Abstract
Background
Genetic variants in GARP (also known as LRRC32) have been reported to have significant associations with asthma and eczema in special populations, but little is known about allergic rhinitis. This study purposes to evaluate the association of single nucleotide polymorphisms (SNPs) in GARP with house dust mite (HDM)-sensitized persistent allergic rhinitis (PER) in a population of Han Chinese.
Methods
In this hospital-based case–control study, 534 HDM-sensitized PER patients and 451 healthy controls were recruited from East China. In this population, six SNPs in GARP were identified. Serum total and specific IgE levels were measured with ImmunoCAP. Secondary structure and minimum free energy were predicted by RNAfold.
Results
rs79525962 was associated with the risk of HDM-sensitized PER (P < 0.05). The individuals with CT+TT genotype demonstrated a higher risk of HDM-sensitized PER than those with CC genotype (adjusted OR = 1.393, 95% CI = 1.019–1.904). The homozygous genotype CC of rs3781699 rendered a lower risk of HDM-sensitized PER than the wild-type genotype AA (adjusted OR = 0.646, 95% CI = 0.427–0.976); however, the genotype and allele frequencies of rs3781699 demonstrated no associations with HDM-sensitized PER (P > 0.05). rs79525962 increased the risk of HDM-sensitized PER in the subgroup aged ≥16 years (adjusted OR = 1.745, 95% CI = 1.103–2.760), and this high risk was also found in the females (adjusted OR = 1.708, 95% CI = 1.021–2.856). The G-C haplotype of rs1320646-rs3781699 rendered a lower risk of HDM-sensitized PER than the common haplotype G-A (adjusted OR = 0.819, 95% CI = 0.676–0.993). The secondary structure of GARP altered in response to different genotypes of rs79525962 and rs3781699.
Conclusion
SNP rs79525962 in the GARP gene marks a risk locus of HDM-sensitized PER in Chinese Hans.
Keywords: allergic rhinitis, mites, GARP, LRRC32, single nucleotide polymorphism, genetic association studies
Introduction
Immunoglobulin E (IgE) mediates inflammation to evoke allergic rhinitis (AR), featuring nasal symptoms such as nasal itching, sneezing, rhinorrhea, and nasal congestion.1,2 The prevalence of allergic diseases, such as AR and asthma, has kept increasing markedly worldwide over the past 50 years.1 Currently, AR prevalence ranges from 9.8% to 23% in China adults.3 Epidemiologic studies have consistently shown that asthma and rhinitis often prevail in the same population throughout the world.4–6 As the upper and lower airway inflammatory responses present physiological similarities, rhinitis is considered as a risk factor for asthma at the pediatric age.7,8
AR involves an intricate cooperation between genetic predisposition and environmental factors, with implicated genes being intensely studied.9 Genomic searches have verified the roles of chromosomes 2, 3, 4, and 99. Some AR-related single nucleotide polymorphisms (SNPs) have also been discovered10–13. Genome-wide association studies have found that through biological pathways involved in the immunosuppression of regulatory T lymphocytes (Tregs),14 a common variant on chromosome 11q13.5 near the glycoprotein A repetitions predominant gene (GARP, also known as leucine-rich repeat containing 32, LRRC32) increases the susceptibility to asthma,15 AR,16 and atopic dermatitis.17 Disease-associated genetic polymorphisms within 11q13.5 can regulate GARP expression on human Treg cells.18 In a recent study, by using a model of conditional Garp-deficient mice, Lehmkuhl et al19 confirmed that the decreased GARP expression on Tregs increased susceptibility to inflammatory diseases. They also found that as Foxp3 protein acetylation diminished, Garp deficiency led to an unstable Treg phenotype, suggesting that GARP plays a central role in maintaining immune stability.
Genetic variants in GARP have been reported to have significant associations with asthma and eczema in special populations,20 but little is known about AR. Here, we genotyped and analyzed six GARP SNPs in a Chinese Han population to manifest the role of GARP polymorphisms in AR induced by house dust mites (HDM), and its clinical phenotypes.
Materials and Methods
Subjects
This is a hospital-based, case–control study of the association between genetic variations in GARP and susceptibility to AR. This study included 534 unrelated cases (356 males, 178 females) with persistent allergic rhinitis (PER) ages 2 to 66 years (median age [quartiles] of 16.0 [10.0–27.0] years) and 451 unrelated healthy controls (278 males, 173 females) ages 3 to 63 years (median age [quartiles] of 16.0 [10.0–29.0] years). All these subjects were enrolled from Chinese Hans in Jiangsu and Anhui provinces in East China, who had received medical care in the First Affiliated Hospital of Nanjing Medical University between 2008 and 2014. PER was diagnosed according to the criteria described in Allergic Rhinitis and its Impact on Asthma (ARIA) 2008 Update.1 A questionnaire was used to obtain data about disease history, family history, symptoms and concurrent diseases. The controls were recruited from the hospital seeking health care or routine health examinations and were frequency-matched with the cases in age (±5 years) and sex. The selection criteria for controls:12,13,21 (1) no symptoms and medical history of AR and nasal diseases; (2) no symptoms and medical history of other allergic diseases, such as asthma, eczema and urticaria; (3) negative blood test for serum allergen-specific IgE in the phadiatop assay (Phadia, Uppsala, Sweden); (4) no history of AR or other allergic diseases in the immediate family. The exclusion criteria for all subjects: (1) complicated with acute upper respiratory tract infection, severe deviation of nasal septum, rhinosinusitis with or without nasal polyps, neoplasms in paranasal sinuses and nasal cavity; (2) combined with other immune diseases and systemic diseases; (3) use of corticosteroids during previous 4 weeks and antihistamines/antileukotrienes during previous 2 weeks. The research protocol complying with the Declaration of Helsinki was approved by the Ethics Committee of Nanjing Medical University (20080305), and written informed consent was obtained from all participants.
Clinical Parameters
A visual analogue scale (VAS) was used to assess the severity of nasal symptoms. 0 was a score meaning “not at all bothersome”, and 10 meaning “extremely bothersome”. VAS score ≤5 indicated mild AR and VAS score >5 indicated moderate-to-severe AR.22
After interview, about 5 mL of peripheral blood was collected from each subject for in vitro allergy testing. The ImmunoCAP assays (Phadia, Uppsala, Sweden) were carried out to measure the levels of serum total IgE and specific IgE antibodies to common inhalant allergens, including Dermatophagoides pteronyssinus (Der p, d1), Dermatophagoides farinae (Der f, d2), cat dander (e1), dog dander (e5), Blatella germanica (i6), Alternaria alternata (m6), Ambrosia elatior (w1), and Artemisia vulgaris (w6). Specific IgE ≥0.35 kUA/L indicated positivity. AR cases were sensitized with HDM (Der p and/or Der f), and 139 of them (26.0%) were further sensitized with other aeroallergens.
