Abstract
Background
The arrival of the Delta variant of SARS-CoV-2 was associated with increased transmissibility and illness of greater severity. Reports of nosocomial outbreaks of Delta variant COVID-19 in acute care hospitals have been described but control measures varied widely.
Aim
Epidemiological investigation of a linked two-ward COVID-19 Delta variant outbreak was conducted to elucidate its source, risk factors, and control measures.
Methods
Investigations included epidemiologic analysis, detailed case review serial SARS-CoV-2 reverse transcriptase–polymerase chain reaction (RT–PCR) testing of patients and healthcare workers (HCWs), viral culture, environmental swabbing, HCW-unaware personal protective equipment (PPE) audits, ventilation assessments, and the use of whole genome sequencing (WGS).
Findings
This linked two-ward outbreak resulted in 17 patient and 12 HCW cases, despite an 83% vaccination rate. In this setting, suboptimal adherence and compliance to PPE protocols, suboptimal hand hygiene, multi-bedded rooms, and a contaminated vital signs cart with potential fomite or spread via the hands of HCWs were identified as significant risk factors for nosocomial COVID-19 infection. Sudden onset of symptoms, within 72 h, was observed in 79% of all Ward 2 patients, and 93% of all cases (patients and HCWs) on Ward 2 occurred within one incubation period, consistent with a point-source outbreak. RT–PCR assays showed low cycle threshold (CT) values, indicating high viral load from environmental swabs including the vital signs cart. WGS results with ≤3 SNP differences between specimens were observed.
Conclusion
Outbreaks on both wards settled rapidly, within 3 weeks, using a `back-to-basics' approach without extraordinary measures or changes to standard PPE requirements. Strict adherence to recommended PPE, hand hygiene, education, co-operation from HCWs, including testing and interviews, and additional measures such as limiting movement of patients and staff temporarily were all deemed to have contributed to prompt resolution of the outbreak.
Keywords: SARS-CoV-2, Point source, COVID-19, Outbreak, Delta, Personal protective equipment, Whole genome sequencing
Introduction
The arrival of the Delta variant of SARS-CoV-2 was associated with increased transmissibility, reported to be as high as 97% compared to the ancestral lineages, and causing illness of greater severity [[1], [2], [3]]. There have been reports of nosocomial outbreaks of Delta variant COVID-19 in acute care hospitals but control measures varied widely [[4], [5], [6], [7]]. We describe a two-ward acute care hospital outbreak of Delta variant where the probable modes of transmission were elucidated using epidemiologic, laboratory and virologic investigations, environmental investigations, and whole genome sequencing (WGS).
Methods
The outbreak investigation and reporting followed the ORION guidelines, with the exception of the exact start and finish dates, to protect patient and healthcare worker (HCW) confidentiality [8].
Outbreak setting and epidemiologic investigations
Ward 1
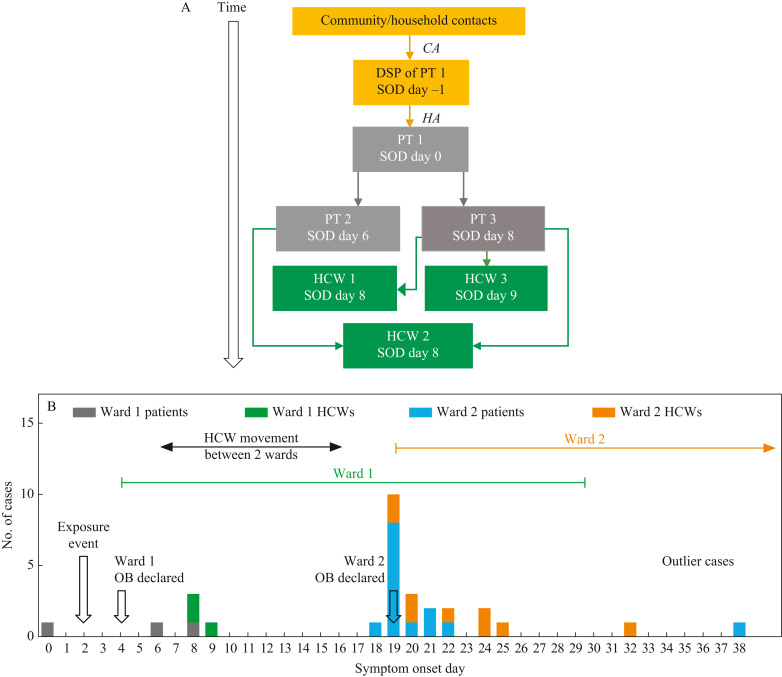
Entry of the Delta variant occurred in an 1100-bed tertiary acute care facility in the spring of 2021 via a designated support person with community COVID-19 acquisition and secondary transmission via close contact, to the index patient (Figure 1A ). The entry of the virus to Ward 1 and the subsequent outbreak occurred during a time of relatively low community transmission in our jurisdiction (active cases: 153 per 100,000; https://covid-tracker.chi-csm.ca/) during an interwave period and just before the Delta wave emergence in our jurisdiction. Further to the first case, two additional hospital-acquired (HA) patient cases (Supplementary Appendix: HA cases require the absence of any epidemiologic evidence to support a community or household exposure) in separate rooms on the same ward were identified following respiratory symptom onset four and six days later, respectively (Figure 1A, B). Three HCWs who became infected, all of whom developed respiratory symptoms, had worked directly with at least one of the affected patients. It was noted that all three infected patients had high dependency needs. Nasopharyngeal (NP) swabs were obtained from all patients on the ward and were requested from all HCWs who attended Ward 1 (patient census = 45; Ward staff = 140; physicians = 25; medical ward). Patients were tested serially (q3 days), with SARS-CoV-2 reverse transcriptase–polymerase chain reaction (RT–PCR) until closure of the outbreak [9].
