1.68×10−3 | 8.34×10−3 | 0.012 |
| Methylobacterium | 1.63×10−3 | 2.20×10−3 | 4.07×10−4 | 2.85×10−3 | −5.56×10−4 | 4.97×10−3 | 0.031 |
| Stenotrophomonas | 7.89×10−4 | 1.95×10−3 | 6.14×10−4 | 9.64×10−4 | 1.00×10−3 | 2.90×10−3 | 0.001 |
| Delftia | 7.67×10−4 | 1.91×10−3 | 5.60×10−4 | 9.74×10−4 | 9.03×10−4 | 2.91×10−3 | 0.001 |
| Massilia | 6.32×10−4 | 1.63×10−3 | 2.39×10−4 | 1.02×10−3 | 7.78×10−4 | 2.48×10−3 | 0.002 |
| Acidovorax | 5.90×10−4 | 1.25×10−3 | 4.27×10−4 | 7.54×10−4 | 7.33×10−4 | 1.76×10−3 | 0.001 |
| Rhizobium | 2.53×10−4 | 1.98×10−3 | 1.92×10−4 | 3.15×10−4 | −6.14×10−4 | 4.57×10−3 | 0.000 |
| Clavibacter | 1.00×10−4 | 2.12×10−3 | 5.68×10−5 | 1.44×10−4 | −1.80×10−5 | 4.25×10−3 | 0.028 |
| Gemmiger | 3.04×10−4 | 9.69×10−4 | 1.26×10−4 | 4.82×10−4 | −1.56×10−4 | 2.09×10−3 | 0.003 |
| Phascolarctobacterium | 3.88×10−4 | 5.37×10−4 | 7.24×10−5 | 7.04×10−4 | 1.76×10−4 | 8.98×10−4 | 0.023 |
Abbreviations: AC, acne; HC, healthy controls; CI, confidence intervals.
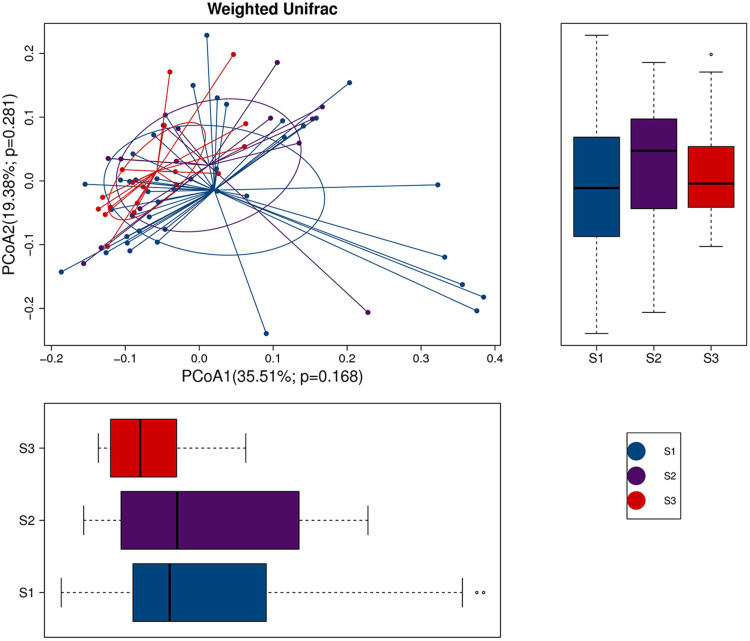
Moreover, we assessed the correlation of skin microbes with the disease severity of acne vulgaris. The PCoA plot did not show a trend in microbe isolation among the mild, moderate, and severe patient groups (Figure 3).
Figure 3.
The PCoA plot with different relative abundances of OTUs among the mild, moderate, and severe acne patient groups. The severity of mild, moderate, severe acne was defined as S1 (score: 1–18), S2 (score: 19–30), and S3 (score: ≥31).
