Abstract
Streptococcus salivarius 20P3 produces a 22-amino-acid residue lantibiotic, designated salivaricin A (SalA), that inhibits the growth of a range of streptococci, including all strains of Streptococcus pyogenes. Lantibiotic production is associated with the sal genetic locus comprising salA, the lantibiotic structural gene; salBCTX genes encoding peptide modification and export machinery proteins; and salYKR genes encoding a putative immunity protein and two-component sensor-regulator system. Insertional inactivation of salB in S. salivarius 20P3 resulted in abrogation of SalA peptide production, of immunity to SalA, and of salA transcription. Addition of exogenous SalA peptide to salB mutant cultures induced dose-dependent expression of salA mRNA (0.2 kb), demonstrating that SalA production was normally autoregulated. Inactivation of salR encoding the response regulator of the SalKR two-component system led to reduced production of, and immunity to, SalA. The sal genetic locus was also present in S. pyogenes SF370 (M type 1), but because of a deletion across the salBCT genes, the corresponding lantibiotic peptide, designated SalA1, was not produced. However, in S. pyogenes T11 (M type 4) the sal locus gene complement was apparently complete, and active SalA1 peptide was synthesized. Exogenously added SalA1 peptide from S. pyogenes T11 induced salA1 transcription in S. pyogenes SF370 and in an isogenic S. pyogenes T11 salB mutant and salA transcription in S. salivarius 20P3 salB. Thus, SalA and SalA1 are examples of streptococcal lantibiotics whose production is autoregulated. These peptides act as intra- and interspecies signaling molecules, modulating lantibiotic production and possibly influencing streptococcal population ecology in the oral cavity.
Streptococcus salivarius is a primary colonizer of neonatal oral mucosal surfaces and a predominant component of the human adult oral microbiota and is not associated with disease in healthy individuals (39). Streptococcus pyogenes, on the other hand, persists in the pharynx in a carrier state in approximately 10% of the population, is a common cause of pharyngeal infections, especially in school-aged children, and is usually present in high numbers only during acute infection (1). All S. pyogenes strains tested have been found to be susceptible to growth inhibition by salivaricin A (SalA), a lantibiotic peptide produced by S. salivarius (34, 37), and it has been suggested that growth of S. pyogenes in vivo may be modulated by indigenous SalA-producing S. salivarius.
Lantibiotics are antimicrobial peptides that are produced by, and are active against, closely related gram-positive organisms. These peptides are ribosomally synthesized and then undergo posttranslational modifications, including amino acid dehydration (38) and thioether bridge formation (20). Lantibiotics form two families; type A lantibiotics are linear, and type B lantibiotics have more globular conformations. The peptides are synthesized as prepropeptides consisting of an inhibitor propeptide and a leader region, the features of which have been utilized to group the type A lantibiotics into three subclasses (36). In the members of one of these subclasses, subclass AII, the amino acid (aa) residues Gly-Gly, Gly-Ser, or Gly-Ala immediately precede the site of leader cleavage. These Gly-Gly type cleavage sites are more commonly found in nonmodified bacteriocins or pheromones, such as the ComC peptide responsible for competence induction in Streptococcus pneumoniae (15). The streptococcal pheromones are quorum-sensing molecules, analogous to the N-acyl homoserine lactones produced by gram-negative bacteria (35), that regulate microbial community responses.
SalA (22 aa residues) is a subclass AII lantibiotic produced by S. salivarius 20P3 via processing of a 48-aa prepropeptide encoded by the salA gene (34). By utilizing salA as a DNA hybridization probe it was shown that all SalA peptide-producing strains of S. salivarius contained salA sequences and that, somewhat surprisingly, 63 of 65 S. pyogenes strains of different M types contained a salA gene homolog designated salA1 (37). We hypothesized that S. pyogenes failed to produce SalA1 inhibitor and was sensitive to inhibition by SalA from S. salivarius because the genetic locus for lantibiotic production and immunity was incomplete or transcriptionally inactive. In the present study, the structure of the sal genetic locus in S. salivarius 20P3 was determined and the genes necessary for regulation of lantibiotic production were identified. In S. pyogenes SF370 (M type 1), the corresponding sal genetic locus had a deletion spanning three genes encoding peptide modification and export proteins, and the salA1 gene was transcriptionally inactive, thus accounting for the lack of SalA1 production. However, the salA gene was actively transcribed in S. pyogenes T11 (M type 4), which contained a complete sal genetic locus, and active SalA1 inhibitor peptide was produced. To the best of our knowledge, this is the first report of an autoregulatory lantibiotic produced by streptococci, and the evidence presented here suggests that SalA-like peptides form a family of signaling factors recognized by different species of streptococci.
MATERIALS AND METHODS
Bacterial strains and media.