Figure 1.
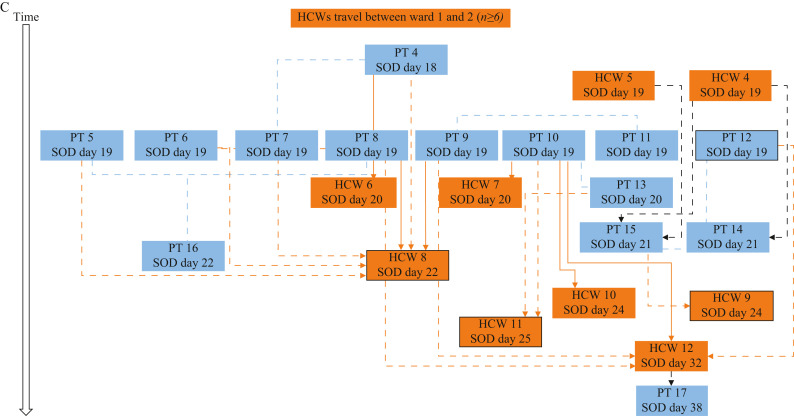
Delta SARS-CoV-2 introduction and transmission between two wards. (A) Origin of SARS-CoV-2 Delta variant on Ward 1. CA, community-acquired; HA, hospital-acquired; SOD, date of core respiratory, core gastrointestinal or expanded COVID symptom onset or the date the case tested positive for COVID-19, whichever is sooner; PT, patient; HCW, healthcare worker; DSP, designated support person. Solid lines: strong epidemiological link based on work assignment, detailed interview and/or any reported personal protective equipment (PPE) breaches. Direction of arrow: direction of transmission. (B) Epidemic curve of Delta SARS-CoV-2 outbreak on two medical wards. Epidemic curve showing cases (patient and HCW) by symptom onset date (SOD). SOD is the date of core respiratory, core gastrointestinal or expanded COVID symptom onset or the date the case tested positive for COVID-19, whichever is sooner. OB, outbreak. Dates of outbreaks on Wards 1 and 2 are indicated by a green and orange line, respectively. Outbreak on Ward 2 closed on day 59 (indicated by forward-facing arrow). Black arrow indicates HCW movement between Wards 1 and 2. Open black arrows indicate key epidemiological events. (C) Introduction and transmission of Delta SARS-CoV-2 on Ward 2. SOD is the date of core respiratory, core gastrointestinal or expanded COVID symptom onset or the date the case tested positive for COVID-19, whichever is sooner. Solid lines denote transmission event with strong epidemiological link based upon work assignment, detailed interview and/or any reported PPE or hand hygiene breaches; dashed black lines denote possible transmission based upon care dates and communicable phase of illness; blue lines indicate patient-to-patient transmission event or patients who were roommates; black lines indicate HCW-to-patient transmission event; orange lines indicate patient-to-HCW transmission event; black frames around HCW cases denote shift overlap with a co-worker. Absence of any line indicates that source of infection is unknown/no specific interaction between patients and/or HCWs identified. In all cases the risk period refers to the communicable phase of illness (defined as 48 h before symptom onset until 14 days after symptom onset).
Ward 2
An initial patient case was identified on Ward 2 (census = 41; medical ward), in a separate building, after onset of fever and cough 10 days following the last identified case on Ward 1. Symptom onset occurred in eight patients within 24 h and, within 72 h, 11 patients had symptoms (Figure 1B). NP swabs were obtained from all patients and were requested from all HCWs who attended Ward 2 (patient census = 41; Ward staff = 109; physicians = 28; medical ward). The patient NP swabs were collected serially (q2–3 days) and tested by RT–PCR until closure of the outbreak [9].
For both wards HCWs were offered SARS-CoV-2 NP testing and it was strongly encouraged. Neither vaccination nor testing was mandatory. Investigations by the Outbreak Management Team were conducted using classic epidemiologic tools, including interviews of infected individuals, review of medical records, review of patient placement, patient and HCW movement between wards and HCW contact tracing using forward and backward contact tracing and a linkage interview, with the aim of determining the transmission routes to patients (Supplementary Appendix) and HCWs and to facilitate outbreak control measures [10].
Environmental investigations
Environmental swabs were collected from multiple sites in patient rooms and from shared medical equipment, including a mobile vital signs cart (VSC). One set of swabs was collected the day the outbreak was declared on Ward 2 and a second set the following day. As a negative control, swabs were taken from the main shower room, which was in a section of the unit separate from where the initial positive patients were identified. Environmental specimen PCR assays were performed according to methods previously described [9,[11], [12], [13]]. Typing of strains was confirmed using Variant testing PCR, as previously described [3].
Laboratory and virologic investigations
Clinical specimens, primarily NP swabs, but in some cases throat swabs, were collected and tested for SARS-CoV-2 using RT–PCR. Serial NP swab COVID-19 testing was done with 13 patients (Ward 2) by experienced HCWs to observe viral kinetics over time in patients using a validated RT–PCR assay, based on an E gene target with internal controls [9]. To confirm the presence of infectious virus, clinical samples from consented patients were cultured using Vero cells as described by Lin et al. [12].
