Abstract
The leucine-responsive regulatory protein (Lrp) binds to three sites centered 252, 216, and 152 bp upstream of the transcription start site of the Escherichia coli glutamate synthase operon (gltBDF) and activates transcription. Activators of ς70-dependent promoters usually bind closer to the −35 hexamer of the core promoter sequence. To study the mechanism by which Lrp-dependent activation occurs over this relatively large distance, the gltBDF upstream region was sequentially replaced with corresponding portions from the well-characterized ς70-dependent promoter lacZYAp. The glt-lac promoter hybrids were placed upstream of lacZ, allowing transcriptional activity to be monitored via β-galactosidase assays. Even replacing all gltBDF sequences downstream of and including the −35 hexamer did not eliminate Lrp-dependent activation of transcription. When a 91-bp region between the −35 hexamer and the proximal Lrp binding site (−48 to −128) was replaced with heterologous DNA of the same length, transcription was reduced nearly 40-fold. Based on the presence of a consensus binding sequence, this region seemed likely to be a binding site for integration host factor (IHF). Experiments to study the effects of a himD mutant on expression of a gltB::lacZ transcriptional fusion, gel mobility shift analyses, and DNA footprinting assays were used to confirm the direct participation of IHF in gltBDF promoter regulation. Based on these results, we suggest that IHF plays a crucial architectural role, bringing the distant Lrp complex in close proximity to the promoter-bound RNA polymerase.
Escherichia coli synthesizes glutamate under ammonia-limiting conditions using the enzymes glutamate synthase, specified by the gltBDF operon, and glutamine synthetase, specified by glnA. The genes gltB and gltD specify the large and small subunits of glutamate synthase, respectively (7), and gltF specifies a protein of unknown function (8, 20). The gltBDF operon is positively regulated by the leucine-responsive regulatory protein (Lrp) (12, 13) and is transcribed at very low levels in the absence of Lrp (12). The coregulator for Lrp is leucine, and exogenous leucine reduces the expression of gltBDF about twofold, although this level is still substantially above baseline transcription in the absence of Lrp. Thus, relative to more responsive operons, gltBDF is a leucine-independent member of the Lrp regulon (12).
There are three Lrp binding sites centered 246, 215, and 152 bp upstream of the gltBDF transcription start site (35); the closest of these sites is ∼110 bp from the −35 hexamer. The presence of such distal regulatory sites is unusual in ς70-regulated promoters, although it has precedence in promoters that are recognized by ς54 (6, 9, 28). In ς54-dependent promoters, upstream elements or activators binding far upstream of the core promoter sequence are brought closer to the RNA polymerase by a looping mechanism that is sometimes assisted by DNA-bending proteins (9), raising the question of whether additional regulatory proteins are required for activation of gltBDF transcription. Upon binding the glt DNA, the Lrp dimer bends the DNA towards itself; occupancy of all three sites results in both phased hypersensitivity of the region from −126 to −264 (35) and compaction of the DNA in this region (D. E. Wiese II, M. Young, R. G. Matthews, and C. Bustamante, unpublished data), suggesting formation of a nucleosome-like structure. It is not known whether Lrp activates transcription by a mechanism involving direct contact with RNA polymerase.
In this study, we have examined the role of the core promoter and downstream sequences in Lrp-dependent activation of the gltBDF operon. We show that Lrp-dependent activation persists even when the entire region downstream of and including the −35 hexamer is replaced by the corresponding region from the lac operon. We also demonstrate that integration host factor (IHF) binds to the region between the proximal Lrp binding site and the −35 hexamer and positively regulates gltBDF transcription. We propose that IHF-induced bending of DNA positions bound Lrp close to RNA polymerase for transcriptional activation of the gltBDF operon.
MATERIALS AND METHODS
Strains and growth conditions.
The strains used in this study are listed in Table 1. Cultures were grown in LB medium (30) or glucose minimal MOPS (morpholinopropanesulfonic acid) medium (25) at 37°C. The antibiotics ampicillin (80 to 100 μg/ml), tetracycline (20 μg/ml), and chloramphenicol (25 μg/ml), were added to the medium as indicated below.
TABLE 1.
Strains of E. coli and primers used in this work
| Strain, lysogen, or primer | Description | Reference or source |
|---|---|---|
| Strains | ||
| BE1 | W3110 lrp-201::Tn10 | 12 |
| DH5α | supE44 Δ(lacZYA-argF)U169 (φ80dlacZΔM15) deoR hsdR17 (rK− mK+) recA1 endA1 gyrA96 thi-1 relA1 λ− F′ | Life Technologies |
| JWD3 | BE1/pJWD2 | 13 |
| PS2209 | W3110 Δlac-169 | F. C. Neidhardt |
| RJ1413 | MC1000 himD::cat | R. Osuna (originally from B. Ely) |
| W3110 | E. coli F− prototroph | F. C. Neidhardt |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| Recombinant λ lysogens of strain PS2209 | ||
| LP1008 | glt-lacZ transcriptional fusion with −406 to +8 of glt fused to +1 of the lac operon | This study |
| LP1023 | glt-lacZ transcriptional fusion with −406 to −23 of glt fused to −24 of the lac operon | This study |
| LP1035 | glt-lacZ transcriptional fusion with −406 to −35 of glt fusion to −36 of the lac operon | This study |
| LP1048 | LP1007 with the region from −48 to −128 replaced with a fragment of equal length from the cat gene | This study |
| LP1000 | gltB::lacZ transcriptional fusion with −406 to +246 of the gltBDF operon | This study |
| LP2000 | LP1000 with the himD::cat allele from strain RJ1413 | This study |
| Primersa | ||
| GLT1 | ggatccgAATTCGGCGATGACCTGGATCAA (−406 to −384) | |
| GLT2 | ACtGCaGATTTCCAACTTATCGGG (complementary to −15 to +8) | |
| GLT3 | cTgCAGGTCAAATGGCAAGCTTAT (complementary to −47 to −23) | |
| GLT4 | AAGCTTATTGGTACAGAAATGTGC (complementary to −62 to −39) | |
| GLT5 | ggatccccgggATCCCTCTCAAGGGATTTATCGTA (complementary to +217 to +246) | |
| GLT6 | tcgcGAAGTCGTTAGAGAAACAGTC (complementary to −148 to −128) | |
| GLT7 | ctgcagCAATAAGCTTGCCATTTGACC (−54 to −28) | |
| GLTFP | GTCGTCAGTTCAAGGCAGGATAAGG (−203 to −179) | |
| LAC1 | CAGCAGGATATCCTGCACCATCGTCTGCTC (complementary to +1175 to +1146) | |
| LAC2 | ctgcagAATTGTGAGCGGATAACAAT (+1 to +20) | |
| LAC3 | ctgcagGCTTCCGGCTCGTATGTTGTGTGGA (−24 to +1) | |
| LAC4 | aaGCTTGCCATTTACACTTTATGCTTCCGG (−40 to −17) | |
| CAT1 | GTCGcgaTATTCACTCCAGAGCGATG (complementary to bp 576 to 601 of pKK-232) | |
| CAT2 | ctgCagTAGTGTTCACCCTTGTTACACC (bp 526 to 550 of pKK-232) |
The restriction enzyme sites present in the primes are underlined, and sequences different from the template are given in lowercase letters. GLT primers correspond to gltBDF operon sequences, LAC primers correspond to the lac operon sequences, and CAT primers correspond to the cat gene sequences. The locations of the GLT and LAC primers are given with respect to the transcription start sites of the glt (26) and lac (23) operons, respectively.