S. salivarius 20P3 (wild type, SalA+) (34), S. pyogenes SF340 (M type 1, SalA−) (41), and S. pyogenes T11 (M type 4, SalA+ clinical isolate) were maintained on Columbia agar plates (Life Technologies Ltd., Paisley, United Kingdom) supplemented with human blood (5%, vol/vol) and CaCO3 (0.1%, wt/vol) in a 5% CO2–air atmosphere at 37°C. Streptococcal strains were routinely cultured in Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.) or in M17 medium (Difco) supplemented with CaCO3 (10 mM) and sucrose (10 mM) (M17CS medium) for maximal SalA peptide production. Escherichia coli DH5α (9) and the highly electrocompetent derivative strain DH10B (14) were maintained on Luria-Bertani (LB) agar (Difco) or were grown in LB broth with aeration at 37°C. The following antibiotic concentrations were used (where appropriate): ampicillin, 100 μg/ml (E. coli); and spectinomycin, 100 μg/ml (E. coli), 60 μg/ml (S. pyogenes), or 600 μg/ml (S. salivarius).
PCR and sequence analysis.
Primers for PCR amplification of salA and the regions downstream of salA in S. salivarius 20P3 were designed on the basis of the corresponding sal locus sequence in S. pyogenes SF370 (University of Oklahoma Advanced Center for Genome Technology; http://www.genome.ou.edu/strep.html). The major primers utilized (and their corresponding target sites within the S. salivarius 20P3 sal locus [10,610 nucleotides; GenBank accession number AY005472]) were SalAF (positions 604 to 629; 5′GATATTTTGAACAATGCTATCGAAGA), DsintF (positions 856 to 876; 5′CAACATCAGTTTTACGAATAC), ORFIR (positions 2340 to 2320; 5′GTGAACTTCAATCTTTCATCG), SalYF (positions 6665 to 6684; 5′GGTCAAGCTCTAGTTATCGC), SalYR (positions 8413 to 8393; 5′GATTATGGATGTCACTAACCG), and SalRterm (positions 10610 to 10590; 5′TCAGAAATCCATAAAATACCC). PCRs were carried out with Taq polymerase (Roche Diagnostics Ltd., Lewes, England) for 30 cycles consisting of denaturation at 95°C for 30 s, annealing at between 55 and 64°C (as appropriate) for 30 s, and extension at 72°C for 1 min per kb of DNA to be amplified; this was followed by a final elongation step at 72°C for 5 min. PCR products were ligated into pGEM-T (Promega, Madison, Wis.) and sequenced with a Perkin-Elmer ABI 377A sequencer. Primary sequence data were collated with SeqEd sequencer software, and sequence alignment, translation, and general analyses were performed by using DNAMAN (Lynnon BioSoft, Vaudreuil, Canada). Deduced amino acid sequences for open reading frames were compared with sequences in protein sequence databases by using the BLAST facilities on the National Center for Biotechnology Information server (www.ncbi.nlm.nih.gov) and the University of Oklahoma server.
DNA manipulation and transformation.
Streptococcal genomic DNA was isolated as described by Upton et al. (43). Briefly, confluent growth from one-half of an agar plate culture was collected in TE buffer (10 mM Tris-HCl [pH 8.0] containing 10 mM EDTA), the cells were harvested by centrifugation (8,000 × g for 10 min) and suspended in 0.3 ml of TE buffer, and 0.3 ml of a lysis mixture (50 mM Tris-HCl [pH 8.0] containing 10 mM EDTA and 2% [vol/vol] Triton X-100) was added. Lysis was achieved by adding 30 U of mutanolysin (Sigma) and then 0.3 mg of pronase (Sigma) and incubating the preparation at 37°C for 2 h. The suspension was extracted once with 0.6 ml of phenol and once with 0.6 ml of phenol-chloroform (0.3 ml of phenol plus 0.3 ml of chloroform-isoamyl alcohol [24:1]). Nucleic acids were precipitated from the aqueous phase with 2 volumes of 100% ethanol at −70°C and collected by centrifugation (12,000 × g for 20 min), and the pellet was washed with 70% (vol/vol) ethanol, air dried, and dissolved in TE buffer. In a number of experiments the solutions were then incubated with 50 μg of RNase A (Sigma)/ml at 37°C for 30 min and extracted with phenol-chloroform, and the DNA was precipitated, washed, and dissolved in TE buffer as described above.
Plasmid DNA was isolated from E. coli by using Qantum Prep (Bio-Rad Laboratories, Hercules, Calif.). For insertional inactivation of streptococcal genes, target fragments were generated by PCR amplification, cloned into pGEM-T, and recovered by restriction digestion with NcoI and NsiI endonucleases. Fragments were gel purified and ligated into vector pFW5 (29) digested with NcoI and NsiI. E. coli strains were transformed with plasmid DNA by using a TransPorator (BTX, San Diego, Calif.) in 0.1-cm cuvettes (Bio-Rad Laboratories, Richmond, Calif.). Following electroporation, bacteria were incubated for 1 h in 1 ml of 2 × YT broth (16 g of Bacto Tryptone per liter, 10 g of Bacto Yeast Extract per liter, 5 g of NaCl per liter; pH 7.0) at 37°C with shaking at 200 rpm before being spread onto LB agar plates containing the appropriate antibiotics (9). S. salivarius and S. pyogenes strains were prepared for electrotransformation by the method of Chen et al. (4); dl-threonine was included in the media at final concentrations of 0.6 and 0.4 M, respectively. Electroporation was carried out in 0.1-cm cuvettes by using a Bio-Rad Gene Pulser II set at 1.8 kV, 25 μF, and 200 Ω, and transformants (frequency, 50 to 100 transformants per μg of DNA) were selected on agar containing spectinomycin.