Correlation Between Microbes and Skin Barrier Parameters
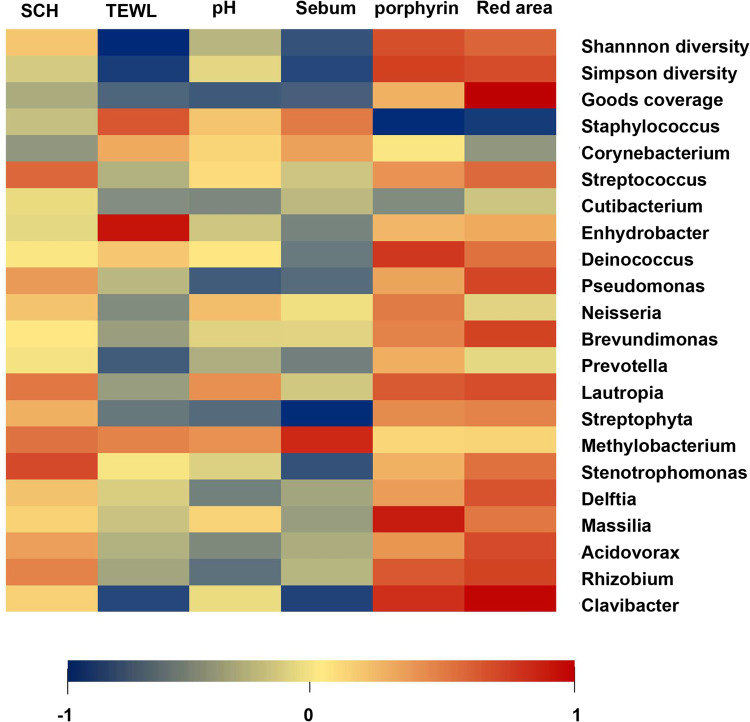
To further analyze the correlation between skin microbes and epidermal barrier function, the microbial diversity index, the top 20 most abundant skin resident microbes, and the top 20 different most abundant microbes between patients and controls were included, which were displayed by heat map (Figure 4). Both TEWL and sebum were negatively associated with the Shannon (R=−0.255, P = 0.014 for TEWL; R=−0.201, P = 0.05 for sebum) and Simpson diversity index (R=−0.229, P = 0.027 for TEWL; R=−0.216, P = 0.038 for sebum). The porphyrins had a negative correlation with Deinococcus (R = 0.265, P < 0.01), Lautropia (R = 0.215, P < 0.039), Massilia (R = 0.036, P < 0.003), Rhizobium (R = 0.217, P < 0.037), and Clavibacter (R = 0.277, P < 0.007), while the red area had a negative correlation with Pseudomonas (R = 0.246, P < 0.017), Brevundimonas (R = 0.25, P < 0.016), Lautropia (R = 0.234, P < 0.024), Delftia (R = 0.225, P < 0.03), Acidovorax (R = 0.238, P < 0.022), Rhizobium (R = 0.248, P < 0.017), and Clavibacter (R = 0.338, P < 0.001). As for sebum, it showed a negative correlation with Clavibacter (R=−0.222, P < 0.033), but a positive association with Methylobacterium (R = 0.288, P < 0.005). For the core microbes, the genus of Staphylococcus showed a positive correlation with porphyrins (R = −0.252, P < 0.015) and red areas (R=−0.232, P < 0.025). The taxa Stenotrophomonas was positively associated with stratum corneum hydration (R = 0.24, P < 0.021), and Enhydrobacter showed a positive correlation with TEWL (R = 0.372, P < 0.001).
Figure 4.
Correlation between bacteria and skin barrier parameters was assessed by Pearson’s correlation test and displayed as a heat map.
Abbreviations: SCH, Stratum Corneum Hydration; TEWL, Transepidermal Water Loss.
