Abstract
The CRISPR/Cas system is widely used for molecular diagnostics after the discovery of trans-cleavage activity, especially now with the COVID-19 outbreak. However, the majority of contemporary trans-cleavage activity-based CRISPR/Cas biosensors exploited standard single-strand DNA (ssDNA) reporters, which were based on the FRET principle from pioneering research. An in-depth comparison and understanding of various fluorescent readout types are essential to facilitate the outstanding analytical performance of CRISPR probes. We investigated various types of fluorescent reporters of Cas12a comprehensively. Results show that trans-cleavage of Cas12a is not limited to ssDNA and dsDNA reporters, but can be extended to molecular beacons (MB). And MB reporters can achieve superior analytical performance compared with ssDNA and ds DNA reporters at the same conditions. Accordingly, we developed a highly-sensitive SARS-CoV-2 detection with the sensitivity as low as 100 fM were successfully achieved without amplification strategy. The model target of ORF1a could robustly identify the current widespread emerging SARS-CoV-2 variants. A real coronavirus GX/P2V instead of SARS-CoV-2 were chosen for practical application validation. And a minimum of 27 copies/mL was achieved successfully. This inspiration can also be applied to other Cas proteins with trans-cleavage activity, which provides new perspectives for simple, highly-sensitive and universal molecular diagnosis in various applications.
Keywords: CRISPR/Cas, Fluorescent reporters, Molecular beacon, SARS-CoV-2, Sensitive detection
Graphical Abstract
1. Introduction
The ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has emerged as a major global healthcare and economic threat. On the time of writing this article, As of 18 July 2022, there have been 557,917,904 confirmed cases globally, reported to WHO.[1], [2] Timely, accurate surveillance and diagnostic tests are necessary to control the current outbreak and prevent transmission. Despite the urgency of the current pandemic, most available diagnostic methods use RT-PCR or sequencing-based technologies to detect nucleic acid-specific to SARS-CoV-2.[3], [4], [5], [6], [7] These methods are limited by the requirement of large laboratory space, long detection time, high reagent costs, and the potential for cross-contamination. Many efforts have been made to develop new technologies that will simplify and speed up the detection process, shorten detection time and improve detection accuracy.[8], [9], [10], [11], [12], [13], [14], [15], [16].
Clustered regularly interspaced short Palindromic Repeats (CRISPR) and CRISPR-associated (Cas) system is an acquired immune system found in most bacteria and archaea.[17], [18] Emerging CRISPR/Cas-assisted SARS-CoV-2 assays are viewed as augment gold-standard RT-PCR-based tests due to their advantages of simple synthesis, convenient use, low cost, and strong specificity.[19], [20] Taking advantages of indiscriminate single-stranded DNase activity of Cas12a[21], [22], extensive Cas12a-aid bioassays with unprecedented analytical performances have been achieved. [23], [24], [25], [26], [27] Amongst them, the fluorescence signal readout played a vital role benefit from its excellent characteristics of qualitative and quantitative easily in a very short time. [28] However, most current fluorescent reporters still used the typical single-stranded DNA (ssDNA) reporter based on the principle of fluorescence resonance energy transfer (FRET) from the pioneering works[22], [29], [30]. The ssDNA reporters have a 5-nt (5′-TTATT-3′) ssDNA or 12-nt (5′-NNNNNNNNNNNN-3′) random sequences labeled by fluorophore and quencher. Upon activation of the Cas12a/crRNA complex with the corresponding target, the ssDNA reporters would be cleaved, resulting in a recovery of fluorescence. The sensitivity is around 1 nM in the absence of amplification for CRISPR probes.[31] Wang et al. adjusted different parameters for experimental condition to find the optimal value for the current experimental condition and using this version as a basis for testing to the next experimental condition. The detection limit, detection speed, and signal background ratio of the unamplified targets were greatly improved.[32].
Molecular beacons (MB) are a class of small, single-stranded oligonucleotide a fluorophore and a quencher at the two termini [27], [33], [34], [35], which has been widely used as hybridization-activated FRET probes, but rarely applied in CRISPR-based assays. [27] FRET is a distance-dependent process of energy transfer between two fluorophores, with one serving as energy donor and the other as acceptor [28], [31], [36]. The length of the fluorophore/quencher labeled ssDNA reporter is an important factor affecting analytical performance. The longer the distance between the fluorophore and the quencher, the lower the fluorescence quenching efficiency, resulting in an increase in background noise. [37] Although the ssDNA reporters are common used for research and applications, limiting selections of signal readout from an application and probe diversification standpoint [13], [25], [38]. The expansion of the substrate reporter library for CRISPR/Cas could be significant for biosensing and biochemistry. On the basis of ssDNA reporter, we herein investigated and compared various fluorescent reporters of CRISPR/Cas systematically. Cas12a, one of Cas enzymes with trans cleavage activity, was selected as model, a diverse set of Cas12a substrates with alternating reporters were designed and studied, including ssDNA, dsDNA, and MB. Moreover, we developed a sensitive SARS-CoV-2 detection, and the analytical performance was enhanced by introducing dual crRNA. We further explored the potential practical application by an assay of real coronavirus GX/P2V instead of SARS-CoV-2, due to its a high degree of similarity in gene sequence with SARS-CoV-2, but no transmissibility to humans. These findings can also apply to other Cas proteins, such as Cas 12b, Cas13, Cas14, which provides perspectives for simple and universal CRISPR designs in various applications.
2. Material and methods
2.1. Materials and Reagents
All synthetic nucleic acid oligonucleotides of Tables S1–S4 diethylpyrocarbonate (DEPC)-treated water were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). Lba Cas12a and buffer 3.1 were purchased from New England Biolabs (Beijing, China). RNase inhibitors were purchased from Takara Biotechnology Co., Ltd. (Dalian, China). The RNase-free environment throughout the experiments using DEPC-treated water and RNase-free tips and tubes. GX/P2V beta coronavirus was isolated by using Vero E6 cells.