SNP Selection and Genotyping
Six SNPs in the GARP gene (rs947998, rs79525962, rs1320646, rs3781699, rs1803627 and rs7685) were selected based on genotype data of Han Chinese in Beijing (CHB) in CSHL-HAPMAP (HapMap Data Rel 27 Phase II+III, Feb09, on NCBI B36 assembly, dbSNP b126), and the allele frequency data from 1000GENOMES (pilot_1_CHB+JPT_mar_2010, b132). For genotyping, the TaqMan SNP Genotyping Assay was performed using the 384-well ABI 7900HT Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Concordance was 100% in more than 15% of the samples randomly selected. The primers and TaqMan probes are shown in Table 1.
Table 1.
Primers and Probes for Genotyping by TaqMan Assay
| NCBI rs No. | Base Change | Primersa | Probesa |
|---|---|---|---|
| rs947998 | G>T | F: GCCTGATCTTTGAAAACACTACACA | G allele: FAM-CCCAGGCCGCAGC-MGB |
| R: CCACACTGCTTCTCCAAAATTAGTT | T allele: HEX-CCCAGGCCTCAGC-MGB | ||
| rs79525962 | C>T | F: GGTTCCCCTGCAGGTTGAG | G allele: FAM-ACACCTTTGCCAATC-MGB |
| R: CTTAAGCCACAATGCCCTGGAG | A allele: HEX-ATACACCTTTACCAATCT-MGB | ||
| rs1320646 | G>A | F: TCTTCTGAGAATGACTTTCAGTCTCTCT | G allele: FAM-TAGGACCGGAAGAGA-MGB |
| R: CCAGATCCGAGACACACTCGTA | A allele: HEX-TTAGGACCAGAAGAGAG-MGB | ||
| rs3781699 | A>C | F: GCCAAGCTGGGTGCAAAA | T allele: FAM-AGAACCAGATATCTAAG-MGB |
| R: GCTGCTGAGCCAGGAGCTAA | G allele: HEX-AGAACCAGATAGCTAAGGT-MGB | ||
| rs1803627 | T>G | F: TCTCTCTGTGCTCTTGCATTCTCT | T allele: FAM-ATTCCCTTTTCCTCTATTGA-MGB |
| R: TCATTCTCTTCCTAAGCCTCAGTTTC | G allele: HEX-TTTTCCTCTATGGAGCAGA-MGB | ||
| rs7685 | T>G | F: GCCTGATCTTTGAAAACACTACACA | G allele: FAM-CCCAGGCCGCAGC-MGB |
| R: CCACACTGCTTCTCCAAAATTAGTT | T allele: HEX-CCCAGGCCTCAGC-MGB |
Note: aThe alleles were arrayed as the location of the primers or probes from 5’ to 3’.
In silico Analysis
HaploReg (http://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) and RegulomeDB (https://www.regulomedb.org/regulome-search/) were used to predict putative functions of SNPs.23 HaploReg could be used to predict the functions of variations, including sequence conservation, regulatory protein binding, expression quantitative trait loci, regulatory motifs, and catalog of variants.24 RegulomeDB score represented different functions of SNPs.25 Changes in secondary structure and minimum free energy (MFE) in different SNP genotypes were predicted by using RNAfold (http://rna.tbi.univie.ac.at/). The alterations of secondary structure and MFE might change the binding affinity to microRNAs.26
Statistical Analysis
A goodness-of-fit χ2 test was first performed to measure allele frequencies against departure from the Hardy-Weinberg equilibrium (HWE). Differences in demographic data, selected variables, and frequencies of genotypes and alleles between PER patients and the controls were evaluated. Two-sided χ2 test was employed for categorical variables, Student’s t-test for continuous variables in normal distribution, and nonparametric test for continuous variables in non-normal distribution. Data about serum total IgE were normalized with a logarithmic model. Percentiles of total IgE measurements (n = 979) were calculated to divide the total serum IgE into “low” (<90th quantile [712.0 kU/L] after logarithmic transformation) and “high” (≥90th quantile [712.0 kU/L] after logarithmic transformation).27 Analysis of covariance and nonparametric tests were conducted to manifest the associations between GARP polymorphisms and clinical phenotypes. Unconditional univariate and multivariate logistic regression models were established to generate crude and adjusted odds ratios (ORs) and 95% confidence intervals (CIs) for the association between GARP polymorphisms and the risk of HDM-sensitized PER. Multivariate analysis was performed after age and gender adjustment. The SNP indicating a positive association with PER was further submitted to stratification analysis of age, gender, concomitant asthma, family history of allergic diseases, VAS score, and serum total IgE levels. The linkage disequilibrium (LD) between SNPs was computed using the normalized measure of allelic association D’ and r2 and illustrated using HaploView 4.2 software.28 Haplotype analysis was performed by HAPSTAT 3.0 (The University of North Carolina at Chapel Hill, NC, USA) with observed genotypes. All statistical analyses were accomplished with SPSS software version 20.0.0 (IBM Corporation, Armonk, NY, USA). P-value <0.05 was considered statistically significant.
Results
Basic Demographic Data
In terms of frequency distributions (Table 2), the cases and controls were well matched in age (P = 0.535) and gender (P = 0.101). The median age of all subjects was 16 years. PER cases showed higher serum total IgE (255.1 [118.9–560.5] kU/L) (P<0.001) than controls (25.2 [10.4–47.2] kU/L). All PER cases were sensitized with HDM (Der p and/or Der f), and 139 of them (26.0%) were further sensitized with other aeroallergens; however, they were not the main allergens causing symptoms. The serum levels of allergen-specific IgE against Der p and Der f in PER cases were 27.4 (6.1–69.5) kUA/L and 23.1 (6.0–63.0) kUA/L, respectively. In PER cases, 208 (45.6%) had mild and 248 (54.4%) had moderate-to-severe AR according to VAS score; 124 (27.5%) reported concomitant asthma and 154 (34.4%) had family history of allergic diseases. These variables were subjected to the multivariate logistic regression analysis to eliminate residual confounding effect.
Table 2.
Distribution of Selected Variables Among Cases and Controls
| Variables | Case (n=534) | Control (n=451) | P | ||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Age (years), median (quartiles) | 16.0 (10.0–27.0) | 16.0 (10.0–29.0) | 0.535a | ||
| Gender | |||||
| Male | 356 | 66.7 | 278 | 61.6 | 0.101a |
| Female | 178 | 33.3 | 173 | 38.4 | |
| Serum total IgE (kU/L), median (quartiles) | 255.1 (118.9–560.5) | 25.2 (10.4–47.2) | < 0.001b | ||
| Specific IgE (kUA/L), median (quartiles) | |||||
| Der p | 27.4 (6.1–69.5) | ||||
| Der f | 23.1 (6.0–63.0) | ||||
| Rhinitis severity (VAS score)c | |||||
| Mild (VAS score ≤ 5) | 208 | 45.6 | |||
| Moderate-to-severe (VAS score > 5) | 248 | 54.4 | |||
| Concomitant asthmac | |||||
| Yes | 124 | 27.5 | |||
| No | 327 | 72.5 | |||
| Family history of allergic rhinitisc | |||||
| Yes | 154 | 34.4 | |||
| No | 294 | 65.6 | |||
Notes: aTwo-sided χ2 test for comparison of discrete variables and nonparametric test for continuous variables with non-normal distribution. bSelective variables were transformed into logarithmic model before unpaired Student’s t-test between cases and controls. cData about VAS score, concomitant asthma, and family history of allergic diseases were not available in some cases.
