Abstract
Chemical-peptide conjugation is the molecular initiating event in skin sensitization. The OECD test guideline uses a high-performance liquid chromatography/ultraviolet (HPLC/UV) detection method to quantify chemical-peptide conjugation in a direct peptide reactivity assay (DPRA), which measures the depletion of two synthetic peptides containing lysine or cysteine residues. To improve assay throughput, sensitivity and accuracy, an automated 384-well plate-based RapidFire solid-phase extraction (SPE) system coupled with tandem mass spectrometry (MS/MS) DPRA was developed and validated in the presence of a newly designed internal standard. Compared to the HPLC/UV-based DPRA, the automated SPE-MS/MS-based DPRA improved throughput from 16 min to 10 s per sample, and substrate peptides usage was reduced from 100 mM to 5 μM. When implementing the SPE-MS/MS-based DPRA into a high-throughput platform, we found 10 compounds that depleted lysine peptide and 23 compounds that depleted cysteine peptide (including 7 unreported chemicals from 55 compounds we tested) in a concentration-response manner. The adduct formation between cysteine and cinnamic aldehyde and ethylene glycol dimethacrylate were further analyzed using high-performance liquid chromatography time-of-flight mass spectrometry (HPLC-TOF-MS) to confirm the conjugation. Overall, the automated SPE-MS/MS-based platform is an efficient, economic, and accurate way to detect skin sensitizers.
Keywords: Direct peptide reaction assay (DPRA), RapidFire-MS/MS, high throughput mass spectrometry
1. Introduction
Allergic contact dermatitis (ACD) is a delayed hypersensitivity reaction caused by allergens (Saint-Mezard et al., 2003). Sensitizers are typically small molecules with allergenic potential found in drugs, cosmetics, environmental chemicals, or even heavy metals. ACD induced by sensitizers is a common environmental and occupational health problem. The animal test of contact sensitization risk in the cosmetic industry has been gradually replaced by in vitro assays (Basketter et al., 2013). Currently, a battery of in vitro testing assays has been developed based on the adverse outcome pathway (AOP) of skin sensitization (MacKay et al., 2013; OECD, 2012). Four key events in AOP include covalent modification of self-proteins (also known as haptenation by sensitizers), activation of keratinocytes, presentation of new antigens (hapten/carrier complexes) by dendritic cells, and inflammatory response of lymphocytes (Tollefsen et al., 2014). The molecular initiating event (MIE) of skin sensitization involves chemicals that covalently bind with an endogenous protein. The direct peptide reactivity assay (DPRA), an in chemico skin sensitization assay, addresses this MIE and has been well accepted in industrial and regulatory applications (Van Loveren et al., 2008). The DPRA uses two synthetic peptides with one containing lysine and the other containing cysteine in order to predict a chemical’s conjugating ability with each peptide (cysteine peptide Ac-RFAACAA-COOH, and lysine peptide Ac-RFAAKAA-COOH) (Gerberick et al., 2009; Gerberick et al., 2004). The Organization for Economic Co-operation and Development (OECD) test guideline adopted the DPRA using a high-performance liquid chromatography (HPLC) with UV detector to quantify the covalent conjugation between chemicals and peptides (No, 2015).
The current OECD TG 442C DPRA requires high concentrations of peptides and chemicals whose absorption properties often interfere with the UV detection method, causing unexpected false positive or false negative results (Gerberick et al., 2004). DPRA HPLC-MS based protocols could provide useful quantitative information to determine whether the chemicals truly bind to peptides based on the molecular weight change after the binding. However, use of the HPLC-MS method for analyzing a sample is labor-intensive and time-consuming. Compared to the DPRA HPLC-MS method, HPLC-MS/MS with fragment information of the analyte avoids UV absorbance interference and demands less product for detection, while demonstrating higher sensitivity and selectivity. In addition, the HPLC-MS/MS assay described previously by Zhang et al. (2018) gives more information about the structure of a chemical-peptide conjugate than the HPLC-MS method (Natsch and Gfeller, 2008), which detects adduct formation of the chemical with the peptide. In the HPLC-MS/MS method, the different product ions generated from the parental conjugated peptide and unconjugated peptide can be used as a “signature” to evaluate whether the original peptide remains. Unfortunately, chromatography is time-consuming with the elution time varying from compound to compound. Since animal tests have been banned for cosmetics product development in European Union, Israel, Turkey, India, Taiwan, South Korea, New Zealand and Guatemala (Hartung et al., 2003), there is an urgent need to develop a high-throughput screening method that can quickly detect sensitizers with minimal data processing effort by an analyst. The RapidFire system, an automated high-throughput solid-phase extraction (SPE) platform, conducts high speed solid phase extraction (sampling, loading, washing, and injection) through multiple pumps running in concert and direct coupling to an MS/MS instrument provides peptide and other analyte quantitative analysis (Asano et al., 2019; Clausse et al., 2019; Highkin et al., 2011; Hutchinson et al., 2012; Leveridge et al., 2016; Leveridge et al., 2012; Lowe et al., 2014; Lu et al., 2016; Meng et al., 2015; Plant et al., 2011; Veach et al., 2017a). Compared to 10–20 minutes per sample in the HPLC method, the RapidFire method reduces sample cycle time to 7–10 seconds per injection (Veach et al., 2017b).
Herein, we describe the implementation of a modified DPRA using a RapidFire-MS/MS method, which could significantly improve throughput and screening efficiency. In addition, to reduce the potential for false positive results, we introduced a new alanine peptide (Ac-RFAAAAA-COOH) as the internal standard control. To make the method universal and applicable to screening a large compound library, the multiple reaction monitoring (MRM) method measures the amount of unreacted cysteine or lysine compared to alanine peptide. Ratios of free cysteine peptide or lysine peptide compared to alanine peptide were used to quantify the percent depletion of free cysteine or lysine peptide. We miniaturized the reaction into a 384-well plate format to increase testing throughput and decrease peptide usage dramatically. The results showed that this modified RapidFire-MS/MS protocol is a reliable and sensitive method to distinguish sensitizers and non-sensitizers through the percent depletion of free cysteine and lysine peptides, while dramatically improving throughput.
2. Materials and methods
2.1. Peptides
Cysteine and lysine containing hepa-peptides, as well as the internal standard alanine peptides (> 95% purity) were purchased from New England Peptide (Gardner, MA). The amino acid sequences were (1) cysteine: Ac-RFAACAA-COOH, (2) lysine: Ac-RFAAKAA-COOH, and (3) alanine: Ac-RFAAAAA-COOH. According to manufacture instructions, the peptide powder was dissolved in water at 10 mM and stored at −20 °C.
2.2. Chemicals, reagents, and solvents
Dimethyl sulfoxide (DMSO), ammonium hydroxyl solution, hydrogen chloride (HCl), acetic acid, formic acid, HPLC/MS grade water, and acetonitrile were purchased from Sigma-Aldrich (St. Louis, MO, USA). Phosphate-buffered saline (PBS) was purchased from ThermoFisher Scientific (Waltham, MA, USA). All compounds used in this study were obtained from Sigma-Aldrich. The compound plates were prepared by NCATS (National Center for Advancing Translational Sciences, National Institutes of Health) compound management group. The ACQUITY UPLC BEH C18 column (2.1×150 mm, particle size 1.7 μm) was purchased from Waters Corporation (Milford, MA, USA)
2.3. HPLC-TOF-MS
High-performance liquid chromatography was performed using an Agilent 1290 Infinity II LC system (Agilent Technologies, Wilmington, DE, USA) equipped with a Diode-Array Detection (DAD), binary pump, multicolumn thermostat, and autosampler. The mobile phases used for the separation were MS-grade water with 0.1% formic acid (solvent A) and MS-grade acetonitrile with 0.1% formic acid (solvent B). Gradient elution was performed at a flow rate of 0.4 mL/min, and the mobile phase was 15% solvent B for 3 min, then from 15% to 45% solvent B for 12 min, 45% solvent B to 90% solvent B for 0.5 min and maintained at 90% solvent B for 3 min, followed by 3.5 min for column re-equilibration. Separations were performed at a column temperature of 60 °C with a total run time of 22 min. A varying volume of sample was injected onto the column for each experimental run.
The HPLC-TOF-MS experiments were conducted on an Agilent 6230 TOF system (Agilent Technologies, Wilmington, DE, USA), equipped with a DUAL Jet Stream electrospray ionization (AJS ESI) source operating in positive ion mode. MS spectra were acquired from m/z 150 to 1700 at the scan rate of 1 spectrum per second. The ESI source parameters were used as follows: Gas Temp: 325 °C; Gas Flow: 11 L/min; Nebulizer: 35 psi; Vcap: 3500 V; Nozzle V: 1000; Fragmentor: 175V.