Discussion
The skin microflora maintains homoeostasis by cross-talk with the host immune system. Host–microbiome interactions that affect both innate and adaptive immune homeostasis appear to be a central factor in skin diseases.12 As a future perspective, examining the skin microbiome could be seen as a potential new diagnostic and therapeutic target in skin disorders. With the advances in sequencing technology, such as 16S ribosomal RNA (rRNA) gene sequencing, there have been increasing insights into the skin microbiome. Previous literature has confirmed that the bacterial composition differs according to the body site and that C. acnes is the most prevalent and abundant microbe in sebaceous sites, such as on the forehead, nose, and back, in both acne patients and healthy people.13,14 However, our results suggested that the facial microbiota is dominated by Staphylococcus, which may be because the skin microbiome of only the cheek region was detected in this study. In fact, there are differences in the properties of the stratum corneum and also of the biophysical properties of the forehead, cheek, nose, and perioral regions. The T-zone of the face, comprising the forehead and nose, has previously been reported to have higher levels of sebum than other facial regions.15–18 The microbial data of this study reflected the skin flora only of the cheek regions for both healthy people and acne patients.
It was believed that the diversity of the human skin microbiome may have the potential to influence pathogenic microbes and cause skin disease, eg atopic dermatitis.12 Some previous studies reported an increased diversity of cutaneous bacteria in patients with acne, compared to that of healthy controls,8,19 and loss of microbial diversity could indeed lead to chronic inflammatory skin diseases, including acne vulgaris.7,20 In this study, the diversity of skin microbes of acne patients was significantly less than that of healthy controls, which decreased with the disease severity. It was suggested that the reduced microbe diversity on acne patients’ skin may cause the overgrowth of pathogens, which might influence the pathogenesis in acne.
Additionally, the majority of skin microbes populate the stratum corneum, the outermost layer of the skin. The stratum corneum has a thin texture in the facial region, with some unique functional characteristics to provide the hydrated skin surface with relatively poor barrier function. The stratum corneum in the epidermal barrier acts in the homeostatic control of water and assists in the recognition and neutralization of microorganisms.21 Hence, the microbiome composition changes as a result of barrier disruption of the stratum corneum (for example, changes in pH, moisture, and sebum content).22 The direct evidence has been provided that quantitative levels of sebum and hydration are significantly associated with the distribution of the microbiome in facial skin.23 This study revealed that acne patients have a significantly increased skin pH and sebum level, compared with healthy controls, which was consistent with previous studies.24–26 To further explain how the modification of microorganisms accompanies the damage of the epidermal barrier, the association between them was analyzed, which can help in understanding more about the disease and finding better treatment options.
There were a few studies reporting the association of a certain bacteria with deterministic skin barrier parameters, although the pattern of the colonization of skin microbes has been thought to be mainly determined by skin physiology; for example, Staphylococcus and Corynebacterium are most likely to reside in moist sites, but Propionibacterium and Malassezia usually dominate sebaceous areas.12 However, we did not find any association of Corynebacterium or Staphylococcus with either sebum or hydration levels of the facial skin, as previous studies reported.23 Instead, we found that Enhydrobacter is positively associated with TEWL, which has been linked to cutaneous inflammation.27 Meanwhile, the taxa Stenotrophomonas was found to be positively associated with stratum corneum hydration; it is a commensal and also emerging pathogen previously noted in broad-spectrum