Replacement of the gltBDF upstream region with the lac operon sequences and construction of λ lysogens.
Fusions of the glt and lac operon regions (Fig. 1) were constructed from fragments of glt and lac operons obtained by PCR amplification using the primers listed in Table 1. The glt regions were amplified from plasmid pBE10 (13), and the lac regions were amplified from E. coli strain W3110 genomic DNA purchased from Sigma Aldrich (St. Louis, Mo.).
FIG. 1.
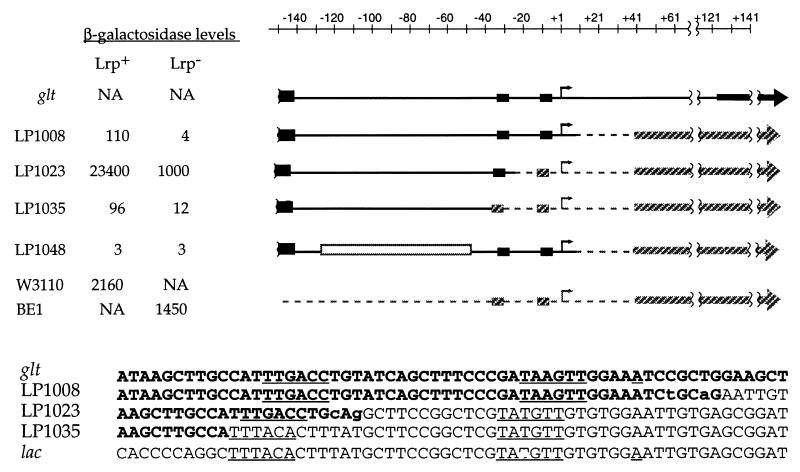
Maps and β-galactosidase activities of glt-lac chimeric operon fusions. The chimeric promoters were introduced as a single copy into the attB site of E. coli strain PS2209 (W3110 Δlac). The gltBDF sequences are shown with a black line, and the lac sequences are shown by a gray hatched line. The base pairs that were changed to introduce restriction sites in the chimeras are indicated in lowercase letters in the sequences shown at the bottom. The region from −48 to −128 in the gltBDF operon that was replaced with a fragment from the cat gene in strain LP1048 is shown as an open rectangular box. The box on the far left of each construct indicates the 3′ portion of the proximal Lrp binding site that extends from −142 to −161. The −35 and −10 hexamers are also represented by boxes and are shown underlined in the chimeric sequences at the bottom; arrows and the underlined A indicate the start sites of transcription determined for the native gltBDF (26) or lacZYA (23) promoters. The β-galactosidase activity from each chimeric strain in the presence or absence of Lrp is presented on the left side of the construct diagrams. The β-galactosidase activities from the chromosomal lacZ gene in E. coli strain W3110 (lrp+) and isogenic strain BE1 (lrp::Tn10) are also shown for comparison. NA, not applicable.
The PCR mixtures contained 2 ng of template plasmid DNA or 200 ng of E. coli genomic DNA, 1 U of Vent polymerase (New England Biolabs, Beverly, Mass.), 200 mM concentrations of each primer, and 200 μM concentrations of each deoxynucleoside triphosphate (Life Technologies, Rockville, Md.) in thermopol reaction buffer [10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris-HCl (pH 8.8), 2 mM MgSO4, and 0.1% Triton X-100] in a total volume of 100 μl. The reaction was carried out for 20 cycles of 96°C for 30 s, 60°C minus 0.5°C/cycle for 1 min, and 72°C for 1.5 min, followed by 10 cycles of 96°C for 30 s, 50°C for 1 min, and 72°C for 1.5 min. The PCR products were purified using a Qiaquick PCR purification kit (Qiagen Inc., Valencia, Calif.). A single deoxyadenosine was added to the 3′ end of the products by incubating them at 72°C for 20 min with 5 U of Taq DNA polymerase (Life Technologies) and 200 μM dATP in 20 mM Tris-HCl (pH 8.4) containing 50 mM KCl and 2 mM MgCl2. The products were resolved on a 1.0% agarose gel, purified using a Qiaquick gel extraction kit (Qiagen), and cloned into E. coli strains XL-1 Blue or DH5α using the pGEM-T vector (Promega, Madison, Wis.). The sequences of the cloned fragments were verified using the ABI PRIZM dye terminator cycle sequencing method (Applied Biosystems, Foster City, Calif.) at the University of Michigan DNA Sequencing Core Facility.