2.2. Signal Readout properties of different fluorescent reporters
To a series of Eppendorf tubes, 0.5 μL of 250 nM Cas12a enzyme, 0.5 μL of 500 nM, 1 μL of 500 nM reporters (ssDNA, dsDNA, MB reporters), 15 μL of Buffer 3.1 and 1 μL of RNase inhibitor were transferred into the tubes. And 2 μL of different concentration targets were added into the a serious of tubes. Finally, the resulting mixture was put into the real-time fluorescent PCR instrument and kept at 37 ℃ for 1 h.
2.3. Quantitative Assay of with Single or dual crRNA
1 μL of 20 μM crRNA (1 μM), 4 μL of 10 μM Cas12a (2 μM), 1 μL of 10 μM MB reporter (500 nM), 2 μL of target at different concentrations (100 nM, 10 nM, 1 nM, 100 pM, 10 pM, 5 pM, 2 pM, 1 pM, 0), 11 μL of Buffer 3.1, and 1 μL of RNase inhibitor were added to a series of 1.5-mL tubes. Simultaneously, 1 μL 20 μM crRNA ORF1a (1 μM), 1 μL 20 μM crRNA-2 (1 μM), 4 μL 10 μM Cas12a (2 μM), 1 μL 10 μM MB reporter (500 nM), 2 μL target at different concentrations (1 nM, 100 pM, 50 pM, 20 pM, 10 pM, 1 pM, 100 fM, 0), 10 μL of buffer 3.1 and 1 μL of RNase inhibitor were added to a series of 1.5-mL tubes. Finally, the obtained mixture was put into a real-time fluorescence PCR instrument at 47 °C for 1 h to collect the fluorescence signal.
3. Results and discussion
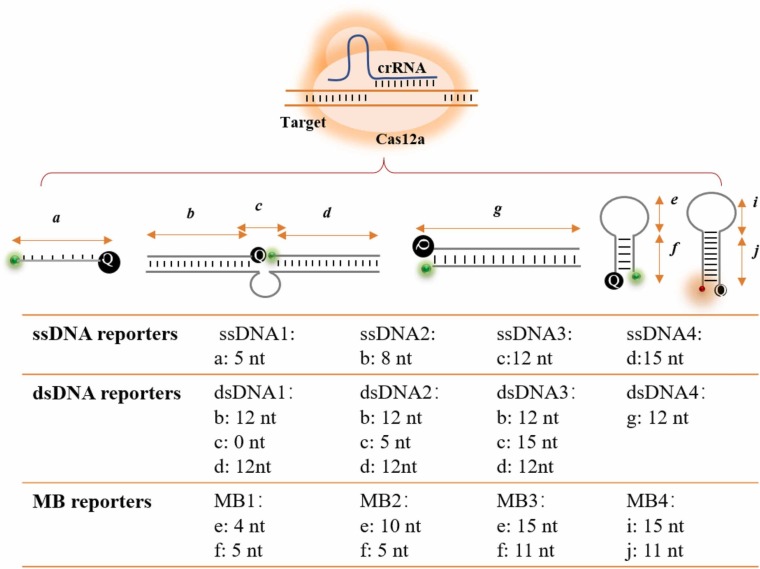
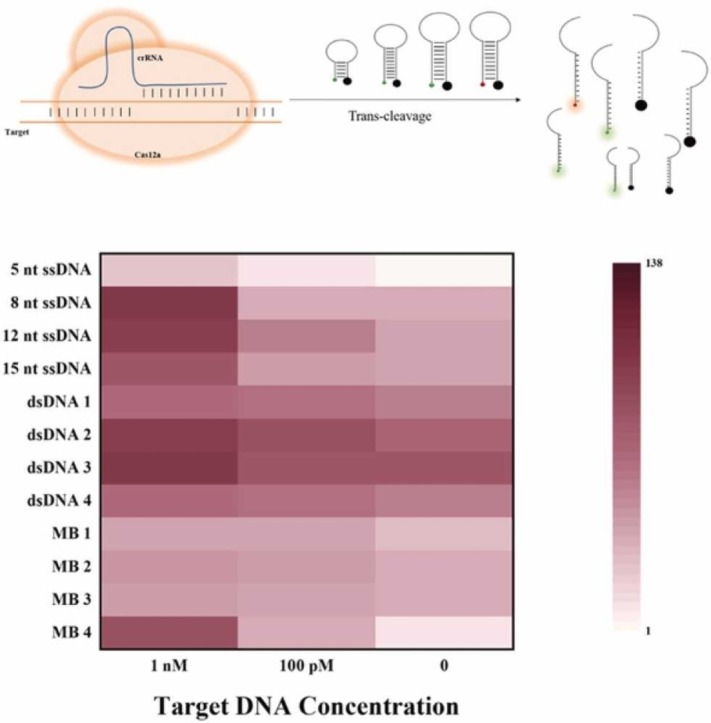
Extensive CRISPR/Cas12a-based bioassays with outstanding analytical performance have been developed. However, most fluorescent reporters are limited by ssDNA templated substrates from pioneering works [22], [29], [30]. Although ssDNA reporters have been shown to be effective for diagnostic applications, limiting selections to short ssDNA may restrict its application and probe diversification. The expansion of the DNA substrate library for probing Cas12a activity is essential in diagnostics. However, only a few reporters were concerned and thoroughly investigated the fluorescence signal read-out.[13] In this work, the cleavage properties of Cas12a with a variety of fluorescent substrates were studied and compared. Scheme 1 illustrates the different reporter formulations, upon activation of the Cas12a/crRNA complex with the target, the FRET reporters would be cleaved resulting in a recovery of fluorescence. FRET pairs labeled 5(6)-carboxyfluorescein (FAM) or Texas Red as donor and BHQ1 or BHQ2 as quenchers. The ssDNA reporters are designed based on the length between fluorophores and quencher with 5 nt, 8 nt,12 nt and 15 nt ssDNA. The dsDNA reporters are designed based on the spacers between fluorophores and quenchers at secondary structure after annealing. The MB reporters were designed based on the number of bases on the stem and loop. Signal readout properties of various fluorescent reporters including the real-time fluorescence curves, background noise, signal-to-background ratio, fluorescence recovery rate and cleavage rate of all fluorescent reporters were compared under the same conditions.
