Summary
Discovery of efficacious antiviral agents targeting SARS-CoV-2 main protease (Mpro) is of the highest importance to fight against COVID-19. Here, we describe a simple protocol for high-throughput screening of Mpro inhibitors using a robust fluorescence polarization (FP) assay. Candidate Mpro inhibitors from large compound libraries could be rapidly identified by monitoring the change of millipolarization unit value. This affordable FP assay can be modified to screen antiviral agents targeting virus protease.
For complete details on the use and execution of this protocol, please refer to Li et al. (2022), Yan et al. (2021), and Yan et al. (2022c).
Subject areas: Biophysics, Cell-based assays, High throughput screening, Protein biochemistry, Protein expression and purification
Graphical abstract
Highlights
-
•
Production of SARS-CoV-2 main protease (Mpro) in E. coli cells
-
•
Measurement of Mpro activity using the fluorescence resonance energy transfer assay
-
•
A robust fluorescence polarization (FP) assay for rapid screening of Mpro inhibitors
-
•
Discovery of anacardic acid as an inhibitor targeting Mpro using this FP assay
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Discovery of efficacious antiviral agents targeting SARS-CoV-2 main protease (Mpro) is of the highest importance to fight against COVID-19. Here, we describe a simple protocol for high-throughput screening of Mpro inhibitors using a robust fluorescence polarization (FP) assay. Candidate Mpro inhibitors from large compound libraries could be rapidly identified by monitoring the change of millipolarization unit value. This affordable FP assay can be modified to screen antiviral agents targeting virus protease.
Before you begin
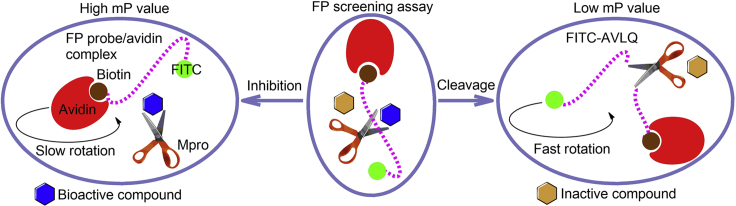
This protocol details an in vitro sandwich-like fluorescence polarization (FP) assay for high-throughput screening (HTS) of SARS-CoV-2 main protease (Mpro) inhibitors. The FP approach is based on the positive correlation between millipolarization unit (mP) value and apparent molecular weight of a fluorescent moiety in the solution (Hall et al., 2016; Hendrickson et al., 2020). In this FP assay, a synthetic peptide is conjugated with a fluorescein isocyanate (FITC) fluorophore and biotin to generate the FP probe, FITC-AVLQSGFRKK-Biotin, and this probe is used as the preferred substrate of Mpro. Subsequently, Mpro cleaves the FP probe to release a small fluorescent fragment FITC-AVLQ, resulting in a low mP value due to a fast rotation. In contrast, in the presence of a bioactive compound that inhibits Mpro activity, the binding of this intact FP probe to avidin leads to formation of a large complex, resulting in a high mP value due to a slow rotation (Figure 1). Therefore, candidate compounds targeting Mpro from large compound libraries could be rapidly identified by measuring the change of mP value (Yan et al., 2021). This FP assay mainly includes candidate compounds, active Mpro, FP probe, and avidin, which looks like a sandwich. This step-by-step protocol can be easily used to screen and assess candidate Mpro inhibitors in vitro. Recently, we also utilized a modified FP approach to identify novel antiviral agents targeting SARS-CoV-2 papain-like protease (PLpro) from a natural product library (Yan et al., 2022b).
Figure 1.
The principle of the developed FP screening assay for discovery of Mpro inhibitors
In this FP assay, incubation of FP probes (dashed purple line) with active Mpro (the scissors) and subsequent addition of avidin (red crescent) will produce a FP signal that is proportional to the relative amount of cleaved and uncleaved FP probes. In the presence of a bioactive compound (blue hexagon) that inhibits Mpro activity, the uncleaved FP probe produces a high mP value upon binding to avidin, whereas in the presence of an inactive compound (yellow hexagon), the cleaved fragment FITC-AVLQ produces a low mP value because of the failure for avidin binding. This adapted graphic is produced with permission (Yan et al., 2022b).
Prior to the experiment, active Mpro, FP assay buffer, stock solutions of a natural product library, GC-376 (positive control), a fluorescence resonance energy transfer (FRET) substrate, the FP probe, and avidin should be properly prepared to start an enjoyable screening journey.
Prepare active SARS-CoV-2 Mpro
Timing: 2 weeks
-
1.Expression of active Mpro.
-
a.Ligate the synthetic codon-optimized DNA fragment encoding Mpro (GenBank: YP_009725301.1) into a pET-21a(+) vector with Nde I and Xho I restriction sites to carry an extra C-terminal 6×His tag.
 CRITICAL: According to the principle of codon bias, the codon-optimized sequence of Mpro gene is generated using JAVA Codon Adaption Tool (JCat, www.jcat.de). The services of Mpro gene synthesis and plasmid construction are provided by Anhui Gene Universal Corporation Limited (Chuzhou, China), and the incorporated Mpro gene is further confirmed by DNA sequencing.
CRITICAL: According to the principle of codon bias, the codon-optimized sequence of Mpro gene is generated using JAVA Codon Adaption Tool (JCat, www.jcat.de). The services of Mpro gene synthesis and plasmid construction are provided by Anhui Gene Universal Corporation Limited (Chuzhou, China), and the incorporated Mpro gene is further confirmed by DNA sequencing. -
b.Transform the E. coli DH5α competent cells with the recombinant plasmids.
-
i.Thaw the competent cells on ice.
-
ii.Transfer 200 ng plasmids to 50 μL aliquot of competent cells.Note: Mix by gently swirling the pipette tip.
-
iii.Incubate for 30 min on ice.
-
iv.Heat the tube for 90 s at 42°C.
-
v.Cool for 2 min on ice.
-
vi.Add 200 μL of LB medium (10 g tryptone, 10 g NaCl, 5 g yeast extract for 1 L). Gently shake for 1 h at 37°C in a bacterial incubator.
-
vii.Transfer 100 μL of the transformation mix onto the LB agar plate containing 100 μg/mL ampicillin and 1.5% agar.
 CRITICAL: Ampicillin must be added after the LB agar cooling down as it could degrade at a high temperature.
CRITICAL: Ampicillin must be added after the LB agar cooling down as it could degrade at a high temperature. -
viii.Incubate the LB agar plate for 10 h at 37°C.