In addition, HCWs were offered voluntary prevalence testing on site to increase uptake and convenience of testing. Testing was highly recommended every five days for ‘on unit’ and ‘off unit’ staff who attended the ward in the 14 days prior to and since the start of the outbreak. Based upon the numbers of tests completed, it was learned that HCW testing was incomplete and that the number of HCWs tested decreased substantially after the initial round of prevalence testing.
HCW symptom screening
A daily ‘Fit-for-Work’ symptom review was required for all HCWs. A HCW was deemed fit for work if asymptomatic, as well as having no discrete COVID-19 exposure risk. HCWs were required to show their fit-for-work status on arrival at work and mid-way through shift. If they developed COVID-19 symptoms while at work they were directed to leave, isolate, and test for COVID-19.
PPE compliance based on covert observations
Anonymous ‘HCW unaware’ audits of adherence to PPE and hand hygiene had been ongoing in the hospital for several months and stored on an accessible database. Multiple audits were completed just prior to the outbreak on the affected ward and were available for comparison with the designated COVID-19 wards across the health region. These covert audits were performed by clinical nurse educators from other hospital wards and hence were unknown to the HCWs on the audited ward. A standardized audit tool was utilized and loaded into a RedCap database. The audit tool addressed the individual components for each step of PPE use including the PPE environment (i.e. availability of PPE materials), donning of all components of the PPE and the doffing of all PPE components, including the gloves, gowns, eye protection and masks plus the required hand hygiene for each doffing step. Separate from the audits, any infected HCWs were asked via questionnaire about the individual elements of PPE and to self-describe their doffing procedures and hand hygiene technique as part of the contact tracing interviews.
Whole genome sequencing
WGS of patient and HCW samples was done retrospectively. The full genome of SARS-CoV-2 strains obtained from the NP swabs of HCWs and patients between the two wards was amplified by multiplex PCR according to the LoCost ARTIC protocol (https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye) using the Freed oligos as 1200 bp amplicons with sequencing done using Oxford Nanopore (Oxford, UK) or Illumina (San Diego, CA, USA) sequencing technology [14,15]. Consensus sequences were aligned with mafft and visualized using snipit (https://github.com/aineniamh/snipit) [16]. The full protocol was completed as outlined previously [11].
Ventilation assessments
Ventilation was measured in air exchanges per hour (AEH), by the Facilities, Maintenance, and Engineering department of the hospital, at the end of the outbreak. Values were interpreted relative to the Canadian Safety Association standards for Heating, Ventilation, and Air Conditioning (HVAC) Systems in Health Care Facilities (CSA-Z317.2–15).
Outbreak control measures
Multiple measures were employed concomitantly for control of the outbreak, including isolation of patients, frequent clinical monitoring using a comprehensive COVID-19 symptom/sign monitoring tool, serial testing for SARS-CoV-2, selected transfer of infected patients (to designated COVID-19 wards), implementation of enhanced environmental cleaning and review of all cleaning and disinfection practices for the patient environment and shared medical equipment, staff and visitor restrictions, review of ventilation parameters, and coached adherence to contact and droplet PPE by all HCWs (medical masks, goggles/face shields, gowns, gloves) [17]. All break and lunch rooms were reviewed for compliance with the restrictions, occupancy limits and segregated physical distancing limits implemented by Site Administration at the beginning of the pandemic.
Statistical analysis
Statistical analysis was performed using χ2-test or Student's t-test for categorical and continuous variables as appropriate; P < 0.05 was considered significant.
Ethics approval
This investigation was conducted as part of a formal Epidemiologic Investigation under Public Health in the Province of Alberta. It also was assessed using the ARECCI (A Project Ethics Community Consensus Initiative) tool which scored this project as fitting with a quality improvement project for which ethics approval is not required. The consenting and culturing of virus from the affected patients was approved by the University of Calgary Conjoint Research Ethics Board (REB20-0444).
Results
Descriptive epidemiology: Wards 1 and 2
Following initial entry of the Delta variant via a designated support person as described above, epidemiologic investigations identified that transmission to patients 2 and 3 occurred when patient 1, with known difficulty masking from underlying medical conditions, and having core respiratory symptoms, including frequent coughing, was inadvertently seated directly beside patients 2 and 3 (masking status unknown) in a treatment waiting area elsewhere in the facility on day 2 (Figure 1A). The three HCWs who became positive, despite continuous masking (Figure 1A) were considered as patient-to-HCW transmission based on contact tracing, patient assignments and the high dependency needs of the patients. Prospective serial PCR testing (q3 days) of all patients and of HCWs who were tested revealed no other positive cases on Ward 1.
Symptom screening and serial testing identified a total of 14 SARS-CoV-2-infected patients on Ward 2 (Figure 1B, C; designated patients 4–17). One outlier patient case and one HCW case were identified >7 days from outbreak declaration. All Ward 2 patients (N = 14) had core respiratory symptoms with or without low-grade fever based on the symptom screening tool employed, regardless of their vaccination status. The frequency of vaccinated and unvaccinated patients and HCWs is presented in Table I . For patients and HCWs, vaccination rates were 77% and 92%, respectively, for an overall rate of 83%. The 14 SARS-CoV-2-infected patients had mobility scores indicating that they were at higher risk of falls and had higher dependency needs.
Table I.