Scheme 1.
The various fluorescent reporter structures of the assay, upon activation of the Cas12a/crRNA complex with the target, the FRET reporters would be cleaved resulting in fluorescence recovery.
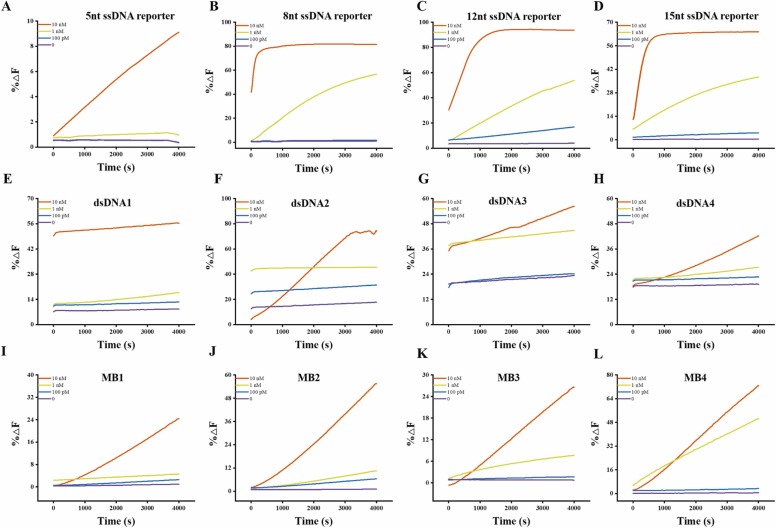
Take COVID-19 ORF 1a gene 13328–13437 sequence as input targets, we first evaluated the analytical performance of Cas12a on several lengths of ssDNA reporters. As shown in Figs. 1A and 2, the most commonly used 5 nt ssDNA reporter (5'-FAM-TTATT-BHQ1) with low background noise and high signal-to-background ratio. The sensitivity as low as 1 nM was achieved, the differences at concentrations of < 100 pM were found not to be statistically significant (Fig. S1), which was comparable with the available literature.[31] When the length of ssDNA increased from 5 nt to 8 nt, the lowest detectable concentration is reduced to 100 pM. When continuing to increase the base length until 15 nt, the lowest detectable concentration did not decrease with it. In comparison to the 5 nt ssDNA reporters at same conditions, the longer distance between the fluorophores and quenchers leads to higher background noise. The fluorescence recovery of 5 nt ssDNA reporters was about 37% at 10 nM, while it achieved about 82% and 94% for 8 nt ssDNA reporters and 12 nt reporters, respectively. The maximum shearing rates were calculated to be 0.01645, 0.275, 0.162 and 0.135 s−1 at 10 nM for 5 nt, 8 nt, 12 nt and 15 nt ssDNA reporters, which first increased and then decreased with the length raised. In contrast, the rate of fluorescence recovery and shearing rates of 5 nt ssDNA reporter is minimum, which might attribute the steric hindrance limited the shearing activity, but further structural research is needed.
Fig. 1.
The fluorescent curves of different types of reporters of Cas12a detection, the target at 10 nM, 1 nM, 100 pM and 0, respectively for all cases.
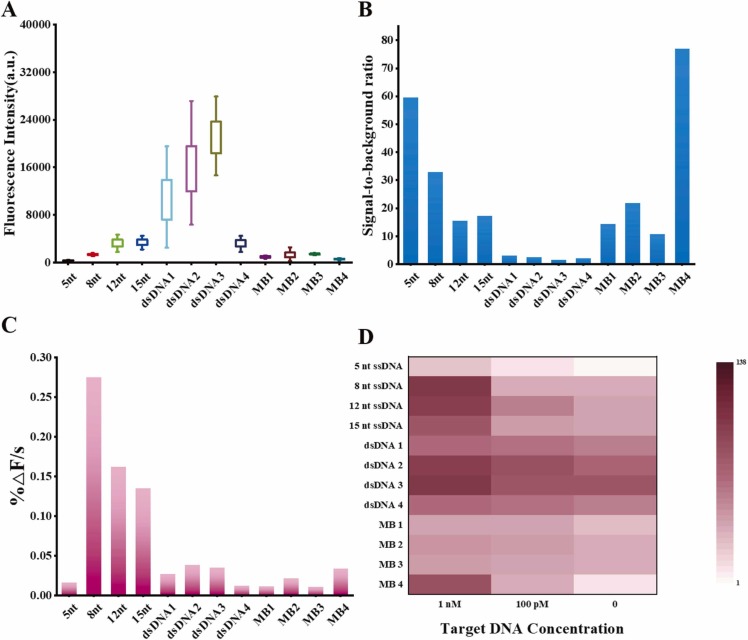
Fig. 2.
(A) The background fluorescence, (B) the signal-to-background ratio at 10 nM and (C) the fluorescence recovery rate at 10 nM of all fluorescent reporters. (D) Heat map comparison of endpoint fluorescence ratio with the different types of reporters of the Cas12a detection. Target DNA at 1 nM, 100 pM and 0, respectively for all cases (t = 4000 s).