Generation of mutants.
To generate S. salivarius UB1309 salB::pMU1011 Spr, an internal 1.48-kb salB fragment (Fig. 1), PCR amplified from 20P3 DNA by using primers DsintF and ORF1R, was cloned into pFW5 to produce pMU1001, and this plasmid was then transformed onto the S. salivarius 20P3 chromosome. S. pyogenes UB1308 salB::pMU1016 Spr was generated in the same way by transforming pMU1016 (pFW5 carrying a 1.48-kb salB fragment from T11) onto the S. pyogenes T11 chromosome. S. salivarius UB1310 salR::pMU1002 Spr was generated by PCR amplifying a 1.16-kb fragment from the salKR gene region (Fig. 1) of 20P3 DNA with primers SalKF (positions 9249 to 9269; 5′GTTGGATTGTACTCATGAAGG) and SalRR (positions 10411 to 10391; 5′TCAACATAATCCTGAGATTCG), cloning this fragment into pFW5 to produce pMU1002, and transforming this plasmid into wild-type strain 20P3 as described above. Integration of pFW5 plasmid constructs onto the streptococcal chromosome was confirmed by PCR amplification with pFW5-specific primers pFW5F (5′GATCAGGAGTTGAGAGTGGAC) and pFW5R (5′TGGAGAAGATTCAGCCACTGC).
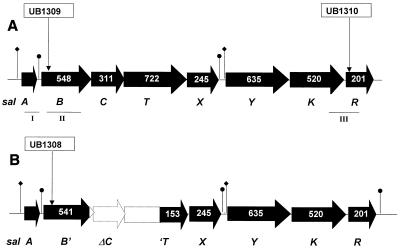
FIG. 1.
Genetic structures of the sal loci in S. salivarius 20P3 (A) and S. pyogenes SF370 (M type 1) (B). Open reading frames are depicted by arrows which show the numbers of aa residues in the deduced polypeptides. The sites of insertional inactivation with plasmid pFW5 constructs used to generate strains UB1309, UB1310, and UB1308 are shown. PCR-amplified segments of DNA used for hybridization probes or cloning in pFW5 are indicated as follows: I, 0.3-kb salA probe; II, 1.48-kb salB fragment; III, 1.16-kb salKR fragment. Symbols: ⧫, putative promoter; ●, inverted repeat.
RNA analysis.
To prepare total RNA from streptococci, cells were harvested by centrifugation from M17CS medium, suspended in 0.2 ml of spheroplasting buffer (20 mM Tris-HCl [pH 6.8] containing 10 mM MgCl2 and 26% [wt/vol] raffinose) containing 500 U of mutanolysin/ml and 0.1 mg of spectinomycin/ml, and incubated 37°C for 30 min. Spheroplasts were collected by centrifugation, and RNA was extracted by using a Qiagen RNAeasy kit and a Qiashredder column (Qiagen Ltd., Crawley, England) as recommended by the manufacturer. RNA samples (10 μg per lane) were subjected to electrophoresis through 1% (wt/vol) agarose in 1× MOPS (morpholinepropanesulfonic acid) buffer (0.2 M MOPS containing 50 mM sodium acetate and 10 mM EDTA; pH 7.0) supplemented with 1% (wt/vol) formaldehyde at 80 V for 3 h and were vacuum blotted onto a nylon membrane. For Northern analysis of sal transcripts, a PCR product generated with primers SalAF and SalAR (positions 901 to 883; 5′AGAAGTATCTAGTATGTCG) was radioactively labeled with [32P]dATP by using Prime-a-Gene (Promega) as directed by the manufacturer. Hybridization reactions were carried out at 65°C for 18 h as described elsewhere (5). For reverse transcription (RT)-PCR analysis, DNase-treated RNA that had been extracted from exponentially growing cells was utilized for cDNA synthesis by using the SalRterm oligonuclotide primer and avian myeloblastosis virus reverse transcriptase (First Strand cDNA synthesis kit; Roche Diagnostics). PCR amplifications were then performed by utilizing the Expand Long Template PCR system (Roche Diagnostics) with primers SalAF and SalXR (positions 6510 to 6490; 5′CTCTCCTCTATCGGATAAAGC), primers SalY2S (positions 6571 to 6591; 5′CTATTTTCTAGCTACGATCGG) and SalRterm, or primers SalAF and SalRR.
SalA production and expression.
SalA peptide was purified from S. salivarius 20P3 M17CS broth culture supernatant as previously described (34). To test for SalA induction of transcription, streptococcal strains were grown for 18 h in M17CS medium with or without SalA peptide. Cells were harvested by centrifugation (16,000 × g for 20 min at 4°C), and the cell-free supernatants were collected in sterile tubes. For medium swap experiments the bacterial pellets were suspended in the appropriate spent culture supernatant and incubated at 37°C for 4 h before RNA was extracted as described above. Levels of inhibitor production and of immunity to inhibition were determined by using a deferred antagonism assay performed on Columbia blood agar plates supplemented with 0.1% (wt/vol) CaCO3 as described elsewhere (42). Levels of lantibiotic production by producer strains and growth inhibition of test strains were scored as 0 (no inhibition zone), 1 (narrow inhibition zone that was 1 to 2 mm wide), 2 (medium zone of inhibition that was 2 to 5 mm wide), 3 (larger zone of inhibition that was 5 to 8 mm wide), or 4 (inhibition zone that was more than 8 mm wide).