Number of Delta SARS-CoV-2 cases and vaccination status on Wards 1 and 2
| Ward | Healthcare workers |
Patients |
||||||
|---|---|---|---|---|---|---|---|---|
| n = 0 | n = 1 | n = 2 | Total | n = 0 | n = 1 | n = 2 | Total | |
| 1 | 0 | 1 | 2 | 3 | 0 | 3 | 0 | 3 |
| 2 | 1 | 3 | 5 | 9 | 4 | 4 | 6 | 14 |
| No. of cases | 1 | 4 | 7 | 12 | 4 | 7 | 6 | 17 |
n refers to the number of vaccine doses.
All 17 patient cases were considered hospital-acquired. The median number of days from admission to symptom onset was 14 days; only four patient cases had an interval <10 days, one of whom was the index patient (PT1, Figure 1C) with a known designated support person-to-patient transmission event and the other three patient cases (patients 7, 13, 14; Figure 1C) had a least one negative SARS-CoV-2 NP test on admission and/or in the three to five days prior to symptom onset.
There were 140 and 109 primary HCWs (including registered nurses, licensed practical nurses, healthcare aides, unit clerks and allied health staff including physiotherapists/occupational therapists) who were assigned to Wards 1 and 2, respectively, and there were 25 and 28 physicians who worked at various times on Wards 1 and 2, respectively. There was high uptake of SARS-CoV-2 NP prevalence testing during this outbreak with 91.4% of the HCWs and physicians undertaking NP prevalence testing at least once between the two units, based on public health records of individuals tested. A total of 551 NP RT–PCR tests were completed between the two wards over the course of the outbreak. Some HCWs had multiple tests done. Of the 12 PCR-positive HCWs detected between the two wards, some of whom were identified as exposed contacts of cases, none initially reported ‘exposure’ or PPE breaches on initial contact tracing interview, but post-diagnosis questionnaires and interviews conducted on all 12 cases with a detailed selection of additional interview questions, including backwards contact tracing, found six out of 12 (50%) reported breaches related to PPE (eye protection or masking), hand hygiene, or environmental exposure or to difficulty accessing wipes for shared computer workstations. All HCWs were asked about COVID-19 within their households and none had a household contact prior to their own onset of COVID-19 symptoms. Three of the HCWs reported suspected onward transmission of infections to their household contacts following their occupationally acquired COVID-19. Although initially only one HCW was found to have worked between the two wards, more comprehensive and collaborative investigations between ward managers, WHS, IPC, and hospital administration identified at least six HCWs who had worked between both wards during the 14 days prior to the Ward 2 outbreak, not all of whom were confirmed to have been tested for SARS-CoV-2. It was also learned that many HCWs frequently and simultaneously aided with at least one of the patients reported to have had frequent forceful coughing and who had very high care and dependency needs, particularly for positioning and toileting.
Multi-bedded rooms with shared bathrooms
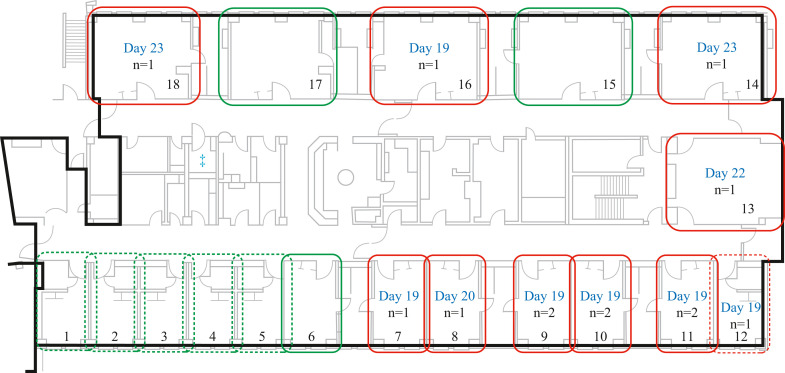
All but one Ward 2 case occurred in multi-bedded rooms (two, three, or four beds with a shared bathroom) (Figure 2 ). A significant association with infection (P = 0.04) was seen in patients who were in multi-bedded rooms with a shared bathroom (nine out of 12) versus those in a private room. No aerosol-generating medical procedures (AGMPs) were performed in multi-bedded rooms [18].
Figure 2.
Room assignment of positive cases. Room number is indicated for each room (1–14) and shower room is indicated by blue double-dagger symbol. Symptom onset date (SOD) is indicated by day in blue text and the number of patients that were positive in each room is indicated in italics (n = x). SOD is the date of core respiratory, core gastrointestinal or expanded COVID symptom onset or the date the case tested positive for COVID-19, whichever is sooner. Solid red boxes indicate shared rooms where positive cases were found; dotted red box indicates private room where positive case was found; solid green boxes indicate shared rooms where no positive cases were found; dotted green box indicates private rooms where no positive cases were found.
Environmental investigations
SARS-CoV-2 PCR positivity was found on 10/10 swabs from high-touch surfaces on the VSC (Table II ) with C T values (N gene) between 17.14 and 19.70, which is in the range highly predictive of infectious virus [12]. All samples, including those from the VSC, were confirmed to be Delta strain by RT–PCR [11]. COVID-19-positive cases were significantly associated with rooms (10/13 vs 0/5; P = 0.007) on Ward 2 where the VSC was used by HCWs as opposed to rooms with wall-mounted vital signs equipment. No record or log of cleaning the mobile VSC was found.
Table II.