Although dsDNA reporters are widely used in biosensors, few were found in CRISPR probes. Unlike short ssDNA probes applying the FRET principle, dsDNA can provide more DNA sequence programmability as well as the combination and arrangement of fluorophore and quencher. Christopher et al. developed a variety of dsDNA fluorescent probes to detect the cleavage activity of Cas12a[13]. When the target triggers the cleavage activity of the Cas12a/crRNA complex, the dsDNA probe can be cleaved like the ssDNA reporters, and the FRET groups are separated and released fluorescent signals for detection, which giving a new direction to expand the DNA probe library. We observed that steric hindrance effect would increase reaction rate and signal in the ssDNA reporters. To test whether such a phenomenon exists in dsDNA reporters, we designed 0, 5, and 15-nucleotide enzymatic cleavage sites in dsDNA reporters, respectively. We labeled the fluorophores and quenchers at two ends of the ssDNA and formed a dsDNA reporter through complementary base pairing. As shown in Figs. 1E-1 G and 2, the fluorescence recovery values reached equilibrium in a very short time. The sensitivity of dsDNA probes is not significantly improved with about 100 pM, but the background noise increased dramatically in contrast to ssDNA reporters, leading to low signal-to-background ratio. The fluorescence background noise increased and the signal-to-background ratio reduced with the spacers enlarged between the fluorophore and quencher, while the cleavage rate increased and then decreased, indicating that the steric hindrance might works, which is consistent with the phenomenon of ssDNA reporters. Moreover, we added a dsDNA5 group for verification with equal distance between fluorophores and quencher as dsDNA2 reporters, but increasing the number of base pairs in the double strand makes the DNA structure more stable (Fig. S2), the background signals and fluctuation of dsDNA5 is smaller and more stable, which is consistent with the FRET principle. On the other hand, the FRET pairs in the middle or end of the dsDNA will affect the quenching effect of dsDNA reporters. We explored whether fluorophores and quenchers labeled in the middle or end of the dsDNA could be used for Cas12a detection. Results showed that fluorophore labeled at the ends of dsDNA producing lower fluorescence background noise and shearing rate. These findings were beneficial for Cas12a biosensor applications.
Molecular beacons (MBs) are specifically designed DNA hairpin structures that are commonly used as FRET probes but are rarely used in CRISPR-based fluorescent assays. We first investigated the feasibility of Cas12a trans-cleave on MB reporters. Following the formation of the Cas12a/crRNA complex, the addition of the targets activated the cleavage activity, resulting in a dramatic recovery of fluorescence of the hairpin structure. Inspired by ss- and ds-DNA reporters, stem-loop structure of the MB inevitably influenced Cas12a nuclease activity (Figs. 1I-L and 2). Specifically, we designed a hairpin structure with a 4-nt loop, and 5-bp stem named MB1, the ΔΔG° calculated by NUPACK is − 4.43 kcal/mol.[39] The fluorescent signal can be significantly distinguished from the blank with sensitivity as low as 100 pM (Fig. 1I). We increased the number of bases in the loop region to verify the steric hindrance of the Cas12a cleavage named MB2 with a 10-nt loop, and 5-bp stem, as shown in Fig. 1J, the ΔΔG° calculated by NUPACK is − 3.16 kcal/mol. [39] The sensitivity is 100 pM, and the background noise and rate fluorescence recovery and cleavage rate rise obviously in contrast to MB1. We further designed a stable hairpin structure with a 15-nt loop and a 11-bp stem named MB3 to reduce background signal and steric hindrance of the enzyme cleavage reaction, ΔΔG° calculated by NUPACK is − 10.79 kcal/mol. The background signal and fluctuation were greatly reduced as expected, but the reaction rate decreased meanwhile. It is speculated that the balance between the increase of the stability of the stem region and the decrease of the steric resistance of the loop region in MB reporters (Fig. 1K). In addition, we investigated the impact of fluorophores on CRISPR probes (Fig. 1L). Results show that using Texas Red-labeled hairpin structures has higher sensitivity and lower background than FAM. These findings provide perspectives about choosing appropriate reporters for simple and universal probe designs in various applications.
Heatmaps are used to compare the overall analytical capabilities of all reporters (Fig. 2D). In order to compare under a uniform standard, all fluorescence values were divided by the background fluorescence value of the 5 nt ssDNA reporters to get the fluorescence ratio. The larger the fluorescence ratio, the darker the area where the reporters are located, and vice versa, the lighter the color. By comparing ssDNA, dsDNA, and MB reporters, we found that the 8 nt ssDNA and the MB4 reporters had darker colors at higher concentrations, and even at lower concentrations, the color of this area is also significantly different from the control area. Although 5 nt ssDNA reporter showed a high signal-to-background ratio at 10 nM, the lowest detectable concentration basically was merely 1 nM under the same conditions. Moreover, the MB4 reporters could detect target concentrations as low as 1 pM with excellent signal-to-background ratio while the 8 nt ssDNA reporters was 100 pM with faster cleavage speed. The above results indicate that Cas12a can trans-cleave the MBs reporters with outstanding analytical performance, making it a good candidate for CRISPR-Cas12a probes. Consequently, we developed a highly-sensitive SARS-CoV-2 detection using MB as signal read-out reporters.
To achieve the excellent analytical performance, the experimental conditions are optimized, including the amount of the MB reporters, temperature, and the ratio of Cas12a between crRNA. Collateral cleavage efficiency of the activated Cas12a nuclease represents an ability to cut all reporters around it. We first optimized MB reporter’s concentration because the reporter concentration plays a crucial role in fluorescence readout. We set the concentration of MB reporters at 500 nM and 5 μM respectively. The higher the concentration of reporters, the shorter the threshold time, but the sensitivity was not improved significantly (Fig. S3). To save reagents and costs, the amount of MB reporters was optimized as 500 nM. It is reported that reaction temperature will affect the collateral cleavage activity of Cas12a. Therefore, we investigated the trans-cleavage efficiency of Cas12a at 37 ℃, 42 ℃, 47 ℃, and 52 ℃, respectively. As shown in Fig. S4, a significant fluorescence recovery at all temperatures was found. Taking advantages of stem-loop structures in MB, the fluorescence recovery value increases gradually with the increase in temperature, and the background signal does not increase significantly. It reaches the peak value at 47 ℃, and then decreases gradually. It is likely attributed that the MB reporters are cleaved rapidly by highly activated Cas12a and easily disintegrated at 47 ℃. Therefore, the optimal reaction temperature is 47 ℃ for subsequent experiments. In addition, different proportions of Cas12a and crRNA are essential to the analytical performance of the CRISPR probes. Fixed the target concentration at 10 nM and 100 nM, we explored the fluorescence value when the ratio of Cas12a and crRNA was 0.5, 1, and 2, respectively (Fig. S5). The fluorescence change was remarkable when the ratio of Cas12a to crRNA was 2 in both 10 nM and 100 nM cases. In comparison, fluorescence recovery values quickly reach a saturation state at the target of 100 nM. Because of enough targets activating the trans-cleavage activity of Cas12a, the 500 nM MB reporters were wholly digested. The above results suggested that the optimal ratio of Cas12 to crRNA was 2.