2.4. Peptide reaction condition
The stock solution of cysteine-peptide was mixed with internal standard alanine peptide and diluted in PBS (pH=7.5) to reach 5.0 μM final concentration of each peptide. The stock solution of lysine-peptide was mixed with internal standard alanine peptide and diluted in acetate ammonia buffer (pH=10.2) to reach 0.5 mM as final concentration of each peptide, respectively. Chemicals dissolved in DMSO (DMSO final concentration is < 0.1%) were added to the peptides mixture and incubated at room temperature for 24 h.
2.5. Mass spectrometry platform in high-throughput screening
The high-throughput mass spectrometry system, which conducts high speed solid phase extraction (sampling, loading, washing, and injection) through multiple pumps and valve system, delivers eluted analytes directly to the mass spectrometer (RapidFire-MS/MS) (Clausse et al., 2019). An Agilent RapidFire 360 automated extraction system with three HPLC pumps coupled to an Agilent 6470 triple quadrupole mass spectrometer (Agilent Technologies, Santa Clara, CA, USA) equipped with an AJS ESI interface source was used for the compound screen assay. Agilent RapidFire 4.0 and Agilent MassHunter B.08.00 software were used for instrument control and data acquisition. The RapidFire-MS/MS system was equipped with a C4 Type A solid-phase extraction (SPE) cartridge.
The RapidFire operating procedure and parameters are described here and summarized in Table 1. There are two major solvents for the RapidFire-MS/MS: solvent A (water 0.1% (v/v) formic acid) was used for sample loading and washing, and solvent B (80% acetonitrile, 20% water, 0.1% (v/v) formic acid) was used for sample elution. In brief, samples were first aspirated onto the sample collection loop (10.0 μL) under vacuum directly from 384-well assay plates. A fixed time of 600 ms was defined as the maximum aspiration time with a liquid sensor for detecting whether the loop is full (state 1). The 10.0 μL of sample was loaded onto the C4 cartridge and washed, by pump 1, using the solvent A at a flow rate of 1.5 mL/min for 3,000 ms (state 2). The retained analytes on C4 cartridge were then directly eluted to the mass spectrometer by pump 3, using solvent B at a flow rate of 1.25 mL/min for 3,500 ms (state 4). After elution, the system was re-equilibrated by pump 1, using solvent A at a flow rate of 1.5 mL/min for 2,500 ms (state 5). State 3 was designed for extra wash, which was not required in the optimized method. The sample collection loop was washed with solvent B at a flow rate of 1.25 mL/min for 3,500 ms using pump 2, while sample was under elution (state 4). The entire sampling cycle was approximately 10 s per well, corresponding to approximately 64 min per 384-well plate analysis (Clausse et al., 2019).
Table 1.
RapidFire Operating Parameters
| State Name | Time (ms) | |
|---|---|---|
| State 1 | Aspirate | 600 |
| State 2 | Sample load/wash | 3000 |
| State 3 | Extra wash | 0 |
| State 4 | Sample elute | 3500 |
| State 5 | Re-equilibrate | 2500 |
The Agilent 6470 QQQ mass spectrometer equipped with an AJS ESI was tuned and calibrated in positive mode prior to use. To achieve the best sensitivity and specificity, the mass spectrometer was operated in multiple reaction monitoring (MRM) mode with Q1 resolution set to unit and Q3 resolution set to unit, delta EMV at 200 V. The gas temperature, gas flow, sheath gas temperature, and sheath gas flow were set to 325 °C, 10 L/min, 400 °C, and 11 L/min, respectively. Electrical voltages were optimized for the capillary voltage at +3500 V, nebulizer voltage at +1000 V. The precursor and product ions, collision energies, Fragmentor voltage, collision energy, and cell accelerator voltage for each MRM transition are provided in Table 2.
Table 2.
Parental ion and product ion for free peptide quantification and parameters in RapidFire-MS/MS method
| Peptide Name | Prec Ion | MS1 Res | Prod Ion | MS2 Res | Dwell (ms) | Frag (V) | CE (V) | Cell Acc (V) | Polarity |
|---|---|---|---|---|---|---|---|---|---|
| Cysteine | 751.4 | Unit | 680.2 | Unit | 12 | 205 | 40 | 5 | Positive |
| Cysteine | 751.4 | Unit | 400.2 | Unit | 12 | 205 | 42 | 5 | Positive |
| Cysteine | 751.4 | Unit | 329.1 | Unit | 12 | 205 | 50 | 5 | Positive |
| Alanine | 719.4 | Unit | 648.2 | Unit | 12 | 205 | 40 | 5 | Positive |
| Alanine | 719.4 | Unit | 471.2 | Unit | 12 | 205 | 40 | 5 | Positive |
| Alanine | 719.4 | Unit | 329.1 | Unit | 12 | 205 | 48 | 5 | Positive |
| Lysine | 776.4 | Unit | 705.4 | Unit | 12 | 205 | 44 | 5 | Positive |
| Lysine | 776.4 | Unit | 400.2 | Unit | 12 | 205 | 45 | 5 | Positive |
| Lysine | 776.4 | Unit | 329.1 | Unit | 12 | 205 | 51 | 5 | Positive |
Abbreviations: Prec Ion: Precursor ion; Res: Resolution; Prod Ion: Product ion; Frag: Fragmentor; CE: Collision energy; Cell Acc: Cell accelerator.
RapidFire-MS/MS high-throughput screening was conducted in a 384-well plate. The plate map was designed to fit 48 compounds at 7 concentrations in column 4–24 (Supplemental Fig. 1). Blank, negative, and positive controls were in column 1–3. All the test compounds were dispensed into respective wells. Then 50 μL of the peptide mixture (final concentration of peptide was described in 2.4) was added by multi-drop dispenser to the 384-well plate. The final concentration range of test compounds in cysteine peptide depletion assay was either 0.25 μM to 1 mM, or 12 μM to 50 mM. The final concentration range of test compounds in lysine peptide depletion assay was either 12 μM to 50 mM, or 25 μM to 100 mM. The peptide mixture was diluted to 0.05 μM using loading buffer (3% acetonitrile in water with 0.1% formic acid, v/v) prior to RapidFire-MS/MS analysis.
2.6. Data analysis
Data normalization and concentration–response curve fitting for the data from the cysteine-peptide depletion screening were performed in Prism (GraphPad). Raw plate reads for each titration point were first normalized relative to DMSO-only wells. % Activity = VCompound/VDMSO×100, where VCompound denotes the compound well values, and VDMSO denotes the average values of the DMSO-only wells. Concentration–response titration points for each compound were fitted to a three-parameter Hill equation yielding concentrations of half-maximal inhibitory activity (IC50) and maximal response (efficacy) values calculated by Prism. The mean and SD were calculated by 3 independent replicate plates and reported as % activity of vehicle control.
3. Results and discussion
3.1. RapidFire-MS/MS DPRA assay design
A quantitative high-throughput screen (qHTS) assay based on RapidFire-MS/MS is described in Figure 1. First, we chose appropriate parameters and ion profiles for both parental and product ion in MRM to measure the amount of free cysteine and lysine peptide (Table 2). To reduce the false positive and negative rates, we used an alanine peptide as an internal standard, which had a similar sequence to cysteine/lysine peptide, only replacing cysteine/lysine by alanine to eliminate the reaction sites. The ratios of cysteine or lysine peptide to alanine peptide were used to quantify the percent of depletion of free cysteine or lysine peptide. The absolute signals of alanine peptide dropping could also be an indicator for non-specific reaction for peptides and chemicals, warranting further confirmation by HPLC-TOF-MS experiments.
Figure 1.
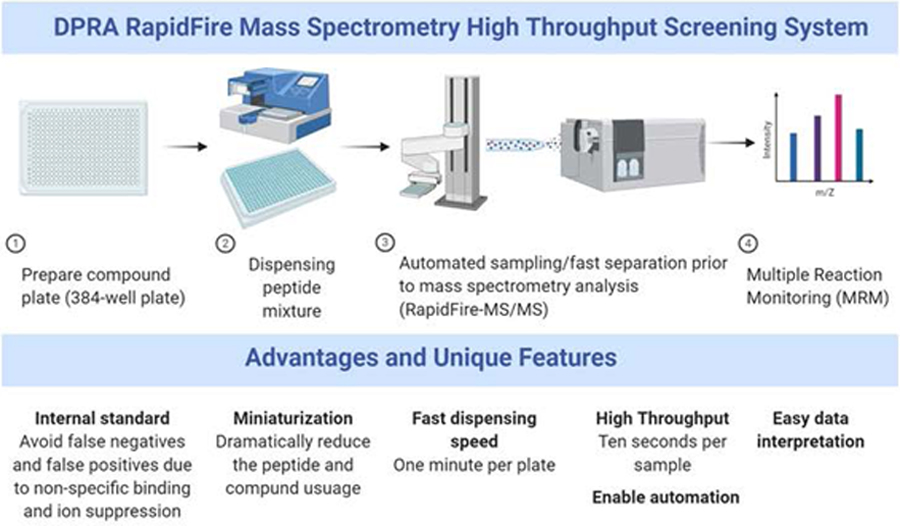
Scheme of the RapidFire-MS/MS DPRA assay high-throughput screening process. The advantages and unique features of each step are listed in the lower panel.