Restriction enzyme sites were incorporated into the primers used for PCR (Table 1), allowing ligation of the glt and lac operon sequences (Fig. 1). All the glt fragments had an EcoRI site at the 5′ end introduced from the primer glt1, and all the lac fragments had an EcoRV site at the 3′ end introduced from the primer lac1. The ligated glt-lac fragments were introduced into plasmid pRS528 (31) cut with EcoRI and EcoRV.
The glt-lac chimera in strain LP1008 was generated using fragments derived by amplification of glt DNA using primers glt1 and glt2 (−406 to +8) and amplification of lac DNA using primers lac2 and lac1 (+1 to +1166); primers glt2 and lac2 each contain a PstI site, allowing ligation of the fragments together. The glt and lac fragments are numbered with respect to their transcription start sites (23, 26).
The glt-lac chimera in strain LP1023 was obtained using the primer pairs glt1 and glt3 (−406 to −23) and lac3 and lac1 (−24 to +1166). Primers glt3 and lac3 each contain a PstI site, which was used to ligate the two fragments together.
The chimera in strain LP1035 was generated using primers glt1 and glt4 (−406 to −35) and lac4 and lac1 (−36 to +1166). Primer glt4 has a HindIII site that is present in the glt sequence, and primer lac4 has a HindIII site upstream of the lac sequence.
The chimera in strain LP1048 was constructed by replacing the glt sequence between −48 and −128 in strain LP1008 with a fragment of the chloramphenicol acetyltransferase gene (cat). The cat fragment was amplified from the plasmid vector pKK232-8 (Pharmacia) using the primers cat1 and cat2. The flanking regions were amplified from pLP1008, which contains the glt-lac chimera introduced into strain LP1008, with primers glt1 and glt6 for the upstream flanking region and glt7 and lac1 for the downstream flanking region. Primer glt6 contains an NruI site, and primer cat1 contains complementary sequence, allowing ligation of the upstream fragment with the cat insert. Primer glt7 contains a PstI site, as does primer cat2, allowing ligation of the downstream fragment to the cat insert.
The glt region of strain LP1000 (Table 1) was generated using primers glt1 and glt5 (−406 to +246), resulting in a fragment with an EcoRI site at the 5′ end and a BamHI site at the 3′ end which was ligated into plasmid pRS415 (31) cut with EcoRI and BamHI.
The glt-lac fusions from the pRS vectors were transferred into the chromosome of E. coli strain PS2209 (W3110 Δlac) at the attB site using λRZ5 (2) according to the procedure described in the work of Weise et al. (35). Single-copy lysogens were identified using the PCR method described by Powell et al. (27).
Transferring lrp and himD mutations into λ lysogens.
An lrp-201::Tn10 allele from the E. coli strain BE1 (13) and a himD::cat allele from E. coli strain RJ1413 (from Robert Osuna, University at Albany, Albany, N.Y.) were transferred into the λ lysogens carrying glt-lacZ fusions by P1 vir transduction (24). The transductants were selected on LB medium-ampicillin plates containing tetracycline or chloramphenicol.
β-Galactosidase assays.
The λ lysogens were grown in glucose minimal MOPS medium supplemented with 0.4 mM isoleucine and 0.6 mM valine (13). IPTG (isopropyl-β-d-thiogalactopyranoside; 0.5 mM) was added to the cultures to avoid any effect of the lac repressor on the expression of glt-lac fusion constructs. Samples for β-galactosidase assays were taken throughout the growth period, and the assays were carried out as described by Ernsting et al. (13). The absorbances of the cultures at 420 nm were plotted versus the β-galactosidase levels. The points were fitted by linear regression, and the slope of the line indicates the β-galactosidase activity of the culture (35). Only samples taken before the cultures reached an A420 of 1.0 were used for the slope determinations. The β-galactosidase levels from the wild-type chromosomal lacZ in Lrp+ (W3110) and Lrp− (BE1) backgrounds were used as controls to account for the effect of Lrp on lacZ transcription.
Gel mobility shift assays.
The DNA fragments used for gel mobility shift assays were amplified by PCR from pBE10 (13). The DNA was labeled at the 5′ end using T4 polynucleotide kinase (Life Technologies) and [γ-32P]ATP (7,000 Ci/mmol; ICN Biomedical Research Products, Costa Mesa, Calif.) at 37°C for 45 min. The unincorporated nulceotides were removed by passing the labeling reaction mixtures through a Biospin 6 gel filtration column (Bio-Rad Laboratories, Hercules, Calif.). Lrp was purified from an overexpressing E. coli strain (JWD3) using the procedure described earlier (13). Purified IHF was a gift from Steven D. Goodman (University of Southern California). The labeled DNA fragment (1.15 nM) was incubated with various concentrations of Lrp and/or IHF in a buffer containing 20 mM Tris-acetate (pH 8.0), 0.1 mM EDTA, 0.1 mM dithiothreitol, 50 mM NaCl, 4 mM Mg acetate, 12.5% glycerol (vol/vol), and 200 ng of poly(dI-dC) · poly(dI-dC) (Amersham Pharmacia Biotech, Pistcataway, N.J.) in a total volume of 20 μl. The mixtures were left at room temperature for 5 min before being incubated at 28°C for 15 min. The samples were loaded directly onto a 6% polyacrylamide gel (8.3 cm wide by 7.3 cm long by 1.0 mm) in 0.5× TBE (45 mM Tris-borate [pH 8.3], 0.1 mM EDTA) and electrophoresed at 12 mA and 4°C. The electrophoresis buffer contained 0.5× TBE and 5 mM MgCl2. The gel was fixed in a solution of 10% acetic acid and 10% methanol for 15 min and dried at 80°C. Biomax MS film (Kodak, Rochester, N.Y.) was used for autoradiography. The gels were scanned using a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.), and the intensities of the bands were determined using ImageQuant version 1.2 software. The data were fitted to the Michaelis-Menten equation using Kaleidagraph (Synergy Software, Reading, Pa.).
DNase I footprinting.