Nucleotide sequence accession number.
The nucleotide sequence data for the S. salivarius 20P3 sal locus have been deposited in the GenBank database under accession number AY005472.
RESULTS
Structure of the sal locus in S. salivarius 20P3.
To determine the sequence and genetic structure of the S. salivarius 20P3 sal gene locus, PCR primers were synthesized based on the nucleotide sequence of the approximately 7-kb region downstream of the salA1 gene in S. pyogenes SF370 (Fig. 1). These primers were utilized to PCR amplify segments that comprised the complete sal genetic locus in S. salivarius 20P3. The genetic locus comprised eight open reading frames designated salABCTXYKR (Fig. 1A). These open reading frame designations were assigned on the basis of database homology searches and were in accordance with conventional lantibiotic gene cluster nomenclature (7, 36).
The first gene (salA), encoding the 48-aa precursor lantibiotic peptide, was preceded by a potential ribosome binding sequence (GGGAG) and an AT-rich region containing putative promoter sequences. Downstream of salA, the salB and salC genes (Fig. 1A) are predicted to encode peptides with significant identities to the N-terminal and C-terminal sequences, respectively, of lactococcin biosynthesis protein produced by Lactococcus lactis (32) (Table 1). We identified salB and salC as separate genes because there are multiple stop codons present in all three reading frames in the putative intergenic sequence. SalB contains a number of motifs that are conserved in LanM modification peptides (36), while SalC is predicted to contain at least five membrane-spanning sequences and is probably responsible for the formation of thioether bridges in mature SalA peptide (12, 27). The deduced aa sequences encoded by the next two genes, salT and salX, exhibit significant identities with ATP binding cassette (ABC) transporters (16, 46) (Table 1). For lantibiotics and unmodified bacteriocins with a GG cleavage site, the leader is removed by a cysteine protease activity in the N-terminal region of the LanT polypeptide (16). Retention of the leader peptide might have an immunity function (45). Motifs characteristic of LanT protease regions (36) are present in the N-terminal region of the SalT polypeptide. Collectively, these data suggest that leader peptide cleavage and export of modified SalA are carried out by a SalTX protein complex located within the cell membrane.
TABLE 1.
Levels of homology of S. salivarius sal gene product sequences to peptide database sequences and to corresponding S. pyogenes SF370 sal gene product sequences
| S. salivarius 20P3 gene | Size of precursor (aa) | Most similar sequence
|
% Identity to corresponding sal gene product in S. pyogenes SF370a | |||
|---|---|---|---|---|---|---|
| Molecule (accession no.) | No. of aa residues | Region of homology to sal gene product (residues) | % Identity | |||
| salA | 48 | Salivaricin A, S. salivarius (P36500) | 48 | 1–48 | 100 | 92 |
| salB | 548 | Lacticin 481 biosynthesis protein, L. lactis (P37609) | 922 | 29–470 | 26 | 83 |
| salC | 311 | Lacticin 481 biosynthesis protein, L. lactis (P37609) | 922 | 539–908 | 25 | |
| salT | 722 | ComA transporter, S. pneumoniae (Q03727)b | 717 | 11–697 | 24 | 92 |
| salX | 245 | ABC transporter, B. subtilis (P42423) | 257 | 5–246 | 44 | 99 |
| salY | 635 | No significant database match | 91 | |||
| salK | 520 | ComP sensor kinase, B. subtilis (Q99027) | 769 | 564–754 | 21 | 85 |
| salR | 201 | DegU transcriptional regulator, B. brevis (P54662) | 236 | 10–232 | 27 | 92 |
salC is not present in SF370, while the salB and salT genes encode truncated proteins in SF370.
The salY gene, situated upstream of the salKR genes at the distal end of the locus, encodes a 635-aa polypeptide for which there were no significant database matches. It is proposed that the products of the salKR genes form a two-component sensor kinase-response regulator system (40) based on significant sequence identities between SalK and Bacillus subtilis ComP (47) and between SalR and the Bacillus brevis DegU transcriptional regulator (24) (Table 1). Other features of the sal locus that have relevance for transcriptional regulation include putative promoter regions upstream of the salA and salY genes (Fig. 1A) and inverted repeats with the potential to form stem-loop structures immediately downstream of the salA and salX genes (Fig. 1A). An inverted repeat downstream of the inhibitor peptide structural gene is present in several lantibiotic gene clusters (13, 18, 23, 33). The potential stem-loop structure that is formed functions as a transcriptional attenuator that allows only partial readthrough transcription of the downstream genes.
Structure of the sal locus in S. pyogenes.
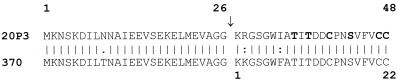
S. pyogenes SF370 (M type 1) contains the salA-like gene designated salA1 (37) but does not produce detectable levels of lantibiotic inhibitor (Table 2). While the deduced aa sequence of the mature SalA1 peptide from strain SF370 has two conserved changes, residue R2 to K and residue I7 to F (Fig. 2), it is predicted that these changes do affect the hydrophobicity of the mature peptide or the maturation process. Indeed, the deduced aa sequence of SalA1 from the lantibiotic inhibitor-producing strain S. pyogenes T11 (M type 4) is identical to that of SF370 SalA1. In deferred antagonism tests the inhibitory profile of S. pyogenes T11 was similar to that of S. salivarius 20P3 (Table 2).