Abbreviations: Der p, Dermatophagoides pteronyssinus; Der f, Dermatophagoides farina; VAS, visual analogue scales.
Association Analysis of Each SNP
The genotype distributions in controls presented similarity with those in HWE (all P > 0.05, Table 3). As shown in Table 4, the genotype (P = 0.034) and allele (P = 0.018) frequencies of rs79525962 were significantly different between the cases and the controls. In addition, genotype CT+TT of rs79525962 brought with a higher risk of HDM-sensitized PER than the wild-type genotype CC (adjusted OR = 1.393, 95% CI = 1.019–1.904). The homozygous genotype CC of rs3781699 brought with a lower risk of HDM-sensitized PER than the wild-type genotype AA (adjusted OR = 0.646, 95% CI = 0.427–0.976); however, the genotype and allele frequencies of rs3781699 showed no significant association with the risk of HDM-sensitized PER (all P > 0.05).
Table 3.
Primary Information of Six SNPs in the GARP Gene
| SNP No. | NCBI rs No. | Chromosome Positiona | Location | Base Change | MAF | P for HWEc | Genotyped (%) | ||
|---|---|---|---|---|---|---|---|---|---|
| Databaseb | Case | Control | |||||||
| 1 | rs947998 | 76672762 | 5’ near gene | G>T | 0.305 | 0.310 | 0.317 | 0.290 | 98.1 |
| 2 | rs79525962 | 76660374 | exon | C>T | 0.092 | 0.129 | 0.095 | 0.274 | 99.2 |
| 3 | rs1320646 | 76659003 | 3ʹUTR | G>A | 0.111 | 0.093 | 0.109 | 0.893 | 99.3 |
| 4 | rs3781699 | 76658741 | 3ʹUTR | A>C | 0.407 | 0.326 | 0.362 | 0.392 | 99.8 |
| 5 | rs1803627 | 76658300 | 3ʹUTR | T>G | 0.061 | 0.087 | 0.081 | 0.051 | 99.2 |
| 6 | rs7685 | 76657595 | 3ʹUTR | T>G | 0.350 | 0.332 | 0.355 | 0.548 | 99.2 |
Notes: aSNP position in NCBI dbSNP (http://www.ncbi.nlm.nih.gov/snp). bMAF for CHB from the HapMap databases (http://hapmap.ncbi.nlm.nih.gov) or NCBI dbSNP (http://www.ncbi.nlm.nih.gov/snp). cHWE P-value in the control group using a goodness-of-fit χ2 test.
Abbreviations: SNP, single nucleotide polymorphism; GARP, glycoprotein A repetitions predominant; MAF, minor allele frequencies; HWE, Hardy-Weinberg equilibrium; UTR, untranslated region; CHB, Han Chinese in Beijing, China.
Table 4.
Genotypes and Allele Frequencies in GARP Polymorphisms Among Cases and Controls
| SNP No. | NCBI rs No. | Genotypes | Case | Control | Crude OR (95% CI) | Adjusted OR (95% CI)a | Pb | ||
|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | ||||||
| 1 | rs947998 | n=531 | n=435 | ||||||
| GG | 249 | 46.9 | 198 | 45.5 | 1.000 (reference) | 1.000 (reference) | 0.911 | ||
| GT | 235 | 44.3 | 198 | 45.5 | 0.944 (0.724–1.231) | 0.937 (0.718–1.225) | |||
| TT | 47 | 8.9 | 39 | 9.0 | 0.958 (0.603–1.524) | 0.988 (0.620–1.574) | |||
| GT+TT | 282 | 53.1 | 237 | 54.5 | 0.946 (0.734–1.220) | 0.946 (0.733–1.221) | 0.670 | ||
| T allele | 0.310 | 0.317 | 0.725 | ||||||
| 2 | rs79525962 | n=533 | n=444 | ||||||
| CC | 406 | 76.2 | 362 | 81.5 | 1.000 (reference) | 1.000 (reference) | 0.034 | ||
| CT | 117 | 22.0 | 80 | 18.0 | 1.304 (0.949–1.792) | 1.319 (0.959–1.814) | |||
| TT | 10 | 1.9 | 2 | 0.5 | 4.458 (0.970–20.481) | 4.225 (0.918–19.452) | |||
| CT+TT | 127 | 23.8 | 82 | 18.5 | 1.381 (1.011–1.886) | 1.393 (1.019–1.904) | 0.042 | ||
| T allele | 0.129 | 0.095 | 0.018 | ||||||
| 3 | rs1320646 | n=532 | n=446 | ||||||
| GG | 442 | 83.1 | 354 | 79.4 | 1.000 (reference) | 1.000 (reference) | 0.169 | ||
| GA | 81 | 15.2 | 87 | 19.5 | 0.746 (0.534–1.041) | 0.742 (0.531–1.037) | |||
| AA | 9 | 1.7 | 5 | 1.1 | 1.442 (0.479–4.340) | 1.460 (0.484–4.405) | |||
| GA+AA | 90 | 16.9 | 92 | 20.6 | 0.783 (0.568–1.082) | 0.781 (0.565–1.079) | 0.138 | ||
| A allele | 0.093 | 0.109 | 0.250 | ||||||
| 4 | rs3781699 | n=534 | n=449 | ||||||
| AA | 240 | 44.9 | 187 | 41.6 | 1.000 (reference) | 1.000 (reference) | 0.151 | ||
| AC | 240 | 44.9 | 199 | 44.3 | 0.940 (0.719–1.229) | 0.932 (0.713–1.220) | |||
| CC | 54 | 10.1 | 63 | 14.0 | 0.668 (0.443–1.007) | 0.646 (0.427–0.976) | |||
| AC+CC | 294 | 55.1 | 262 | 58.4 | 0.874 (0.679–1.127) | 0.863 (0.670–1.114) | 0.299 | ||
| C allele | 0.326 | 0.362 | 0.093 | ||||||
| 5 | rs1803627 | n=534 | n=443 | ||||||
| TT | 446 | 83.5 | 377 | 85.1 | 1.000 (reference) | 1.000 (reference) | 0.574 | ||
| TG | 83 | 15.5 | 60 | 13.5 | 1.169 (0.816–1.675) | 1.171 (0.817–1.678) | |||
| GG | 5 | 0.9 | 6 | 1.4 | 0.704 (0.213–2.326) | 0.701 (0.212–2.320) | |||
| TG+GG | 88 | 16.5 | 66 | 14.9 | 1.127 (0.796–1.595) | 1.128 (0.797–1.597) | 0.500 | ||
| G allele | 0.087 | 0.081 | 0.645 | ||||||
| 6 | rs7685 | n=532 | n=445 | ||||||
| TT | 237 | 44.5 | 188 | 42.2 | 1.000 (reference) | 1.000 (reference) | 0.492 | ||
| TG | 237 | 44.5 | 198 | 44.5 | 0.949 (0.726–1.242) | 0.942 (0.719–1.233) | |||
| GG | 58 | 10.9 | 59 | 13.3 | 0.780 (0.518–1.175) | 0.757 (0.501–1.143) | |||
| TG+GG | 295 | 55.5 | 257 | 57.8 | 0.911 (0.706–1.174) | 0.899 (0.697–1.161) | 0.470 | ||
| G allele | 0.332 | 0.355 | 0.281 | ||||||
Notes: aAdjusted for age and gender in multivariate logistic regression model. Bold values denote statistical significance at the P < 0.05 level. bTwo-sided χ2 test for the distributions of genotype and allele frequencies.