Footprinting assays were carried out as described earlier (5). The gltBDF DNA fragment used for footprinting of the template strand was amplified by PCR from plasmid pBE10 (13) using primers glt1 and glt2 and digested with the restriction enzyme HincII, resulting in a fragment from −176 to +8. For footprinting of the nontemplate strand, DNA was amplified with primers gltFP and glt5 and digested with Bsp1286I, resulting in a fragment from −203 to +161. The DNA fragments were labeled at the glt2 and gltFP ends for the template and nontemplate strands, respectively, using T4 polynucleotide kinase and [γ-32P]ATP as described above for gel mobility shift assays. Various concentrations of IHF were incubated with the labeled DNA in a buffer containing 10 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 1 mM CaCl2, 2 mM dithiothreitol, 50 μg of bovine serum albumin per ml, and 2 μg of poly(dI-dC) · poly(dI-dC) per ml in a total volume of 20 μl. The mixtures were incubated at 28°C for 30 min, after which 0.05 U of RNase-free DNase I (amplification grade from Life Technologies) was added to each tube. DNase I digestion was stopped after 2 min by adding 700 μl of a stop solution (645 μl of 100% ethanol, 5 μl of saturated ammonium acetate, and 5 μg of yeast tRNA) (5). The precipitated DNA fragments were resolved on a 6% acrylamide gel containing 8 M urea (30). A dideoxy sequencing reaction of the gltBDF upstream region with the primer corresponding to the labeled end, carried out using a T7 Sequenase version 2.07-deaza-dGTP kit (Amersham Pharmacia Biotech), was used to generate the ladder.
RESULTS
Effect of replacing the gltBDF promoter regions with those of the lac promoter on Lrp-dependent activation.
To study the role of the DNA sequences downstream of the proximal Lrp binding site centered at −152 in the regulation of the glutamate synthase (gltBDF) operon of E. coli, these downstream sequences were systematically replaced with the corresponding regions of the lacZYA operon. Single copies of the chimeric constructs were introduced into E. coli strain PS2209 (W3110 Δlac) at the attλ site by lambda integration. Transcription from these hybrid constructs was monitored using lacZ as the reporter gene. Figure 1 illustrates the sequence replacements and the β-galactosidase activities of each construct. As described below, replacing the gltBDF region downstream of and including the −35 hexamer with the corresponding regions of the lac operon did not abolish Lrp-dependent regulation. Thus, Lrp is clearly able to regulate the well-characterized ς70-dependent promoter lacZYAp from its binding sites more than 110 bp upstream of the −35 hexamer.
In strain LP1008, the region downstream of +8 of the gltBDF operon was replaced by the lac operon sequence. Bases at +4 and +7 were changed (C to T and T to A, respectively) to introduce a PstI site, which was used for ligating the glt and lac fragments. In strain LP1008, lacZ was still under the control of Lrp and showed a nearly 30-fold reduction in β-galactosidase levels in an Lrp− background relative to the wild-type strain.
Strain LP1023 has a chimeric promoter, with the −35 hexamer being from gltBDF and the −10 hexamer being from the lac operon. The sequence in the spacer region derived from glt was changed in two positions (T to G at −23 and T to C at −25) to generate a PstI site for preparing the construct, and the spacer sequence downstream of −23 was from the lac operon. The length of the spacer sequence is 17 bp as in glt; the wild-type lac spacer region is 18 bp. This construct showed an extremely high level of β-galactosidase activity: 200-fold higher at both the basal and activated levels compared to levels in strain LP1008. Nonetheless transcription from this chimeric promoter was activated over 20-fold by Lrp.
In strain LP1035 the −35 hexamer and the entire region downstream of it have been replaced with the corresponding sequences from the lac operon. The distance between the proximal Lrp binding site and the −35 hexamer was 1 bp shorter than in the other constructs (111 versus 110 bp). The β-galactosidase level from this construct was reduced sevenfold in an Lrp− background, but the basal level of transcription was higher than in strain LP1008.
In the gltBDF operon, the proximal Lrp binding site centered at −152 and the RNA polymerase recognition sequence at −34 are farther apart (more than 110 bp) than is usually seen in other ς70-dependent promoters. To see if the intervening region is involved in transcriptional regulation of the glt operon, the region from −48 to −128 was replaced with a portion of the chloramphenicol acetyltransferase (cat) gene of the same length (LP1048). This replacement had no effect on basal transcription but completely abolished Lrp-mediated activation.
IHF is required for positive regulation of transcription from the gltBDF promoter.
In preliminary experiments we used surface-enhanced laser desorption and ionization (SELDI) ProteinChip technology (Ciphergen Biosystems, Fremont, Calif.) to identify proteins that might bind to the upstream region of the gltBDF operon. In this approach, biotin-labeled DNA fragments of the glt region (from −406 to +132) were attached to streptavidin-coated chips and incubated with cell extracts. The masses of proteins that bound to the DNA fragments were determined using mass spectroscopy. This binding experiment was carried out under nonstringent conditions, and there were multiple peaks corresponding to various proteins that bound the DNA nonspecifically. Even though the results of these experiments were not conclusive, they suggested that two polypeptides with molecular masses of 10.6 and 11.2 kDa bound to the gltBDF upstream region. The molecular masses of these polypeptides matched those of the two subunits of IHF. These preliminary results led us to focus our study on the possible role of IHF in gltBDF transcription.
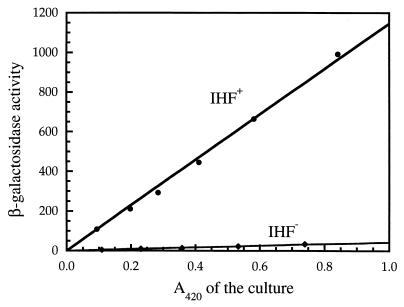
Upstream of the gltBDF promoter, within the region replaced by the cat fragment in strain LP10048, there are two regions resembling the consensus IHF binding sequence WATCAANNNNTTR, where W is A or T and R is A or G (10, 17). The region from −95 to −83 (TTTCAGTCATTTA) has two mismatches and the overlapping region from −91 to −79 (AGTCATTTAATAA) has three mismatches to the consensus sequence. An insertion in the himD gene encoding the β subunit of IHF (himD::cat) was transferred by P1 vir transduction into strain LP1000, which contains a gltB::lacZ transcriptional fusion. The IHF− strain had a β-galactosidase level over 30-fold lower than that of the IHF+ strain (Fig. 2).