3.2. Peptide stability, linearity, and reproducibility test
According to the OECD test guideline 442C, the DPRA assay requires a standard curve to quantify the amount of free peptide by the HPLC/UV method. In the present study, we used a standard curve of the cysteine or lysine peptide consisting of 10 concentrations ranging from 0.1 to 50 μM in the presence of 5 μM alanine peptide. The integrated area of different concentrations of the cysteine (Figure 2A) or lysine peptide (Figure 2B) were plotted and excellent linearity with an R2 > 0.99 calculated by simple linear regression model for both peptides.
Figure 2.
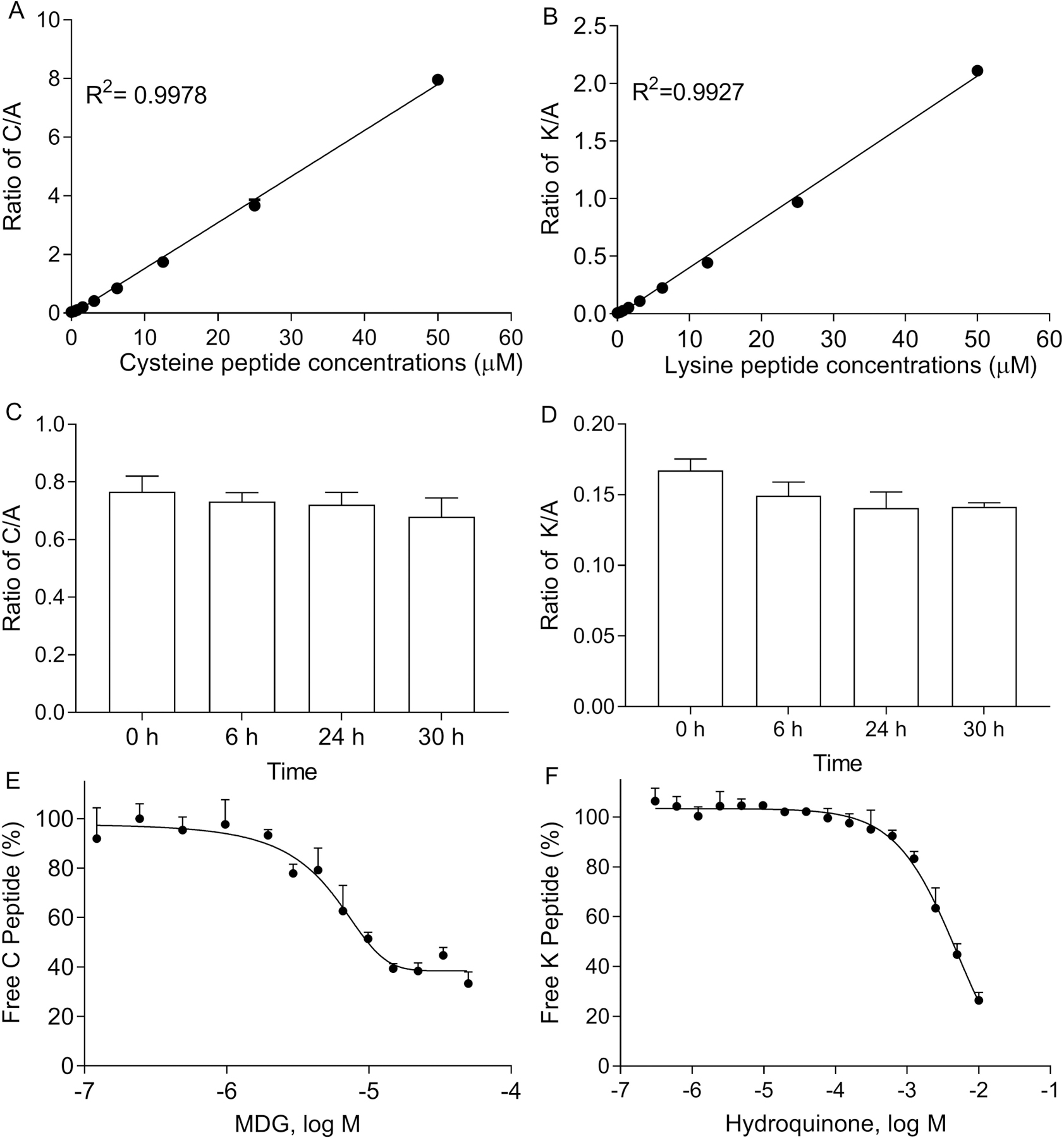
Validation of the cysteine and lysine peptide depletion assays. Linear response of cysteine (A) and lysine (B) peptide calibration standards detected by the RapidFire-MS/MS system. Stability of cysteine (C) /lysine (D) peptide mixed alanine peptide up to 30 h (n=6). Concentration response curves of (E) MDG- and (F) hydroquinone-induced cysteine/lysine peptide depletion. The percentage of free cysteine/lysine peptide was normalized based on positive and negative controls and plotted as average ± SD (n=3).
The DPRA assay was measured after 24 h incubation with compounds at room temperature. To demonstrate the stability of cysteine/lysine when mixed with alanine peptides, we measured the stability of the 5 μM peptide mixtures at 6, 24, or 30 h. As shown in Figure 2C and 2D, no significant changes were found in the ratio of cysteine or lysine to alanine up to 30 h.
Methyldibromo glutaronitrile (MDG), a known skin sensitizer (Basketter, 2010), causes cysteine peptide depletion (Gerberick et al., 2009). Hydroquinone depletes 83.3% of the lysine peptide at 0.5 mM (Gerberick et al., 2004). Both MDG and hydroquinone were selected as the positive controls in this study to ensure the reproducibility of the method for the cysteine peptide and lysine peptide, respectively. The reproducibility of the assay was tested in three independent runs. The concentration response curve of MDG-induced cysteine peptide depletion (Figure 2E), and hydroquinone-induced lysine peptide depletion (Figure 2F) exhibited IC50 values of 4.7 μM and 5 mM, respectively.
Overall, use of the RapidFire-MS/MS system can quantitatively measure the cysteine peptide depletion assay. This method demonstrated high reproducibility and sensitivity to detect peptide-chemical conjugation.
3.3. Assay performance in a 384-well plate format
To reduce peptide usage and improve assay throughput, the RapidFire-MS/MS DPRA assay was developed and optimized in a 384-well plate format. The plate map was designed to fit 48 compounds at 7 concentrations in column 4–24, while blank, negative, and positive controls were in column 1–3 (shown in Supplemental Figure 1). The assay was validated by testing the positive control, MDG, in a 384-well plate format. The signal to basal ratio (S/B) was 4.42, which is calculated based on the average of 8 wells of positive controls over 8 wells of DMSO vehicle controls. The Z’ factor was 0.7, which is calculated based on 3 replicate plates. Subsequently, 55 compounds comprised of 28 known sensitizers, 20 known non-sensitizers, and 7 not previously tested compounds were screened by incubating with cysteine/lysine mixed with alanine peptides. For the cysteine peptide depletion assay, 22 out of 28 known sensitizers were detected and 18 out of 20 known non-sensitizers remained negatives, according to the threshold defined by OECD test guideline 442C. The sensitivity and specificity of this assay were 78.5% and 90%, respectively. The IC50 and efficacy values of each compound are reported in Table 3. The concentration-response curves of the compounds showing cysteine depletion are reported in Figure 3. The 6 known sensitizers which were not detected in the cysteine peptide depletion assays are imidazolidinyl urea, 2-propenoic acid, ethyl acrylate, phenylacetaldehyde, dodecyl gallate, and palmitoyl chloride (labeled with * in Table 3). Palmitoyl chloride, was a strong sensitizer that almost completely depleted lysine peptide (No, O.T., 2015). However, we observed immediate precipitation when adding 100 mM palmitoyl chloride into the lysine peptide buffer. We believe that due to poor solubility, the palmitoyl chloride does not react with peptides (cLogp = 7.5). A similar observation was reported in a previous study (Yamamoto et al.) which discussed precipitation of palmitoyl chloride in DPRA peptide buffer.
Table 3.