Abbreviations: GARP, glycoprotein A repetitions predominant; SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Stratification Analysis of SNP rs79525962 in PER Subgroups
Considering that rs79525962 showed a significant association with HDM-sensitized PER, we evaluated the distributions of rs79525962 among PER subgroups. PER patients were stratified into six subgroups based on age, gender, concomitant asthma, family history of allergic diseases, rhinitis severity and serum total IgE level. As shown in Table 5, in contrast to wild-type genotype CC, genotype CT+TT of rs79525962 brought with a higher risk of HDM-sensitized PER in the subgroup of age ≥16 years (adjusted OR = 1.745, 95% CI = 1.103–2.760), and this high risk was also found in the females (adjusted OR = 1.708, 95% CI = 1.021–2.856). No evident associations were found between rs79525962 with other PER-related variables (all P > 0.05).
Table 5.
Stratification Analysis of rs79525962 in the Dominant Model in PER Subgroups
| Variables | Subcategory | Case | Control | Adjusted OR (95% CI)a |
|---|---|---|---|---|
| Age (years) | n=533 | n=444 | ||
| < 16 | 257 | 225 | 1.076 (0.693–1.671) | |
| ≥ 16 | 276 | 219 | 1.745 (1.103–2.760) | |
| Gender | n=533 | n=444 | ||
| Male | 356 | 273 | 1.241 (0.836–1.840) | |
| Female | 177 | 171 | 1.708 (1.021–2.856) | |
| Concomitant asthma | n=450 | |||
| No | 326 | 1.000 (reference) | ||
| Yes | 124 | 0.936 (0.565–1.552) | ||
| Family history of allergic diseases | n=447 | |||
| No | 293 | 1.000 (reference) | ||
| Yes | 154 | 0.987 (0.620–1.571) | ||
| Rhinitis severity (VAS score) | n=455 | |||
| VAS score ≤ 5 | 207 | 1.000 (reference) | ||
| VAS score > 5 | 248 | 0.909 (0.585–1.411) | ||
| Serum total IgEb | n=529 | n=442 | ||
| Lower level | 431 | 442 | 1.000 (reference) | |
| Higher level | 98 | 1.144 (0.696–1.880) |
Notes: aAdjusted for age and gender in multivariate logistic regression model. Bold values denote statistical significance at the P < 0.05 level. bLower level: below the 90th percentile of logarithmic total IgE; higher level: above the 90th percentile of logarithmic total IgE.
Abbreviations: Dominant model: MW+MM/WW; MW: heterozygotes; MM: mutation homozygotes; WW: wild homozygotes. PER, persistent allergic rhinitis; VAS, visual analogue scale; OR, odds ratio; CI, confidence interval.
Association Between GARP Polymorphisms and AR-Related Phenotypes
As shown in Table 6, the genotypes of rs947998, rs79525962, rs1320646, rs3781699, rs1803627 and rs7685 in the GARP gene were not linked with the susceptibility to PER with or without asthma in the dominant model (all P > 0.05). Given the significant difference in serum total IgE levels between cases and controls, the associations of serum total IgE with six SNPs were evaluated separately. As showed in Table 7, both total IgE levels demonstrated no significant difference among individuals classified according to genotypes of six SNPs (all P > 0.05). No obvious associations were observed between different SNP genotypes and other AR-related phenotypes, including VAS score and specific IgE levels against Der p and Der f (all P > 0.05).
Table 6.
Genotype Frequencies of GARP Polymorphisms in the Dominant Model in PER Patients with and without Asthma
| SNP No. | NCBI rs No. | Case with Asthma vs Control | Case without Asthma vs Control |
|---|---|---|---|
| Adjusted OR (95% CI)a | Adjusted OR (95% CI)a | ||
| 1 | rs947998 | 0.670 (0.445–1.008) | 1.140 (0.853–1.525) |
| 2 | rs79525962 | 1.249 (0.765–2.041) | 1.410 (0.993–2.003) |
| 3 | rs1320646 | 0.948 (0.570–1.576) | 0.714 (0.490–1.041) |
| 4 | rs3781699 | 0.919 (0.612–1.380) | 0.951 (0.712–1.271) |
| 5 | rs1803627 | 0.950 (0.536–1.684) | 1.199 (0.813–1.768) |
| 6 | rs7685 | 1.060 (0.703–1.598) | 0.945 (0.707–1.263) |
Note: aAdjusted for age and gender in multivariate logistic regression model.
Abbreviations: Dominant model, MW+MM/WW; MW, heterozygotes; MM, mutation homozygotes; WW, wild homozygotes; GARP, glycoprotein A repetitions predominant; PER, persistent allergic rhinitis; SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Table 7.
Associations Between GARP Polymorphisms and AR-Related Phenotypes
| SNP No. | NCBI rs No. | P for Serum Total IgEa | P for Specific IgEb | P for VAS Scorec | ||
|---|---|---|---|---|---|---|
| Case | Control | Der p | Der f | |||
| 1 | rs947998 | 0.782 | 0.780 | 0.179 | 0.213 | 0.139 |
| 2 | rs79525962 | 0.608 | 0.270 | 0.820 | 0.800 | 0.524 |
| 3 | rs1320646 | 0.713 | 0.442 | 0.543 | 0.244 | 0.518 |
| 4 | rs3781699 | 0.173 | 0.938 | 0.803 | 0.434 | 0.736 |
| 5 | rs1803627 | 0.944 | 0.232 | 0.874 | 0.678 | 0.717 |
| 6 | rs7685 | 0.361 | 0.956 | 0.977 | 0.628 | 0.529 |
Notes: aSelective variables were transformed into logarithmic model to normalize the distribution before analysis of covariance adjusted for age and gender. bNonparametric test for selective variables with non-normal distribution. cAnalysis of covariance adjusted for age and gender for selective variables with normal distribution.
Abbreviations: GARP, glycoprotein A repetitions predominant; AR, allergic rhinitis; SNP, single nucleotide polymorphism; Der p, Dermatophagoides pteronyssinus; Der f, Dermatophagoides farinae.