life-threatening infections among the vulnerable but more recently as a pathogen in immunocompetent individuals.28 C. acnes have the ability to maintain an acidic pH of the skin surface by hydrolyzing triglycerides in the sebum and promoting the release of fatty acids.29 In this study, we did not find a correlation between Propionibacterium and pH. In clinical practice, porphyrins and red areas are known as microscopic manifestations of porphyrins, produced by C. acnes and hemoglobin, indicating the severity of inflammation. In this study, porphyrins and red areas were negatively correlated with Deinococcus, Lautropia, Massilia, Rhizobium, Clavibacter, Pseudomonas, Brevundimonas, Delftia, and Acidovorax; however, they were positively correlated with Staphylococcus, which was obviously increased in acne patients compared with that in healthy controls. Staphylococcus epidermidis was recently considered to play a role in acne prevention by inhibiting the growth of C. acnes via releasing succinic acid, secreting polymorphic toxins, and generating staphylococcal lipoteichoic acid.30–33 A recent article and this study both reported that Staphylococcus is more prevalent in acne patients, compared with healthy controls, which could be due to the Staphylococcus epidermidis using fermentation as a way to inhibit C. acnes growth.34 However, this hypothesis needs further testing. Additionally, a low skin pH contributes to blocking pathogens (ie, Staphylococcus aureus and Streptococcus) from colonizing the skin, and the increased pH in facial regions of acne patients promoted the growth of Staphylococcus.35
In summary, this study explored the differences in skin microbial diversity and composition, evaluated the epidermal barrier function of acne patients and healthy controls in the cheek region, and analyzed their association. We again verified that there is a loss of diversity of the skin microbial community and increased abundance of Staphylococcus in patients with acne vulgaris, compared with healthy controls. Additionally, acne patients had increased TEWL, sebum levels, pH values, porphyrins, and red areas. Finally, the taxa Enhydrobacter, Stenotrophomonas, and Staphylococcus were positively associated with TEWL, stratum corneum hydration, porphyrins, and red areas. There are still several limitations in this study. The sample size was not large, and we only detected the skin microbiota in cheek regions. However, previous studies have focused more on the skin microbiota of the forehead, nose, and back regions for acne vulgaris, and the cheek region has been ignored.36–38 It was reported that the sebum concentration of the cheek, not the forehead, is significantly correlated with microbial composition and diversity.23 Most importantly, this was the first investigation to discuss the link between the skin microbiota and epidermal barrier function as it relates to acne vulgaris. Although the causal relationship between the integrity of the skin barrier, microbiota of the epidermis, and sebaceous gland in the pathophysiology of acne was not clarified, the results could help in understanding more about the disease and finding better treatments.
Acknowledgments
The authors thank Ms Tingting Zhao and Mr Yinghao Pan for their assistance with patient recruitment.
Funding Statement
This study was funded by the major special project for social development of the Sichuan Science and Technology Department (2019YFS0253), the joint project of the Southwest Medical University and Luzhou Science and Technology Bureau (2021LZXNYD-Z04), and the joint project of the Southwest Medical University and Suining people’s Hospital (NO.2021SNXNYD01, 2021SNXNYD04) and General Project of Southwest Medical University (Grant Nos. 2021ZKMS027 and 2021ZKMS030).
Ethics Approval
The entire study was conducted in accordance with the Declaration of Helsinki and permitted by the ethical review board of the hospital affiliated with Southwest Medical University, China (20211116-007).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
None of the authors have conflicts of interest to declare.
References