FIG. 2.
Effect of IHF on gltB-lacZ expression. The β-galactosidase activity of the gltB-lacZ transcriptional fusion, integrated as a single copy into the attB site of isogenic IHF+ and IHF− (himD::cat) strains (LP1000 and LP2000, respectively) constructed from E. coli strain PS2209 (W3110 Δlac), is shown. The cultures were grown in glucose minimal MOPS medium, and samples were taken at intervals during the growth. The optical density of the cultures was measured at 420 nm. The β-galactosidase activities for the cultures were calculated from the slopes of the lines and were 1,180 for strain LP1000 and 39 for strain LP2000.
Purified IHF binds to the upstream region of the gltBDF promoter.
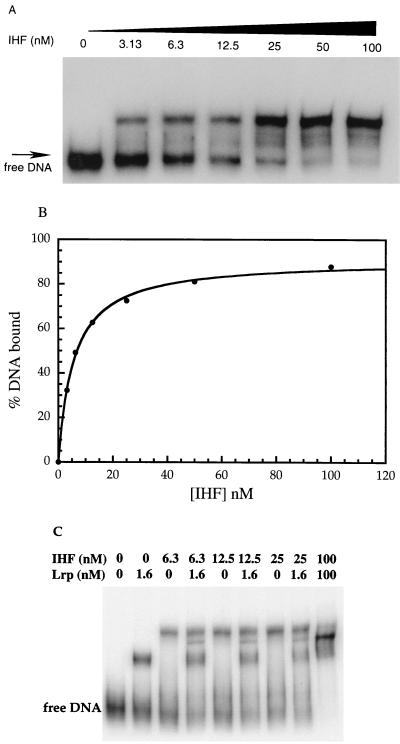
To see if IHF affects gltBDF transcription directly, gel mobility shift assays were carried out using purified IHF and the region upstream of the gltB translational initiation codon. Assays with both the region from −406 to +246 that included the first 30 bp of the coding region of gltB and a shorter fragment (−406 to +8) showed similar band shift patterns. A gel mobility shift assay with the shorter fragment is shown in Fig. 3A. Migration of the DNA fragment was retarded in the presence of IHF. IHF binds to the gltBDF upstream region with an apparent Kd of 5.7 ± 0.3 nM (Fig. 3B). A DNA fragment from −406 to +246 but with the region from −48 to −128 replaced with an equivalent length of the cat gene sequence did not bind IHF under identical binding conditions (not shown).
FIG. 3.
Gel mobility shift analysis of IHF binding to glt DNA. (A) The assays used a DNA fragment containing the region from −406 to +8 of the gltBDF promoter region and various amounts of purified IHF. The reaction mixtures contained 1.15 nM DNA and 0 to 100 nM IHF. (B) The free DNA in the above gel mobility shift assay was quantitated using a phosphorImager and used to calculate the bound DNA. The data were fitted using the Michaelis-Menten equation. The Kd of IHF for the gltBDF upstream region was 5.7 ± 0.3 nM. (C) Lrp and IHF binding to gltBDF DNA (−324 to +246). The PCR fragment from −406 to +246 was digested with the restriction enzyme NsiI to obtain the above-described fragment. The reaction mixtures contained 1.6 to 100 nM Lrp and 6.3 to 100 nM IHF as noted above the lanes. In the presence of both Lrp and IHF in the reaction mixture, we observed an additional band corresponding to the ternary complex, which was not present when either Lrp or IHF alone was used in the reactions; at saturating concentrations of Lrp and IHF, most of the DNA migrated in this band.
When both Lrp and IHF were present in the gel mobility shift assays, we observed an additional band that was not present in assays with either IHF or Lrp alone. The position of the shifted DNA fragment corresponding to the ternary complex depended on the distance of the IHF and Lrp binding sites from the ends of the DNA fragment used for the assay (37). When the glt DNA fragment from −324 to +246 was used, the band corresponding to the ternary complex was seen between those corresponding to IHF or Lrp alone (Fig. 3C). In the presence of saturating amounts of Lrp and IHF, most of the DNA was bound to both IHF and Lrp.