Depletion of cysteine peptide measured by RapidFire-MS/MS method
| Sample Name | CASRN | IC50 (M) | Efficacy (% peptide depletion) |
|---|---|---|---|
| p-Benzoquinone | 106-51-4 | 4.84E-06 | −119.4 |
| (E)-2-Hexenal | 6728-26-3 | 5.58E-05 | −28.06 |
| (S)-4-Isopropenyl-1-cyclohexene-1-carboxaldehyde | 18031-40-8 | 1.16E-04 | −108.8 |
| 1-Butanola | 71-36-3 | Inactive | Inactive |
| 2,3-Butanedione | 431-03-8 | 4.11E-05 | −95.45 |
| 2-Mercaptobenzothiazole | 149-30-4 | 1.31E-04 | −97.27 |
| 2-Methyl-4-isothiazolin-3-one hydrochloride | 26172-54-3 | 3.63E-06 | −101.9 |
| 2-Propenoic acid, 2-methyl-, monoester with 1,2-propanedioa,* | 923-26-2 | Inactive | Inactive |
| 3-(Dimethylamino)propylaminea | 109-55-7 | Inactive | Inactive |
| 3,4-Dihydrocoumarin | 119-84-6 | 6.95E-05 | −56.7 |
| 4-Hydroxybenzoic acid | 99-96-7 | 1.27E-04 | −14.37 |
| 4-Methoxyacetophenone | 100-06-1 | Inactive | Inactive |
| 5-Chloro-2-methyl-3(2H)-isothiazolone | 26172-55-4 | 1.44E-05 | −122.3 |
| 6-Methyl coumarina | 92-48-8 | Inactive | Inactive |
| Benzisothiazolin-3-one | 2634-33-5 | 8.65E-07 | −110.5 |
| Benzoyl peroxide | 94-36-0 | 9.53E-05 | −107 |
| Benzyl alcohola | 100-51-6 | Inactive | Inactive |
| Benzyl benzoatea | 120-51-4 | Inactive | Inactive |
| Benzylideneacetone | 122-57-6 | 2.12E-005 | 97.67 |
| Chlorobenzenea | 108-90-7 | Inactive | Inactive |
| Cinnamic aldehyde | 104-55-2 | 1.05E-03 | −124.4 |
| Cyclamen aldehyde | 103-95-7 | 2.91E-05 | −14.77 |
| Diethyl maleate | 141-05-9 | 3.00E-04 | −108 |
| Diethyl phthalatea | 84-66-2 | Inactive | Inactive |
| Dimethyl isophthalatea | 1459-93-4 | Inactive | Inactive |
| Dinitrochlorobenzene | 97-00-7 | 3.19E-04 | −198.8 |
| Diphenylcyclopropenone | 886-38-4 | 2.38E-05 | −43.84 |
| Dodecyl gallatea,* | 1166-52-5 | Inactive | Inactive |
| Ethyl acrylatea,* | 140-88-5 | Inactive | Inactive |
| Ethylene glycol dimethacrylateb | 97-90-5 | N/A | −60.29 |
| Farnesalb | 19317-11-4 | N/A | −17.4 |
| Formaldehydeb | 50-00-0 | N/A | −11.6 |
| Glutaraldehyde | 111-30-8 | 1.03E-03 | −119.3 |
| Glycerola | 56-81-5 | Inactive | Inactive |
| Hexyl cinnamic aldehydea | 165184-98-5 | Inactive | Inactive |
| Hydroxycitronellala | 107-75-5 | Inactive | Inactive |
| Hydroxyethyl acrylate | 818-61-1 | 5.82E-05 | −132.1 |
| Imidazolidinyl ureaa,* | 39236-46-9 | Inactive | Inactive |
| Lactic acida | 50-21-5 | Inactive | Inactive |
| m-Aminophenol | 591-27-5 | 8.66E-06 | −26.38 |
| Methyl 2-nonynoate | 111-80-8 | 3.51E-04 | −39.99 |
| Methyl salicylatea | 119-36-8 | Inactive | Inactive |
| Methyldibromo glutaronitrile | 35691-65-7 | 1.01E-06 | −102.5 |
| n-Hexanea | 110-54-3 | Inactive | Inactive |
| Octanoic acid | 124-07-2 | 3.31E-04 | −25.36 |
| Oxalic acida | 144-62-7 | Inactive | Inactive |
| Oxazolone | 15646-46-5 | 9.09E-06 | −95.43 |
| Palmitoyl chloridea,* | 112-67-4 | Inactive | Inactive |
| Penylcinnamaldehyde | 122-40-7 | 2.92E-04 | −104.8 |
| Phenylacetaldehydea,* | 122-78-1 | Inactive | Inactive |
| Phthalic anhydridea | 85-44-9 | Inactive | Inactive |
| Propyl parabena | 94-13-3 | Inactive | Inactive |
| Protectol PP (Lilial) | 80-54-6 | 3.58E-05 | −15.36 |
| Trimellitic anhydridea | 552-30-7 | Inactive | Inactive |
| Vinyl pyridinea | 100-69-6 | Inactive | Inactive |
Inactive indicated no concentration-response inhibition.
N/A indicates the curve is too wide to calculate IC50, and efficacy values are the span of % peptide depletion between the lowest and highest concentrations of compound treatment.
False negative compounds
Figure 3.
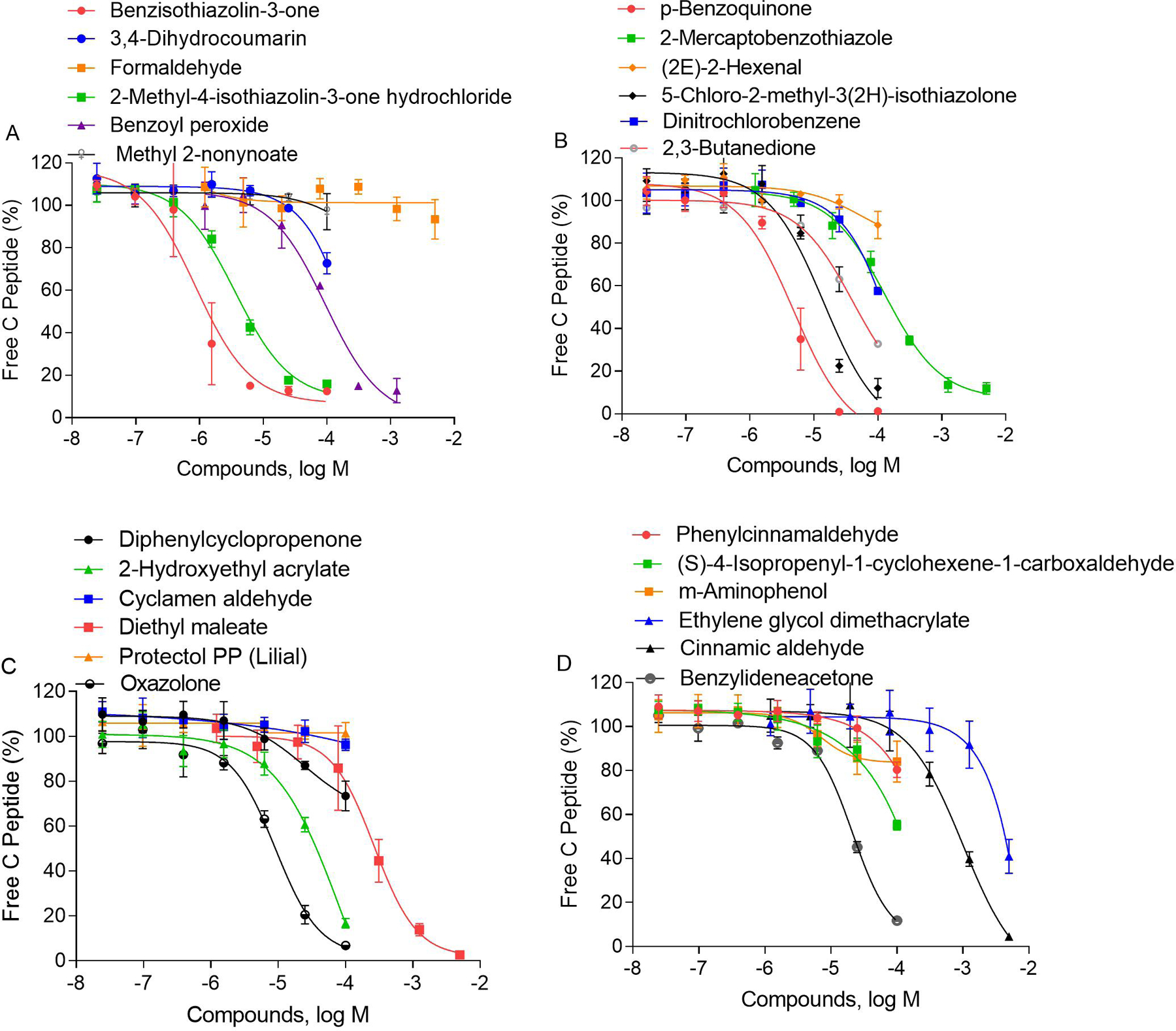
Concentration responses of identified compounds induced depletion of free cysteine peptide. The percent of free cysteine peptide was normalized based on negative contros and plotted as average ± SD (n=3).