TABLE 2.
SalA production and immunity profiles for S. salivarius and S. pyogenes strains as determined by deferred cross-antagonism testing
| Indicator organism | Activities with the following producersa:
|
|||||
|---|---|---|---|---|---|---|
|
S. salivarius strains
|
S. pyogenes strains
|
|||||
| 20P3 | UB1309 salB | UB1310 salR | SF370 | T11 | UB1308 salB | |
| S. salivarius strains | ||||||
| 20P3 wild type | 0 | 0 | 0 | 0 | 0 | 0 |
| UB1309 salB | 4 | 0 | 2 | 0 | 2 | 0 |
| UB1310 salR | 3 | 0 | 0 | 0 | 1 | 0 |
| S. pyogenes strains | ||||||
| SF370 wild type | 4 | 1 | 4 | 0 | 3 | 0 |
| T11 wild type | 3 | 0 | 3 | 0 | 0 | 0 |
| UB1308 salB | 4 | 0 | 3 | 0 | 3 | 0 |
The activities of SalA in producer strains against indicator strains were scored from 4 (strongest activity) to 0 (no activity) as described in Materials and Methods.
FIG. 2.
Comparison of the inferred aa sequences of the precursor SalA peptide in S. salivarius 20P3 and SalA1 peptide in S. pyogenes SF370. The arrow indicates the site of proteolytic cleavage (after residue 26) to generate the 22-aa SalA or SalA1 propeptides. The residues in boldface type are those likely to be involved in thioether bridge formation.
A comparison of the sal locus sequence present in S. pyogenes SF370 with that in S. salivarius 20P3 showed that a 3.2-kb deletion occurred in strain SF370, which was predicted to result in truncation of the salB gene product and abrogation of SalC and SalT production (Fig. 1B). Thus, if the salA1 gene was to be transcribed and translated normally, the prepropeptide could not be modified or exported. Apart from the deleted region, the genetic structure of the sal locus in S. pyogenes SF370 (Fig. 1B) was similar to that in S. salivarius 20P3 (Fig. 1A), and the corresponding polypeptide sequence identities ranged from 85 to 99% (Table 1). These comparisons were based on the available GAS genomic sequence (University of Oklahoma), and the results were confirmed by sequencing several PCR products generated for the salYKR genes that allowed minor corrections to be made to the SF370 genomic sequence in this region. By utilizing PCR amplification with primers DsintF and SalYR to screen a number of S. pyogenes strains, we have shown that only M type 4 (including S. pyogenes T11) and M type 57 strains carry the complete salBCT coding region.
Transcriptional analysis of the sal locus in S. pyogenes and S. salivarius.
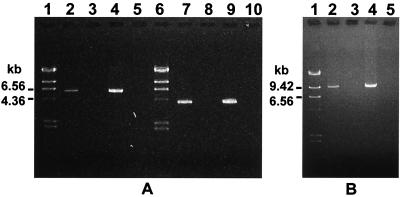
For Northern analysis of salA mRNA transcript levels, nitrocellulose blots of streptococcal RNAs were incubated with a PCR-generated 32P-labeled 0.3-kb DNA fragment (salA) (Fig. 1). The salA fragment probe reacted strongly with a single mRNA transcript approximately 200 bases long in S. salivarius 20P3 and less strongly with a band of similar size in S. pyogenes T11; no significant signal was obtained with mRNA extracts of S. pyogenes SF370 (Fig. 3 and 4). The relative salA mRNA levels corresponded well to the levels of inhibitor produced by these strains, as assayed by deferred antagonism on agar (Table 2). To confirm the identities of the other predicted transcripts of the locus, mRNA from S. salivarius 20P3 cells was subjected to RT-PCR analysis. A 5.9-kb product was generated with primers SalAF and SalXR (Fig. 5A), supporting the suggestion that the salABCTX genes were cotranscribed. In addition, amplification of a 4-kb fragment with primers SalYS and SalRterm demonstrated that there was transcriptional linkage of the salYKR genes (Fig. 5A). By utilizing primers SalAF and SalRR we were also able to demonstrate the presence of a 9.3-kb mRNA transcript corresponding to salABCTXYKR (Fig. 5B). In all these experiments no PCR products were obtained from RNA samples that had not been incubated with reverse transcriptase (Fig. 5).
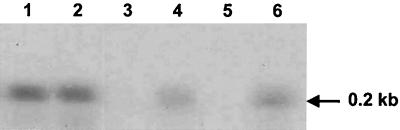
FIG. 3.
Northern analysis of SalA1 gene transcription in S. pyogenes probed with a salA1 fragment corresponding to salA fragment I (Fig. 1B). Lane 1, S. pyogenes T11 grown in M17CS medium; lane 2, S. pyogenes T11 grown in S. pyogenes UB1308 salB culture medium; lane 3, S. pyogenes SF370 grown in M17CS medium; lane 4, S. pyogenes SF370 grown in strain T11 culture medium; lane 5, S. pyogenes UB1308 salB grown in M17CS medium; lane 6, S. pyogenes UB1308 salB grown in strain T11 culture medium. The position of the salA1 mRNA transcript (0.2 kb) is indicated by an arrow. The lanes contained equivalent amounts of total RNA (10 μg).