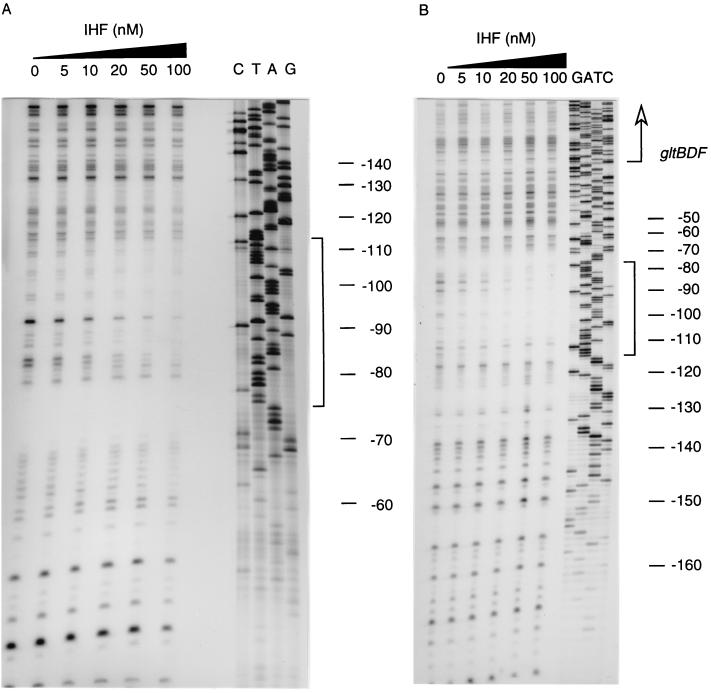
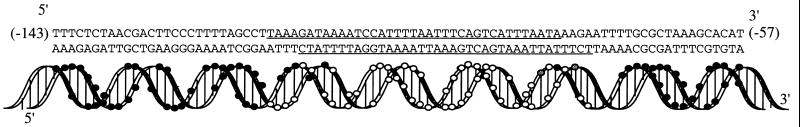
The binding site of IHF on the gltBDF upstream DNA was determined using DNase I footprinting assays of both strands of the DNA (Fig. 4). IHF protected a 41-bp region from −115 to −75 on the nontemplate strand and a 38-bp region from −113 to −76 on the template strand (Fig. 5). The bases −78 on the nontemplate strand and −82 on the template strand were made hypersensitive to DNase I digestion by IHF binding. Upstream of the IHF binding consensus site there are two short tracts of adenine (−106 to −109 and −113 to −115), both of which were protected in the DNase I footprinting assays. High-affinity IHF binding sites are often preceded by a poly (A) tract (21), whose narrow minor groove helps optimize IHF-DNA contacts (29). In Fig. 6, the sequences upstream of the gltBDF operon of E. coli are compared with those in other genera of Enterobacteriaceae: Salmonella enterica serovar Typhimurium LT2, Klebsiella pneumoniae, and Yersinia pestis. The approximate distance between the proximal Lrp binding site and the IHF site is conserved among the different genera, but the sequence of the intervening region is not highly conserved except for the poly (A) region. Though the sequence of the IHF binding site is not conserved among the four genera, the cytosine that is present in all known natural IHF binding sites (16) is conserved. An adenine following the cytosine is also generally conserved (17), but this residue is a T rather than an A in K. pneumoniae.
FIG. 4.
DNase I footprinting analysis of IHF binding to the glt DNA. DNA fragments extending from −176 to +8 of the template strand (the strand complementary to the mRNA sequence) (A) or from −203 to +161 of the nontemplate strand (B) of the gltBDF operon were footprinted in the presence of 0 to 100 nM IHF. Dideoxy sequencing reactions of the gltBDF promoter region with the primer glt2 (for the template strand) and gltFP (for the nontemplate strand) were used to generate the ladders. IHF protected the region (shown in brackets) from −113 to −76 on the template strand and the region from −115 to −75 on the nontemplate strand against DNase I digestion. The positions of the sequences are given with respect to the transcription start site.
FIG. 5.
Diagramatic representation of the region upstream of the gltBDF promoter that was protected against DNase I digestion by IHF binding. Open circles indicate protection against cleavage in the presence of IHF, and the filled circles indicate unchanged cleavage. Where no circles are shown, the DNA was not cleaved by DNase I in the absence of IHF. The two gray circles indicate hypersensitivity to DNase I digestion due to IHF binding. The underlined sequences indicate the region protected from DNase I digestion.
FIG. 6.
Comparison of sequences upstream of the gltBDF operon from E. coli (4) with corresponding sequences from related genera: S. enterica serovar Typhimurium LT2 (WUGSC 99287/stmlt2-contig1496), K. pneumoniae (WUGSC 573/kpneumo B KPN.contig272), and Y. pestis (Sanger 632/Y pestis Contig850). The sequences of the related genera were obtained from unfinished genomes using a BLAST search (1), and the alignment was generated using CLUSTAL W (33). Bases conserved in all four genera are shown in bold, and the shaded regions indicate the proximal Lrp binding site, the poly(A) region upstream of the IHF binding site, the consensus IHF binding site (−95 to −83), and the −35 and −10 hexamers. The non-E. coli sequences are prepublication communications from the Genome Sequencing Center at Washington University and The Sanger Centre.
DISCUSSION
glt::lac hybrid promoters are activated by Lrp.
To study the minimum DNA sequence required for Lrp-dependent regulation, the gltBDF operon sequence downstream of and including the −35 hexamer was sequentially replaced with the corresponding regions of the lacZYA operon. The effects of these substitutions on transcription were studied by monitoring the activity of the reporter gene lacZ (Fig. 1).
The gltBDF sequences downstream of −23, including the −10 hexamer, are not required for Lrp-dependent activation of transcription. Strain LP1023 has the −35 hexamer of gltBDF, the −10 hexamer of the lac operon, and a hybrid spacer region, yet Lrp-dependent activation (23.4-fold) was similar to that of strain LP1008 (27.5-fold), in which the gltBDF core promoter regions were intact. β-Galactosidase expression in strain LP1023 was >200-fold higher than in strain LP1008 and over 10-fold higher than the IPTG-induced level from the native chromosomal lacZ gene in W3110. Apparently, the combination of the −35 and the −10 hexamers or the hybrid spacer region affected the strength of the promoter. A study using random sequences in the spacer region of Lactococcus lactis promoters in L. lactis and E. coli backgrounds has shown that the sequence of the spacer region can affect promoter strength up to 400-fold (22).
Lrp was able to activate transcription even when the gltBDF core promoter sequence (the −35 hexamer and downstream sequence) was completely replaced with that of the ς70-dependent lac promoter in strain LP1035. The lac promoter normally requires catabolite activator protein for activation. Dove et al. (11) have shown that any DNA binding protein that contacts RNA polymerase can activate transcription by strengthening the interaction of RNA polymerase with the promoter region. The extent of Lrp-dependent activation was lower in LP1035 than in strain LP1023 with the chimeric promoter or in strain LP1008 with the gltBDF core promoter. It is possible that the longer spacer region between the −35 and −10 hexamers or the shorter region between the proximal Lrp binding site and the −35 hexamer in strain LP1035 affected transcriptional activation. Alternatively, there may be something about the gltBDF −35 hexamer that makes it more amenable to Lrp-dependent activation. In the E. coli fis promoter, changes in the −35 hexamer, but not in the −10 hexamer, reduce the response to stringent control (34).