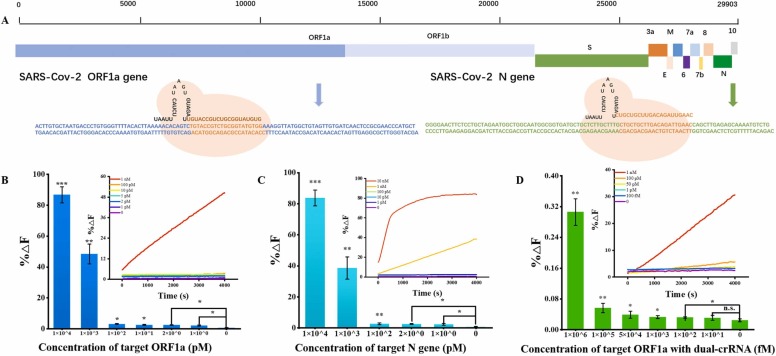
With the optimized conditions of MB reporter (500 nM), incubation temperature (47 ℃), and the ratio of Cas12a to crRNAs (2:1), we established a CRISPR biosensor for SARS-COV-2 detection with MB fluorescent reporters. Taking the ORF1a gene fragment as model target, as shown in Fig. 3A, with the increase of the concentration, the fluorescence recovery value gradually increased, which was positive with the target concentration. The sensitivity as low as 1 pM were successfully achieved without amplification strategy. To improve the accuracy of clinical diagnosis, it is usually select two genes as targets for COVID-19 practical applications. The Chinese CDC recommends ORF1ab and the N gene fragment as targets. We further evaluated the proposed probes by targeting the N gene from28881 to 28979 in the SARS-CoV-2 genome map (Fig. 3B). There is a good relationship between the fluorescence signal and the target concentration. The limit of detection was achieved at 1 pM without DNA amplification. The above results strongly demonstrated that the proposed method could realize the unbiased highly sensitive detection of SARS-CoV-2, superior for practical applications.
Fig. 3.
(A)The ORF1a gene and the N gene in the SARS-CoV-2 genome map used in this work. The real-time of fluorescence intensity and fluorescence value of ORF1a gene(B) and N gene fragment at 47 ℃ and 4000 s with using MB4 reporters. (C) The real-time of fluorescence intensity and Fluorescence value of ORF1a gene at 47 ℃ and 4000 s with the use of MB reporters with dual crRNAs when the ratio of Cas12a to crRNAs at 2. n = 3 replicates, bars represent mean ± S.D. *P < 0.05, * *P < 0.01, * ** *P < 0.0001, n.s. not significant.
It is reported that the introduction of dual crRNAs enables highly sensitive detection [19], [40]. We investigated the feasibility of dual crRNAs using MB reporters inspired by this. The crRNA1 was designed with the PAM site (5′-TTTG-3′) limitation, and crRNA2 was not. With the conditions of MB reporters at 500 nM, incubation temperature at 47 ℃, and the ratio of Cas12a to crRNAs at 0.5, fluorescence curves of dual crRNAs showed a faster fluorescence response than single crRNA1 and crRNA2 (Fig. S6). However, the sensitivity was not improved. It’s likely attributed that crRNAs could not combine with limited Cas12a to exert trans-cleavage activity when the concentration of Cas 12a is too low. Under the conditions of the ratio of Cas12a (2 μM), dual crRNA (1 μM), MB reporter (500 nM), and incubation temperature (47 ℃), The sensitivity as low as 100 fM was successfully achieved for the ORF1a gene, increasing 10 times over single crRNA1 and 1000 times (Fig. S7) over single crRNA2 without amplification as expected (Fig. 3C). The approximate 100 fM sensitivity achieved with the proposed method is by far among the highest without involving the amplification strategy or signal enhancement for CRISPR fluorescent probes (Table S5).
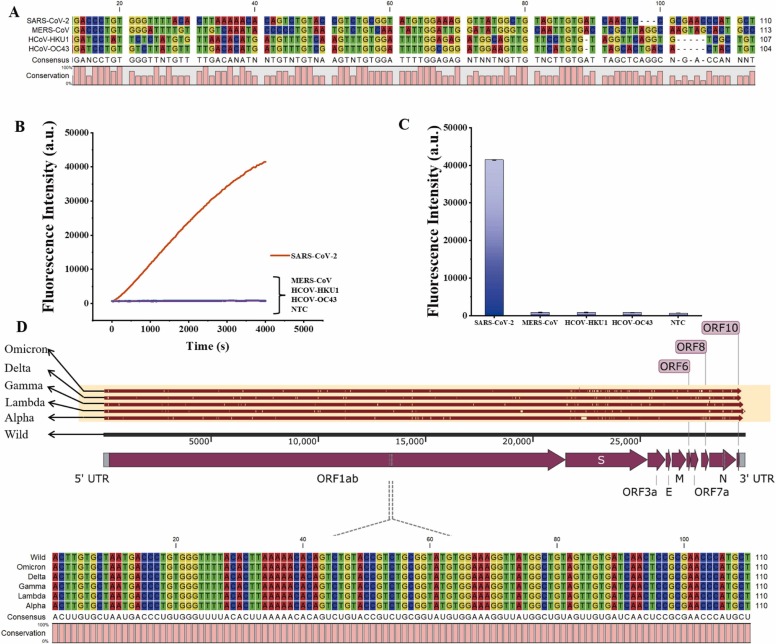
Since the SARS-COV-2 presents a similar syndrome to the influenza virus and coronavirus, which may complicate the diagnosis and clinical management of these respiratory infectious diseases, especially during flu season. Sequence alignment of SARS-CoV-2 with corresponding sites in related beta coronaviruses that cause middle east respiratory syndrome (MERS-CoV), Human coronavirus (HCoV-HKU1) and human coronaviruses OC43 (HCoV-OC43) were shown in Fig. 4 and S8. To evaluate the detection specificity, we tested the proposed method using these three viruses and SARS-CoV-2. Only SARS-CoV-2 showed a significantly increased fluorescence signal which resulted from the cleaved MB reporters due to activated Cas12a/crRNA complex. In contrast, only weak fluorescence signals were observed without noticeable difference between the control group. Moreover, we found that the target region of ORF1a could robustly identify multiple emerging SARS-CoV-2 variants by nucleotide sequence alignment of wild SARS-CoV-2 with five emerging variants. The alignment was performed using CLC Genomics Workbench 9 and nucleotide sequence of wild and variants of SARS-CoV-2 are downloaded from NCBI. Alignment of target sequence showed that no mutations were identified for all variants in target region. Results verified that the proposed method shows a high specificity with SARS-CoV-2 detection.