Environmental specimens from Ward 2 taken on days 1 and 2a
| Environmental specimens | RT–PCR |
|---|---|
| RT–PCR (Ward 2, day 1)b | CT (E gene) |
| Patient room 11 (two-bed) | |
| Call bell | 32.28 |
| Commode | 34.82 |
| Bed rail | 33.92 |
| Table top | 31.98 |
| A random blood pressure monitor, thermometer, bladder scanner and O2 monitor and shower room (N = 5) | Negative |
| Room 13 specimens (call bell, bed rail, light switch, table top (N = 4)) | Negative |
| RT–PCR (Ward 2, day 2) | CT (N gene) |
| Room 13 (overcapacity space) | |
| Vital signs cart 1: thermometer 1 | 19.14 |
| Vital signs cart 1: metal temperature probe | 18.27 |
| Vital signs cart 1: pulse oximeter | 19.15 |
| Vital signs cart 1: push handle | 17.14 |
| Commode: under seat and armrests | 19.26 |
| Patient room 14 (four-bed) | |
| Vital signs cart 2: thermometer 2 | 18.1 |
| Vital signs cart 2: pulse oximeter 2 | 18.8 |
| Vital signs cart 2: push handle and monitor buttons | 20.14 |
| Vital signs cart 2: stethoscope | 18.06 |
| Patient room 11 (two-bed) | |
| Commode: under seat, armrests | 16.92 |
| Patient room 9 (two-bed) | |
| Commode | 15.2 |
| Bed rail | 19.4 |
| Call bell | 20.08 |
| Stethoscope hanging on door | 19.7 |
| Patient room 12 (private) | |
| Room sink taps and vanity counter | 18.48 |
| Call bell (on bedrail only) | 18.5 |
| Vitals monitor buttons, thermometer, pulse oximeter | 18.53 |
| Computer on wheels between rooms 10 and 11 | |
| Computer keyboard and mouse | 17.23 |
| Shared equipment stored across from room 5 | |
| Bladder scanner: push handle, wand, monitor buttons, gel bottle | 16.96 |
| Masimo vital cart: push handles, temp probe, monitor buttons, pulse oximeter | 17.44 |
Laboratory and virologic investigations
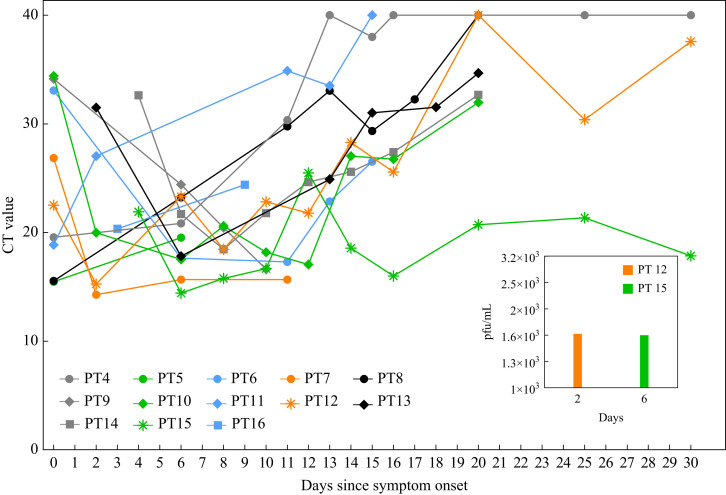
Viral kinetics studies were done on 13 patients up to 30 days post symptom onset (Figure 3 ) (N = 6 fully immunized; N = 7 not fully immunized, see Supplementary Appendix for definitions). On illness day 6, 11 out of 11 patients (three unvaccinated) had C T values ≤25 (E-gene) and six out of six patients (one unvaccinated) continued to have C T values ≤25 up to day 10. Viral cultures for two consented patients (both fully immunized) revealed 1.60 × 103 plaque-forming units (pfu)/mL (C T N gene 18.09) at day 2 and 1.58 × 103 pfu/mL (C T N gene 17.25) at day 6 of infection (Figure 3) [12].
Figure 3.
Transmission of Delta SARS-CoV-2 cases on Ward 2. Serial CT values of patients over time. pfu, plaque-forming units (per mL); CT, cycle threshold value for the Envelope (E) gene; PT, patients (for Ward 2 patients numbering starts at PT 4, as per numbering in Figure 1C). Data represent 13 out of 14 patients; PT 17 was not tested for serial CT. Median number of serial CT values per patient is 6 (range: 2–11).
Monitoring of PPE compliance based on covert observations
There was a highly significant association with sub-optimal adherence to doffing, hand hygiene and order of doffing on Ward 2 (55% and 64% of the time, respectively) compared to four adult-designated COVID-19 wards (78% and 89%, respectively; P = 0.007; N = 54 HCW-unaware audits) in the three-month period prior to the outbreak. Regarding specific components of the PPE doffing, which may place HCWs at risk of mucous membrane inoculation of SARS-CoV-2, in the three months prior to the outbreak, lack of hand hygiene after glove and gown removal was identified in 27.2% and 36.3% of audits, and improper doffing of eye protection and masks in 27.2% and 36.3% of audits, respectively. In addition, it is noteworthy that in the four weeks following the declaration of the outbreak, the lack of hand hygiene after glove and gown removal was markedly reduced, being found in only 2.7% and 5.4% of audits, respectively, while improper doffing of eye protection and masks was noted in only in 13.5% and 16.2% indicating a marked improvement in overall adherence. There was no difference in the type of PPE used (medical mask, googles or face shields, gowns, and gloves) between COVID-19 wards and general wards.