Association Between GARP Haplotypes and PER
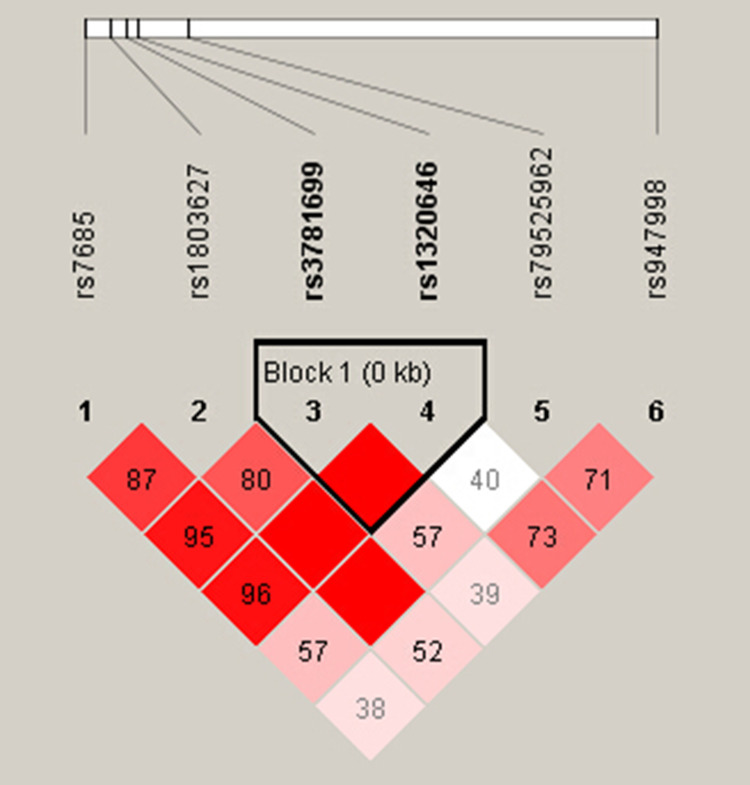
As shown in Figure 1, the LD between rs1320646 and rs3781699 was quite strong (D′ = 1.0, r2 = 0.058). We then identified rs1320646-rs3781699 as a potential GARP haplotype and evaluated its association with PER risk among cases and controls. As shown in Table 8, compared with the common haplotype G-A of rs1320646-rs3781699, the G-C haplotype exhibited a lower risk of HDM-sensitized PER (adjusted OR = 0.819, 95% CI = 0.676–0.993).
Figure 1.
Linkage disequilibrium (LD) of six SNPs in the GARP gene. LD of six SNPs was determined using the solid spine of LD option of Haploview 4.2. D’ values are displayed in the squares. Empty red squares have a pairwise D’ of 1.0. Red squares indicate strong pairwise LD, gradually coloring down to white squares of weak pairwise LD.
Table 8.
Associations Between PER Risk and the Frequencies of Haplotypes
| Haplotype | Haplotype Frequencies | Adjusted OR (95% CI)a | |
|---|---|---|---|
| rs1320646-rs3781699 | Case | Control | |
| G-A | 0.581 | 0.529 | 1.000 (reference) |
| G-C | 0.326 | 0.362 | 0.819 (0.676–0.993) |
| A-A | 0.093 | 0.109 | 0.777 (0.573–1.050) |
Notes: aDerived from logistic regression model performed by HAPSTAT 3.0 based on observed genotypes. Bold values denote statistical significance at the P < 0.05 level.
Abbreviation: PER, persistent allergic rhinitis; OR, odds ratio; CI, confidence interval.
Functions of SNP rs1320646, rs3781699 and rs79525962
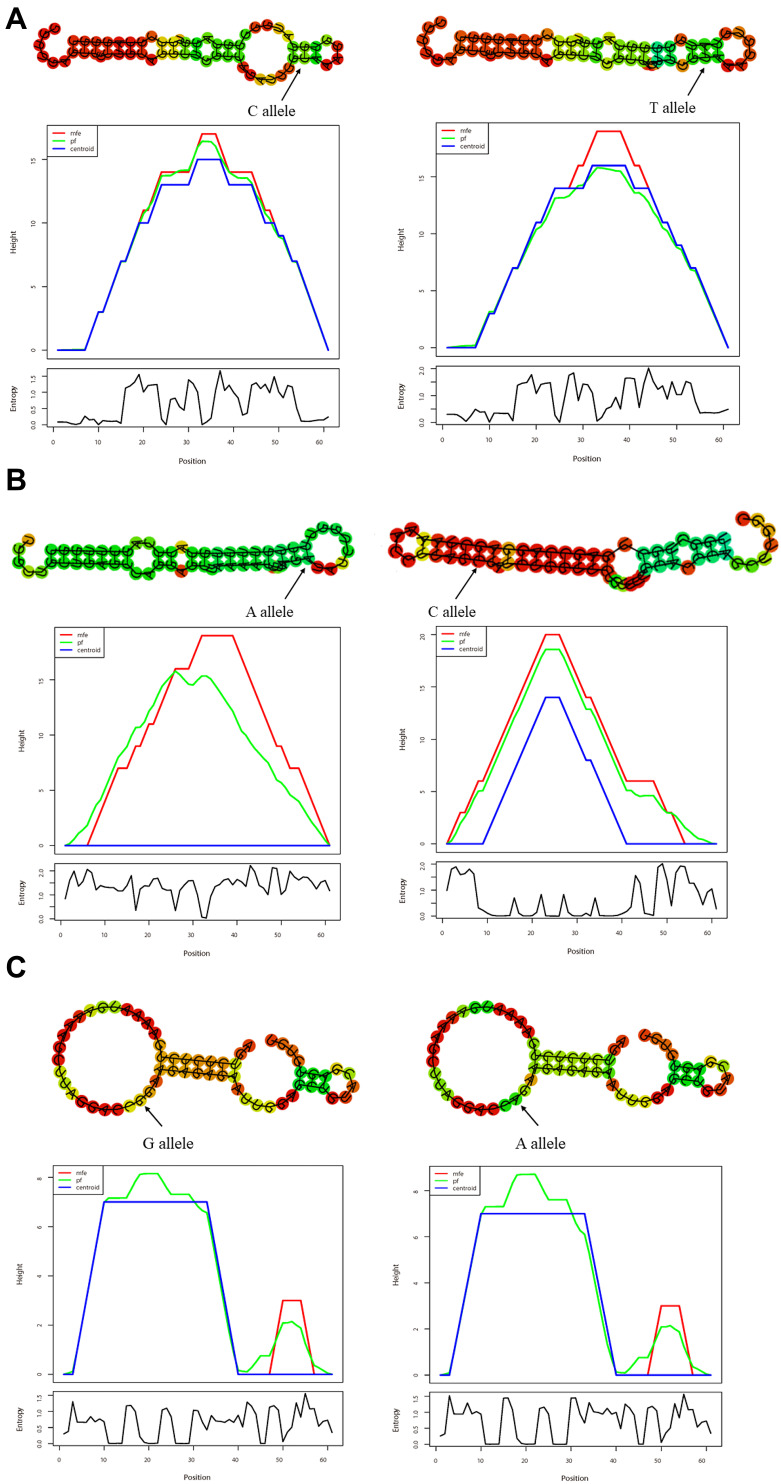
Using HaploReg, we observed that rs1320646, rs3781699 and rs79525962 possessed enhancer histone marks, altered motifs and selected expression quantitative trait locus (eQTL) hits. Besides, GRASP QTL hits were rich in rs1320646 and rs79525962. The RegulomeDB scores of rs1320646, rs3781699 and rs79525962 were 1f, 3a and 4, respectively (Table 9). RNAfold predicted that rs79525962 C to T substitution led to the alteration in GARP secondary structure, with MFE increasing from −21 kcal/mol to −19.1 kcal/mol (Figure 2A), indicating that the C allele had a higher binding affinity to microRNAs than the T allele. We also observed that rs3781699 A to C substitution changed the secondary structure of GARP, with MFE decreasing from −16.4 kcal/mol to −21.6 kcal/mol, suggesting that the C allele had a higher binding affinity to microRNAs than the A allele (Figure 2B). However, no similar results were found in rs1320646 (Figure 2C).
