Abstract
The luxS gene of quorum-sensing Vibrio harveyi is required for type 2 autoinducer production. We identified a Porphyromonas gingivalis open reading frame encoding a predicted peptide of 161 aa that shares 29% identity with the amino acid sequence of the LuxS protein of V. harveyi. Conditioned medium from a late-log-phase P. gingivalis culture induced the luciferase operon of V. harveyi, but that from a luxS insertional mutant did not. In P. gingivalis, the expression of luxS mRNA was environmentally controlled and varied according to the cell density and the osmolarity of the culture medium. In addition, differential display PCR showed that the inactivation of P. gingivalis luxS resulted in up-regulation of a hemin acquisition protein and an arginine-specific protease and reduced expression of a hemin-regulated protein, a TonB homologue, and an excinuclease. The data suggest that the luxS gene in P. gingivalis may function to control the expression of genes involved in the acquisition of hemin.
Quorum sensing, the density-dependent regulation of gene expression, is widespread among both gram-negative and gram-positive bacteria. Quorum sensing involves the synthesis and detection of extracellular signaling molecules termed autoinducers (AIs) (2, 13). Quorum sensing in gram-negative bacteria was first described for the marine symbiotic organism Vibrio fischeri. The number of acyl homoserine lactone (HSL) AI molecules in a given culture of V. fischeri increases as the cell density increases, and once a critical concentration of AI is reached, a signal transduction cascade that leads to the production of bioluminescence by cells is initiated (15). Components of this system include LuxI, an acyl HSL synthase that directs synthesis of 3-oxo-hexanoyl-HSL (V. fischeri AI-1); AinS, an acyl HSL synthase that catalyzes the synthesis of octanoyl-HSL (V. fischeri AI-2); and LuxR, a transcriptional activator necessary for responses to V. fischeri AI-1 (10). Homologues of the luxI and luxR genes of V. fischeri have been described now for a range of gram-negative bacteria and are responsible for the density-dependent regulation of quite diverse physiological functions (1, 2, 13, 30, 41). Light production by Vibrio harveyi is similarly under the control of quorum-sensing systems, however the bioluminescence genes are not regulated by homologues of the V. fischeri LuxI and LuxR proteins (3, 4). Rather, in V. harveyi, quorum sensing involves two parallel regulatory systems. Signaling system 1 is dependent on two genes, luxL and luxM, for the synthesis of N-3-hydroxybutanoyl-l-HSL (V. harveyi AI-1), and signal detection is mediated by the sensor kinase LuxN (3, 26). LuxM shows sequence homology to V. fischeri AinS (10). Signaling system 2 requires the luxS gene for the synthesis of V. harveyi AI-2, a non-HSL AI, the structure of which is unknown (41, 42). The primary sensor for V. harveyi AI-2 is thought to be LuxP, and the LuxP–AI-2 complex interacts with LuxQ to initiate signal transduction (4, 26). Signals from both LuxN and LuxQ feed into the LuxU phosphorelay protein that then transmits the signal to the response regulator LuxO (4, 26). Whereas the V. harveyi AI-1 quorum-sensing circuit is species specific, the AI-2 system can be used for interspecies cell-cell signaling and may confer upon bacterial cells the ability to monitor the total bacterial density of mixed populations (2, 40).
luxS-based signaling has recently been described for Escherichia coli, Salmonella enterica serovar Typhimurium, Helicobacter pylori, and Shigella flexneri (9, 12, 21, 42). In S. enterica serovar Typhimurium, the expression of the luxS gene is controlled by environmental factors. AI production and signaling activity increase at high osmolarity and low pH levels and during the mid-to-late-exponential-growth phase. Since these conditions are relevant to S. enterica serovar Typhimurium as an enteric pathogen, the luxS gene is thought to play an important role in the virulence of the organism (41). However, the full extent of the role of the luxS gene in different organisms is still a matter of conjecture.
Porphyromonas gingivalis, a gram-negative anaerobe, is an etiologic agent of severe adult periodontitis (38). The environmental niche of this organism is within a mixed-species biofilm that exists in the gingival crevice, an area that experiences fluctuations in temperature, pH, osmolarity, and nutrient availability (17, 43). Additionally, P. gingivalis can invade, replicate, and persist at high density within gingival epithelial cells (5, 23). Thus, there is a potential role for density-dependent gene regulation in P. gingivalis. In this study, we identified a gene of P. gingivalis encoding a peptide exhibiting 29% identity with LuxS of V. harveyi. We also show that conditioned culture medium of P. gingivalis 33277, but not that of a luxS insertional mutant, induced luciferase expression in V. harveyi. Inactivation of luxS also influenced the expression of several genes involved in hemin uptake, suggesting that LuxS may play a role in the acquisition of hemin by P. gingivalis.
MATERIALS AND METHODS
Bacteria and culture conditions.
P. gingivalis 33277 and its derivative (see below) were grown from frozen stocks in Trypticase soy broth (TSB) (BBL) supplemented with 1 mg of yeast extract per ml, 5 μg of hemin per ml, and 1 μg of menadione per ml. For AI assays, P. gingivalis was cultured in AI bioassay (AB) medium (16) modified by the addition of 0.5 mg of yeast extract per ml, 2.5 μg of hemin per ml, and 0.5 μg of menadione per ml. P. gingivalis was grown under anaerobic conditions (85% N2, 10% H2, 5% CO2) at 37°C. E. coli strains were grown in Luria-Bertani (LB) broth (Difco) under aerobic conditions at 37°C. Streptococcus gordonii DL1 was grown in Trypticase-peptone broth supplemented with 5 mg of yeast extract per ml and 0.5% glucose under aerobic conditions at 37°C. V. harveyi reporter strain BB170 (sensor 1−, sensor 2+) was kindly provided by B. Bassler (Princeton University) and was grown in AB medium overnight at 30°C.