FIG. 4.
Autoinduction of SalA in S. salivarius 20P3: Northern analysis of salA mRNA transcripts from streptococci grown at 37°C for 30 min in M17CS medium (lane 1) and in M17CS medium containing purified SalA peptide at concentrations of 0.05 pmol/ml (lane 2), 0.5 pmol/ml (lane 3), and 5 pmol/ml (lane 4). The lanes contained equivalent amounts of total RNA (10 μg).
FIG. 5.
RT-PCR analysis of sal locus transcripts from S. salivarius 20P3. cDNA was generated from mRNA by using the oligonucleotide SalRterm. (A) Lanes 2 to 5 PCRs performed with primers SalAF and SalXR; lanes 7 to 10, PCRs performed with primers SalY2S and SalRterm; lanes 2 and 7, cDNA template generated by RT; lanes 3 and 8, RNA controls (no RT); lanes 4 and 9, chromosomal DNA template; lanes 5 and 10, no-template controls. (B) PCRs performed with primers SalAF and SalRR. Lane 2, cDNA template generated by RT; lane 3, RNA control; lane 4, chromosomal DNA; lane 5, no template. The molecular mass markers (panel A, lanes 1 and 6; panel B, lane 1) were λ DNA HindIII fragments (23.1, 9.41, 6.56, 4.36, 2.32, and 2.02 kb).
Transcription of salA depends upon SalA modification and export.
In order to test the hypothesis that the salBCT gene products were essential for production of SalA inhibitor, the salB gene in S. salivarius 20P3 was inactivated by inserting integrational plasmid pMU1011 in the salB coding region (Fig. 1A). A Northern blot analysis of mRNA extracted from UB1309 salB::pMU1011 in which the salA probe was used demonstrated that transcription of the salA gene was abrogated (Fig. 6, lane 1). S. salivarius UB1309 was phenotypically SalA− as determined by an agar inhibition assay and did not significantly inhibit growth of S. pyogenes (Table 2). The analogous salB gene inactivation experiment was then carried out with S. pyogenes T11 by integrating plasmid pMU1016 in the salB chromosomal gene (Fig. 1B). The resulting isogenic mutant, UB1308, did not produce a 0.2-kb salA1 transcript (Fig. 3, lane 5) and was also inhibitor production negative (SalA1−) as determined by the agar inhibition assay (Table 2). These results confirmed that the salBCT gene products were essential for production of SalA and SalA1 peptides and suggested that SalA and SalA1 lantibiotics modulated salA and salA1 gene transcription.
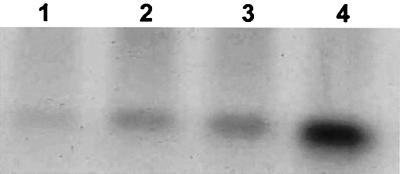
FIG. 6.
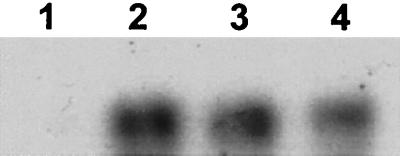
Northern analysis of salA gene transcription in S. salivarius UB1309 salB probed with salA fragment I (Fig. 1A). mRNAs were extracted from S. salivarius UB1309 cells grown at 37°C for 4 h. Lane 1, cells grown in their own cell-free spent culture medium; lane 2, cells grown in fresh M17CS medium containing 5 pmol of purified SalA/ml; lane 3, cells grown in cell-free S. salivarius 20P3 spent culture medium; lane 4, cells grown in cell-free S. pyogenes T11 spent culture medium.
Expression of SalA is autoregulated.
To test the hypothesis that salA expression depended upon the presence of active SalA, S. salivarius 20P3 cells were removed from a late-exponential-phase culture in M17CS medium. The spent medium was inoculated with S. salivarius UB1309 salB cells; the control consisted of spent medium from a UB1309 culture. The cultures were incubated at 37°C for 4 h, and mRNAs were then extracted, used for Northern analysis, and probed with the salA gene fragment as described above. Following incubation in the S. salivarius 20P3 spent culture medium, the UB1309 salB cells produced a 0.2-kb salA mRNA transcript (Fig. 6, lane 3) that was absent from the control cells (Fig. 6, lane 1).