-
i.
-
c.Inoculate 5 mL of LB medium containing 100 μg/mL ampicillin with a single colony of transformed bacteria, and further incubate the culture for 10 h at 37°C with vigorous shaking.
-
d.Prepare the pure plasmids using an EasyPure® Plasmid MiniPrep Kit.Note: The kit user guide is accessible at https://www.transgen.com.cn/plasmid/337.html.
-
e.Transform the E. coli Rosetta (DE3) competent cells with the pure plasmids as described above.
 Pause point: LB agar plate with colonies can be stored at 4°C for 1 week.
Pause point: LB agar plate with colonies can be stored at 4°C for 1 week. CRITICAL: The fresh competent cells and plasmids should be used for this transformation.
CRITICAL: The fresh competent cells and plasmids should be used for this transformation. -
f.Inoculate a single colony into 5 mL of LB medium with 100 μg/mL ampicillin and grow the cell culture at 37°C for 10 h.
-
g.Inoculate 2 mL of the cell culture into 1 L of LB medium with 100 μg/mL ampicillin until the culture has reached mid-log phase (A600=0.6–0.8).
-
h.Induce Mpro expression by adding 1 M isopropyl-β-D-thiogalactoside (IPTG, 238.3 g/mol) solution to a final concentration of 0.2 mM, and further incubate the culture at 30°C for additional 8 h for the bulk production.
-
i.Transfer an aliquot of the slurry to a 15 mL polypropylene tube.
-
j.Centrifuge the tubes at 514 × g for 10 min at 4°C. Discard the supernatant to harvest the bacterial pellets.
 Pause point: After induction by IPTG, the E. coli cell pellets can be stored at −20°C for 1 week.
Pause point: After induction by IPTG, the E. coli cell pellets can be stored at −20°C for 1 week. CRITICAL: It is important to monitor the density of the E. coli cell culture before IPTG induction. For a high yield of Mpro, the E. coli cells are induced by addition of 0.2 mM IPTG at mid-log phase, and the entire length of induction should not exceed 12 h. We strongly recommend increasing the volume of cell culture to several liters to increase yield.
CRITICAL: It is important to monitor the density of the E. coli cell culture before IPTG induction. For a high yield of Mpro, the E. coli cells are induced by addition of 0.2 mM IPTG at mid-log phase, and the entire length of induction should not exceed 12 h. We strongly recommend increasing the volume of cell culture to several liters to increase yield.
-
a.
-
2.Purification of active Mpro.
-
a.Resuspend the E. coli cell pellets in 40 mL of lysis buffer (pH8.0).Note: The lysis buffer contains 50 mM tris (hydroxymethyl) aminomethane (Tris, 121.14 g/mol), 150 mM NaCl (58.44 g/mol), pH8.0.
-
b.Disrupt E. coli cells by a cell sonicator on ice. Alternate 3 s of sonication at 35% amplitude with 7 s rest periods for a total of 45 min sonication.
-
i.Remove cell debris by centrifugation at 12,857 × g for 30 min at 4°C.
-
ii.Carefully decant the supernatant (40 mL) to a new tube and store on ice until the next step.Note: During the sonication, always put the supernatant on ice! After sonication, the solution should be more translucent than the starting solution. If needed, repeat sonication for a complete lysis.
-
i.
-
c.Filter the supernatant by the syringe filters (0.45 μM, Millipore).
-
d.Load the supernatant onto a HisTrap chelating column (5 mL, Cytiva) in binding buffer A (pH8.0).Note: The binding buffer A contains 25 mM Tris (121.14 g/mol), 0.5 M NaCl (58.44 g/mol), 5 mM imidazole (68.08 g/mol), pH8.0.
-
e.Wash the column with 50 mL of binding buffer A to elute unbound proteins.Note: The N-terminal methionine residue can be removed by addition of E. coli methionine aminopeptidase in the supernatant to generate the N-terminal free Mpro, but additional polyhistidine tag at the C terminus of Mpro is not detrimental to its enzyme activity (Sacco et al., 2020).Based on our experience, direct loading without adding methionine aminopeptidase could still retain expected Mpro activity via our described procedure (Chen et al., 2021a).
 CRITICAL: Before sample loading, the supernatant and buffers must be filtered by 0.45 μM pore-size filters. Otherwise, the particles will block the HisTrap column. Once the supernatant is prepared, the sample must be loaded as soon as possible.
CRITICAL: Before sample loading, the supernatant and buffers must be filtered by 0.45 μM pore-size filters. Otherwise, the particles will block the HisTrap column. Once the supernatant is prepared, the sample must be loaded as soon as possible. -
f.Apply a linear 5–250 mM imidazole gradient to the column using 0%–50% elution buffer B (pH8.0) in 10 column volumes (CV).Note: The elution buffer B contains 25 mM Tris (121.14 g/mol), 0.5 M NaCl (58.44 g/mol), 0.5 M imidazole (68.08 g/mol), pH8.0.
-
g.Monitor the change of A280 value for each fraction.
-
h.Collect 3 fractions in the elution peak of A280, which likely yield 20 mL sample of Mpro.
-
i.Wash the column with 5 CV of 100% elution buffer B. Stop collecting fractions.
-
j.Analyze the protein samples by SDS-PAGE.
-
i.Run 12% SDS-PAGE gel for the fractions with A280 peak.
-
ii.Stain SDS-PAGE gel with coomassie brilliant blue R250 to confirm which fractions contain pure Mpro.
- iii.
-
i.
-
k.Combine fractions and dialyze.
-
i.Dialyze 20 mL combined sample in a dialysis sac (molecular weight cut-off of 3.5 kDa) for 10 h at 4°C against 2 L dialysis buffer (pH8.0).Note: The dialysis buffer contains 25 mM Tris (121.14 g/mol), 150 mM NaCl (58.44 g/mol), 1 mM ethylenediaminetetraacetic acid tetrasodium salt tetrahydrate (EDTA tetrasodium salt, 452.23 g/mol), pH8.0.
-
ii.Concentrate the pure Mpro using a centrifugal filter unit (Amicon® Ultra-15 Ultracel-3, Millipore) by centrifugation at 1,157 × g for 5 h at 4°C to generate 15 mL sample.
-
iii.Determine pure Mpro concentration by a Pierce BCA protein assay kit.Note: The kit user guide is accessible at https://www.thermofisher.cn/order/catalog/product/23225?SID=srch-srp-23225.