Autoinducer assay.
Cell-free culture supernatants from P. gingivalis parent and mutant strains (see below) were prepared by centrifugation (10,000 × g for 10 min) and filtration (filter pore size, 0.22 μm) and tested for the induction of signaling system 2 in V. harveyi BB170 by the previously described luminescence assay (16, 40). Briefly, an overnight culture of BB170 was diluted 1:2,000 in AB medium, and 100 μl of cell-free P. gingivalis culture fluid was added to 900 μl of diluted V. harveyi cells. Cell-free culture fluid of V. harveyi BB170 was included as a positive control, and sterile medium was included as a negative control. The reaction was carried out at 30°C, and light production was monitored with a Bio-Orbit 1251 luminometer.
Oligonucleotides and PCR conditions for the luxS gene.
The oligonucleotides for luxS PCR were luxS1 (5′-CCGTCGCTACATCGAGTACC-3′) and luxS2 (5′-CGAGGCATATATGTCTCCCG-3′ the antisense primer). The oligonucleotides used for testing the cotranscription of luxS and the upstream open reading frame (ORF) were luxpro1 (5′-GAGGATCTTCTCGCCCTTTT-3′) and luxS2. For testing cotranscription with the downstream ORF, the primers luxS1 and luxdwn2 (5′-GTGCCGTCTGATTCACATT-3′) were used. Reverse transcription was performed in the presence of 2 μg of total RNA, 50 ng of antisense primer, 50 U of reverse transcriptase (RT) (Ambion), 13 U of RNase inhibitor, 10 mM deoxynucleoside triphosphate (dNTP), and 1× RT buffer. Annealing of the primer and template was carried out at 72°C for 2 min and then at 48°C for 1 h. Controls without RT were included in all experiments. The resulting cDNA was amplified, with each 100 μl of PCR mixture containing 1× PCR buffer, 3 μl of cDNA, 1.5 mM MgCl2, 10 mM dNTP, 100 ng of each primer, and 2.5 U of Taq DNA polymerase. The amplification conditions were denaturation at 94°C for 30 s, annealing at 45°C for 30 s, and elongation at 72°C for 2 min for 35 cycles.
Construction of a luxS mutant.
An insertional mutation of the luxS gene was constructed using standard recombinant DNA technology (34). The plasmids used are listed in Table 1. Plasmid DNA was prepared by using the Wizard Plus Miniprep kit (Promega) according to the manufacturer's instructions. The 409-bp PCR product containing the luxS gene was cloned into the BamHI-XbaI sites of plasmid pCRII-TOPO using the TOPO TA cloning kit (Invitrogen). The 502-bp BamHI-XbaI region of the resulting pCR-LUX was then cloned into the BamHI-XbaI sites of suicide plasmid pVA3000 carrying the erythromycin resistance gene cassette ermAM-ermF to create pLR409. E. coli DH5α containing R751 was transformed with pLR409 to create the donor strain for mating with P. gingivalis. An overnight culture of the donor was used to inoculate LB medium and cultured aerobically for 2 to 3 h, until the culture reached an A600 of 0.2. An overnight culture of the recipient, P. gingivalis 33277, was used to inoculate TSB and cultured anaerobically for 6 h, until the culture reached an A600 of 0.3. The donor and the recipient were mixed at a ratio of 1 to 5 and spotted onto HAWP filters (pore size, 0.45 μm; Millipore). The mating was performed initially under aerobic conditions for 16 h and then under anaerobic conditions for 8 h. Transconjugants were selected on Trypticase-soy-blood plates supplemented with erythromycin (20 μg/ml) and gentamicin (100 μg/ml).
TABLE 1.
Plasmids used and constructed in this study
| Plasmid | Descriptiona | Source or reference |
|---|---|---|
| pCRII-TOPO | 3.9-kb cloning vector | Invitrogen |
| pCR-LUX | 409-bp luxS PCR product cloned into pCRII-TOPO | This study |
| pVA3000 | Suicide vector for Bacteroides; Emr | 25 |
| R751 | IncP plasmid used to mobilize vectors from E. coli to Bacteroides recipient; Tpr, Tra+ | 29 |
| pLR409 | pVA3000 containing an insertion of BamHI-XbaI fragment from pCR-LUX | This study |
Abbreviations: Emr, erythromycin resistant; Tpr, trimethoprim resistant; Tra+, self-transferable.
Confirmation of integration events.
To ensure that the correct fusion had occurred on the P. gingivalis chromosome, a Southern blot analysis was performed. Chromosomal DNA from six transconjugants was digested sequentially with BamHI, PvuI, HindIII, and SstI and probed with the PCR-amplified luxS that was biotin labeled with the Bionick labeling kit (Gibco BRL). The hybridized probe was detected by using the avidin peroxidase detection system (KPL). Failure to produce luxS mRNA by mutant strains was confirmed by RT-PCR using the primers lxmut1 (5′-CAGCACTTGTGCTTCTCCAA-3′) and lxmut2 (5′-GAGGAGCAGGACTTTGTTCG-3′) under the conditions described above. One transconjugant with the appropriate chromosomal integration and with loss of mRNA production was designated PLM1 and selected for further study. The growth rates of the parent and PLM1 mutant strains were comparable.