Table 9.
Functional Annotation of 3 SNPs in silico Analysis
| Chr: Positiona | Variant | Scoreb | Enhancer Histone Marks | Motifs Changed | GRASP QTL Hits | Selected eQTL Hits | dbSNP Func Annot |
|---|---|---|---|---|---|---|---|
| 11: 76659003 | rs1320646 | 1f | 7 tissues | 7 altered motifs | 2 hits | 3 hits | 3ʹUTR |
| 11: 76658741 | rs3781699 | 3a | 8 tissues | Hltf | – | 3 hits | 3ʹUTR |
| 11: 76660374 | rs79525962 | 4 | 7 tissues | 7 altered motifs | 2 hits | 3 hits | Missense |
Notes: aBased on NCBI build 38 of the human genome. bBased on RegulomeDB (https://www.regulomedb.org/regulome-search/).
Abbreviation: SNP, single nucleotide polymorphisms; GRASP, Genome-Wide Repository of Associations Between SNPs and Phenotypes; eQTL, expression quantitative trait locus; UTR, untranslated region.
Figure 2.
In silico prediction of secondary structures and minimum free energy (MFE) changes corresponding to GARP SNPs. (A) The MFE changed from −21 kcal/mol to −19.1 kcal/mol caused by rs79525962 (C > T), indicating a stronger binding affinity of microRNAs to the C allele. (B) The MFE changed from −16.4 kcal/mol to −21.6 kcal/mol caused by rs3781699 (A > C), indicating a stronger binding affinity of microRNAs to the C allele. (C) No change was found in MFE caused by rs1320646.
Abbreviation: GARP, glycoprotein A repetitions predominant.
Discussion
In this study, we report SNP rs79525962 in GARP appears to mark a genuine HDM-sensitized PER risk locus and the GARP gene may contribute to susceptibility of HDM-sensitized PER in this Chinese Han population. Moreover, we predict a putative function of the SNPs. Current literature suggests our research unprecedented in exploring the relationship between GARP variations and AR risk in Chinese population.
GARP protein, encoded by the GARP gene localized in the 11q13.5 chromosomal region, is a 80-kDa transmembrane protein with its extracellular region primarily composed of 20 leucine-rich repeats.14 GARP protein is highly expressed on the surface of activated Tregs and increases the immune-suppressing function of Tregs.29 In addition, GARP, functioning as a carrier and a cell surface receptor for latent transforming growth factor (TGF)-β, binds directly to the latent TGF-β, then modifies latent TGF-β into active TGF-β.14 Tregs can regulate immune tolerance and inflammatory responses,30,31 which in turn makes it a potential target in the treatment of allergic diseases. TGF-β is a pleiotropic cytokine critical to the generation of Treg cells.32 It plays crucial roles in the remodeling and immunosuppression of allergic inflammatory airway diseases33 and mediates pro- and anti-inflammatory responses.34
Our recent studies have found that a TGFB1 promoter polymorphism may be associated with the susceptibility and the severity of PER,12 and SNPs in microRNA target sites of TGF-β signaling pathway genes may be associated with risk of HDM-sensitized PER stratified by age and gender in Chinese Hans.13 Studies have demonstrated that GARP promotes TGF-β secretion and activation, then increases the suppressive function of Tregs through TGF-β.35–37 Moreover, GARP/latent TGF-β1 complexes can induce Th17 differentiation.14 Studies have also reported that human B lymphocytes, upon stimulation, produce active TGF-β1 from surface GARP/latent TGF-β1 complexes and induce IgA isotype switching.38,39 As GARP mediates the immune-suppressing activity of Treg cells,36,37,40,41 and allergen-dependent inflammation is suppressed by Tregs via soluble GARP,29 we proposed that GARP might be implicated in the pathogenesis of AR.
The present study is the first to evaluate the genetic association of GARP polymorphisms with AR in Chinese population. The results suggested that SNP rs79525962 was significantly correlated with HDM-sensitized PER risk, and genotype CT+TT with a high risk of HDM-sensitized PER, compared with the wild-type genotype CC. The C/T polymorphism of rs79525962 located within the exon in the GARP gene yields missense coding sequence. This polymorphism drives the transition of alanine (C allele) into threonine (T allele), consequently compromising the conformation and function of GARP protein and might affect the suppressive role of Tregs. Moreover, we observed that rs79525962 C to T substitution altered the secondary structure of GARP. Previous study reported that structural changes affect the stability and translation of mRNA.42 Hence, rs79525962 might regulate the translation of GARP and subsequent Tregs’ function by this way. Manz et al17 have shown a significant excess of LRRC32 variants, such as rs79525962, in patients with atopic dermatitis. Structural protein modeling and bioinformatic analysis have revealed that protein transport is disrupted upon these variants. Deficient Tregs responses are associated with various allergic and autoimmune diseases,43,44 suggesting that rs79525962 polymorphism of the GARP gene may have a role in these diseases.
In the stratification analysis of rs79525962 in PER subgroups, we found rs79525962 exhibited a significant association with HDM-sensitized PER risk in the subgroups of age ≥16 years and females. Therefore, age of ≥16 years and female may be factors for susceptibility to PER caused by GARP variants. This susceptibility may arise from the immature immunity in adolescents and the cycle-mediated endocrine function in women. On the other hand, lack of association was found between rs79525962 and AR concomitant asthma. We speculate that GARP polymorphisms increase the risk of allergic sensitization which, in turn, increases the risk of subsequent development of AR and asthma. Ferreira et al45 found a genetic variant (rs7130588) in GARP was significantly associated with atopic status among asthmatics, suggesting that it is a risk factor for allergic but not non-allergic asthma, which was consistent with our results.
Furthermore, as various SNPs may co-work to raise the risk of AR, we identified rs1320646-rs3781699 as a potential haplotype. Compared with G-A of rs1320646-rs3781699, the G-C haplotype decreased the susceptibility to HDM-sensitized PER. Genetic variant rs3781699 was located in the 3ʹUTR region of GARP. At the post-transcriptional level, microRNAs bind to the 3ʹUTR of their target messenger RNA to regulate GARP expression.46 Notably, the mutation (A to C allele) in rs3781699 decreased MFE from −16.4 kcal/mol to −21.6 kcal/mol, indicating that the C allele has a higher binding affinity to microRNAs than the A allele.26 Thus, we speculated that the G-C haplotype of rs1320646-rs3781699 might regulate the expression of GARP through this pathway, therefore decreasing the susceptibility to HDM-sensitized PER.