Fig. 4.
(A) Sequence alignment of the SARS-CoV-2 target region (ORF1a gene) and the corresponding regions on other human coronaviruses. (B) Real-time fluorescence intensity of specificity analysis for SARS-CoV-2. (C) The Histogram of specificity of Cas12a assay at 4000 s for detection of the SARS-CoV-2 ORF1a target. n = 3 replicates, bars represent mean ± S.D. (D) Sequence alignment of the SARS-CoV-2 target region (ORF1a gene) and variants.
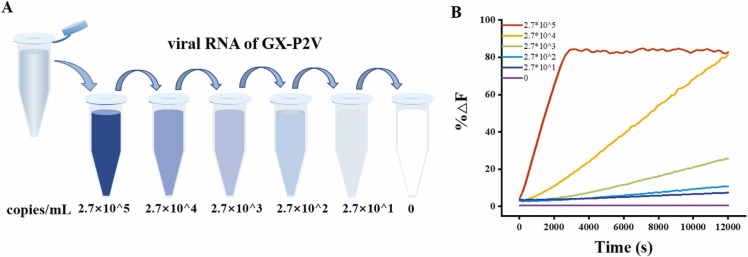
Given the outstanding performance, we further explored the feasibility of SARS-CoV-2 assay in real samples. Since GX/P2V shares over 85.8% of gene similarity with SARS-COV-2 and has no pathogenicity to humans, GX/P2V was used for clinical validation for the safety of experimental operators.[41], [42] 110 bp gene sequence at the location from 26719 to 26828 in the GX/P2V genome map were selected as model target. GX/P2V viral RNA was extracted from 200 μL of the cell supernatant using an equal volume of lysis buffer and 15 μL of 25 mg/mL magnetic beads. Then the obtained composite was directly used as a template to amplify the target DNA. As shown in Fig. 5, fluorescence intensity is proportional to the concentration of the target, and as low as 27 copies/mL was successfully achieved. These results firmly demonstrated the potential of the proposed method in practical applications, which provides perspectives for simple and universal CRISPR probes and assay kits in various application scenarios.
Fig. 5.
(A) Extracted GX/P2V viral RNA. (B) Real-time fluorescence intensity of GX P2Vgene fragment at 47 ℃.
4. Conclusions
In summary, we discovered that the trans-cleavage of Cas12a is not limited to ssDNA, dsDNA reporters but can be extended to MB reporters. The properties of various types of fluorescent reporters were systematically investigated and compared. Cas12a biosensors based on MB reporters showed outstanding analytical performance compared with ssDNA and ds DNA reporters. The high sensitivity of 100 fM achieved was by far among the highest level without involving the amplification strategy or signal enhancement. Moreover, the model target region of ORF1a could robustly identify the current widespread emerging SARS-CoV-2 variants by nucleotide sequence alignment. A minimum of 27 copies/mL viral RNA of GX/P2V showing the great potential for practical applications. This work expands the choices of CRISPR-based fluorescent biosensor and should find potential applications in clinical diagnosis.
CRediT authorship contribution statement
Sitong Liu: Methodology, Investigation, Data curation. Tie Xie: Methodology, Investigation, Data curation. Zhaohe Huang: Writing – review & editing. Xiaojing Pei: Writing – original draft, Writing – review & editing, Supervision. Shujing Li: Supervision, Writing – review & editing. Yifan He: Supervision, Writing – review & editing. Yigang Tong: Conceptualization, Resources, Supervision. Guoqi Liu: Conceptualization, Resources, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 22004005), General items of Beijing Municipal Education Commission (No. KM202110011011), High-end Foreign Experts Recruitment Plan of China (No. G2021003005L).
Declaration of Competing Interests
Sitong Liu and Xiaojing PEI declare that a patent relative the study has been filed. The remaining authors declare no competing interests in this study.
Biographies
Sitong Liu is a Master's degree candidate majoring in chemistry at Beijing Technology and Business University. He graduated with a bachelor’s degree in 2021 from Beijing Technology and Business University. His current research interest is CRISPR probes on nucleic acid detection.
Tie Xie is a Master's degree candidate majoring in chemistry at Beijing University of Chemical Technology. He graduated with a bachelor’s degree in 2016 from Beijing University of Chemical Technology. His current research interest is CRISPR probes on nucleic acid detection.
Zhaohe Huang is a Master's degree candidate majoring in light chemical engineering at Beijing Technology and Business University. He graduated with a bachelor’s degree in 2022 from Beijing Technology and Business University. His current research interest is cosmetic formulations and DNA nanomaterials.
Xiaojing Pei received her Ph.D. in analytical chemistry in 2019 from Peking University. She graduated with a bachelor’s degree in 2014 from Shandong Normal University. She is now associate professor of the College of chemistry and materials engineering at Beijing Technology and Business University. Her current research interest is developing novel optical biosensors and CRISPR probes for biomarkers detection and applications.