In similar experiments, induction of salA1 transcription was observed in S. pyogenes UB1308 salB cells when these cells were grown in spent culture supernatant from S. pyogenes T11 cultures (SalA1+) (Fig. 3, lane 6) but not when they were grown in fresh broth or in spent culture supernatant from their own cultures (Fig. 3, lane 5). In addition, S. pyogenes SF370 cells, which did not normally express salA1 mRNA (Fig. 3, lane 3), were induced to express the salA1 mRNA transcript when they were grown in T11 culture supernatant (Fig. 3, lane 4). These data strongly suggested that production of SalA and production of SalA1 were autoregulated in S. salivarius 20P3 and S. pyogenes T11, respectively. To confirm this, S. salivarius 20P3 cells were grown to the mid-exponential phase in M17CS medium, harvested, washed, and suspended in fresh medium containing 0 to 5 pmol of purified SalA peptide per ml. After incubation of the cultures at 37°C for 4 h, mRNAs were extracted and subjected to Northern analysis. In the absence of exogenous SalA, no salA mRNA transcript was detected (Fig. 4, lane 1), but there was a dose-dependent increase in salA mRNA levels as the SalA concentration increased up to 5 pmol/ml (Fig. 4). Purified SalA peptide was also effective in inducing salA transcription in S. salivarius UB1309 salB (Fig. 6, lane 2).
SalA and SalA1 are intra- and interspecies signaling molecules.
To investigate the specificity of autoinduction by SalA and SalA1, medium swap experiments were carried out with the SalA1-producing organism S. pyogenes T11 and the non-SalA-producing organism S. salivarius UB1309 salB. In the latter strain, salA mRNA transcription was clearly induced by the S. pyogenes SalA1 peptide present in T11 culture medium (Fig. 6, lane 4), and the level was similar to that induced by the S. salivarius peptide SalA (Fig. 6, lane 3).
Since gene regulatory activities of other lantibiotics and peptide pheromones are mediated through two-component systems, it seemed likely that SalA and SalA1 function through the salKR gene products, sensor kinase and response regulator proteins. To test this hypothesis, we attempted to inactivate the salK gene in S. salivarius 20P3, but we were not successful. Attempts to inactivate the salY gene in S. salivarius 20P3 or the salK and salY genes in S. pyogenes SF370 also were unsuccessful, but we were able to obtain an insertion of pMU1022 in the salR gene of S. salivarius 20P3 (Fig. 1A). Strain UB1310 expressed salA mRNA at a level that was about 50% of the level present in wild-type 20P3 (data not shown). In deferred antagonism assays strain UB1310 was much less inhibitory to growth of S. salivarius UB1309 (Table 2), providing evidence that SalR played a role in expression of SalA.
The inhibitory activities of the various strains and their cross-immunity profiles are shown in Table 2. S. salivarius 20P3 produced the highest SalA inhibitor activity of all the strains, and growth of wild-type 20P3 cells was not inhibited by any of the other strains. S. pyogenes T11 had an inhibitory and immunity profile similar to that of S. salivarius 20P3, but it was weaker. By contrast, S. pyogenes SF370 and UB1309 and S. salivarius UB1308 did not produce inhibitory activities (Table 2). These strains were also sensitive to growth inhibition by SalA and SalA1 peptides produced by S. salivarius 20P3 and UB1310 salR and S. pyogenes T11. S. salivarius UB1310 salR, which exhibited a moderate level of inhibitory activity against S. pyogenes T11 and UB1308, was sensitive only to inhibition by S. salivarius 20P3 and S. pyogenes T11. Thus, streptococci that produce SalA or SalA1 peptides have increased immunity to both lantibiotics compared with SalA-negative or SalA1-negative streptococci.
DISCUSSION
It is now well established that gram-positive bacteria, especially lactic acid bacteria, produce an array of extracellular peptides with diverse functions. These molecules may act as highly specific intercellular signals for controlling competence development, mating responses, and virulence factor production in bacterial communities or may act as bacteriocin-like inhibitors of the growth of other neighboring species (10). Salivaricin A was one of the first bacteriocin-like inhibitors that were purified, sequenced, and characterized (34). This lantibiotic is produced by S. salivarius, and homologous SalA1 peptides are produced by M type 4 strains of S. pyogenes (37). In this study we extended our understanding of the production and function of this lantibiotic through comparative genetic analysis of the sal genetic loci in S. salivarius and S. pyogenes.
The genetic organization of the sal locus in strain 20P3 resembles that of other lantibiotic synthesis gene clusters (36). Transcriptional analyses suggested that there are at least two sal mRNA transcripts, a major (inducible) salA mRNA transcript approximately 200 bases long and a salABCTXYKR readthrough transcript. RT-PCR data are consistent with the proposal that the salBCTX genes are cotranscribed with salA, but the presence of an inverted repeat sequence downstream of the salA structural gene would be predicted to form a hairpin loop and attenuate transcriptional readthrough from salA into salBCTX. This would allow production of higher levels of SalA compared with the levels of SalBCTX, the modification and export machinery, and is a control feature found in a number of other lantibiotic synthesis gene operons (18, 23, 33). From the sequence analysis, we predicted that transcription of salYKR may also be initiated from a promoter immediately upstream of salY, but transcriptional start point mapping experiments would be necessary to confirm this. The significance of the different transcripts to lantibiotic production and control is not yet understood, but it is worth noting that among the transcripts from the L. lactis nisin lantibiotic gene locus, a transcript corresponding to the entire locus has been reported (31).
Comparison of the polypeptide sequences encoded by the sal gene loci in S. salivarius and S. pyogenes with the sequences in protein sequence databases allowed us to assign polypeptide functions. One unusual feature of the locus was the presence of two genes (salB and salC) encoding lantibiotic modification enzymes, since subclass AII lantibiotics with the characteristic Gly-Gly motif are usually modified by the product of a single gene (lanM) (36). The only gene for which a possible function of the product could not be inferred from a database match was salY. Since most lantibiotic gene clusters carry a self-immunity protein gene (36), it is possible that the 635-aa SalY protein is associated with self-protection against SalA. However, it is also possible that genetic determinants required for immunity are located elsewhere on the chromosome (2, 11).