-
iv.Divide Mpro sample into 50 μL aliquots and store in an ultra-low temperature freezer at −80°C before use.Note: The volume of Mpro stock should be appropriate based on the required amount of Mpro for desired experiments to minimize freeze-thaws. The final concentration of pure Mpro is 1.6 mg/mL (47 μM), and the stock solution is stable for 6 months at −80°C.
-
i.
-
a.
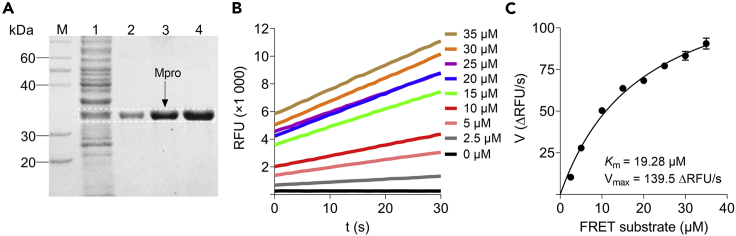
Figure 2.
Production of active SARS-CoV-2 main protease (Mpro) in E. coli
(A) Bacterial expression and purification of Mpro. The SDS-PAGE gel was stained with coomassie brilliant blue R250. M: Protein ladder; 1: Total cell extract; 2–4: Pure Mpro (34 kDa).
(B) Measurement of the initial velocity of Mpro enzyme reaction. All initial rates were obtained by fitting the linear portion of the curves to a straight line.
(C) Determination of the Km value. The Michaelis-Menten equation was plotted using GraphPad Prism 8.0 according to the initial rates, and the Km and Vmax values were recorded as 19.28 μM and 139.5 ΔRFU/s, respectively. This assay was performed in triplicate.
Prepare the FP assay buffer
Timing: 30 min
The FP assay buffer contains 10 mM Tris (121.14 g/mol), 50 mM NaCl (58.44 g/mol), 1 mM EDTA tetrasodium salt (452.23 g/mol), 1 mM dithiothreitol (DTT, 154.25 g/mol), pH8.0.
-
3.
Dissolve 1.21 g Tris, 2.92 g NaCl, and 0.45 g EDTA tetrasodium salt in 800 mL of ultrapure water by stirring in a beaker.
-
4.
Adjust pH to 8.0 by drop-wise addition of 1 M HCl.
-
5.
Adjust volume to 1 liter with ultrapure water and label it as the Tris buffer.
Note: This Tris buffer should be filtered with the 0.45 μM pore-size filters.
-
6.
Divide into 50 mL aliquots and store at −20°C before use.
-
7.Dissolve 3.08 g DTT in 20 mL of ultrapure water.
-
a.Sterilize by filtration to prepare 1 M DTT stock solution and divide into aliquots of 1 mL.
-
b.Store at −20°C before use.
-
a.
-
8.Add 50 μL of 1 M DTT stock solution to 50 mL of Tris buffer to generate the FP assay buffer for natural product screening.
-
a.Mix well by vortexing.
-
b.Freshly prepared before use.
-
a.
CRITICAL: Considering the stringent caution of DTT addition for the reliability in HTS, it is recommended to perform this FP assay in the presence of 1 mM DTT (Ma et al., 2020). For best results, freshly prepared assay buffer is appreciated in HTS experiments. Moreover, addition of EDTA could efficiently prevent the possible interference of metal ions with Mpro activity (Kozak et al., 2020). Because the theoretical pI value of native Mpro is 6.2, this FP assay buffer is preferred to use in HTS experiments.
Prepare natural product library plates
Timing: 2 weeks
-
9.
Dissolve 1 mg natural product library powder in 100 μL dimethyl sulfoxide (DMSO) to generate fresh stock solutions at 10 mg/mL in barcoded 96-well plates with a V-conical bottom.
Note: The natural product library contains 3,000 natural molecules extracted from traditional Chinese medicine. This stock solution of a natural product library can be stably stored at −80°C for 2 years. Make sure that all the compounds are soluble before use.
-
10.Add 18 μL DMSO to each well of another barcoded 96-well plate with a V-conical bottom and transfer 2 μL stock solution to all wells to generate a final concentration of 1 mg/mL.
-
a.Label this plate as a working library plate for the primary screening.
-
b.All plates should be carefully sealed to avoid evaporation.
-
a.
-
11.
The working library plates are stored at −20°C before use.
Prepare the stock solution of GC-376 (positive control)
Timing: 5 min
-
12.
Dissolve 5 mg GC-376 (507.53 g/mol) in 492.58 μL DMSO to generate a stock solution at 20 mM and divide into 20 μL aliquots as stock solution.
-
13.
Store at −20°C before use.
Note: The final working concentration of GC-376 is 1 μM in HTS experiments. This stock solution is stable for 1 year.
Prepare the stock solution of FRET substrate
Timing: 5 min
-
14.
Dissolve 1 mg FRET substrate peptide (1,514.66 g/mol) in 330.1 μL DMSO to generate a stock solution at 2 mM.
-
15.
Divide into 10 μL aliquots.
-
16.
Store at −20°C and protect from light.
Note: The FRET substrate sequence is MCA-AVLQSGFRK(Dnp)-K-NH2 (λex/λem = 320/405 nm), and the purity is more than 95.0%. This sequence is derived from the N-terminal auto-cleavage sequence of SARS-CoV-2 main protease (Jin et al., 2020). This stock solution is stable for 2 months.
Prepare the stock solution of FP probe
Timing: 5 min
-
17.
Dissolve 1 mg FP probe (1,862.21 g/mol) in 268.5 μL DMSO to generate a stock solution at 2 mM.
-
18.
Divide into 10 μL aliquots.
-
19.
Store at −20°C and protect from light.
Note: The FP probe sequence is FITC-AVLQSGFRKK-Biotin (λex/λem = 485/535 nm), and the purity is more than 95.0%. The working concentration of FP probe is 60 nM in HTS experiments. As described above, the stock solution of FITC-AVLQ peptide (2 mM) is also prepared at the same time. This peptide could be used as a background control in HTS experiments. These stock solutions are stable for 2 months, but the peptide powders are stable for 1 year if stored frozen.
Prepare the stock solution of avidin
Timing: 5 min
-
20.
Dissolve 5 mg avidin in 2.78 mL FP assay buffer to generate a stock solution at 30 μM.
-
21.
Divide into 20 μL aliquots.
-
22.
Store at −20°C before use.