The native gltBDF promoter is presumed to be recognized by ς70, but evidence for this is not definitive. The promoter is recognized in vitro by purified ς70 and core polymerase (B. R. Ernsting, unpublished data), and the sequence and spacing of the −35 and −10 hexamers relative to the transcription start site defined by primer extension are consistent with those of moderately expressed ς70-dependent promoters. The experiments reported here provide definitive evidence that Lrp dimers, bound more than 110 bp upstream of the −35 hexamer, can activate transcription of a ς70-dependent promoter, since the lac operon is controlled by what is probably the best-categorized such promoter.
IHF binding is required for Lrp-dependent activation of the gltBDF operon.
The mechanism by which the leucine-responsive regulatory protein (Lrp) regulates the genes under its control is not completely understood. In some operons, e.g., gcv, Lrp exerts its effect in tandem with other regulatory proteins (32); in others, such as ilvIH, Lrp is sufficient for regulation (36). Lrp has been reported to play an architectural role in the transcription of the gcv operon (32). Although the gltBDF operon of E. coli is positively regulated by Lrp (13), in vitro transcription with purified Lrp produced minimal activation of transcription from the gltBDF promoter (B. R. Ernsting, unpublished results). This observation suggested that additional factors might be involved in the transcriptional activation of the gltBDF operon of E. coli.
The 110-bp distance between the proximal Lrp binding site (centered at −152) and the −35 hexamer of the gltBDF operon is unusual for a ς70-dependent promoter. When a 91-bp sequence in this region (−128 to −48) was replaced with a fragment of the cat gene (strain LP1048), transcription was reduced almost 40-fold. Results of SELDI ProteinChip experiments suggested that IHF might bind to the gltBDF upstream sequence. The presence of a consensus IHF binding sequence in this region further supported this idea. In vivo assays using a himD mutant (Fig. 2) confirmed that IHF is required for activation of transcription from the gltBDF promoter.
While this work was in progress, a global transcript analysis of a himA mutant was published (3). Mutation in himA led to a 7.1-fold reduction in gltD transcript levels in microarrays. This effect was not as drastic as the approximately 30-fold effect seen in our assays with the himD mutant or with a strain where the IHF binding region was replaced by heterologous DNA (strain LP1048). It is possible that functional himD homodimers were formed in the himA mutant used for the microarray assay, resulting in activation of the gltBDF operon transcription to a certain extent (3, 38).
We confirmed that IHF binds to the gltBDF upstream region using gel mobility shift and footprinting assays (Fig. 3 to 5). The two overlapping matches to the consensus IHF binding sequence in the glt upstream region (−95 to −83 and −91 to −79) are protected in footprinting assays. Site-specific mutational studies have to be conducted to identify the site actually recognized by IHF. The activation of the gltBDF operon by Lrp is totally dependent on the presence of IHF, since introduction of an Lrp mutation in strain LP1048 did not lead to a further decrease in β-galactosidase level.
IHF has been shown to be involved in transcriptional regulation of many operons on its own or in concert with additional activator proteins (14, 18). The crystal structure of IHF bound to DNA shows that this protein bends DNA at an ∼160° angle (29). This facilitates looping of DNA, allowing an upstream element or upstream-bound activator protein to come in close contact with RNA polymerase. Transcriptional activation of the gltBDF operon depends completely on the presence of both proteins, suggesting that a DNA looping mechanism may be involved in gltBDF regulation.
The proposed IHF-induced bend in the gltBDF upstream region would bring the proximal Lrp binding site close to the region just upstream of the −35 hexamer, a position where activators of ς70-dependent promoters usually bind (19). So far, activation of transcription by direct contacts between Lrp and RNA polymerase has not been documented for any promoter. The effect of IHF on transcription from the gltBDF promoter is phase dependent: transcription from the gltBDF promoter was reduced fivefold when the IHF binding site was taken out of phase with respect to the −35 hexamer and the Lrp binding sites by 5-bp insertions at −40 and −120 (D. E. Weise II, unpublished results).
It is not known at present whether IHF acts as a direct regulator of gltBDF transcription or whether it plays a structural role in bringing the transcriptional machinery together. When IHF plays an architectural role, it can be replaced by an intrinsically bent region of DNA (15). Such an experiment remains to be done for the gltBDF operon. Since the IHF binding site and the Lrp binding sites upstream of the glt promoter are conserved in S. enterica, K. pneumoniae and Y. pestis, the glutamate synthase operons of enteric bacteria appear to be regulated by a common mechanism.
Our results suggest an exception to the general rule that transcriptional activators of ς70-dependent promoters bind close to the −35 hexamer (19). When a confirmed binding site for IHF is present, activators may bind farther upstream and retain function. Since the proximal Lrp binding site in gltBDF operon is positioned 110 bp upstream of the −35 hexamer, an intermediate IHF binding site appears to be essential to Lrp-mediated activation. Future experiments will probe for evidence of direct contact between Lrp and/or IHF and RNA polymerase.
ACKNOWLEDGMENTS
We thank Steven D. Goodman (University of Southern California) for providing us with purified IHF, William S. Brusilow (Wayne State University) for λRZ5, Robert Osuna (University at Albany) for strain RJ1413, and Ruth Van Bogelen (Pfizer, Ann Arbor, Mich.) and Lisa Bradbury (Ciphergen Biosystems) for their assistance with the SELDI ProteinChip experiment. We thank the Genome Sequencing Center, Washington University, St. Louis, Mo., and The Sanger Centre for communication of DNA sequence data prior to publication.