Shujing Li graduated from technical Institute of Physics and Chemistry, Chinese Academy of Sciences with a doctorate degree in organic chemistry from 2002 to 2007. She is a postdoctoral fellow in polymer science, School of Science, Osaka University, Japan from 2008 to 2010. From 2018 to present, she has been working as a professor in the Department of Chemistry, School of Science, Beijing Technology and Business University.
Yifan He graduated from the University of New South Wales with a degree in biomedical engineering. He focuses on the research of medical devices and human compatibility and safety.
Yigang Tong is now a professor of College of Chemistry and Materials Engineering, Beijing University of Chemical Technology Beijing, China. He is an expert in the field of novel coronavirus and SARS-COV-2 and published more than 300 papers, including Nature, PNAS, Lancet Infectious Disease with a total of more than 3000 citations. He is the chief expert of the National Major Special Project on Infectious Diseases and the chief expert of the National Key Special Project of "Synthetic Biology". Chief scientist of China's aid to Africa to fight Ebola epidemic (the third batch to Sierra Leone). Consultant of the working group of the Ministry of Science and Technology of the Ministry of Science and Technology of New Coronavirus Traceability, and the leader of the Chinese "Animal and Environment" group of the WHO-China joint research on new coronavirus traceability. He has published three research papers in the main journal of Nature, among which the research paper on the evolution of Ebola virus was rated as one of the top ten scientific advances in China in 2015. The research group discovered the new coronavirus SADS-CoV at the first time, and traced it to the bat host. The article has attracted widespread attention at home and abroad after it was published in Nature
Guoqi Liu is Ph.D. from Nankai University and post-doctoral fellow at Vanderwaal University in the United States. He was a professor at the University of Colorado. He is currently the chief scientist of Biotechnovo (Beijing) Co.,LTD. He is an immunology expert of Northwestern Polytechnical University, a visiting professor of Beijing University of Chemical Technology. In 2021, the Ministry of Science and Technology of China's high-level talent program (high-end foreign experts) was selected. In 2022, Hebei Province will be selected for the introduction of foreign intelligence projects, selected as experts of Hebei Province Talent Introduction Demonstration Base, and winner of Hebei Provincial Science and Technology Plan Project. He committed to the application of biopharmaceuticals and the CRISPR molecular detection technology in clinical diagnosis.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.snb.2022.132746.
Appendix A. Supplementary material
Supplementary material
.
Data Availability
Data will be made available on request.
References
- 1.https://covid19.who.int/.
- 2.Zhang J.C.L., He M., Su X. Exploration. 2022;2:20210265. doi: 10.1002/EXP.20210265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mattioli I.A., Hassan A., Oliveira O.N., Jr, Crespilho F.N. On the challenges for the diagnosis of SARS-CoV-2 based on a review of current methodologies. ACS Sens. 2020;5:3655–3677. doi: 10.1021/acssensors.0c01382. [DOI] [PubMed] [Google Scholar]
- 4.Drobysh M., Ramanaviciene A., Viter R., Ramanavicius A. Affinity sensors for the diagnosis of COVID-19. Micromachines. 2021;12:19. doi: 10.3390/mi12040390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dronina J., Samukaite-Bubniene U., Ramanavicius A. Advances and insights in the diagnosis of viral infections. J. Nanobiotechnol. 2021;19:23. doi: 10.1186/s12951-021-01081-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drobysh M., Ramanaviciene A., Viter R., Chen C.F., Samukaite-Bubniene U., Ratautaite V., et al. Biosensors for the determination of SARS-CoV-2 virus and diagnosis of COVID-19 infection. Int J. Mol. Sci. 2022;23:25. doi: 10.3390/ijms23020666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drobysh M., Liustrovaite V., Baradoke A., Viter R., Chen C.F., Ramanavicius A., et al. Determination of rspike protein by specific antibodies with screen-printed carbon electrode modified by electrodeposited gold nanostructures. Biosensors. 2022;12:16. doi: 10.3390/bios12080593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park J.S., Hsieh K., Chen L., Kaushik A., Trick A.Y., Wang T.-H. Digital CRISPR/Cas-assisted assay for rapid and sensitive detection of SARS-CoV-2. Adv. Sci. 2021;8:2003564. doi: 10.1002/advs.202003564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang D., Yan Y., Que H., Yang T., Cheng X., Ding S., et al. CRISPR/Cas12a-mediated interfacial cleaving of hairpin DNA reporter for electrochemical nucleic acid sensing. ACS Sens. 2020;5:557–562. doi: 10.1021/acssensors.9b02461. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J., Liu C. CRISPR-powered DNA computing and digital display. ACS Synth. Biol. 2021;10:3148–3157. doi: 10.1021/acssynbio.1c00431. [DOI] [PubMed] [Google Scholar]
- 11.Luo X., Xue Y., Ju E., Tao Y., Li M., Zhou L., et al. Digital CRISPR/Cas12b-based platform enabled absolute quantification of viral RNA. Anal. Chim. Acta. 2022;1192 doi: 10.1016/j.aca.2021.339336. [DOI] [PubMed] [Google Scholar]
- 12.Wu X., Chan C., Springs S.L., Lee Y.H., Lu T.K., Yu H. A warm-start digital CRISPR/Cas-based method for the quantitative detection of nucleic acids. Anal. Chim. Acta. 2022;1196 doi: 10.1016/j.aca.2022.339494. [DOI] [PubMed] [Google Scholar]