Note: References have cited the molecular mass for native avidin as 60 kDa because it contains four 15 kDa subunits. This stock solution is stable for 5 months, but the dry powder is stable at least for 3 years if stored frozen. The working concentration of avidin is 0.3 μM in HTS experiments.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli DH5α competent cell | TransGen Biotech, Beijing | Cat# CD201-01 |
| E. coli Rosetta (DE3) competent cell | TransGen Biotech, Beijing | Cat# CD801-02 |
| Chemicals, peptides, and recombinant proteins | ||
| Nde Ⅰ | TransGen Biotech, Beijing | Cat# JN201-01 |
| Xho Ⅰ | TransGen Biotech, Beijing | Cat# JX201-01 |
| EasyPure® Plasmid MiniPrep Kit | TransGen Biotech, Beijing | Cat# EM101-01 |
| Yeast extract | Thermo Fisher Scientific | Cat# LP0021B |
| Tryptone | Thermo Fisher Scientific | Cat# LP0042B |
| NaCl | Sinopharm, Shanghai | Cat# 10019318 |
| Agar | Aladdin, Shanghai | Cat# A109142 |
| IPTG | Aladdin, Shanghai | Cat# I104812 |
| Ampicillin | Aladdin, Shanghai | Cat# A102048 |
| Brilliant blue R250 | Aladdin, Shanghai | Cat# B105005 |
| Protein ladder II | TransGen Biotech, Beijing | Cat# DR201-01 |
| DMSO | Sigma-Aldrich | Cat# V900090 |
| HCl | Sinopharm, Shanghai | Cat# 10011018 |
| Tris | Sigma-Aldrich | Cat# S4762 |
| EDTA | Aladdin, Shanghai | Cat# T161517 |
| DTT | Sigma-Aldrich | Cat# V900830 |
| Imidazole | Sigma-Aldrich | Cat# V900153 |
| FRET substrate | GL Biochem, Shanghai | N/A |
| FP probe | GL Biochem, Shanghai | N/A |
| FITC-AVLQ peptide | GL Biochem, Shanghai | N/A |
| Streptavidin | Sigma-Aldrich | Cat# S4762 |
| Natural product library | TargetMol, Shanghai | Cat# L6000 |
| GC-376 | TargetMol, Shanghai | Cat# T5188 |
| Anacardic acid | TargetMol, Shanghai | Cat# T6389 |
| Pierce BCA protein assay kit | Thermo Fisher Scientific | Cat# 23227 |
| Recombinant DNA | ||
| SARS-CoV-2 Mpro construct | In-house production | N/A |
| Software and algorithms | ||
| JAVA Codon Adaption Tool, JCat | A web server, Germany | www.jcat.de |
| GraphPad Prism 8.0 | GraphPad Software, San Diego, USA | www.graphpad.com/scientific-software/prism/ |
| Other | ||
| Electronic balance | Sartorius, Germany | Cat# Secura324-1CN |
| Clean bench | AIRTECH, Suzhou | Cat# BLB-1600 |
| pH meter | OHAUS, USA | Cat# AB23PH |
| Cell sonicator | Scientz, Ningbo | Cat# JY92-IIDN |
| Bacterial incubator | Zhichu, Shanghai | Cat# ZCLY-180ES |
| Shaker | Kylin-Bell, Haimen | Cat# ZD-9550 |
| Ice machine | Xueke, Changshu | Cat# IMS-25 |
| Vertical electrophoresis cell | Bio-Rad, USA | Cat# Mini-PROTEAN Tetra |
| Culture dish | Aladdin, Shanghai | Cat# D1799-03-100EA |
| CLINX imager | Clinx, Shanghai | Cat# GenoSens 2000 |
| Centrifuge | Eppendorf, Germany | Cat# 5804R |
| Syringe filter | Millipore, USA | Cat# SLHV033NB |
| Dialysis sac | Solarbio, Beijing | Cat# YA1078 |
| Amicon® Ultra-15 | Merck, USA | Cat# UFC900308 |
| Ultrapure water purifier | Millipore, USA | Cat# Milli-Q Biocel |
| Opaque black 96-well microplate | Corning, USA | Cat# 3686 |
| Microplate reader | BioTek, USA | Cat# Cytation 5 |
| HisTrap HP, 5 mL | Cytiva, Sweden | Cat# 17524701 |
| AKTA pure | Cytiva, Sweden | Cat# 25 M |
Materials and equipment
LB medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Tryptone | N/A | 10 g |
| NaCl | N/A | 10 g |
| Yeast extract | N/A | 5 g |
| ddH2O | N/A | Up to 1 L |
Store at room temperature (RT) within 2 weeks.
Note: Autoclave LB medium at 121°C for 20 min before storage.
In this protocol, the term of RT indicates that the ambient temperature in the laboratory is between 25°C and 27°C.
Binding buffer A (pH8.0)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 25 mM | 3.03 g |
| NaCl | 0.5 M | 29.22 g |
| Imidazole | 5 mM | 0.34 g |
| ddH2O | N/A | Up to 1 L |
Store at 4°C within 2 weeks.
Note: Pre-warm the binding buffer A for 1 h at RT before use.
Elution buffer B (pH8.0)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 25 mM | 3.03 g |
| NaCl | 0.5 M | 29.22 g |
| Imidazole | 0.5 M | 34.04 g |
| ddH2O | N/A | Up to 1 L |
Store at 4°C within 2 weeks.
Note: Pre-warm the elution buffer B for 1 h at RT before use.
Dialysis buffer (pH8.0)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 25 mM | 3.03 g |
| NaCl | 150 mM | 8.77 g |
| EDTA | 1 mM | 0.45 g |
| ddH2O | N/A | Up to 1 L |
Store at 4°C within 2 weeks.
Tris buffer (pH8.0)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 10 mM | 1.21 g |
| NaCl | 50 mM | 2.92 g |
| EDTA | 1 mM | 0.45 g |
| ddH2O | N/A | Up to 1 L |
Store at −20°C within 2 months.
Note: The Tris buffer should be divided into 50 mL aliquots and stored at −20°C before use. The FP assay buffer is freshly prepared by adding 1 M DTT to a final concentration of 1 mM. All used buffers must be filtered by 0.45 μM pore-size filters before use.
CRITICAL: Avoid contact and inhalation when handling DTT and other chemical reagents. Do not get in eyes, on skin, or on clothing. Wash thoroughly after handling. Wear protective gloves, protective clothing, and eye protection.