Biofilm formation.
Biofilm formation by the parent and mutant strains of P. gingivalis with S. gordonii was determined as described previously (7). S. gordonii DL1 cells (107 cells/ml) were labeled with hexidium iodide and passed over a saliva-coated glass slide in a flow chamber for 4 h at a flow rate of 2 ml/h. Following the deposition of streptococci, P. gingivalis cells (107 cells/ml) were labeled with fluorescein and passed through the flow cell at 2 ml/h for 4 h. The P. gingivalis-streptococcal biofilm was examined with a confocal microscope (Bio-Rad MRC600). Fluorescent optical sections were collected, and confocal assistant software was used to format and merge images (7).
Invasion of epithelial cells.
Invasion of P. gingivalis strains was quantitated by the standard antibiotic protection assay, as previously described (23). Primary cultures of gingival epithelial cells were obtained from gingival explants and maintained in tissue culture in keratinocyte growth medium (Clonetics). P. gingivalis cells were reacted with gingival epithelial cells at a multiplicity of infection of 100 for 90 min. External, adherent bacteria were killed by incubation for 1 h with gentamicin (300 μg/ml) and metronidazole (200 μg/ml), and internal bacteria were released by lysis of the cells in sterile distilled water for 20 min and enumerated by plate counting.
RNA isolation and DD-PCR.
Total RNA was isolated from the parent and mutant strains cultivated in TSB by using a total RNA isolation kit (Totally RNA; Ambion) and then was subjected to reverse transcription. The reaction mixture, containing 2 μg of RNA, 1 μl of 10 mM dNTP, and 100 pmol of random hexamers, was incubated at 80°C for 10 min and put on ice. The enzyme mixture, containing 40 U of Moloney murine leukemia virus RT (Ambion), 1× RT reaction buffer, and 1 μl of anti-RNase (Ambion), was added to a final volume of 20 μl. The reaction was performed at 42°C for 1 h, followed by inactivation of the enzyme at 92°C for 10 min. Differential display PCR (DD-PCR) was performed using 5 μl of the synthesized cDNA in 100 μl of a solution containing 1 U of Taq DNA polymerase (Promega), 1.5 mM MgCl2, 0.2 mM dNTP, and 100 pmol of arbitrary primers. The arbitrary primers used were act1 (5′-GGCATGGGTCAGAAGGATT-3′), act2 (5′-CTCAAGTTGGGGGACAAAAA-3′), kgp1 (5′-CGGAACAGCTTCTTCCAATC-3′), and kgp2 (5′-AATCTTGCTCCGCCCTTATT-3′). The thermal cycling parameters were 50 cycles of 94°C for 1 min, 34°C for 1 min, and 72°C for 2 min. Differentially expressed PCR products were excised from the gel and cloned into pCRII-TOPO, and DNA sequencing was done by the University of Washington DNA Sequencing Service. The DD-PCR results were further investigated by RT-PCR using RNA preparations identical to those described above and primers derived from the sequences of cloned products. The primers used were Exinuc1 (5′-TACAAGGAGCACGCAGACAG-3′), Exinuc2 (5′-TCCCGTGGACGATATGTAGG-3′), Hemreg1 (5′-TACCGCTGTACCATTGACGA-3′), Hemreg2 (5′-TAACACTCCTCTCGCCGACT-3′), OMP1 (5′-ATACGGAGGAGGTGAGCGTA-3′), OMP2 (5′-AGTGATGCAATGCTCTGACG-3′), RGP1 (5′-TGTTCGGTTCTGCAGTTGTC-3′), RGP2 (5′-TAATCGCTTCCACCACCTTC-3′), TonB1 (5′-CGGCCAAATCTGTCTTGACT-3′), and TonB2 (5′-ACCGTCGTTCATACCCGTAG-3′).
RESULTS
Presence of lux homologues in P. gingivalis.
A BLAST search of the P. gingivalis genomic database of The Institute for Genomic Research (http://www.tigr.org) revealed several predicted open reading frames with significant identity to various Lux proteins of V. harveyi. In particular, an ORF of P. gingivalis exhibited 29% identity (49 of 167 amino acid residues were conserved) and 49% similarity (82 of 167 amino acid residues were similar) with the LuxS protein of V. harveyi. The sequences were also compared to those of the human pathogens (S. enterica serovar Typhimurium and H. pylori) that have been shown to produce a functional signaling molecule. Regions of identity occurred within those portions of LuxS that demonstrated the greatest conservation among species (Fig. 1). In addition, similar regions of identity with the E. coli LuxS protein that is 100% homologous to LuxS of S. flexneri were observed (9). Moreover, a direct comparison of the P. gingivalis LuxS protein with the Borrelia burgdorferi LuxS protein (which has yet to be demonstrated to be functional) showed 50% identity.
FIG. 1.
Alignment of the deduced P. gingivalis LuxS sequence (obtained from the database of the The Institute for Genomic Research [http: //www.tigr.org]) with deduced LuxS sequences from other bacteria. Sequences from V. harveyi (GenBank accession no. AAD17292), E. coli (GenBank accession no. P45578), S. enterica serovar Typhimurium (GenBank accession no. AAF73475), and H. pylori (GenBank accession no. AAD07175) were aligned using the ClustalW algorithm. The amino acid residues of these sequences that are identical appear in boldface. Symbols: ∗, identity; :, strong similarity; ., weak similarity.