Whole genome sequencing
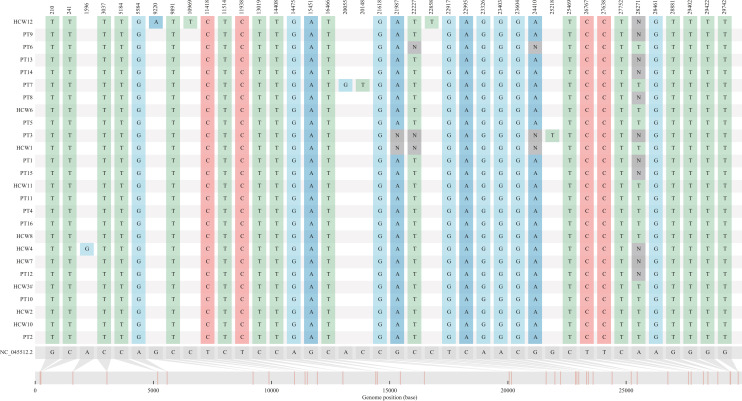
WGS revealed ≤3 single nucleotide polymorphism (SNP) differences in the strains found in all HCWs and patients from Wards 1 and 2, including individuals who had worked between both wards. The outbreak strain was markedly different (mean 17.5 SNPs) than community Delta strains, demonstrating a unique strain was responsible for both outbreaks (Figure 4 ).
Figure 4.
Whole genome sequencing of healthcare workers (HCWs) and patient cases from Wards 1 and 2. PT, patient; HCW, healthcare worker. NC_045512.2 is the genome reference sequence number for SARS-CoV-2 (Wuhan genome). Please refer to Figure 1, which indicates which HCWs and patients are linked to either Ward 1 (Figure 1A) or Ward 2 (Figure 1C).
Ventilation parameters
The testing of air exchanges per hour (AEH) immediately following the outbreak for the rooms on Ward 2 revealed that they exceeded Canadian safety standards, with variance between 6.9 and 9.5 AEH, with 100% outside air. The bathrooms were negative pressure with respect to the patient rooms; the patient rooms were positive with respect to the hallway.
Infection prevention and control measures and outbreak closure
Measures initiated concomitantly with declaration of the outbreak included serial SARS-CoV-2 PCR prevalence testing of all patients and >90% of the HCWs enabling rapid detection of cases, moving some positive patients to the designated COVID-19 Ward to allow all remaining patients to be in single-bedded rooms on Ward 2, a temporary suspension of all new admissions, restricting HCW movement on multiple wards or sites, purchasing of additional VSCs, enhanced environmental cleaning of surfaces and mobile medical equipment, and strict adherence/compliance to our standard PPE measures. Enhanced cleaning by Environmental Services (twice as opposed to once daily) of the patient environment with a hospital-approved cleaning product, in addition to standard cleaning at the time of discharge or transfer of a patient to another ward or whenever any visible soling was present. Cleaning and disinfection of any patient-related mobile or shared medical equipment was the responsibility of the Ward staff rather than Environmental Services and was to be done after use on any patient and at discharge or transfer. A review of the medical equipment cleaning and disinfection revealed verbal reports of staff not following the cleaning and disinfection process. No logs of the cleaning and disinfection of the shared medical equipment were kept. Reviews of all break rooms, lunch rooms, and the cafeteria and WHS investigations with the HCWs identified no breaches in compliance but nonetheless these measures were reinforced. Outbreaks on both wards settled rapidly, within three weeks, without extraordinary measures or changes to PPE other than improved adherence to all measures for PPE donning, doffing, and hand hygiene [19].
Discussion
Patient-to-patient and patient-to-HCW transmission on Ward 1 followed by transmission from Ward 1 to Ward 2 via HCWs who worked between both wards was considered most likely. The two wards were in separate buildings. For Ward 2, with 84% of symptomatic patients presenting within a 72 h period, as well as a significant association with the use of a mobile VSC and the presence of environmental SARS-CoV-2 RNA contamination with very low C T values, a point-source transmission from the VSC was considered a plausible explanation based on the epidemiologic and environmental findings. There is evidence that SARS-CoV-2 from clinical sources (cough droplets, saliva, nasal secretions) can be readily cultured from human hands and may persist on common medical surfaces for many hours, including stethoscope diaphragms, pulse oximeters and plastic surfaces, all of which are basic components of the VSCs [12,13]. The lack of systematic cleaning and disinfection of the VSC and its components would have permitted continued growth of the virus on its high-touch surfaces. The suboptimal hand hygiene and improper doffing of eyewear and masks in the immediate pre-outbreak period which we documented through the covert audits on the Ward would have provided opportunities for contact transmission of the virus to the mucous membranes of the HCWs. It is also possible that additional HCWs were infected and at work communicable to others unbeknownst to the outbreak response teams since testing was not mandatory. Transmission may have occurred in association with close contact between patients who were in multi-bedded rooms with a single shared bathroom, but it does not explain how eight patients became symptomatic and were laboratory-confirmed COVID-19 on the same day but were in different rooms and no one HCW had assignments to all these patients. The lack of finding of any transmission events associated with patients in single rooms, with the one exception where the mobile VSC was used, and the ventilation param-eters exceeding CSA standards do not support long-range airborne transmission across either of the wards. It is possible that transmission occurred via infected HCWs travelling with the cart during their interactions with patients but this possibility is not compatible with the timing.