Alternatives: In this study, we have used a BioTek microplate reader (Cytation 5) for HTS, but other devices equipped by a FP module are also suitable for this study. For the inhibition analysis, a dose-response curve is plotted by a GraphPad Prism 8.0 software to produce a half-maximum inhibitory concentration (IC50) value. Both OriginPro software and Microsoft Excel work well for this purpose.
Alternatives: Because the use of opaque black microplates could reduce the background signal noise, the measurement should be performed using these microplates. Any commercial opaque black microplates with a flat bottom are also suitable for this study, but the measurement program needs to be tuned according to the types of microplates used.
Step-by-step method details
Measurement of SARS-CoV-2 Mpro activity
Timing: 1 h
This section describes a procedure to measure Mpro activity using the FRET assay, which has been widely used to analyze Mpro activity and screen Mpro inhibitors (Zhu et al., 2020). In this FRET assay, a quenched peptide containing the Mpro cleavage site is used as the FRET substrate. After cleavage by active Mpro, the cleaved fragment MCA-AVLQ is released, resulting in a high value of relative fluorescence unit (RFU) (Yan et al., 2022a).
-
1.
Prepare 10 mL FP assay buffer containing 1 mM DTT.
-
2.
Before addition of the FRET substrate and active Mpro, pre-warm the solution for 30 min at RT.
Note: To get reliable results, the preparation of pre-warmed assay buffer is necessary.
CRITICAL: All regular steps are carried out at RT.
-
3.Add 10.6 μL sample containing active Mpro (1.6 mg/mL) into 2 mL FP assay buffer for a final concentration of 8.48 μg/mL (0.25 μM).
-
a.Gently mix by pipetting.
-
b.Divide into 50 μL aliquots.
-
c.Add 50 μL aliquots into a black 96-well microplate.
-
a.
Note: Use a fresh Mpro solution and keep the solution on ice when not in use.
-
4.Dilute the FRET substrate solution (2 mM) by 50 μL aliquots to generate the analyte with FRET substrate concentrations at 0, 2.5, 5, 10, 15, 20, 25, 30, or 35 μM, respectively.
-
a.Gently mix by pipetting to generate a homogenous solution.
-
b.Protect from light.
-
a.
-
5.Proceed to measure the RFU value immediately (troubleshooting 1).
-
a.Place the microplate containing analyte in a microplate reader.
-
b.After setting the gain value of 58, measure the RFU value from the top with the excitation wavelength of 320 nm and emission wavelength of 405 nm every second for 30 s at RT.
-
c.Export the obtained data as RFU values versus time (seconds).
-
a.
Note: Except for RFU value, the intensity of fluorescence can be expressed in arbitrary units (AU) or simply abbreviated as FL, and this value is device-dependent.
CRITICAL: The prepared analyte must be protected from light.
Calculation of the Michaelis constant of SARS-CoV-2 Mpro
Timing: 30 min
As described above, the initial velocity (V) of Mpro enzyme reaction is calculated by a linear regression for the first 30 s. According to the initial velocity, the Michaelis constant (Km) value is determined using a Michaelis-Menten equation to assess Mpro activity in vitro. This equation can be inferred using GraphPad Prism 8.0.
-
6.
Plot RFU values versus time (seconds) using a scatter plot.
-
7.
Select at least 30 continuous points covering the first 30 s in an initial reaction curve.
-
8.
Plot a straight line to infer a curve equation and R-squared value on chart (troubleshooting 2).
Note: For an expected result, the R-squared value should be more than 0.98.
-
9.
Record a slope value as an initial velocity (V=ΔRFU/s) in the indicated FRET substrate concentration (Table 1, Figure 2B).
-
10.Plot the initial velocity against the FRET substrate concentration to generate a Michaelis–Menten equation using GraphPad Prism 8.0 (Figure 2C).
-
a.Program option: Michaelis–Menten equation.
-
b.Export the Km value.
-
a.
Note: The Km value is a measure of the kinetics of an enzyme reaction, and it is equivalent to the concentration of substrate at which the reaction takes place at one half its maximum rate.
Table 1.
The initial velocity of Mpro enzyme reaction in the FRET assay
| FRET substrate (μM) | Linear equations | Avg. velocity (ΔRFU/s) | ||
|---|---|---|---|---|
| 35 | y=88.32x+5829.9 | y=93.12x+5766.6 | y=93.33x+5618.1 | 91.59 |
| 30 | y=85.91x+4913.4 | y=83.4x+5033.3 | y=81.4x+4999.8 | 83.57 |
| 25 | y=75.83x+4126.6 | y=78.59x+4019.9 | y=77.7x+4126.6 | 77.37 |
| 20 | y=69.21x+3699.2 | y=67.5x+3814.5 | y=67.8x+3644.9 | 68.17 |
| 15 | y=64.34x+3610.4 | y=63.21x+3673.2 | y=63x+3553.5 | 63.52 |
| 10 | y=49.26x+2882.2 | y=51.41x+2008.7 | y=51.53x+2698.7 | 50.73 |
| 5 | y=25.63x+1486.9 | y=27.81x+1402.8 | y=27x+1396.2 | 26.81 |
| 2.5 | y=10.66x+683.5 | y=10.5x+608.9 | y=10x+576.1 | 10.39 |
| 0 | 0 | 0 | 0 | 0 |
FP assay operation
Timing: 1 h
This section describes a step-by-step FP assay procedure for the discovery of Mpro inhibitors (Figure 3A). In this FP assay, the change of mP value is dependent on the molecular weight of a fluorescent moiety provided that temperature and viscosity remain constant. Candidate compounds targeting Mpro can be easily identified by monitoring the change of mP value in a pilot screening using this FP assay.
-
11.Prepare 4 mL solution containing 0.4 μM Mpro as working buffer using an aliquot of the FP assay buffer.
-
a.Pre-warm the fresh FP assay buffer for 30 min at RT before use.
-
b.Dilute 34 μL aliquot of Mpro solution in 4 mL FP assay buffer to generate a final concentration of 0.4 μM.
-
c.Gently mix by pipetting.
-
a.
-
12.
Dispense 29 μL aliquots into a black 96-well microplate using a multichannel pipette (troubleshooting 3).
Note: Make sure that bubble formation is minimal in this operation.
-
13.Add 1 μL sample of a natural product (1 mg/mL) to the Mpro-containing mixture using a multichannel pipette. Use new pipette tips for each well.
-
a.Designate the positive (GC-376, 1 μM, A12-C12), negative (DMSO, D12-F120), and background (FITC-AVLQ peptide, 20 nM, G12-H12) wells in each plate (Figure 3B).
-
b.Gently mix using a shaker.