This work was supported by a grant (MCB 9807237) from the National Science Foundation to R.G.M. and R.M.B.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angov E, Brusilow W S A. Use of lac fusions to measure in vivo regulation of expression of Escherichia coli proton-translocating ATPase (unc) genes. J Bacteriol. 1988;170:459–462. doi: 10.1128/jb.170.1.459-462.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arfin S M, Long A D, Ito E T, Tolleri L, Riehle M M, Paegle E S, Hatfield G W. Global gene expression profiling in Escherichia coli K12. The effects of integration host factor. J Biol Chem. 2000;275:29672–29684. doi: 10.1074/jbc.M002247200. [DOI] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett III G I, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N, Kirkpatrick W H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Brenowitz M, Senear D F, Kingston R E. DNA-protein interactions. In: Chanda V B, editor. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1994. pp. 12.4.1–12.4.11. [Google Scholar]
- 6.Buck M, Miller S, Drummond M, Dixon R. Upstream activator sequences are present in the promoters of nitrogen fixation genes. Nature. 1986;320:374–378. [Google Scholar]
- 7.Castano I, Bastarrachea F, Covarrubias A A. gltBDF operon of Escherichia coli. J Bacteriol. 1988;170:821–827. doi: 10.1128/jb.170.2.821-827.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castano I, Flores N, Valle F, Covarrubias A A, Bolivar F. gltF, a member of the gltBDF operon of Escherichia coli, is involved in nitrogen-regulated gene expression. Mol Microbiol. 1992;6:2733–2741. doi: 10.1111/j.1365-2958.1992.tb01450.x. [DOI] [PubMed] [Google Scholar]
- 9.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craig N L, Nash H A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984;39:707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- 11.Dove S L, Joung J K, Hochschild A. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature. 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 12.Ernsting B R, Atkinson M R, Ninfa A J, Matthews R G. Characterization of the regulon controlled by the leucine-responsive regulatory protein in Escherichia coli. J Bacteriol. 1992;174:1109–1118. doi: 10.1128/jb.174.4.1109-1118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernsting B R, Denninger J W, Blumenthal R M, Matthews R G. Regulation of the gltBDF operon of Escherichia coli: how is a leucine-insensitive operon regulated by the leucine-responsive regulatory protein? J Bacteriol. 1993;175:7160–7169. doi: 10.1128/jb.175.22.7160-7169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman D I. Integration host factor: a protein for all reasons. Cell. 1988;55:545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- 15.Goodman S D, Nicholson S C, Nash H A. Deformation of DNA during site-specific recombination of bacteriophage λ: replacement of IHF protein by HU protein or sequence directed bends. Proc Natl Acad Sci USA. 1992;89:11910–11914. doi: 10.1073/pnas.89.24.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodman S D, Velten N J, Gao Q, Robinson S, Segall A M. In vitro selection of integration host factor binding sites. J Bacteriol. 1999;181:3246–3255. doi: 10.1128/jb.181.10.3246-3255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodrich J A, Schwartz M L, McClure W R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF) Nucleic Acids Res. 1990;18:4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goosen N, van de Putte P. The regulation of transcription initiation by integration host factor. Mol Microbiol. 1995;16:1–7. doi: 10.1111/j.1365-2958.1995.tb02386.x. [DOI] [PubMed] [Google Scholar]
- 19.Gralla J D, Collado-Vides J. Organization and function of transcription regulatory elements. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1232–1245. [Google Scholar]
- 20.Grassl G, Bufe B, Muller B, Rosel M, Kleiner D. Characterization of the gltF gene product of Escherichia coli. FEMS Microbiol Lett. 1999;179:79–84. doi: 10.1111/j.1574-6968.1999.tb08711.x. [DOI] [PubMed] [Google Scholar]
- 21.Hales L M, Gumport R I, Gardner J F. Examining the contribution of a dA+dT element to the conformation of Escherichia coli integration host factor-DNA complexes. Nucleic Acids Res. 1996;24:1780–1786. doi: 10.1093/nar/24.9.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen P R, Hammer K. The sequence of the spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl Environ Microbiol. 1998;64:82–87. doi: 10.1128/aem.64.1.82-87.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maizels N M. The nucleotide sequence of the lactose messenger ribonucleic acid transcribed from the UV5 promoter mutant of Escherichia coli. Proc Natl Acad Sci USA. 1973;70:3585–3589. doi: 10.1073/pnas.70.12.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 25.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver G, Gosset G, Sanchez-Pescador R, Lozoya E, Ku L M, Flores N, Becerril B, Valle F, Bolivar F. Determination of nucleotide sequence of glutamate synthase structural genes of Escherichia coli K-12. Gene. 1987;60:1–11. doi: 10.1016/0378-1119(87)90207-1. [DOI] [PubMed] [Google Scholar]
- 27.Powell B S, Court D L, Nakamura Y, Rivas M P, Turnbough C L., Jr Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reitzer L J, Magasanik B. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell. 1986;45:785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- 29.Rice P A, Yang S, Mizuuchi K, Nash H A. Crystal structure of an IHF-DNA complex: a protein induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T, editors. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 32.Stauffer L T, Stauffer G V. Role for the leucine-responsive regulatory protein (Lrp) as a structural protein in regulating the Escherichia coli gcvTHP operon. Microbiology. 1999;145:569–576. doi: 10.1099/13500872-145-3-569. [DOI] [PubMed] [Google Scholar]
- 33.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker K A, Atkins C L, Osuna R. Functional determinants of the Escherichia coli fis promoter: roles of −35, −10, and transcription initiation regions in the response to stringent control and growth phase-dependent regulation. J Bacteriol. 1999;181:1269–1280. doi: 10.1128/jb.181.4.1269-1280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weise D E, II, Ernsting B R, Blumenthal R M, Matthews R G. A nucleoprotein activation complex between the leucine-responsive regulatory protein and DNA upstream of the gltBDF operon in Escherichia coli. J Mol Biol. 1997;270:152–168. doi: 10.1006/jmbi.1997.1057. [DOI] [PubMed] [Google Scholar]
- 36.Willins D A, Calvo J M. In vitro transcription from the Escherichia coli ilvIH promoter. J Bacteriol. 1992;174:7648–7655. doi: 10.1128/jb.174.23.7648-7655.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu H-M, Crothers D M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984;308:509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- 38.Zulianello L, de la Gorgue de Rosny E, van Ulsen P, van de Putte P, Goosen N. The HimA and HimD subunits of integration host factor can specifically bind to DNA as homodimers. EMBO J. 1994;13:1534–1540. doi: 10.1002/j.1460-2075.1994.tb06415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]