Several limitations of this study should be addressed. First, the homozygous genotype CC of rs3781699 was associated with a low risk of HDM-sensitized PER, compared with the wild-type genotype AA; however, no evident associations were established between the genotype and allele frequencies of rs3781699 and HDM-sensitized PER, which might be explained by that our sample size was not large enough and the effect of rs3781699 on AR pathogenesis was mild. Therefore, further larger-scale studies should be required in Chinese population. Second, the cases were collected from one single tertiary hospital setting in East China, and they may not be representative enough for the whole Han Chinese. Last but not least, GARP-related genes were not analyzed in the present study.
Conclusions
SNP rs79525962 in the GARP gene marks a risk locus of HDM-sensitized PER, suggesting that GARP polymorphisms might drive the development of HDM-sensitized PER in this population of Han Chinese. Further functional studies are required to demonstrate their molecular mechanisms in PER, and more detailed environmental exposure data are required to verify the effect of gene–environment interaction on HDM-sensitized PER.
Acknowledgments
We are grateful to the participants in this study and all staff involved in the study through the years. We also thank associate professor Yong-Ke Cao at the College of Foreign Languages of Nanjing Medical University for professional English-language proofreading of the manuscript.
Funding Statement
This work was supported by grants from the Health Promotion Project of Jiangsu Province (XK200719) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (JX10231801), People’s Republic of China.
Abbreviations
AR, allergic rhinitis; ARIA, Allergic Rhinitis and its Impact on Asthma; CHB, Han Chinese in Beijing; CIs, confidence intervals; Der f, Dermatophagoides farinae; Der p, Dermatophagoides pteronyssinus; eQTL, expression quantitative trait locus; GARP, glycoprotein A repetitions predominant; GRASP, Genome-Wide Repository of Associations Between SNPs and Phenotypes; HWE, Hardy-Weinberg equilibrium; IgE, Immunoglobulin E; LD, linkage disequilibrium; LRRC32, leucine-rich repeat containing 32; MAF, minor allele frequency; MFE, minimum free energy; ORs, odds ratios; PER, persistent allergic rhinitis; SNPs, single nucleotide polymorphisms; TGF-β, transforming growth factor-β; Treg, regulatory T lymphocytes; UTR, untranslated region; VAS, visual analogue scale.
Ethics Approval and Informed Consent
The research protocol was approved by the Ethics Committee of Nanjing Medical University (20080305), and written informed consent was obtained from all participants.
Consent for Publication
We have obtained the informed consent from all patients or their legal guardians.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas. All authors took part in drafting, revising, or critically reviewing the article; have agreed on the journal to which the article has been submitted; gave final approval for the version to be published; and agreed to be accountable for the contents of the article.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bousquet J, Khaltaev N, Cruz AA, et al. Allergic Rhinitis and its Impact on Asthma (ARIA) 2008 update (in collaboration with the World Health Organization, GA(2)LEN and AllerGen). Allergy. 2008;63(Suppl 86):8–160. doi: 10.1111/j.1398-9995.2007.01620.x [DOI] [PubMed] [Google Scholar]
- 2.Brożek JL, Bousquet J, Agache I, et al. Allergic Rhinitis and its Impact on Asthma (ARIA) guidelines-2016 revision. J Allergy Clin Immunol. 2017;140(4):950–958. doi: 10.1016/j.jaci.2017.03.050 [DOI] [PubMed] [Google Scholar]
- 3.Cheng L, Chen J, Fu Q, et al. Chinese Society of Allergy guidelines for diagnosis and treatment of allergic rhinitis. Allergy Asthma Immunol Res. 2018;10(4):300–353. doi: 10.4168/aair.2018.10.4.300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samitas K, Carter A, Kariyawasam HH, et al. Upper and lower airway remodelling mechanisms in asthma, allergic rhinitis and chronic rhinosinusitis: the one airway concept revisited. Allergy. 2018;73(5):993–1002. doi: 10.1111/all.13373 [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Zhang Y, Li B, et al. Asthma and allergic rhinitis among young parents in China in relation to outdoor air pollution, climate and home environment. Sci Total Environ. 2021;751:141734. doi: 10.1016/j.scitotenv.2020.141734 [DOI] [PubMed] [Google Scholar]
- 6.Nordeide Kuiper I, Svanes C, Markevych I, et al. Lifelong exposure to air pollution and greenness in relation to asthma, rhinitis and lung function in adulthood. Environ Int. 2021;146:106219. doi: 10.1016/j.envint.2020.106219 [DOI] [PubMed] [Google Scholar]
- 7.Bousquet J, Schünemann HJ, Samolinski B, et al. Allergic Rhinitis and its Impact on Asthma (ARIA): achievements in 10 years and future needs. J Allergy Clin Immunol. 2012;130(5):1049–1062. doi: 10.1016/j.jaci.2012.07.053 [DOI] [PubMed] [Google Scholar]
- 8.Marseglia GL, Caimmi S, Marseglia A, et al. Rhinosinusitis and asthma. Int J Immunopathol Pharmacol. 2010;23(1 Suppl):29–31. [PubMed] [Google Scholar]
- 9.Laulajainen-Hongisto A, Lyly A, Hanif T. Genomics of asthma, allergy and chronic rhinosinusitis: novel concepts and relevance in airway mucosa. Clin Transl Allergy. 2020;10(1):45. doi: 10.1186/s13601-020-00347-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang L, Chen Y, Xiang Q, et al. The association between IL18, FOXP3 and IL13 genes polymorphisms and risk of allergic rhinitis: a meta-analysis. Inflamm Res. 2020;69(9):911–923. doi: 10.1007/s00011-020-01368-4 [DOI] [PubMed] [Google Scholar]
- 11.Gao Y, Li J, Zhang Y, et al. Replication study of susceptibility variants associated with allergic rhinitis and allergy in Han Chinese. Allergy Asthma Clin Immunol. 2020;16:13. doi: 10.1186/s13223-020-0411-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu XJ, Lu MP, Chen RX, et al. Polymorphism -509C/T in TGFB1 promoter is associated with increased risk and severity of persistent allergic rhinitis in a Chinese population. Am J Rhinol Allergy. 2020;34(5):597–603. doi: 10.1177/1945892420913441 [DOI] [PubMed] [Google Scholar]
- 13.Chen RX, Lu WM, Lu MP, et al. Polymorphisms in microRNA target sites of TGF-β signaling pathway genes and susceptibility to allergic rhinitis. Int Arch Allergy Immunol. 2021;182(5):399–407. doi: 10.1159/000511975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shevach EM. Garp as a therapeutic target for modulation of T regulatory cell function. Expert Opin Ther Targets. 2017;21(2):191–200. doi: 10.1080/14728222.2017.1275568 [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Chen Q, Wu C, et al. Genetic variants of the C11orf30-LRRC32 region are associated with childhood asthma in the Chinese population. Allergol Immunopathol (Madr). 2020;48(4):390–394. doi: 10.1016/j.aller.2019.09.002 [DOI] [PubMed] [Google Scholar]