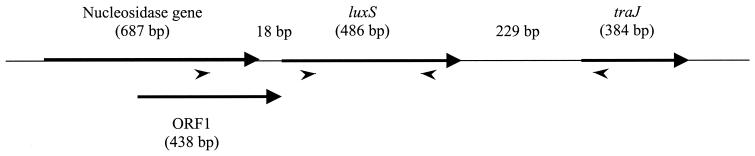
Interrogation of the P. gingivalis genome database revealed that luxS was separated by only 18 bp from an upstream in-frame ORF with homology to the 5-methylthioadenosine nucleosidase–S-adenosylhomocysteine nucleosidase gene (Fig. 2). RT-PCR data indicated that these two genes are cotranscribed (data not shown). The region upstream of the putative 5-methylthioadenosine nucleosidase–S-adenosylhomocysteine nucleosidase gene contained prokaryote promoter consensus sequences identified using the Promoter Predictions search tool of the Berkeley Drosophila Genome Project (http://www.fruitfly.org) and by visual comparison with a set of predicted P. gingivalis consensus promoter sequences (20). Thus, transcription of luxS may require the promoter of the upstream ORF. In addition, a 438-bp ORF with no identifiable homology spans the intergenic region overlapping both the LuxS and 5-methylthioadenosine nucleosidase–S-adenosylhomocysteine nucleosidase genes (Fig. 2). Downstream (229 bp) of luxS is an ORF with homology to the traJ gene of Bacteroides thetaiotaomicron that comprises the transfer region of a conjugative transposon. RT-PCR revealed that this gene is not cotranscribed with luxS (data not shown).
FIG. 2.
Schematic arrangement of the luxS ORF and those upstream (nucleosidase) and downstream (traJ) of it. The ORFs themselves are indicated by arrows, with the direction of the arrow indicating the direction of transcription. The genes for LuxS and the nucleosidase are cotranscribed and encompass an additional ORF in a different reading frame. The primers used for RT-PCR are indicated with arrowheads.
P. gingivalis also possesses putative ORFs that demonstrate significant sequence identity with V. harveyi proteins LuxN and LuxQ, the sensor kinases of signaling systems 1 and 2, respectively, and with LuxO (Table 2). Lux O of V. harveyi is thought to be a negative regulator of luminescence and to integrate sensory inputs from AI-1 and AI-2 signaling systems. The presence of homologues to luxS and luxQ on the P. gingivalis chromosome suggests that P. gingivalis possesses a signaling system similar to the AI-2 circuit in V. harveyi. Some differences between the AI-2 systems in P. gingivalis and V. harveyi can be expected, however, as homologues of LuxP (the primary AI-2 sensor) and LuxU (the phosphorelay protein for AI-2 and AI-1) were not present in P. gingivalis. The absence of homologues of LuxU and of LuxLM indicates that P. gingivalis does not possess a functional homoserine lactone-dependent signaling pathway similar to the V. harveyi AI-1 circuit. In addition, P. gingivalis does not appear to possess a quorum-sensing pathway similar to either the AI-1 or the AI-2 circuit of V. fischeri, as homologues of LuxI, LuxR, and AinS were not detected.
TABLE 2.
Identification of P. gingivalis ORFs that share significant sequence identitya with Lux proteins of V. harveyi
| Lux proteinb | Function | Identity of P. gingivalis ORFc |
|---|---|---|
| LuxS (AAD17292) | AI-2 synthase | 49/167 (29) |
| LuxQ (AA20838) | AI-2 sensor | 107/411 (26) |
| LuxO (S49540) | Response regulator | 167/469 (35) |
| LuxN (S37350) | AI-1 sensor | 94/356 (26) |
e value, <10−10.
GenBank accession numbers appear in parentheses.
Each value is the number of identical amino acid residues/the total number of amino acid residues. Values in parentheses are percentages.
AI-2 activity in P. gingivalis.
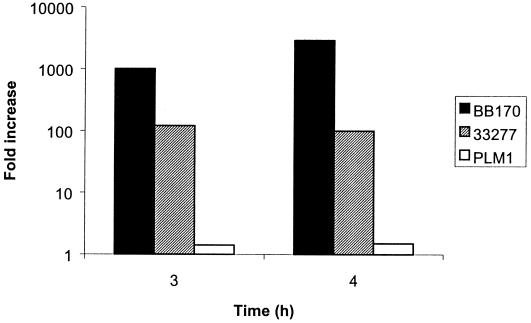
To test whether P. gingivalis exhibits functional signaling activity related to the V. harveyi AI-2 system, cell-free culture media from late-log-phase cultures of P. gingivalis 33277 and PLM1 (in which luxS was insertionally inactivated) were assayed for induction of luminescence in V. harveyi BB170. After 3 h of incubation, conditioned medium from P. gingivalis 33277 induced luminescence by 120-fold in BB170 (Fig. 3). In contrast, luminescence induced by the PLM1 mutant was less than twofold higher than the level induced by media only (Fig. 3). Comparable results were found after 4 h of incubation (Fig. 3). Plate counts showed that the growth of the V. harveyi reporter was similar whether stimulated with supernatant from the parent or with that from the mutant (data not shown). The level of induction by P. gingivalis was almost one log unit lower than that induced by control V. harveyi culture supernatants. This may indicate that the AI molecule of P. gingivalis differs structurally from that of V. harveyi, resulting in less efficient recognition. Alternatively, or additionally, there may be less AI in P. gingivalis culture supernatants.