We selected 10 chemicals that are known to deplete lysine peptides and tested them in 384-well plates. As shown in Figure 4, they all depleted the lysine peptide in a concentration-dependent manner with IC50 values listed in Table 4. Among these compounds, those previously reported as sensitizers (Natsch et al., 2013), including hydroquinone, propyl gallate, 2-hydroxyethyl acrylate, ethyl acrylate and glutaraldehyde, depleted more than 50% of the lysine peptide at the highest tested concentrations.
Figure 4.
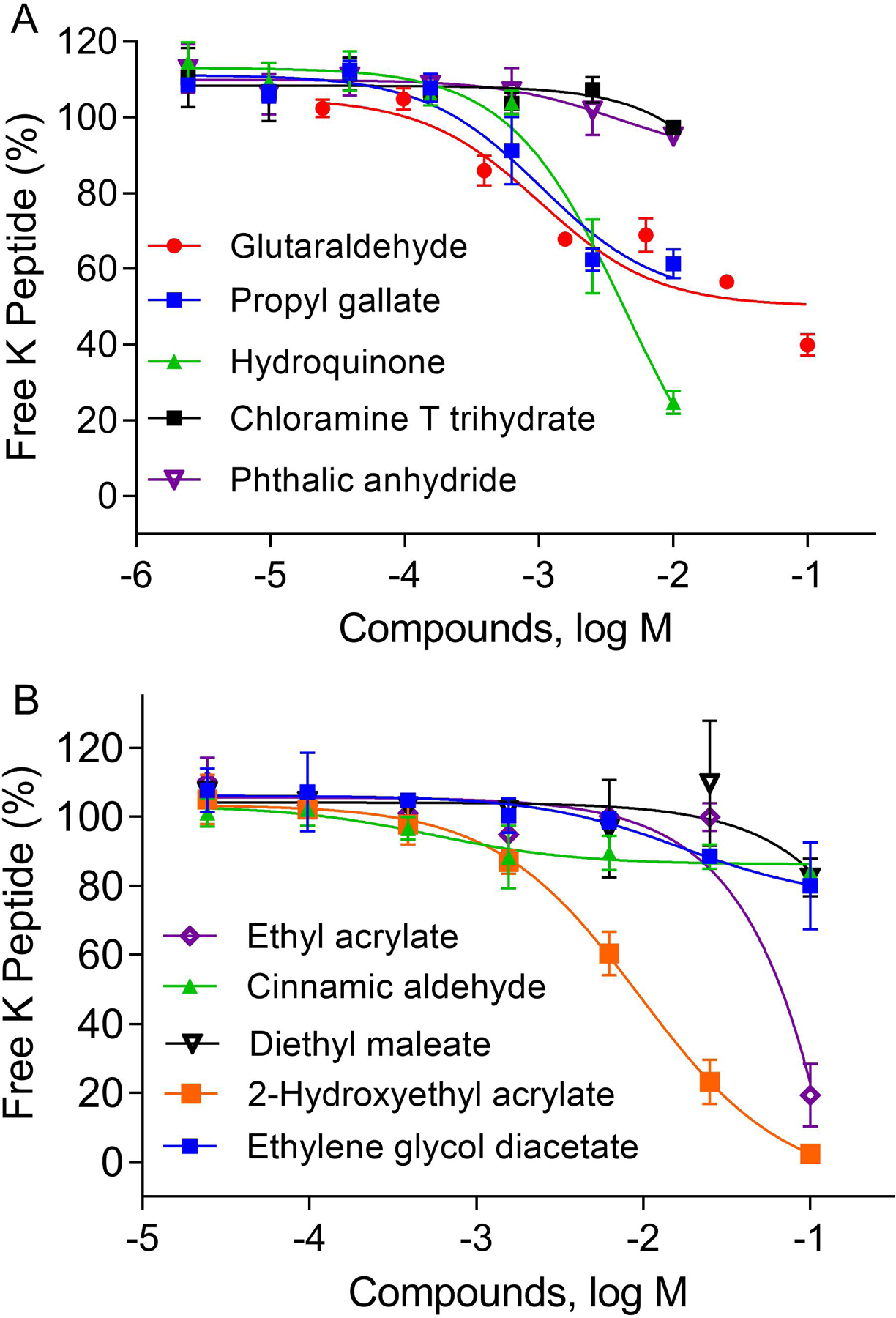
Concentration responses of identified compounds induced depletion of free lysine peptide. The percent of free lysine peptide was normalized based on negative control and plotted as average ± SD (n=3).
Table 4.
Depletion of lysine peptide measured by RapidFire-MS/MS method
| Sample Name | CASRN | IC50 (M) | Efficacy (% peptide depletion) |
|---|---|---|---|
| Glutaraldehyde | 111-30-8 | 1.02E-03 | −55.08 |
| Propyl gallate | 121-79-9 | 1.04E-03 | −59.18 |
| Hydroquinone | 123-31-9 | 4.44E-03 | −128.6 |
| Chloramine T trihydratea | 7080-50-4 | N/A | −13.28 |
| Phthalic anhydride | 85-44-9 | 3.84E-03 | −20.65 |
| Ethylene glycol diacetate | 111-55-7 | 1.707E-02 | −30.2 |
| Cinnamic aldehyde | 104-55-2 | 5.5E-04 | −16.97 |
| 2-Hydroxyethyl acrylate | 818-61-1 | 9.51E-03 | −110.6 |
| Diethyl maleatea | 141-05-9 | N/A | −24.88 |
| Ethyl acrylatea | 140-88-5 | N/A | −90.99 |
N/A indicates the curve is too wide to calculate IC50, and efficacy values are the span of % peptide depletion between the treatment of lowest and highest compound concentration.
3.4. Internal standard improves the assay accuracy
In the current study, we found that some chemicals were very reactive with strong ionization capacity. These compounds caused ion suppression or non-specific binding, resulting in false positive or false negative predictions. Two chemicals, cinnamic aldehyde and glutaraldehyde, were analyzed as examples. The results from RapidFire-MS/MS were compared to the results from the HPLC-TOF-MS method with a 22 min gradient elution by using a 150 mm length C18 column. Both chemicals were tested in a 7-point concentration range. RapidFire-MS/MS showed a decrease in signals of the alanine (Figure 5A) and lysine peptides (Figure 5B) (> 60% depletion) at high concentrations of cinnamic aldehyde treatment (above 1 mM). However, HPLC-TOF-MS data only showed a slight decrease of the lysine peptide (Figure 5B) at 100 mM, and no changes were observed in the alanine peptide (Figure 5A). We speculate that the decrease in both the lysine and alanine peptides for RapidFire-MS/MS at higher concentration (> 10 mM) may be due to a potential ionization suppression. RapidFire-MS/MS ionizes the chemical compound and peptides at a similar time if the chemical is hydrophobic and co-elutes with the peptides. In the absence of the alanine peptide, cinnamic aldehyde could be wrongly identified as a strong sensitizer due to the lysine peptide depletion at higher concentration. With the use of the internal standard in the assay, the ratio of lysine/alanine peptide truly represents the amount of free lysine peptide. As shown in Figure 5C, a very slight change (<15%) was observed across the entire concentration range with the RapidFire-MS/MS method. This matches well with the HPLC-TOF-MS data and shows the benefit of using the internal standard to reduce false positives caused by the potential ion suppression. We further investigated the data for potential ionization, and among the 55 compounds screened in the cysteine or lysine peptide depletion assay, only four exhibited a greater than 15% drop in the alanine peptide signal. Generally, the alanine peptide is very stable and demonstrates the ability to monitor ion suppression or non-specific binding.
Figure 5.