- 16.Li J, Zhang Y, Zhang L. Discovering susceptibility genes for allergic rhinitis and allergy using a genome-wide association study strategy. Curr Opin Allergy Clin Immunol. 2015;15(1):33–40. doi: 10.1097/ACI.0000000000000124 [DOI] [PubMed] [Google Scholar]
- 17.Manz J, Rodríguez E, ElSharawy A, et al. Targeted resequencing and functional testing identifies low-frequency missense variants in the gene encoding GARP as significant contributors to atopic dermatitis risk. J Invest Dermatol. 2016;136(12):2380–2386. doi: 10.1016/j.jid.2016.07.009 [DOI] [PubMed] [Google Scholar]
- 18.Nasrallah R, Imianowski CJ, Bossini-Castillo L, et al. A distal enhancer at risk locus 11q13.5 promotes suppression of colitis by Treg cells. Nature. 2020;583(7816):447–452. doi: 10.1038/s41586-020-2296-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehmkuhl P, Gentz M, Garcia de Otezya AC, et al. Dysregulated immunity in PID patients with low GARP expression on Tregs due to mutations in LRRC32. Cell Mol Immunol. 2021;18(7):1677–1691. doi: 10.1038/s41423-021-00701-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marenholz I, Esparza-Gordillo J, Rüschendorf F, et al. Meta-analysis identifies seven susceptibility loci involved in the atopic march. Nat Commun. 2015;6:8804. doi: 10.1038/ncomms9804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen RX, Dai MD, Zhang QZ, et al. TLR signaling pathway gene polymorphisms, gene-gene and gene-environment interactions in allergic rhinitis. J Inflamm Res. 2022;15:3613–3630. doi: 10.2147/JIR.S364877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bousquet PJ, Combescure C, Neukirch F, et al. Visual analog scales can assess the severity of rhinitis graded according to ARIA guidelines. Allergy. 2007;62(4):367–372. doi: 10.1111/j.1398-9995.2006.01276.x [DOI] [PubMed] [Google Scholar]
- 23.Chen M, Zheng R, Li F, et al. Genetic variants in Hippo pathway genes are associated with house dust mite-induced allergic rhinitis in a Chinese population. Clin Transl Allergy. 2021;11(10):e12077. doi: 10.1002/clt2.12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–81. doi: 10.1093/nar/gkv1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. doi: 10.1101/gr.137323.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W, Yang C, Nie H, et al. LIMK2 acts as an oncogene in bladder cancer and its functional SNP in the microRNA-135a binding site affects bladder cancer risk. Int J Cancer. 2019;144(6):1345–1355. doi: 10.1002/ijc.31757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian HQ, Chen XY, Lu Y, et al. Association of VDR and CYP2R1 polymorphisms with mite-sensitized persistent allergic rhinitis in a Chinese population. PLoS One. 2015;10(7):e0133162. doi: 10.1371/journal.pone.0133162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 29.Miyazono K, Katsuno Y, Koinuma D, et al. Intracellular and extracellular TGF-β signaling in cancer: some recent topics. Front Med. 2018;12(4):387–411. doi: 10.1007/s11684-018-0646-8 [DOI] [PubMed] [Google Scholar]
- 30.Meyer-Martin H, Hahn SA, Beckert H, et al. GARP inhibits allergic airway inflammation in a humanized mouse model. Allergy. 2016;71(9):1274–1283. doi: 10.1111/all.12883 [DOI] [PubMed] [Google Scholar]
- 31.Grant CR, Liberal R, Mieli-Vergani G, et al. Regulatory T-cells in autoimmune diseases: challenges, controversies and–yet–unanswered questions. Autoimmun Rev. 2015;14(2):105–116. doi: 10.1016/j.autrev.2014.10.012 [DOI] [PubMed] [Google Scholar]
- 32.Iizuka-Koga M, Nakatsukasa H, Ito M, et al. Induction and maintenance of regulatory T cells by transcription factors and epigenetic modifications. J Autoimmun. 2017;83:113–121. doi: 10.1016/j.jaut.2017.07.002 [DOI] [PubMed] [Google Scholar]
- 33.DeVries A, Vercelli D. Of pleiotropy and trajectories: Does the TGF-β pathway link childhood asthma and chronic obstructive pulmonary disease? J Allergy Clin Immunol. 2018;141(6):1992–1996. doi: 10.1016/j.jaci.2018.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ojiaku CA, Yoo EJ, Panettieri RA Jr. Transforming growth factor β1 function in airway remodeling and hyperresponsiveness. The missing link? Am J Respir Cell Mol Biol. 2017;56(4):432–442. doi: 10.1165/rcmb.2016-0307TR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Metelli A, Salem M, Wallace CH, et al. Immunoregulatory functions and the therapeutic implications of GARP-TGF-β in inflammation and cancer. J Hematol Oncol. 2018;11(1):24. doi: 10.1186/s13045-018-0570-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salem M, Wallace C, Velegraki M, et al. GARP dampens cancer immunity by sustaining function and accumulation of regulatory T cells in the colon. Cancer Res. 2019;79(6):1178–1190. doi: 10.1158/0008-5472.CAN-18-2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vermeersch E, Liénart S, Collignon A, et al. Deletion of GARP on mouse regulatory T cells is not sufficient to inhibit the growth of transplanted tumors. Cell Immunol. 2018;332:129–133. doi: 10.1016/j.cellimm.2018.07.011 [DOI] [PubMed] [Google Scholar]
- 38.Dedobbeleer O, Stockis J, van der Woning B, et al. Cutting Edge: Active TGF-β1 released from GARP/TGF-β1 complexes on the surface of stimulated human B lymphocytes increases class-switch recombination and production of IgA. J Immunol. 2017;199(2):391–396. doi: 10.4049/jimmunol.1601882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wallace CH, Wu BX, Salem M, et al. B lymphocytes confer immune tolerance via cell surface GARP-TGF-β complex. JCI Insight. 2018;3(7):e99863. doi: 10.1172/jci.insight.99863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lodyga M, Hinz B. TGF-β1 - A truly transforming growth factor in fibrosis and immunity. Semin Cell Dev Biol. 2020;101:123–139. doi: 10.1016/j.semcdb.2019.12.010 [DOI] [PubMed] [Google Scholar]
- 41.de Streel G, Bertrand C, Chalon N, et al. Selective inhibition of TGF-β1 produced by GARP-expressing Tregs overcomes resistance to PD-1/PD-L1 blockade in cancer. Nat Commun. 2020;11(1):4545. doi: 10.1038/s41467-020-17811-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma Y, Miladi M, Dukare S, et al. A pan-cancer analysis of synonymous mutations. Nat Commun. 2019;10(1):2569. doi: 10.1038/s41467-019-10489-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noval Rivas M, Chatila TA. Regulatory T cells in allergic diseases. J Allergy Clin Immunol. 2016;138(3):639–652. doi: 10.1016/j.jaci.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eggenhuizen PJ, Ng BH, Ooi JD. Treg enhancing therapies to treat autoimmune diseases. Int J Mol Sci. 2020;21(19):7015. doi: 10.3390/ijms21197015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferreira MA, Matheson MC, Duffy DL, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378(9795):1006–1014. doi: 10.1016/S0140-6736(11)60874-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ali Syeda Z, Langden SSS, Munkhzul C, et al. Regulatory mechanism of microRNA expression in cancer. Int J Mol Sci. 2020;21(5):1723. doi: 10.3390/ijms21051723 [DOI] [PMC free article] [PubMed] [Google Scholar]