FIG. 3.
Induction of V. harveyi BB170 luminescence by cell-free supernatants of 33277 (parent strain), PLM1 (mutant), and V. harveyi BB170. Activation was measured by comparing the level of luminescence induced by the test strain to that induced by sterile medium.
Environmental control of luxS gene expression in P. gingivalis.
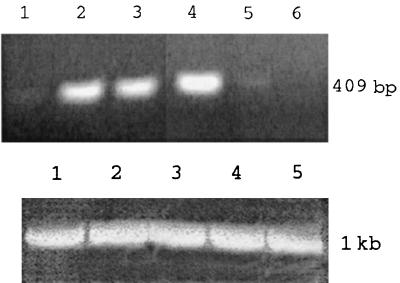
Growth phase-dependent LuxS expression is a feature of the AI-2 systems of bacteria other than P. gingivalis (12, 40, 41). To investigate whether luxS expression in P. gingivalis is controlled by environmental cues, luxS transcripts were examined by RT-PCR using RNA from cells grown under various conditions (Fig. 4). As a control, the P. gingivalis fimA gene was also amplified under the same conditions, using primers described previously (44). The levels of luxS mRNA (quantitated by NIH Image software) in P. gingivalis grown to late log phase were over three times higher than those in the same strain grown to early log phase. The NaCl concentration of the growth media also affected expression of luxS mRNA which was highest at the normal osmolarity of TSB (80 mM, approximately half physiological). At NaCl concentrations of 160 and 240 mM, luxS expression was significantly reduced and absent, respectively (Fig. 4). Expression of luxS was affected neither by growth temperature nor by pH, which was tested between pH 6.5 and 8.5 (data not shown).
FIG. 4.
RT-PCR using RNA from cells grown under various conditions. (Top) RT-PCR of mRNA of luxS of P. gingivalis grown under various conditions. Lanes: 1, early log growth; 2, late log growth; 3, late log growth at 34°C; 4, late log growth at 80 mM NaCl; 5, late log growth at 160 mM NaCl; 6, late log growth at 240 mM NaCl. (Bottom) RT-PCR of mRNA of fimA of P. gingivalis grown under various conditions (as a control for total RNA levels). Lanes: 1, early log growth; 2, late log growth; 3, late log growth at 80 mM NaCl; 4, late log growth at 160 mM NaCl; 5, late log growth at 240 mM NaCl. Note that no constitutively expressed P. gingivalis gene that would be a more appropriate control has been reported and that fimA mRNA levels vary according to growth temperature (44).
Functional role of P. gingivalis LuxS.
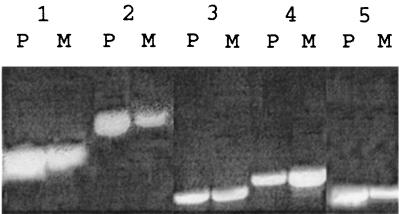
P. gingivalis can form a mixed-species biofilm with S. gordonii and can invade gingival epithelial cells. Both processes could require that P. gingivalis assess the local environment through a quorum-sensing system. Therefore, we examined wild-type and mutant strains for biofilm formation in conjunction with S. gordonii and for invasion of gingival epithelial cells. However, no differences in either biofilm structure or invasion efficiency were observed (data not shown). To determine if luxS plays a role in regulating gene expression in P. gingivalis, parent and mutant strains were analyzed by DD-PCR. Five amplification products that were differentially present or absent in parent and mutant samples were sequenced and identified by a BLAST search of the GenBank database (http://www.ncbi.nlm.nih.gov). As shown in Table 3, genes encoding two previously described P. gingivalis proteins were identified: the expression of the gene encoding hemin-regulated protein (HemR) (22) was reduced in the luxS knockout strain PLM1, whereas that of the gene encoding arginine-specific protease (RgpA) (31) was increased. In addition, a gene homologous to an outer membrane hemin acquisition protein of Pseudomonas fluorescens (19) was up-regulated in PML1; while the expression of genes homologous to those encoding TonB (33) and excinuclease ABC (6) was down-regulated. RT-PCR confirmed the differential expression of these genes with the exception of rgpA (Fig. 5). Discrepancies between the results obtained by DD-PCR and RT-PCR are frequently reported and may be due to differences in the dynamic ranges of the two techniques (11). Moreover, in this case, a further degree of variability could result from the inability of the RT-PCR primers to distinguish between rgpA and the closely related gene kgp, which encodes a lysine-specific protease.
TABLE 3.
Characterization of genes differentially regulated in P. gingivalis PLM1
| Gene product (GenBank accession no.) | Homologous proteina (reference) | Function | Expression in luxS mutant |
|---|---|---|---|
| HemR (AAC44980) | P. gingivalis hemin-regulated outer membrane protein (22) | TonB-dependent receptor | Absent |
| Excinuclease ABC homologue (P14951) | B. subtilis excision nuclease (6) | DNA repair | Absent |
| Outer membrane protein (BAA88494) | P. fluorescens heme acquisition protein (19) | Hemin uptake | Present |
| RgpA (A55426) | P. gingivalis arginine-specific protease (31) | Multiple, including provision of hemin by degradation of hemin-sequestering proteins | Present |
| TonB homologue (E82955) | Pseudomonas siderophore acquisition protein (33) | Energy transducer | Absent |
Determined by comparison with sequences of the GenBank database.