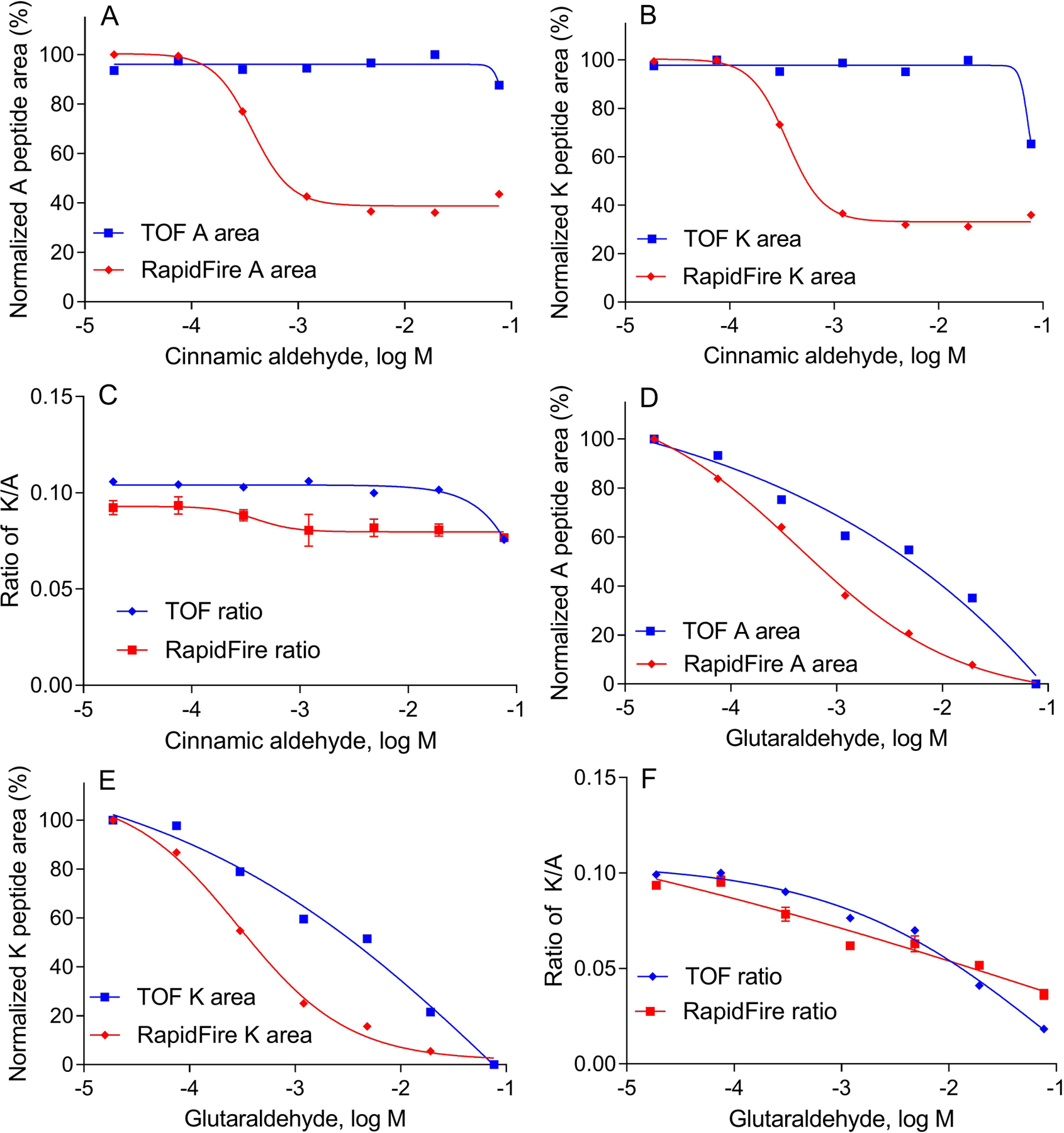
Comparison of the cysteine/lysine depletion assay results in the RapidFire-MS/MS DPRA and HPLC-TOF-MS methods. (A) Depletion of the alanine peptide with cinnamic aldehyde treatment. The percent integrated alanine peptide (A peptide) area was normalized to the DMSO negative control. (B) Depletion of the lysine peptide with cinnamic aldehyde treatment. The percent integrated lysine peptide (K peptide) area was normalized to the DMSO negative control. (C) Ratio of lysine to alanine peptide area with cinnamic aldehyde treatment. (D) Depletion of the alanine peptide with glutaraldehyde treatment. The percent integrated alanine peptide area was normalized to the DMSO negative control. (E) Depletion of the lysine peptide with glutaraldehyde treatment. The % integrated lysine peptide (K peptide) area was normalized to the DMSO negative control. (F) Ratio of lysine to alanine peptide area with glutaraldehyde treatment.
Interestingly, we also observed that glutaraldehyde caused non-specific binding to both lysine and alanine peptides. The signal of alanine (Figure 5D) and lysine peptides (Figure 5E) decreased in a concentration-dependent manner in both the HPLC-TOF-MS and RapidFire-MS/MS methods. In Figure 5F, ratios of the lysine to alanine peptides were used to calculate the IC50, and similar results were obtained in HPLC-MS and RapidFire-MS/MS methods. The IC50 of glutaraldehyde-induced lysine depletion would be much lower (0.3 mM) if it only measured the lysine peptide depletion in the RapidFire-MS/MS method (Figure 5E). Therefore, the use of an internal standard (alanine peptide) is necessary to reduce the false negative results.
3.5. Confirmation of conjugated peptides in HPLC-TOF-MS
To verify peptide chemical conjugation in a 384-well plate is like the test tube-based assay, we utilized HPLC-TOF-MS to conduct a detailed spectral analysis of several chemical-peptide mixtures. The HPLC-TOF-MS allows us to postulate the chemical-peptide adduct formation and explains the mechanisms of peptide modification by sensitizers. We found that several chemicals exhibited the same conjugation pattern with the cysteine peptide as previously reported. As shown in Figure 6A and 6B, cinnamic aldehyde (MW 132.16) and ethylene glycol dimethacrylate (MW 198.2) formed adduct peak with cysteine peptide (C peptide adduct). As shown in Figure 6C, the mass of the cinnamic aldehyde formed C peptide adduct peak (m/z 883.4096) matches the theoretical mass of the cysteine-cinnamic aldehyde adduct (883.6) as reported by Roberts and Natsch (2009). In addition, the dimer of cysteine peptide observed in cinnamic aldehyde sample which had a retention time of 6.851 min, showed a m/z value of 750.3457 for charge state 2 corresponding to a MW of 1498.69 (Figure 6D). A weak sensitizer, ethylene glycol dimethacrylate (MW 198.2), was identified in RapidFire-MS/MS (Figure 3D). It was found to form monomers and dimers with cysteine showing peaks m/z 475.2245 and 949.4418 in Figure 6E, which matches reported values (949.8 and 475.4) in Urbisch et al. (2016), but differs from the 1:1 adduct formation (949.4) reported in Roberts and Natsch (2009).
Figure 6.
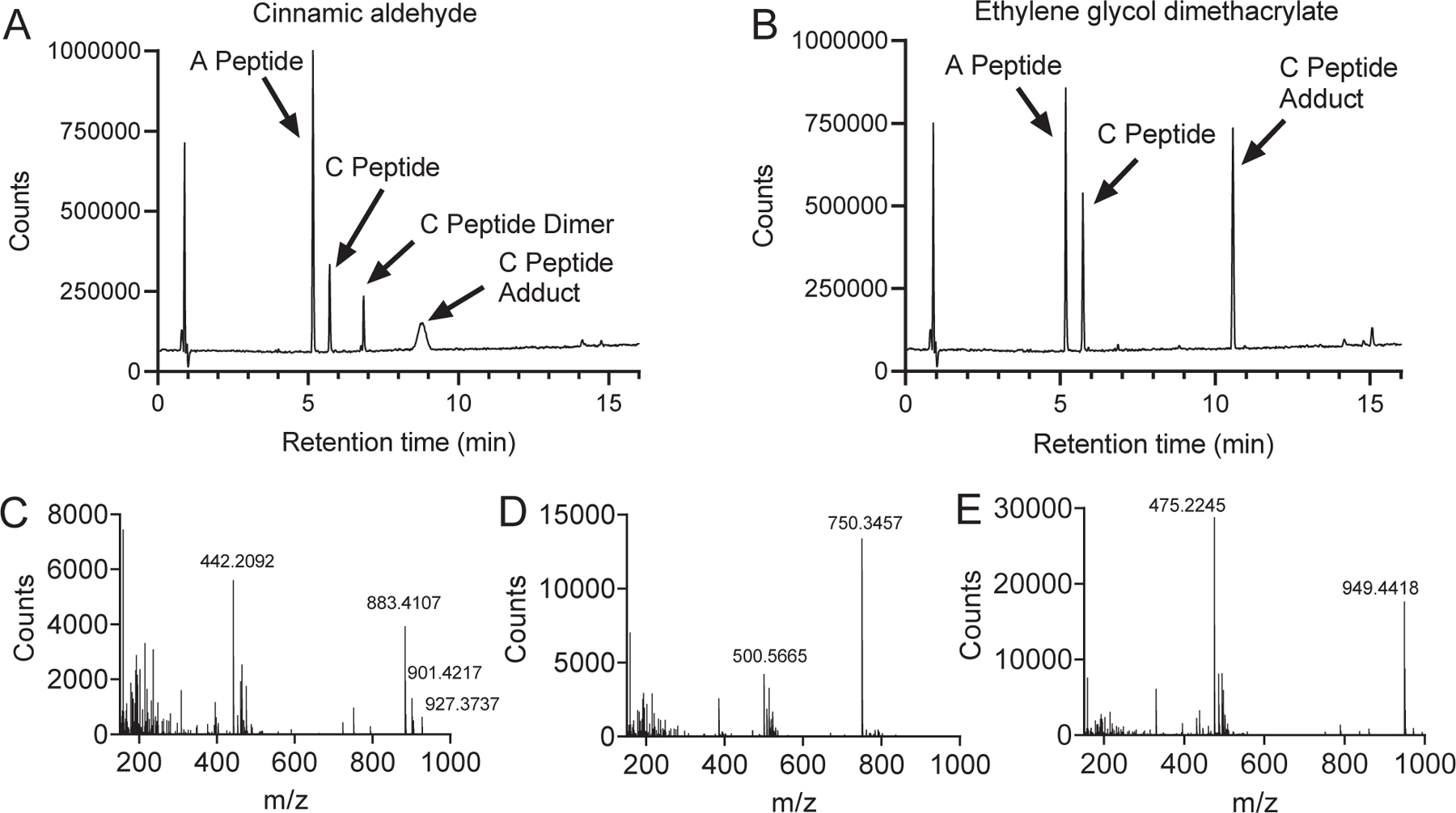
HPLC-TOF-MS spectrum of mixture of chemical-cysteine peptide mixtures. (A) Total Ion Chromatogram (TIC) of the alanine peptide (A peptide) and cysteine peptide (C peptide) mixture with cinnamic aldehyde after 24 h incubation. (B) TIC of the alanine peptide (A peptide), cysteine peptide (C peptide) mixture with ethylene glycol dimethacrylate (EGD). (C) Mass spectra of a selected peak of cinnamic aldehyde adduct with C peptide at the retention time of 8.765 min. (D) Mass spectra of a selected peak of C peptide dimer induced by cinnamic aldehyde at the retention time of 6.851 min. (E) Mass spectra of a selected peak of EGD adduct with C peptide at the retention time of 10.573 min.