- 1.Baurecht H, Rühlemann MC, Rodríguez E., et al. Epidermal lipid composition, barrier integrity, and eczematous inflammation are associated with skin microbiome configuration. J Allergy Clin Immunol. 2018;141(5):1668–1676. doi: 10.1016/j.jaci.2018.01.019 [DOI] [PubMed] [Google Scholar]
- 2.Moradi TS, Makrantonaki E, Ganceviciene R, Dessinioti C, Feldman SR, Zouboulis CC. Acne vulgaris. Nat Rev Dis Primers. 2015;1:15029. doi: 10.1038/nrdp.2015.29 [DOI] [PubMed] [Google Scholar]
- 3.Lukaviciute L, Navickas P, Navickas A, Grigaitiene J, Ganceviciene R, Zouboulis CC. Quality of life, anxiety prevalence, depression symptomatology and suicidal ideation among acne patients in Lithuania. J Eur Acad Dermatol Venereol. 2017;31(11):1900–1906. doi: 10.1111/jdv.14477 [DOI] [PubMed] [Google Scholar]
- 4.Szegedi A, Dajnoki Z, Bíró T, Kemény L, Törőcsik D. Acne: transient arrest in the homeostatic host-microbiota dialog? Trends Immunol. 2019;40(10):873–876. doi: 10.1016/j.it.2019.08.006 [DOI] [PubMed] [Google Scholar]
- 5.Oh J, Conlan S, Polley EC, Segre JA, Kong HH. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 2012;4(10):77. doi: 10.1186/gm378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dréno B, Pécastaings S, Corvec S, Veraldi S, Khammari A, Roques C. Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: a brief look at the latest updates. J Eur Acad Dermatol Venereol. 2018;32(Suppl 2):5–14. doi: 10.1111/jdv.15043 [DOI] [PubMed] [Google Scholar]
- 7.Szabó K, Erdei L, Bolla BS, Tax G, Bíró T, Kemény L. Factors shaping the composition of the cutaneous microbiota. Br J Dermatol. 2017;176(2):344–351. doi: 10.1111/bjd.14967 [DOI] [PubMed] [Google Scholar]
- 8.Barnard E, Shi B, Kang D, Craft N, Li H. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci Rep. 2016;6:39491. doi: 10.1038/srep39491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doshi A, Zaheer A, Stiller MJ. A comparison of current acne grading systems and proposal of a novel system. Int J Dermatol. 1997;36(6):416–418. doi: 10.1046/j.1365-4362.1997.00099.x [DOI] [PubMed] [Google Scholar]
- 10.Goldsberry A, Hanke CW, Hanke KE. VISIA system: a possible tool in the cosmetic practice. J Drugs Dermatol. 2014;13(11):1312–1314. [PubMed] [Google Scholar]
- 11.Lee SH, Kang HJ, Lee YH, et al. Monitoring bacterial community structure and variability in time scale in full-scale anaerobic digesters. J Environ Monit. 2012;14(7):1893–1905. doi: 10.1039/c2em10958a [DOI] [PubMed] [Google Scholar]
- 12.Fyhrquist N, Salava A, Auvinen P, Lauerma A. Skin Biomes. Curr Allergy Asthma Rep. 2016;16(5):40. doi: 10.1007/s11882-016-0618-5 [DOI] [PubMed] [Google Scholar]
- 13.Findley K, Oh J, Yang J, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498(7454):367–370. doi: 10.1038/nature12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bek-Thomsen M, Lomholt HB, Kilian M. Acne is not associated with yet-uncultured bacteria. J Clin Microbiol. 2008;46(10):3355–3360. doi: 10.1128/JCM.00799-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee MR, Nam GW, Jung YC, et al. Comparison of the skin biophysical parameters of Southeast Asia females: forehead-cheek and ethnic groups. J Eur Acad Dermatol Venereol. 2013;27(12):1521–1526. doi: 10.1111/jdv.12042 [DOI] [PubMed] [Google Scholar]
- 16.Wendling PA, Dell’Acqua G. Skin biophysical properties of a population living in Valais, Switzerland. Skin Res Technol. 2003;9(4):331–338. doi: 10.1034/j.1600-0846.2003.00040.x [DOI] [PubMed] [Google Scholar]
- 17.Tagami H. Location-related differences in structure and function of the stratum corneum with special emphasis on those of the facial skin. Int J Cosmet Sci. 2008;30(6):413–434. doi: 10.1111/j.1468-2494.2008.00459.x [DOI] [PubMed] [Google Scholar]
- 18.Lopez S, Le Fur I, Morizot F, Heuvin G, Guinot C, Tschachler E. Transepidermal water loss, temperature and sebum levels on women’s facial skin follow characteristic patterns. Skin Res Technol. 2000;6(1):31–36. doi: 10.1034/j.1600-0846.2000.006001031.x [DOI] [PubMed] [Google Scholar]
- 19.Coughlin CC, Swink SM, Horwinski J, et al. The preadolescent acne microbiome: a prospective, randomized, pilot study investigating characterization and effects of acne therapy. Pediatr Dermatol. 2017;34(6):661–664. doi: 10.1111/pde.13261 [DOI] [PubMed] [Google Scholar]