-
c.Incubate the assay mixture for 30 min at RT.
-
d.Prepare 3 mL FP probe solution containing 60 nM FP probe using the remaining aliquot of the FP assay buffer before next step.
-
a.
Note: The working solution of FP probe must be freshly prepared before use due to the instability of a fluorogenic peptide in the solution. According to the real needs, properly prepare the working solutions prior to the next step. The final concentration of a natural product is about 30 μM in the primary screening, and keep the incubation time within 35 min.
CRITICAL: In this test, the wells containing GC-376 (1 μM), DMSO, and FITC-AVLQ peptide (20 nM) are used as positive, negative, and background controls, respectively. These wells should be properly arranged in each microplate as described in Figure 3B.
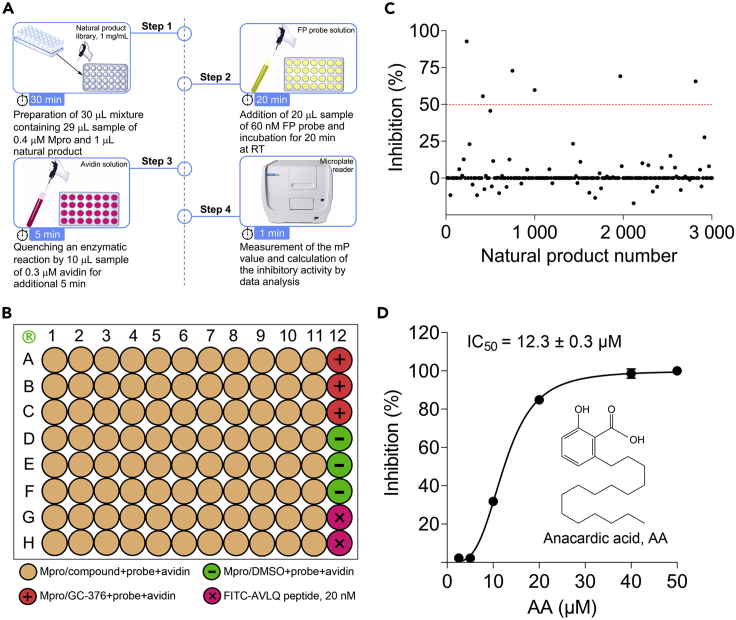
Figure 3.
Application of the FP screening assay for discovery of Mpro inhibitors
(A) The 4-step procedure of the FP assay for rapid screening of Mpro inhibitors. Step 1: 29 μL Mpro solution (0.4 μM) was incubated with 1 μL sample of a natural product (1 mg/mL) for 30 min at RT in a black 96-well microplate. Step 2: 20 μL FP probe solution (60 nM) was added and incubated for 20 min at RT. Step 3: The enzyme reaction was quenched by adding 10 μL avidin solution (0.3 μM) followed by 5 min incubation. Step 4: The mP value was measured by a microplate reader for data analysis.
(B) The microplate layout in a large-scale screening. The positive (GC-376, 1 μM, A12-C12), negative (DMSO, D12-F12), and background (FITC-AVLQ peptide, 20 nM, G12-H12) wells were designated in each plate. These schematic diagrams are adapted with permission (Yan et al., 2022b).
(C) The illustration for the primary screening cycle of a natural product library using this FP assay. The red dotted line indicated a baseline that is equivalent to 50% inhibition, and 6 candidate compounds were identified in a pilot screening.
(D) The inhibitory effect of anacardic acid (AA) on Mpro activity in the FP assay. The chemical structure and IC50 value of AA are shown. Data are represented as mean ± SD.
It should be noted that the actual working concentration of DMSO used in this assay is less than 5% to achieve a reliable screening for natural products (Yan et al., 2021).
-
14.Add 20 μL sample of 60 nM FP probe to the mixture containing natural products using a multichannel pipette (troubleshooting 4).
-
a.Gently mix using a shaker.
-
b.Proceeding for 20 min at RT.
-
c.Protect from light.
-
d.Prepare 1.5 mL solution containing 0.3 μM avidin using the remaining aliquot of the FP assay buffer before next step.
-
a.
Note: The final concentration of FP probe is 20 nM in HTS experiments. Make sure that an equal volume of FP probe solution is dispensed into the assay mixture to initiate an enzyme reaction. During this process, the microplate must be protected from light.
-
15.Add 10 μL sample of 0.3 μM avidin to the FP probe-containing mixture using a multichannel pipette (troubleshooting 5).
-
a.Gently mix using a shaker.
-
b.Incubate the reaction mixture for 5 min at RT to adequately quench this reaction.
-
a.
Note: The final concentration of avidin is 50 nM in HTS experiments.
-
16.Measure the mP value immediately (troubleshooting 6, 7, 8).
-
a.Load the microplate onto the tray of a microplate reader and initiate the FP program by clicking "OK".
-
b.Record the mP value from the top with the excitation wavelength at 485 nm and emission wavelength at 535 nm.
-
a.
-
17.Export the data for inhibition analysis of tested natural products (Figure 3C).
-
a.The inhibitory activity of a tested compound is calculated using the Equation 1.
-
b.The candidate compounds (≥50% inhibition) are repeated in triplicate to confirm the inhibition against Mpro activity at the screening concentration.
-
a.
Note: The final volume in all wells is 60 μL in HTS experiments.
CRITICAL: Considering the instability of the FP probes and proteins, all used buffers must be freshly prepared before use. Importantly, these continuous steps must be completed as quickly as possible.
For a rapid and reliable screening of natural products, each screening microplate should be arranged as shown in Figure 3B, and the positive (GC-376), negative (DMSO), and background controls must be included in each screening cycle.
The evaluation of Mpro inhibition by anacardic acid
Timing: 1 h
This section details a protocol for the inhibition evaluation of anacardic acid against Mpro activity using the FP assay. This procedure could be regarded as an inhibition test of candidate compounds targeting Mpro using the FP assay. In general, this assay will have broad applications for the screening and evaluation of candidate Mpro inhibitors in vitro.
-
18.
Prepare 4 mL solution containing 0.4 μM Mpro using an aliquot of the FP assay buffer as described in step 11.
-
19.Dissolve 5 mg anacardic acid (348.52 g/mol) in 717.3 μL DMSO to prepare 20 mM stock solution.
-
a.Vortex to dissolve.
-
b.Divide into 50 μL aliquots.
-
c.Store at −20°C before use.
-
a.
Note: The stock solution of anacardic acid is stable for 1 year when stored at −20°C.