The prolonged presence of low C T values in patients (Figure 3) that correlate with cultivatable virus, combined with a stronger binding avidity to ACE-2-bearing receptor cells, provides a potential explanation as to why the Delta variant strain was so transmissible [12,20]. This outbreak occurred despite mRNA vaccination in a large number of HCWs and patients (Table I), which corroborates other study findings [4,5].
This outbreak also occurred in the setting of continuous masking by HCWs but thorough investigations led to discovery of well-recognized risk factors for transmission: suboptimal adherence and compliance to PPE protocols, suboptimal hand hygiene, risks associated with multi-bedded rooms, and a contaminated VSC with potential fomite or indirect spread via the attendant HCWs. An underlying impression of a ‘veil of protection’ feeling among vaccinated HCWs, illustrative of the bias of purity risk ritual, may have contributed [21].
Despite the rigour of investigation and the strength of our findings, there are limitations to this study including imprecision due to not testing all HCWs, recall bias, non-responder bias, and inability to capture every patient–HCW interaction and care being provided without documented patient assignment. Nonetheless, the findings underscore the importance of ‘shoe leather’ epidemiology, supplemented with molecular epidemiology and adherence to fundamental IPC principles and rigorous investigations to ascertain modes of transmission during a COVID-19 outbreak.
Unlike other outbreak reports where N95 respirators were employed as a part of the response strategy and considered a necessary component, this outbreak settled rapidly without any change to PPE recommendations [6]. Rather, our approach focused on a back-to-basics approach emphasizing PPE adherence and hygienic practices. This approach is similar to the one utilized by Susky et al. and supports results seen in recent systematic reviews and a large matched case–control study in France where there was no difference demonstrated in a multivariate analysis of whether HCWs who acquired COVID-19 were wearing surgical masks or N95 respirators [7,[22], [23], [24]].
Acknowledgements
We thank P. Dieu, C. Ferrato, K. Gill, R. Ma, and J. Thayer from Genomics and Bioinformatics, Alberta Public Health Laboratory – South, Calgary, AB, Canada. We acknowledge the Public Health Agency and the Government of Canada for their generous assistance to provide personnel, reagents, and consumables to support genome sequencing of SARS-CoV-2. We also acknowledge support from the Canadian COVID-19 Genomics Network (CanCOGeN). We also wish to thank the Workplace Health and Safety occupational health nurses, and Infection Prevention and Control and Public Health team members who facilitated rapid contact tracing and epidemiological investigations related to the outbreak. We would like to thank Drs S. Chacko and A. Lewis for their assistance and astute observations during the outbreak period and for their physician leadership during the course of this investigation. The hospital site leadership team and AHS communication team members were also instrumental in facilitating a highly coordinated and effective outbreak response. Finally, the authors would like to acknowledge the outstanding cooperation of the healthcare workers who had experienced infection themselves, and their affected patients and families, and all of the healthcare workers (both clinical and non-clinical staff members and physicians), who with support from the site leadership and hospital administrators, helped to maintain excellent patient care delivery during the outbreak period.
Footnotes
This work was presented as a late breaker in abstract form (#4654/L0510) at the 32nd ECCMID meeting, Lisbon, Portugal, April 23rd–29th, 2022.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhin.2022.09.019.
Author contributions
H.M.O.: conceptualization, data curation, formal analysis, investigation, methodology, writing, original draft, and review and editing; R.H.: conceptualization, data curation, investigation, methodology, writing, original draft, and review and editing; K.S., L.T., P.Y., L.W., A.F.: data curation, investigation, validation, writing – review and editing; P.J., A.W., J.C.: data curation, validation, resources, writing – review and editing; T.L., M.C., V.L., K.P., A.W., H.Y.Z., T.D., K.H., B.M.B., K.F.: data curation, validation, writing – review and editing; Y.C.L., D.E.: data curation, formal analysis, resources, methodology, writing – review and editing; J.M.C.: conceptualization, data curation, formal analysis, investigation, methodology, resources, writing, funding, original draft, and review and editing.
Conflict of interest statement
None declared.