- 20.Sanchez DA, Nosanchuk JD, Friedman AJ. The skin microbiome: is there a role in the pathogenesis of atopic dermatitis and psoriasis? J Drugs Dermatol. 2015;14(2):127–130. [PubMed] [Google Scholar]
- 21.Baroni A, Buommino E, De Gregorio V, Ruocco E, Ruocco V, Wolf R. Structure and function of the epidermis related to barrier properties. Clin Dermatol. 2012;30(3):257–262. doi: 10.1016/j.clindermatol.2011.08.007 [DOI] [PubMed] [Google Scholar]
- 22.Zeeuwen PL, Boekhorst J, van den Bogaard EH, et al. Microbiome dynamics of human epidermis following skin barrier disruption. Genome Biol. 2012;13(11):R101. doi: 10.1186/gb-2012-13-11-r101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukherjee S, Mitra R, Maitra A, et al. Sebum and hydration levels in specific regions of human face significantly predict the nature and diversity of facial skin microbiome. Sci Rep. 2016;6:36062. doi: 10.1038/srep36062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rocha MA, Bagatin E. Skin barrier and microbiome in acne. Arch Dermatol Res. 2018;310(3):181–185. doi: 10.1007/s00403-017-1795-3 [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto A, Takenouchi K, Ito M. Impaired water barrier function in acne vulgaris. Arch Dermatol Res. 1995;287(2):214–218. doi: 10.1007/BF01262335 [DOI] [PubMed] [Google Scholar]
- 26.Prakash C, Bhargava P, Tiwari S, Majumdar B, Bhargava RK. Skin surface pH in acne vulgaris: insights from an observational study and review of the literature. J Clin Aesthet Dermatol. 2017;10(7):33–39. [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SH, Oh DH, Jung JY, Kim JC, Jeon CO. Comparative ocular microbial communities in humans with and without blepharitis. Invest Ophthalmol Vis Sci. 2012;53(9):5585–5593. doi: 10.1167/iovs.12-9922 [DOI] [PubMed] [Google Scholar]
- 28.Adegoke AA, Stenström TA, Okoh AI. Stenotrophomonas maltophilia as an emerging ubiquitous pathogen: looking beyond contemporary antibiotic therapy. Front Microbiol. 2017;8:2276. doi: 10.3389/fmicb.2017.02276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gribbon EM, Cunliffe WJ, Holland KT. Interaction of Propionibacterium acnes with skin lipids in vitro. J Gen Microbiol. 1993;139(8):1745–1751. doi: 10.1099/00221287-139-8-1745 [DOI] [PubMed] [Google Scholar]
- 30.Lee YB, Byun EJ, Kim HS. Potential role of the microbiome in acne: a comprehensive review. J Clin Med. 2019;8(7):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Dai A, Huang S, et al. Propionic acid and its esterified derivative suppress the growth of methicillin-resistant Staphylococcus aureus USA300. Benef Microbes. 2014;5(2):161–168. doi: 10.3920/BM2013.0031 [DOI] [PubMed] [Google Scholar]
- 32.Christensen GJ, Scholz CF, Enghild J, et al. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. Bmc Genomics. 2016;17:152. doi: 10.1186/s12864-016-2489-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia X, Li Z, Liu K, Wu Y, Jiang D, Lai Y. Staphylococcal LTA-induced miR-143 inhibits Propionibacterium acnes-mediated inflammatory response in skin. J Invest Dermatol. 2016;136(3):621–630. doi: 10.1016/j.jid.2015.12.024 [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Kuo S, Shu M, et al. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: implications of probiotics in acne vulgaris. Appl Microbiol Biotechnol. 2014;98(1):411–424. doi: 10.1007/s00253-013-5394-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christensen GJ, Brüggemann H. Bacterial skin commensals and their role as host guardians. Benef Microbes. 2014;5(2):201–215. doi: 10.3920/BM2012.0062 [DOI] [PubMed] [Google Scholar]
- 36.Fitz-Gibbon S, Tomida S, Chiu BH, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol. 2013;133(9):2152–2160. doi: 10.1038/jid.2013.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pécastaings S, Roques C, Nocera T, et al. Characterisation of Cutibacterium acnes phylotypes in acne and in vivo exploratory evaluation of Myrtacine(®). J Eur Acad Dermatol Venereol. 2018;32(Suppl 2):15–23. doi: 10.1111/jdv.15042 [DOI] [PubMed] [Google Scholar]
- 38.Dagnelie MA, Corvec S, Saint-Jean M, et al. Decrease in diversity of Propionibacterium acnes phylotypes in patients with severe acne on the back. Acta Derm Venereol. 2018;98(2):262–267. doi: 10.2340/00015555-2847 [DOI] [PubMed] [Google Scholar]