- 13.Smith C.W., Nandu N., Kachwala M.J., Chen Y.-S., Uyar T.B., Yigit M.V. Probing CRISPR-Cas12a Nuclease Activity Using Double-Stranded DNA-Templated Fluorescent Substrates. Biochemistry. 2020;59:1474–1481. doi: 10.1021/acs.biochem.0c00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suh J.-S., Kim H.-S., Kim T.-J. Development of a SARS-CoV-2-derived receptor-binding domain-based ACE2 biosensor. Sens Actuators B Chem. 2021;334 doi: 10.1016/j.snb.2021.129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Xue T., Wang M., Ledesma-Amaro R., Lu Y., Hu X., et al. CRISPR-Cas13a cascade-based viral RNA assay for detecting SARS-CoV-2 and its mutations in clinical samples. Sens Actuators B Chem. 2022;362 doi: 10.1016/j.snb.2022.131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z., Jiang S., Wang X., Dong T., Wang Y., Li D., et al. A novel enhanced substrate for label-free detection of SARS-CoV-2 based on surface-enhanced Raman scattering. Sens Actuators B Chem. 2022;359 doi: 10.1016/j.snb.2022.131568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh D., Mallon J., Poddar A., Wang Y., Tippana R., Yang O., et al. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a) Proc. Natl. Acad. Sci. 2018;115:5444–5449. doi: 10.1073/pnas.1718686115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dronina J., Bubniene U.S., Ramanavicius A. The application of DNA polymerases and Cas9 as representative of DNA-modifying enzymes group in DNA sensor design (review) Biosens. Bioelectron. 2021;175:11. doi: 10.1016/j.bios.2020.112867. [DOI] [PubMed] [Google Scholar]
- 19.Fozouni P., Son S., Derby M.Dd.L., Knott G.J., Gray C.N., D'Ambrosio M.V., et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184:323–333. doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.J. Fu, J. Li, J. Chen, Y. Li, J. Liu, X. Su, et al., Ultra-specific nucleic acid testing by target-activated nucleases, Crit Rev Biotechnol, 10.1080/07388551.2021.1983757(2021). [DOI] [PubMed]
- 21.Li S.-Y., Cheng Q.-X., Liu J.-K., Nie X.-Q., Zhao G.-P., Wang J. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018;28:491–493. doi: 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X., Tay J.K., Goh C.K., Chan C., Lee Y.H., Springs S.L., et al. Digital CRISPR-based method for the rapid detection and absolute quantification of nucleic acids. Biomaterials. 2021;274 doi: 10.1016/j.biomaterials.2021.120876. [DOI] [PubMed] [Google Scholar]
- 24.Wu X.L., Tay J.K., Goh C.K., Chan C., Lee Y.H., Springs S.L., et al. Digital CRISPR-based method for the rapid detection and absolute quantification of nucleic acids. Biomaterials. 2021;274 doi: 10.1016/j.biomaterials.2021.120876. [DOI] [PubMed] [Google Scholar]
- 25.Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164 doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim H., Lee S., Yoon J., Song J., Park H.G. CRISPR/Cas12a collateral cleavage activity for simple and rapid detection of protein/small molecule interaction. Biosens. Bioelectron. 2021;194 doi: 10.1016/j.bios.2021.113587. [DOI] [PubMed] [Google Scholar]
- 27.Mao S.Q., Ying Y.C., Wu X.T., Krueger C.J., Chen A.K. CRISPR/dual-FRET molecular beacon for sensitive live-cell imaging of non-repetitive genomic loci. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gkz752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaur A., Dhakal S. Recent applications of FRET-based multiplexed techniques. Trac-Trend Anal. Chem. 2020;123 [Google Scholar]
- 29.Li L., Li S., Wu N., Wu J., Wang G., Zhao G., et al. HOLMESv2: a CRISPR-Cas12b-Assisted Platform for Nucleic Acid Detection and DNA Methylation Quantitation. ACS Synth. Biol. 2019;8:2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- 30.Li S.-Y., Cheng Q.-X., Wang J.-M., Li X.-Y., Zhang Z.-L., Gao S., et al. CRISPR-Cas12a-assisted nucleic acid detection. Cell Disco. 2018;4:20. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsieh K.W., Zhao G.J., Wang T.H. Applying biosensor development concepts to improve preamplification-free CRISPR/Cas12a-Dx. Analyst. 2020;145:4880–4888. doi: 10.1039/d0an00664e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X.T., Mao S.Q., Yang Y.T., Rushdi M.N., Krueger C.J., Chen A.K. A CRISPR/molecular beacon hybrid system for live-cell genomic imaging. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gky304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherrill-Mix S., Hwang Y., Roche A.M., Glascock A., Weiss S.R., Li Y.Z., et al. Detection of SARS-CoV-2 RNA using RT-LAMP and molecular beacons. Genome Biol. 2021;22:169. doi: 10.1186/s13059-021-02387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyagi S., Kramer F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Q., Wang F., Zhang H., Zhang Y., Liu M., Liu Y. Universal Ti3C2 MXenes based self-standard ratiometric fluorescence resonance energy transfer platform for highly sensitive detection of exosomes. Anal. Chem. 2018;90:12737–12744. doi: 10.1021/acs.analchem.8b03083. [DOI] [PubMed] [Google Scholar]
- 37.Silva F.S.R., Erdogmus E., Shokr A., Kandula H., Thirumalaraju P., Kanakasabapathy M.K., et al. SARS-CoV-2 RNA detection by a cellphone-based amplification-free system with CRISPR/CAS-dependent enzymatic (CASCADE) Assay. Adv. Mater. Technol. 2021:2100602. doi: 10.1002/admt.202100602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li T., Hu R., Xia J., Xu Z., Chen D., Xi J., et al. G-triplex: a new type of CRISPR-Cas12a reporter enabling highly sensitive nucleic acid detection. Biosens. Bioelectron. 2021;187 doi: 10.1016/j.bios.2021.113292. [DOI] [PubMed] [Google Scholar]
- 39.http://www.nupack.org/partition/new.
- 40.Ding X., Yin K., Li Z., Lalla R.V., Ballesteros E., Sfeir M.M., et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020;11:4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Y., Wang L., An X., Tong Y. All-in-one in situ colorimetric RT-LAMP assay for point-of-care testing of SARS-CoV-2. Analyst. 2021;146:6026–6034. doi: 10.1039/d1an01043c. [DOI] [PubMed] [Google Scholar]
- 42.Yang B., Zhao Z., Pan Y., Xie J., Zhou B., Li Y., et al. Shear-thinning and designable responsive supramolecular DNA hydrogels based on chemically branched DNA. 2021;13:48414–48422. doi: 10.1021/acsami.1c15494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.