-
20.Dilute the stock solution of anacardic acid using the previous Mpro-containing buffer to generate 100 μL sample at 100, 80, 40, 20, 10, or 5 μM, respectively.
-
a.Gently mix by pipetting.
-
b.Dispense 30 μL sample into a black 96-well microplate using a pipette.
-
c.Incubate for 30 min at RT.
-
a.
Note: A single testing concentration of anacardic acid contains 3 wells. GC-376 (1 μM), DMSO, and FITC-AVLQ peptide (20 nM) are used as positive, negative, and background controls, respectively. The final concentration of anacardic acid is 50, 40, 20, 10, 5, or 2.5 μM in a final volume of 60 μL in each well.
-
21.Add 20 μL sample of 60 nM FP probe to the previous assay mixture using a pipette.
-
a.Gently mix by a shaker.
-
b.Protect from light.
-
c.Incubate for 20 min at RT.
-
a.
-
22.Add 10 μL sample of 0.3 μM avidin to the FP probe-containing mixture using a pipette.
-
a.Gently mix using a shaker.
-
b.Incubate for 5 min at RT.
-
a.
-
23.
Place the microplate in a microplate reader and record the mP values as described in step 16 (Table 2).
-
24.Calculation of an IC50 value of anacardic acid against Mpro activity (Figure 3D).
-
a.Calculate the inhibition value against Mpro activity using the Equation 1.
-
b.Export the inhibition values versus the indicated concentrations of the inhibitor.
-
c.The x-axis presents the inhibitor concentration (μM), and the y-axis presents the relative inhibition (%).
-
d.Draw a dose-response curve using GraphPad Prism 8.0.
-
e.Export an IC50 value.
-
a.
Note: For determination of an IC50 value with the highest accuracy, it is strongly recommended to repeat this measurement in triplicate.
Table 2.
The mP value from a dose-response curve of anacardic acid against Mpro activity in the FP assay
| Compound (μM) | Raw mP values | Avg. mP value | Inhibition (%) | ||
|---|---|---|---|---|---|
| 50 | 193 | 189 | 189 | 190.33 | 102.48 |
| 40 | 192 | 183 | 181 | 185.33 | 99.09 |
| 20 | 164 | 161 | 158 | 161 | 82.58 |
| 10 | 86 | 80 | 82 | 82.67 | 29.41 |
| 5 | 41 | 35 | 40 | 38.67 | 0 |
| 2.5 | 38 | 38 | 41 | 39 | 0 |
| GC-376, 1 μM | 191 | 184 | 185 | 186.67 | 100 |
| DMSO | 39 | 38 | 41 | 39.33 | 0 |
| FITC-AVLQ, 20 nM | 35 | 38 | 38 | 37 | 0 |
Expected outcomes
The active Mpro was highly expressed in E. coli Rosetta (DE3) cells, and successfully purified by a HisTrap chelating column (Figure 2A). Importantly, the pure Mpro exhibited expected enzyme activity with the Km value of 19.28 μM (Figures 2B and 2C). This protocol describes a step-by-step sandwich-like FP assay for rapid screening of Mpro inhibitors. Because only a small amount of FP probe is used (20 nM/well) in this protocol and it takes less than one hour to finish each screening cycle, this FP assay is very cheap, simple, and rapid for laboratory use, which is ideal for a large-scale screening. Through a pilot screening of a natural product library, anacardic acid was identified as an inhibitor targeting Mpro with an IC50 value of 12.3±0.3 μM in vitro (Figure 3). Recent studies have confirmed an expected antiviral potency of anacardic acid using live SARS-CoV-2 viruses by targeting its cysteine proteases (Chen et al., 2021b; Yan et al., 2022b; Yan et al., 2022c; Zhu et al., 2020).
Quantification and statistical analysis
The inhibitory activity of hit compound is calculated using the Equation 1.
| (Equation 1) |
where μHit, μN, and μP represent the average mP values of the hit compound, negative, and positive controls, respectively.
Limitations
For a successful screening, the validation of the prepared Mpro activity is very crucial for the development of a reliable FP assay. In this assay design, the FP probe was added to the Mpro/inhibitor mixture to initiate an enzyme reaction, then the reaction was further quenched by adding avidin. Interestingly, we found that the Mpro fails to cleave the FP probe into a small fragment FITC-AVLQ when the FP probe is first mixed with avidin, which is likely due to the formation of probe-avidin tetramers that blocks the access of Mpro enzyme. As a result, this FP assay procedure must be divided into 4 steps, and it cannot be used for the kinetics study to infer the possible in vitro inhibition mechanism of Mpro inhibitors. However, this FP assay could be widely used for screening of Mpro inhibitors. An in vitro combination of FP and FRET assay should be a powerful strategy for rapid screening of Mpro inhibitors and evaluating their inhibitory mechanisms in the future.
Troubleshooting
Problem 1
Fluorescence signal is unstable in the FRET assay (step 5).
Potential solution
Make sure that the sequence of FRET substrate is correct. All assay buffers should be freshly prepared by ultrapure water. Start the FRET assay using the fresh aliquots of Mpro. Protect from light during the assay. Assembly of the assay mixture containing fresh FRET substrate and active Mpro at RT.
Problem 2
No linearity is found during the measurement of the Km value of active Mpro (step 8).
Potential solution
Make sure that the prepared Mpro from E. coli cells remains active. The measurement must be completed as quickly as possible to monitor enzyme kinetics. Once an obvious pause occurs, repeat this measurement by adjusting the reading settings of the instrument or by using the fresh assay aliquots. The valid data will be produced within the first 30 s. Use the optimum amounts of active Mpro and FRET substrate to begin this measurement.
Problem 3
Formation of bubbles during the dispensation of active Mpro into the microplate wells (step 12).
Potential solution
Make sure to drop the aliquots gently into the flat bottom of each well.
Problem 4
The FP probe cannot be cleaved by active Mpro in the FP assay (step 14).
Potential solution
Make sure that the sequence of FP probe is correct before use. Protect from light after addition of the FP probe. Fresh aliquots should be used.
Problem 5
Avidin often fails to quench an enzyme reaction in the FP assay (step 15).
Potential solution
Make sure that the used avidin is a high-quality reagent. Use fresh aliquots of avidin.
Problem 6
The mP value is unstable during the measurement (step 16).