FIG. 5.
RT-PCR to confirm differential expression of genes of parent strain 33277 (P) and mutant PLM1 (M). Lanes: 1, excinuclease ABC homologue; 2, HemR; 3, RgpA; 4, P. fluorescens hemin acquisition protein homologue; 5, TonB homologue.
DISCUSSION
P. gingivalis will encounter fluctuations in environmental conditions as it traverses the oral fluids, colonizes oral surfaces, and interfaces with oral tissues. Concomitantly, cell density changes will occur as the organism establishes a subgingival infection and thrives in the multispecies biofilm that exists in the periodontal pocket. Therefore, it is possible that P. gingivalis might monitor these diverse environments by quorum sensing. A BLAST search for lux genes in the P. gingivalis genomic sequence database identified ORFs relating to V. harveyi signaling system 2, namely, LuxS, LuxQ, and LuxO. Interestingly, homologues of LuxLM, which are required for the V. harveyi AI-1 pathway, were not found in P. gingivalis, and neither were homologues of the V. fischeri quorum-sensing components LuxI, LuxR, and AinS. Thus, of the described quorum-sensing systems, a pathway related to V. harveyi signaling system 2 would appear to be the only potentially operative circuit in P. gingivalis. Homologues of V. harveyi signaling system 2 components LuxP, LuxU, and LuxR were not detected in P. gingivalis. This could indicate that there are mechanistic differences in the signaling circuits between the two species or that the P. gingivalis functional equivalents have diverged to the extent that they no longer exhibit significant sequence homology with V. harveyi.
The flanking gene arrangements vary among species in which luxS has been identified, and no operon arrangements have been reported (12). In P. gingivalis, the luxS gene was cotranscribed with an upstream nucleosidase homologue. In addition, this mRNA will also encompass a third ORF in a different reading frame. The significance of this arrangement, which appears, thus far, to be unique to P. gingivalis, remains to be investigated. Downstream of luxS, and not cotranscribed, is a gene homologous to the transfer region of a Bacteroides conjugative transposon. Similarly, Yersinia pestis has a gene for a transposase approximately 200 bp upstream of a luxS gene. This design may have implications for horizontal transfer of the luxS gene, which is present in at least 30 species.
As some components of the V. harveyi AI-2 circuit may not be present in P. gingivalis, we initiated a series of experiments to investigate whether the P. gingivalis luxS gene is expressed and is functional. Analysis of mRNA by RT-PCR demonstrated that the luxS gene is transcribed in P. gingivalis. Moreover, conditioned culture medium of P. gingivalis induced bioluminescence in V. harveyi, indicating that the signal molecule produced by P. gingivalis is recognized by the AI-2 receptor of V. harveyi. However, the level of bioluminescence induced by P. gingivalis was significantly lower than the control levels obtained using conditioned broth from overnight cultures of V. harveyi. This suggests that the P. gingivalis signaling molecule may be functionally and structurally distinct from that of V. harveyi. Consistent with this, conditioned broth from Actinobacillus actinomycetemcomitans, which possesses a luxS homologue exhibiting significantly greater similarity to the V. harveyi luxS gene, induced bioluminescence comparable to that of the V. harveyi control (D. R. Demuth, unpublished data).
The production of LuxS-dependent AI-2 in other gram-negative organisms, such as E. coli, S. enterica serovar Typhimurium, and H. pylori, is regulated by metabolic conditions and environmental stimuli such as growth phase, pH, and osmolarity (12, 41). Similarly, P. gingivalis luxS was expressed at higher levels as the cell density increased, during log-phase growth, or when the osmolarity of the growth medium was reduced to approximately half of the physiological level. Such conditions of low osmolarity may be relevant to growth in the oral cavity since saliva is a very hypotonic fluid. However, unlike the situation with S. enterica serovar Typhimurium, changes in pH did not appear to regulate luxS expression in P. gingivalis. This may simply reflect the different environmental conditions encountered by organisms indigenous to the human oral cavity and gastrointestinal tract.
Quorum-sensing systems serve to regulate a variety of physiological responses (3, 13), the production of light being only one example. Given that P. gingivalis is not capable of bioluminescence, we embarked on a series of studies to define the role of luxS in P. gingivalis. Both biofilm formation and intracellular invasive properties are associated with quorum sensing in other species (8, 39); however, the P. gingivalis luxS mutant was not impaired in these activities in our assay systems. Differential display of mRNA from parent and mutant strains revealed a potential novel role for P. gingivalis LuxS in regulating genes involved in the acquisition of hemin. The loss of LuxS activity resulted in up-regulation of a putative hemin-acquisition protein and the arginine-specific protease, RgpA. Interestingly, the RgpA protease has been suggested to play a role in increasing hemin availability by degradation of host hemin-sequestering proteins (32, 36). The mutant strain also demonstrated down-regulation of a TonB homologue and of HemR, a P. gingivalis outer membrane protein that is negatively regulated by hemin and is TonB-dependent. In P. gingivalis, hemin levels can control expression of various virulence-associated genes (14, 27, 37). In addition, the proteolytic and hemagglutination activities in P. gingivalis are intricately interconnected and affect levels of available hemin (18, 24, 28, 35). This raises the possibility that LuxS is a component of a complex virulence-associated cascade that regulates iron acquisition, which, in turn, influences the expression of specific virulence genes of P. gingivalis. Such an ability to potentially modulate virulence factors through iron acquisition mechanisms sets apart the role of LuxS in P. gingivalis from any other species with a known V. harveyi-like AI-2 signaling pathway.