4. Conclusion
Detection of chemicals with a propensity to covalently conjugate to thiol- or amine-containing peptides is an important unmet need in the general fields of skin product evaluation and in vitro toxicology. The development of an automated SPE-MS/MS DPRA with introduction of an internal standard in a high-throughput platform enables fast and precise detection of skin sensitizers. The advantages of this method are stated in Table 5 when compared with existing detection methods of DPRA. Among these methods listed in Table 5, only our DPRA assay runs in an HTS platform with an internal standard. Continuous monitoring of the cysteine/lysine peptide and chemical conjugation used in the study provides useful information for kinetic profiling of peptides previously developed by Roberts and Natsch (2009). In comparison to their cysteine peptide depletion assay, our method measured depletion of both the cysteine and lysine peptide incubated with chemicals at multiple concentrations, which provides concentration response curves to indicate sensitization potency. Cho et al. reported the development of probe-based spectrophotometric assays for skin sensitization testing which measures both peptides at lower cost (Cho et al., 2014; Cho et al., 2019). The issue of color interference is fundamentally embedded in the detection method. Therefore, we developed the automated SPE-MS/MS DPRA probe free detection method which accurately measures the chemical-peptide conjugation. Also, this assay can detect the compound at concentrations as low as 5 pM, minimizing the usage of the cysteine peptide to 5 μM, compared to the usage of 0.667 mM peptide in OECD test guideline, which is more than a 100-fold peptide usage reduction. The testing speed of sampling to data interpretation is less than 10 seconds, meeting the need for fast screening of skin sensitizers. Most importantly, an internal standard was introduced into this assay for the first time. The use of internal standard provides more accurate measurement of cysteine/lysine depletion compared to the traditional DPRA, which measures the amount of free cysteine or lysine peptide by HPLC. Overall, the automated RapidFire-MS/MS DPRA that utilizes a solid-phase extraction system is an efficient, cost-effective, more accurate, and universal assay to detect sensitizers that is compatible with an HTS format.
Table 5.
Comparison with other DPRA methods
| DPRA Methods (Reference) | Standard DPRA (Gerberick et al., 2004) | qDPRA (Wareing et al., 2017) | Kinetic DPRA (Roberts and Natsch, 2009) | Spectro-DPRA (Cho et al., 2014) | HPLC-MS DPRA (Natsch and Gfeller, 2008) | HPLC-MS/MS DPRA (Zhang et al., 2018) | Automated solid-phase extraction system DPRA |
|---|---|---|---|---|---|---|---|
| Detection Method | HPLC | HPLC | Fluorimetric readout | UV-VIS spectrophotometer | LC-MS | LC-MS/MS | RapidFire-MS/MS |
| Peptide(s) concentration | 100 mM | 100, 10, 1 mM | 0.667 mM | 400 μM (cysteine peptide), 200 μM (lysine peptide) | 0.1 mM | 100 mM | 5 μM (cysteine peptide), 0.5 mM (lysine peptide). |
| Chemical concentration | 1M (cysteine peptide) 5M (lysine peptide) |
10 mM | 20, 9, 6, 3, 1.5 mM | 2 mM | 1 mM | 1M (cysteine peptide) 5M (lysine peptide) |
50 μM – 200 mM |
| Format | Test tube | Test tube | 96-Well plate | 96-Well plate | Test tube | Test tube | 384-Well plate |
| Novelty | First report | 3 Points concentration response | Kinetic profiling, new peptide | The use of fluorescent probe 5,5′-dithiobis-2-nitrobenzoic acid (DTNB) | Detection of conjugation by LC-MS | Detection of modified peptide | HTS, 7 points concentration response, detection of modified peptide by MS/MS, internal standard. |
| Efficiency (time/sample) | 40 min | 40 min | < 1 min | < 1 min | 16 min | 20 min | 10 second per sample |
Supplementary Material
Highlights.
Automated mass spectrometry high throughput screening method enables ultra-fast sampling and accurate data interpretation
Introducing an internal standard (alanine peptide) to monitor and correct false positive or negative sensitizers
Multiple reaction monitoring (MRM) is a highly sensitive method which reduces the usage of peptide from mM to several μM range.
Concentration response curve of peptide depletion gives information in depth to categorize sensitization potential
Acknowledgment
We thank Ms. Zina Itkin for technical support in compound management and solution preparation.
Funding
This work was supported by the Intramural Research Programs of the National Center for Advancing Translational Sciences, National Institutes of Health.
Footnotes
Disclaimer: The views expressed in this article are those of the authors and do not necessarily reflect the views of the National Center for Advancing Translational Sciences, or the United States government. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Electronic Supplementary Material
The supporting information is shown in supplemental figure 1. The plate format was as follows: Column 1, row A – H: blank, row I – P: DMSO. Column 2, row A – H: highest concentration of positive control, row I – P: second highest concentration of positive control. Column 3, concentration-response titration of positive control.
Conflicts of Interest
The authors declare no competing financial interest.