Potential solution
Make sure that the fresh aliquots are used in the FP assay, and the entire procedure should be carried out as described in Figure 3A. For the FP assay buffer, it should not contain glycerine to enhance its viscosity. As a result, a high background noise arises that reduces the sensitivity and reliability of the FP assay. A simple assay buffer is usually preferred. Each component must be properly added to generate a final volume of 60 μL in a homogenous solution. Because the temperature is a major affecting factor in the FP assay, all procedures must be carried out at RT. Keep the constant viscosity and ambient temperature during the FP assay. Shaking during measurement is an option that some equipment offers, which ensures a more homogenous distribution of both enzyme and additives. If the recorded mP value is out of the normal range, repeat this after shaking.
Problem 7
The signal window (SW) value is low in the FP assay (step 16).
Potential solution
The SW value mainly depends on the mP values of positive and negative controls. Make sure that the used positive and negative controls are correct. GC-376 is used as a positive control, and the final working concentration is 1 μM in HTS experiments. The SW value is usually above 140.
Problem 8
The mP values of positive and negative controls are abnormal in the FP assay (step 16).
Potential solution
As described above, make sure that the used positive and negative controls are correct before use.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yunyu Chen (chenyunyu1984@163.com).
Materials availability
The recombinant plasmid encoding active SARS-CoV-2 Mpro in this protocol will be available on request by contacting the lead contact.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 81703546); Natural Science Foundation of Anhui Province, China (No. 1808085QH265); University Natural Science Research Project of Anhui Province, China (Nos. KJ2021A0839, YJS20210549); CAMS Innovation Fund for Medical Sciences, China (No. 2021-I2M-1-054); and The Young Talent Project of Wannan Medical College, China (No. wyqnyx202104).
We thank the staff of Antiviral Research Unit at Wannan Medical College, Wuhu, China. We are grateful to Dr. Dongsheng Li (Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China) for his kind assistance on HTS. We thank Mrs. Jingzhe Yang and Mrs. Liming Zhao (Agilent BioTek Shanghai Representative Office, Shanghai, China) for promoting the application of this FP assay in the field of antiviral agent discovery.
Author contributions
J.Z., H.Y., and G.Y. wrote the manuscript and conducted the experiments. X.L. contributed to reagent support and validation. Y.W. and Y.C. conceived the FP assay, revised the manuscript, and analyzed the experiments. All authors reviewed the results and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Yanchang Wang, Email: yanchang.wang@med.fsu.edu.
Yunyu Chen, Email: chenyunyu1984@163.com.
Data and code availability
This study did not generate datasets/codes.
References
- Chen Y., Fu Z., Yan G., Lin Y., Liu X. Optimization of expression conditions and determination the proteolytic activity of codon-optimized SARS-CoV-2 main protease in Escherichia coli. Chin. J. Biotechnol. 2021;37:1334–1345. doi: 10.13345/j.cjb.200416. [DOI] [PubMed] [Google Scholar]
- Chen Z., Cui Q., Cooper L., Zhang P., Lee H., Chen Z., Wang Y., Liu X., Rong L., Du R. Ginkgolic acid and anacardic acid are specific covalent inhibitors of SARS-CoV-2 cysteine proteases. Cell Biosci. 2021;11:45. doi: 10.1186/s13578-021-00564-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M.D., Yasgar A., Peryea T., Braisted J.C., Jadhav A., Simeonov A., Coussens N.P. Fluorescence polarization assays in high-throughput screening and drug discovery: a review. Methods Appl. Fluoresc. 2016;4:022001. doi: 10.1088/2050-6120/4/2/022001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson O.D., Taranova N.A., Zherdev A.V., Dzantiev B.B., Eremin S.A. Fluorescence polarization-based bioassays: new horizons. Sensors. 2020;20:E7132. doi: 10.3390/s20247132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Kozak J.J., Gray H.B., Garza-López R.A. Structural stability of the SARS-CoV-2 main protease: can metal ions affect function? J. Inorg. Biochem. 2020;211:111179. doi: 10.1016/j.jinorgbio.2020.111179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Yan G., Zhou W., Si S., Liu X., Zhang J., Li Y., Chen Y. Ginkgolic acid and anacardic acid are reversible inhibitors of SARS-CoV-2 3-chymotrypsin-like protease. Cell Biosci. 2022;12:65. doi: 10.1186/s13578-022-00806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Hu Y., Townsend J.A., Lagarias P.I., Marty M.T., Kolocouris A., Wang J. Ebselen, disulfiram, carmofur, PX-12, tideglusib, and shikonin are nonspecific promiscuous SARS-CoV-2 main protease inhibitors. ACS Pharmacol. Transl. Sci. 2020;3:1265–1277. doi: 10.1021/acsptsci.0c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco M.D., Ma C., Lagarias P., Gao A., Townsend J.A., Meng X., Dube P., Zhang X., Hu Y., Kitamura N., et al. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Sci. Adv. 2020;6:eabe0751. doi: 10.1126/sciadv.abe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G., Li D., Lin Y., Fu Z., Qi H., Liu X., Zhang J., Si S., Chen Y. Development of a simple and miniaturized sandwich-like fluorescence polarization assay for rapid screening of SARS-CoV-2 main protease inhibitors. Cell Biosci. 2021;11:199. doi: 10.1186/s13578-021-00720-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G., Li D., Qi H., Fu Z., Liu X., Zhang J., Chen Y. Discovery of SARS-CoV-2 main protease inhibitors using an optimized FRET-based high-throughput screening assay. Chin. J. Biotechnol. 2022;38:2236–2249. doi: 10.13345/j.cjb.210657. [DOI] [PubMed] [Google Scholar]
- Yan H., Liu Z., Yan G., Liu X., Liu X., Wang Y., Chen Y. A robust high-throughput fluorescence polarization assay for rapid screening of SARS-CoV-2 papain-like protease inhibitors. Virology. 2022;574:18–24. doi: 10.1016/j.virol.2022.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Yan G., Qi H., Liu Z., Liu X., Liu X., Li N., Chen Y. Identifying SARS-CoV-2 main protease inhibitors by a novel sandwich-like fluorescence polarization screening assay. Chin. J. Biotechnol. 2022;38:2352–2364. doi: 10.13345/j.cjb.210949. [DOI] [PubMed] [Google Scholar]
- Zhu W., Xu M., Chen C.Z., Guo H., Shen M., Hu X., Shinn P., Klumpp-Thomas C., Michael S.G., Zheng W. Identification of SARS-CoV-2 3CL protease inhibitors by a quantitative high-throughput screening. ACS Pharmacol. Transl. Sci. 2020;3:1008–1016. doi: 10.1021/acsptsci.0c00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate datasets/codes.