Funding sources
The investigation associated with this outbreak was unfunded. The culturing of SARS-CoV-2 was funded in part by the University of Calgary Infectious Diseases Research and Innovation Fund for COVID-19
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., et al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9:1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora P., Sidarovich A., Krüger N., Kempf A., Nehlmeier I., Graichen L., et al. B.1.617 2 enters and fuses lung cells with increased efficiency and evades antibodies induced by infection and vaccination. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.109825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian D., Sun Y., Zhou J., Ye Q. The global epidemic of the SARS-CoV-2 Delta variant, key spike mutations and immune escape. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.751778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim W.-Y., Tan G.S.E., Htun H.L., Phua H.P., Kyaw W.M., Guo H., et al. First nosocomial cluster of COVID-19 due to the Delta variant in a major acute care hospital in Singapore: investigations and outbreak response. J Hosp Infect. 2021;122:27–34. doi: 10.1016/j.jhin.2021.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shitrit P., Zuckerman N.S., Mor O., Gottesman B.-S., Chowers M. Nosocomial outbreak caused by the SARS-CoV-2 Delta variant in a highly vaccinated population, Israel. Euro Surveill. 2021;26(39) doi: 10.2807/1560-7917.ES.2021.26.39.2100822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hetemäki I., Kääriäinen S., Alho P., Mikkola J., Savolainen-Kopra C., Ikonen N., et al. An outbreak caused by the SARS-CoV-2 Delta variant (B.1.617.2) in a secondary care hospital in Finland. Euro Surveill. 2021;26(30) doi: 10.2807/1560-7917.ES.2021.26.30.2100636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Susky E.K., Hota S., Armstrong I.E., Mazzulli T., Kestenberg S., Casaubon L.K., et al. Hospital outbreak of the severe acute respiratory coronavirus virus 2 (SARS-CoV-2) delta variant in partially and fully vaccinated patients and healthcare workers in Toronto, Canada. Infect Control Hosp Epidemiol. 2021:1–4. doi: 10.1017/ice.2021.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stone S.P., Cooper B.S., Kibbler C.C., Cookson B.D., Roberts J.A., Medley G.F., et al. The ORION statement: guidelines for transparent reporting of Outbreak Reports and Intervention studies Of Nosocomial infection. J Antimicrob Chemother. 2007;59:833–840. doi: 10.1093/jac/dkm055. [DOI] [PubMed] [Google Scholar]
- 9.Pabbaraju K., Wong A.A., Douesnard M., Ma R., Gill K., Dieu P., et al. Development and validation of RT–PCR assays for testing for SARS-CoV-2. J Clin Microbiol. 2020;58:e01110–e01120. doi: 10.1128/JCM.01110-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Endo A., Leclerc Q.J., Knight G.M., Medley G.F., Atkins K.E., Funk S., et al. Implication of backward contact tracing in the presence of overdispersed transmission in COVID-19 outbreaks. Wellcome Open Res. 2020;5:239. doi: 10.12688/wellcomeopenres.16344.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zelyas N., Pabbaraju K., Croxen M.A., Lynch T., Buss E., Murphy S.A., et al. Precision response to the rise of the SARS-CoV-2 B.1.1.7 variant of concern by combining novel PCR assays and genome sequencing for rapid variant detection and surveillance. Microbiol Spectr. 2021;9 doi: 10.1128/Spectrum.00315-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin Y.-C., Malott R.J., Ward L., Kiplagat L., Pabbaraju K., Gill K., et al. Detection and quantification of infectious severe acute respiratory coronavirus-2 in diverse clinical and environmental samples. Sci Rep. 2022;12:5418. doi: 10.1101/2021.07.08.21259744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajakumar I., Isaac D.L., Fine N.M., Clarke B., Ward L.P., Malott R.J., et al. Extensive environmental contamination and prolonged severe acute respiratory coronavirus-2 (SARS CoV-2) viability in immunosuppressed recent heart transplant recipients with clinical and virologic benefit with remdesivir. Infect Control Hosp Epidemiol. 2021:1–3. doi: 10.1017/ice.2021.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyson J.R., James P., Stoddart D., Sparks N., Wickenhagen A., Hall G., et al. 2020. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv [Preprint]:2020.09.04.283077. [DOI] [Google Scholar]
- 15.Freed N.E., Vlková M., Faisal M.B., Silander O.K. Rapid and inexpensive whole-genome sequencing of SARS-CoV-2 using 1200 bp tiled amplicons and Oxford Nanopore Rapid Barcoding. Biol Methods Protoc. 2020;5:bpaa014. doi: 10.1093/biomethods/bpaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katoh K., Standley D.M. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics. 2016;32:1933–1942. doi: 10.1093/bioinformatics/btw108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Grady H.M., Dixit D., Khawaja Z., Snedeker K., Ellison J., Erebor J., et al. Asymptomatic severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection in adults is uncommon using rigorous symptom characterization and follow-up in an acute care adult hospital outbreak. Infect Control Hosp Epidemiol. 2022:1–25. doi: 10.1017/ice.2022.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alberta Health Services, Infection Prevention and Control. Aerosol-generating medical procedure guidance tool; n.d. Available at: https://www.albertahealthservices.ca/topics/Page17091.aspx [last accessed October 2022].
- 19.Alberta Health Services, Infection Prevention and Control. Personal protective equipment; n.d. Available at: https://www.albertahealthservices.ca/ipc/page6422.aspx [last accessed October 2022].
- 20.Augusto G., Mohsen M.O., Zinkhan S., Liu X., Vogel M., Bachmann M.F. In vitro data suggest that Indian delta variant B.1.617 of SARS-CoV-2 escapes neutralization by both receptor affinity and immune evasion. Allergy. 2022;77 doi: 10.1111/all.15065. 111–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.VanSteelandt A., Conly J., Ghali W., Mather C. Implications of design on infection prevention and control practice in a novel hospital unit: the medical ward of the 21st century. Anthropol Med. 2015;22:149–161. doi: 10.1080/13648470.2014.1003795. [DOI] [PubMed] [Google Scholar]
- 22.Bartoszko J.J., Farooqi M.A.M., Alhazzani W., Loeb M. Medical masks vs N95 respirators for preventing COVID-19 in healthcare workers: a systematic review and meta-analysis of randomized trials. Influenza Other Respi Viruses. 2020;14:365–373. doi: 10.1111/irv.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jefferson T., Del Mar C.B., Dooley L., Ferroni E., Al-Ansary L.A., Bawazeer G.A., et al. Physical interventions to interrupt or reduce the spread of respiratory viruses. Cochrane Database Syst Rev. 2020;2020 doi: 10.1002/14651858.CD006207.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belan M., Charmet T., Schaeffer L., Tubiana S., Duval X., Lucet J.-C., et al. SARS-CoV-2 exposures of healthcare workers from primary care, long-term care facilities and hospitals: a nationwide matched case–control study. Clin Microbiol Infect. 2022;28:1471–1476. doi: 10.1016/j.cmi.2022.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.