Inactivation of luxS also resulted in down-regulation of a putative ABC excinuclease, a DNA repair enzyme that catalyzes the excision of UV-damaged nucleotide segments (6). Therefore, the luxS gene may also play a role in stress response in P. gingivalis.
In summary, we have identified in P. gingivalis a functional luxS-dependent signaling pathway that can activate a quorum-sensing circuit in V. harveyi. This system is regulated in response to specific growth and environmental stimuli and may play a role in regulating the expression of specific genes involved in hemin acquisition by P. gingivalis.
ACKNOWLEDGMENTS
We thank Bonnie L. Bassler for kindly providing V. harveyi and Guy Cook for confocal microscopy.
This work was supported by NIDCR grants DE11111 and DE12505.
REFERENCES
- 1.Bainton N J, Bycroft B W, Chhabra S, Stead P, Gledhill L, Hill P J, Rees C E D, Winson M K, Salmond G P C, Stewart G S A B, Williams P. A general role for the lux autoinducer in bacterial cell signaling: control of antibiotic biosynthesis in Erwinia. Gene. 1992;116:87–91. doi: 10.1016/0378-1119(92)90633-z. [DOI] [PubMed] [Google Scholar]
- 2.Bassler B L. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol. 1999;2:582–587. doi: 10.1016/s1369-5274(99)00025-9. [DOI] [PubMed] [Google Scholar]
- 3.Bassler B L, Wright M, Showalter R E, Silverman M R. Intercellular signaling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 4.Bassler B L, Wright M, Silverman M R. Multiple signal systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 5.Belton C M, Izutsu K T, Goodwin P C, Park Y, Lamont R J. Fluorescence image analysis of the association between Porphyromonas gingivalisand gingival epithelial cells. Cell Microbiol. 1999;1:215–223. doi: 10.1046/j.1462-5822.1999.00022.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen N Y, Zhang J J, Paulus H. Chromosomal location of the Bacillus subtilis aspartokinase II gene and nucleotide sequence of the adjacent genes homologous to uvrC and trx of Escherichia coli. J Gen Microbiol. 1989;135:2931–2940. doi: 10.1099/00221287-135-11-2931. [DOI] [PubMed] [Google Scholar]
- 7.Cook G S, Costerton J W, Lamont R J. Biofilm formation by Porphyromonas gingivalis and Streptococcus gordonii. J Periodont Res. 1998;33:323–327. doi: 10.1111/j.1600-0765.1998.tb02206.x. [DOI] [PubMed] [Google Scholar]
- 8.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 9.Day W A, Maurelli A T. Shigella flexneri LuxS quorum-sensing system modulates virBexpression but is not essential for virulence. Infect Immun. 2001;69:15–23. doi: 10.1128/IAI.69.1.15-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunlap P V. Quorum regulation of luminescence in Vibrio fischeri. J Mol Microbiol Biotechnol. 1999;1:5–12. [PubMed] [Google Scholar]
- 11.Eckmann L, Smith J R, Housley M P, Dwinell M B, Kagnoff M F. Analysis by high density cDNA arrays of altered gene expression in human intestinal epithelial cells in response to infection with the invasive enteric bacteria Salmonella. J Biol Chem. 2000;275:14084–14094. doi: 10.1074/jbc.275.19.14084. [DOI] [PubMed] [Google Scholar]
- 12.Forsyth M H, Cover T L. Intercellular communication in Helicobacter pylori: luxSis essential for the production of an extracellular signaling molecule. Infect Immun. 2000;68:3193–3199. doi: 10.1128/iai.68.6.3193-3199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 14.Genco C A, Simpson W, Forng R Y, Egal M, Odusanya B M. Characterization of a Tn4351-generated hemin uptake mutant of Porphyromonas gingivalis: evidence for coordinate regulation of virulence factors by hemin. Infect Immun. 1995;63:2459–2466. doi: 10.1128/iai.63.7.2459-2466.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenberg E P. Quorum sensing in Gram-negative bacteria: acylhomoserine lactone signaling and cell-cell communication. In: LeBlanc D J, Lantz M S, Switalski L M, editors. Microbial pathogenesis: current and emerging issues. Indianapolis: Indiana University; 1997. [Google Scholar]
- 16.Greenberg E P, Hastings J W, Ulitzur S. Induction of luciferase synthesis in Beneckea harveyiby other marine bacteria. Arch Microbiol. 1979;120:87–91. [Google Scholar]
- 17.Haffajee A D, Socransky S S, Goodson J M. Subgingival temperature (I). Relation to baseline clinical parameters. J Clin Periodontol. 1992;19:401–408. doi: 10.1111/j.1600-051x.1992.tb00670.x. [DOI] [PubMed] [Google Scholar]
- 18.Hoover C I, Ng C Y, Felton J R. Correlation of haemagglutination activity with trypsin-like protease activity of Porphyromonas gingivalis. Arch Oral Biol. 1992;7:515–520. doi: 10.1016/0003-9969(92)90133-s. [DOI] [PubMed] [Google Scholar]
- 19.Idei A, Kawai E, Akatsuka H, Omori K. Cloning and characterization of the Pseudomonas fluorescensATP-binding cassette exporter, HasDEF, for the heme acquisition protein HasA. J Bacteriol. 1999;181:7545–7551. doi: 10.1128/jb.181.24.7545-7551.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson C A, Hoffmann B, Slakeski N, Cleal S, Hendtlass A J, Reynolds E C. A consensus Porphyromonas gingivalispromoter sequence. FEMS Microbiol Lett. 2000;186:133–138. doi: 10.1111/j.1574-6968.2000.tb09094.x. [DOI] [PubMed] [Google Scholar]