References
- Asano W, Takahashi Y, Kawano M, Hantani Y, 2019. Identification of an Arginase II Inhibitor via RapidFire Mass Spectrometry Combined with Hydrophilic Interaction Chromatography. SLAS Discov 24, 457–465. [DOI] [PubMed] [Google Scholar]
- Basketter D, Alépée N, Casati S, Crozier J, Eigler D, Griem P, Hubesch B, de Knecht J, Landsiedel R, Louekari K, 2013. Skin sensitisation–moving forward with non-animal testing strategies for regulatory purposes in the EU. Regulatory Toxicology and Pharmacology 67, 531–535. [DOI] [PubMed] [Google Scholar]
- Basketter DA, 2010. Methyldibromoglutaronitrile: skin sensitization and quantitative risk assessment. Cutaneous and ocular toxicology 29, 4–9. [DOI] [PubMed] [Google Scholar]
- Cho S-A, Jeong YH, Kim JH, Kim S, Cho J-C, Heo Y, Suh K-D, An S, Shin K, 2014. Method for detecting the reactivity of chemicals towards peptides as an alternative test method for assessing skin sensitization potential. Toxicology Letters 225, 185–191. [DOI] [PubMed] [Google Scholar]
- Cho SA, An S, Park JH, 2019. High-throughput screening (HTS)-based spectrophotometric direct peptide reactivity assay (Spectro-DPRA) to predict human skin sensitization potential. Toxicol Lett 314, 27–36. [DOI] [PubMed] [Google Scholar]
- Clausse V, Tao D, Debnath S, Fang Y, Tagad HD, Wang Y, Sun H, LeClair CA, Mazur SJ, Lane K, Shi ZD, Vasalatiy O, Eells R, Baker LK, Henderson MJ, Webb MR, Shen M, Hall MD, Appella E, Appella DH, Coussens NP, 2019. Physiologically relevant orthogonal assays for the discovery of small-molecule modulators of WIP1 phosphatase in high-throughput screens. J Biol Chem 294, 17654–17668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerberick GF, Troutman JA, Foertsch LM, Vassallo JD, Quijano M, Dobson RL, Goebel C, Lepoittevin J-P, 2009. Investigation of peptide reactivity of pro-hapten skin sensitizers using a peroxidase-peroxide oxidation system. Toxicological Sciences 112, 164–174. [DOI] [PubMed] [Google Scholar]
- Gerberick GF, Vassallo JD, Bailey RE, Chaney JG, Morrall SW, Lepoittevin J-P, 2004. Development of a peptide reactivity assay for screening contact allergens. Toxicological Sciences 81, 332–343. [DOI] [PubMed] [Google Scholar]
- Hartung T, Bremer S, Casati S, Coecke S, Corvi R, Fortaner S, Gribaldo L, Halder M, Roi AJ, Prieto P, 2003. ECVAM’s response to the changing political environment for alternatives: consequences of the European Union chemicals and cosmetics policies. Alternatives to laboratory animals 31, 473–481. [DOI] [PubMed] [Google Scholar]
- Highkin MK, Yates MP, Nemirovskiy OV, Lamarr WA, Munie GE, Rains JW, Masferrer JL, Nagiec MM, 2011. High-throughput screening assay for sphingosine kinase inhibitors in whole blood using RapidFire(R) mass spectrometry. J Biomol Screen 16, 272–277. [DOI] [PubMed] [Google Scholar]
- Hutchinson SE, Leveridge MV, Heathcote ML, Francis P, Williams L, Gee M, Munoz-Muriedas J, Leavens B, Shillings A, Jones E, Homes P, Baddeley S, Chung CW, Bridges A, Argyrou A, 2012. Enabling lead discovery for histone lysine demethylases by high-throughput RapidFire mass spectrometry. J Biomol Screen 17, 39–48. [DOI] [PubMed] [Google Scholar]
- Leveridge M, Collier L, Edge C, Hardwicke P, Leavens B, Ratcliffe S, Rees M, Stasi LP, Nadin A, Reith AD, 2016. A High-Throughput Screen to Identify LRRK2 Kinase Inhibitors for the Treatment of Parkinson’s Disease Using RapidFire Mass Spectrometry. J Biomol Screen 21, 145–155. [DOI] [PubMed] [Google Scholar]
- Leveridge MV, Bardera AI, LaMarr W, Billinton A, Bellenie B, Edge C, Francis P, Christodoulou E, Shillings A, Hibbs M, Fosberry A, Tanner R, Hardwicke P, Craggs P, Sinha Y, Elegbe O, Alvarez-Ruiz E, Martin-Plaza JJ, Barroso-Poveda V, Baddeley S, Chung CW, Hutchinson J, 2012. Lead discovery for microsomal prostaglandin E synthase using a combination of high-throughput fluorescent-based assays and RapidFire mass spectrometry. J Biomol Screen 17, 641–650. [DOI] [PubMed] [Google Scholar]
- Lowe DM, Gee M, Haslam C, Leavens B, Christodoulou E, Hissey P, Hardwicke P, Argyrou A, Webster SP, Mole DJ, Wilson K, Binnie M, Yard BA, Dean T, Liddle J, Uings I, Hutchinson JP, 2014. Lead discovery for human kynurenine 3-monooxygenase by high-throughput RapidFire mass spectrometry. J Biomol Screen 19, 508–515. [DOI] [PubMed] [Google Scholar]
- Lu H, Kopcho L, Ghosh K, Witmer M, Parker M, Gupta S, Paul M, Krishnamurthy P, Laksmaiah B, Xie D, Tredup J, Zhang L, Abell LM, 2016. Development of a RapidFire mass spectrometry assay and a fluorescence assay for the discovery of kynurenine aminotransferase II inhibitors to treat central nervous system disorders. Anal Biochem 501, 56–65. [DOI] [PubMed] [Google Scholar]
- MacKay C, Davies M, Summerfield V, Maxwell G, 2013. From pathways to people: applying the adverse outcome pathway (AOP) for skin sensitization to risk assessment. ALTEX-Alternatives to animal experimentation 30, 473–486. [DOI] [PubMed] [Google Scholar]
- Meng J, Lai MT, Munshi V, Grobler J, McCauley J, Zuck P, Johnson EN, Uebele VN, Hermes JD, Adam GC, 2015. Screening of HIV-1 Protease Using a Combination of an Ultra-High-Throughput Fluorescent-Based Assay and RapidFire Mass Spectrometry. J Biomol Screen 20, 606–615. [DOI] [PubMed] [Google Scholar]
- Natsch A, Gfeller H, 2008. LC-MS–based characterization of the peptide reactivity of chemicals to improve the in vitro prediction of the skin sensitization potential. Toxicological Sciences 106, 464–478. [DOI] [PubMed] [Google Scholar]
- Natsch A, Ryan CA, Foertsch L, Emter R, Jaworska J, Gerberick F, Kern P, 2013. A dataset on 145 chemicals tested in alternative assays for skin sensitization undergoing prevalidation. J Appl Toxicol 33, 1337–1352. [DOI] [PubMed] [Google Scholar]
- No OT, 2015. 442C: In Chemico Skin Sensitisation. OECD Guidelines for the Testing of Chemicals, Section 4
- OECD, 2012. The Adverse Outcome Pathway for Skin Sensitisation Initiated by Covalent Binding to Proteins Part 1: Scientific Evidence. OECD Environment, Health and Safety Publications Series on Testing and Assessment 168, 1–59. [Google Scholar]
- Plant M, Dineen T, Cheng A, Long AM, Chen H, Morgenstern KA, 2011. Screening for lysine-specific demethylase-1 inhibitors using a label-free high-throughput mass spectrometry assay. Anal Biochem 419, 217–227. [DOI] [PubMed] [Google Scholar]
- Roberts DW, Natsch A, 2009. High throughput kinetic profiling approach for covalent binding to peptides: application to skin sensitization potency of Michael acceptor electrophiles. Chemical research in toxicology 22, 592–603. [DOI] [PubMed] [Google Scholar]
- Saint-Mezard P, Krasteva M, Chavagnac C, Bosset S, Akiba H, Kehren J, Nicolas J, Berard F, Kanitakis J, Kaiserlian D, 2003. Afferent and efferent phases of allergic contact dermatitis (ACD) can be induced after a single skin contact with haptens: evidence using a mouse model of primary ACD. Journal of Investigative Dermatology 120, 641–647. [DOI] [PubMed] [Google Scholar]
- Tollefsen KE, Scholz S, Cronin MT, Edwards SW, de Knecht J, Crofton K, Garcia-Reyero N, Hartung T, Worth A, Patlewicz G, 2014. Applying adverse outcome pathways (AOPs) to support integrated approaches to testing and assessment (IATA). Regulatory Toxicology and Pharmacology 70, 629–640. [DOI] [PubMed] [Google Scholar]
- Urbisch D, Becker M, Honarvar N, Kolle SN, Mehling A, Teubner W, Wareing B, Landsiedel R, 2016. Assessment of pre-and pro-haptens using nonanimal test methods for skin sensitization. Chemical research in toxicology 29, 901–913. [DOI] [PubMed] [Google Scholar]
- Van Loveren H, Cockshott A, Gebel T, Gundert-Remy U, De Jong WH, Matheson J, McGarry H, Musset L, Selgrade MK, Vickers C, 2008. Skin sensitization in chemical risk assessment: report of a WHO/IPCS international workshop focusing on dose–response assessment. Regulatory Toxicology and Pharmacology 50, 155–199. [DOI] [PubMed] [Google Scholar]
- Veach BT, Mudalige TK, Rye P, 2017a. RapidFire Mass Spectrometry with Enhanced Throughput as an Alternative to Liquid-Liquid Salt Assisted Extraction and LC/MS Analysis for Sulfonamides in Honey. Anal Chem 89, 3256–3260. [DOI] [PubMed] [Google Scholar]
- Veach BT, Mudalige TK, Rye P, 2017b. RapidFire Mass Spectrometry with Enhanced Throughput as an Alternative to Liquid–Liquid Salt Assisted Extraction and LC/MS Analysis for Sulfonamides in Honey. Analytical chemistry 89, 3256–3260. [DOI] [PubMed] [Google Scholar]
- Wareing B, Urbisch D, Kolle SN, Honarvar N, Sauer UG, Mehling A, Landsiedel R, 2017. Prediction of skin sensitization potency sub-categories using peptide reactivity data. Toxicology in Vitro 45, 134–145. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Tahara H, Usami R, Kasahara T, Jimbo Y, Hioki T, Fujita M, 2015. A novel in chemico method to detect skin sensitizers in highly diluted reaction conditions. J Appl Toxicol 35, 1348–1360. [DOI] [PubMed] [Google Scholar]
- Zhang F, Erskine T, Klapacz J, Settivari R, Marty S, 2018. A highly sensitive and selective high pressure liquid chromatography with tandem mass spectrometry (HPLC/MS-MS) method for the direct peptide reactivity assay (DPRA). Journal of pharmacological and toxicological methods 94, 1–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