- 21.Joyce E A, Bassler B L, Wright A. Evidence for a signaling system in Helicobacter pylori: detection of a luxS-encoded autoinducer. J Bacteriol. 2000;182:3638–3643. doi: 10.1128/jb.182.13.3638-3643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karunakaran T, Madden T, Kuramitsu H. Isolation and characterization of a hemin-regulated gene, hemR, from Porphyromonas gingivalis. J Bacteriol. 1997;179:1898–1908. doi: 10.1128/jb.179.6.1898-1908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamont R J, Chan A, Belton C M, Izutsu K T, Vasel D, Weinberg A. Porphyromonas gingivalisinvasion of gingival epithelial cells. Infect Immun. 1995;63:3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamont R J, Jenkinson H F. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S W, Hillman J D, Progulske-Fox A. The hemagglutinin genes hagB and hagC of Porphyromonas gingivalisare transcribed in vivo as shown by use of a new expression vector. Infect Immun. 1996;64:4802–4810. doi: 10.1128/iai.64.11.4802-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lilley B N, Bassler B L. Regulation of quorum sensing in Vibrio harveyiby LuxO and sigma-54. Mol Microbiol. 2000;36:940–954. doi: 10.1046/j.1365-2958.2000.01913.x. [DOI] [PubMed] [Google Scholar]
- 27.McKee A S, McDermid A S, Bakerville A, Dowsett B, Ellwood D C, Marsh P D. Effect of hemin on the physiology and virulence of Bacteroides gingivalisW50. Infect Immun. 1986;52:349–455. doi: 10.1128/iai.52.2.349-355.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishikata M, Yoshimura F. Characterization of Porphyromonas (Bacteroides) gingivalishemagglutinin as a protease. Biochem Biophys Res Commun. 1991;178:336–342. doi: 10.1016/0006-291x(91)91819-x. [DOI] [PubMed] [Google Scholar]
- 29.Park Y, McBride B C. Characterization of the tpr gene product and isolation of a specific protease-deficient mutant of Porphyromonas gingivalisW83. Infect Immun. 1993;61:4139–4146. doi: 10.1128/iai.61.10.4139-4146.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosavirulence genes requires cell to cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 31.Pike R N, McGraw W, Potempa J, Travis J. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization, and evidence for the existence of complexes with hemagglutinins. J Biol Chem. 1994;269:406–411. [PubMed] [Google Scholar]
- 32.Pike R N, Potempa J, McGraw W, Coetzer H T, Travis J. Characterization of the binding activities of proteinase-adhesin complexes from Porphyromonas gingivalis. J Bacteriol. 1996;178:2876–2882. doi: 10.1128/jb.178.10.2876-2882.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poole K, Zhao Q, Neshat S, Heinrichs D E, Dean C R. The Pseudomonas aeruginosa tonBgene encodes a novel TonB protein. Microbiology. 1996;142:1449–1458. doi: 10.1099/13500872-142-6-1449. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Shah H N, Gharbia S E, Progulske-Fox A, Brocklehurst K. Evidence for independent molecular identity and functional interaction of the haemagglutinin and cysteine proteinase (gingivain) of Porphyromonas gingivalis. J Med Microbiol. 1992;36:239–244. doi: 10.1099/00222615-36-4-239. [DOI] [PubMed] [Google Scholar]
- 36.Shi Y, Ratnayake D B, Okamoto K, Abe N, Yamamoto K, Nakayama K. Genetic analysis of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J Biol Chem. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 37.Smalley J W, Birss A J, McKee A S, Marsh P D. Haemin-restriction influences haemin-binding, haemagglutination and protease activity of cells and extracellular membrane vesicles of Porphyromonas gingivalisW50. FEMS Microbiol Lett. 1991;61:63–67. doi: 10.1016/0378-1097(91)90647-s. [DOI] [PubMed] [Google Scholar]
- 38.Socransky S S, Haffajee A D. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992;63:322–331. doi: 10.1902/jop.1992.63.4s.322. [DOI] [PubMed] [Google Scholar]
- 39.Sperandio V, Mellies J L, Nguyen W, Shin S, Kaper J B. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl Acad Sci USA. 1999;96:15196–15201. doi: 10.1073/pnas.96.26.15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surette M G, Bassler B L. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surette M G, Bassler B L. Regulation of autoinducer production in Salmonella typhimurium. Mol Microbiol. 1999;31:585–595. doi: 10.1046/j.1365-2958.1999.01199.x. [DOI] [PubMed] [Google Scholar]
- 42.Surette M G, Miller M B, Bassler B L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Theilade E. Factors controlling the microflora of the healthy mouth. In: Hills M J, Marsh P D, editors. Human microbial ecology. Boca Raton, Fla: CRC Press; 1990. pp. 1–56. [Google Scholar]
- 44.Xie H, Cai S, Lamont R J. Environmental regulation of fimbrial gene expression in Porphyromonas gingivalis. Infect Immun. 1997;65:2265–2271. doi: 10.1128/iai.65.6.2265-2271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]