Inactivation of the salB gene in S. salivarius 20P3 or S. pyogenes T11 led to a lantibiotic-negative (SalA− or SalA1−) phenotype, confirming that the salB gene product plays an essential role in lantibiotic production. However, inactivation of salB also resulted in abrogation of salA and salA1 transcription. This transcription was restored by exogenous addition of subinhibitory levels of SalA or SalA1 peptides, demonstrating that production of these lantibiotics is autoregulated, like production of nisin and subtilin (21, 23). Interestingly, salB mutants were also rendered sensitive to growth inhibition by SalA or SalA1 peptides, so lantibiotic production and immunity to the lantibiotic are coregulated. These results explain clearly why S. pyogenes strains which contain a deletion across the salBCT genes, such as SF370 (M type 1), are not producers of SalA1 lantibiotic and are also very sensitive to growth inhibition by SalA or SalA1 peptides.
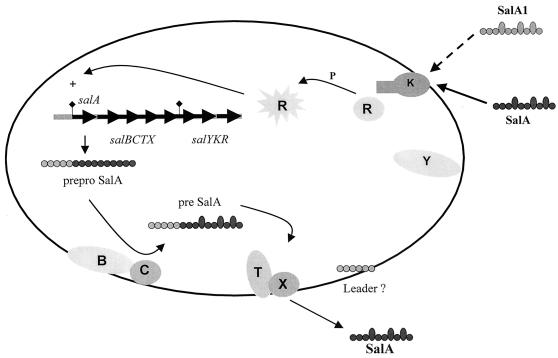
A model for autoregulation of SalA production in S. salivarius is presented in Fig. 7; this model is based on experimental evidence presented here and comparative data for other lantibiotic synthesis systems (7, 8, 20, 21, 24). Transcription of the salABCTX genes is initiated at a promoter upstream of salA. The primary translation products include the 48-aa precursor pro-SalA and the SalB, SalC, SalT, and SalX protein modification and transport machinery. Modification of the precursor peptide occurs with a membrane-associated SalB-SalC enzyme complex, and the membrane-integral SalT-SalX polypeptides then mediate translocation of SalA across the cytoplasmic membrane (Fig. 7). Cleavage of the precursor SalA leader peptide results from cysteine protease activity located in the N-terminal region of SalT, which generates extracellular mature SalA peptide. The fate of the precursor SalA leader peptide is not known.
FIG. 7.
Model for autoregulation of SalA production in S. salivarius and S. pyogenes. The product of the salA gene, preproSalA peptide, is modified by the membrane-associated SalBC polypeptides and then exported with leader peptide cleavage by the membrane-integral SalTX polypeptides. Extracellular lantibiotic peptide from S. salivarius (SalA) or from S. pyogenes (SalA1) is sensed at the cell surface, possibly by the two-component SalKR system, and transcription of the salA promoter is upregulated via a phosphorylated (P) regulatory protein (R). Only the bacteria that respond to extracellular SalA peptide by producing active SalA are immune to the inhibitory effects of the lantibiotic peptide, but the mechanism of immunity is unknown.
The gene clusters encoding production of nisin (12), streptococcin SA-FF22 (26), and subtilin (22) each contain two genes for a two-component (sensor-regulator) system. It has been shown that in the nisin and subtilin systems the mature inhibitor peptide is recognized by the sensor kinase (21, 23, 30). We propose that mature SalA peptide is sensed by the sensor histidine kinase of a two-component system (putatively SalKR), which leads to activation of the response regulator (SalR) and induction of transcription from the salA promoter (Fig. 7). Although we were unable to generate a salK mutant and therefore were unable to formally test the notion that SalK is the cognate sensor for SalA, we generated a salR mutant that contained reduced salA mRNA transcript levels and exhibited reduced SalA inhibitor production. This is consistent with the proposed role for SalR in autoregulation of SalA, but since salA mRNA transcription was not shut down in this mutant, other regulatory pathways must also be operative.
An interesting finding is that SalA production by one streptococcal species may be induced by sensing of the homologous peptide from another streptococcal species. Interspecies signaling between taxonomically diverse streptococcal species has not been demonstrated previously. Most peptide signaling molecules are specific for their cognate sensor-response systems in streptococci (6, 10), but the SalA peptide sensing system apparently does not discriminate between SalA and SalA1 (Fig. 2). Minor modifications to other inhibitor peptides, such as epilancin K7 (44), streptococcin SA-FF22 (19), and subtilin (3), are sufficient to affect their biological activities, but the two conservative changes in SalA1 do not significantly affect the inhibitor activity or profile. Interspecies community sensing could regulate relative population levels of streptococci. The ability of SalA1-producing S. pyogenes strains to respond to SalA from S. salivarius (and vice versa) could provide a selective mechanism for cocolonization of the mucosal epithelium by pathogen and commensal cell populations. For example, SalA1 produced by rapidly multiplying S. pyogenes cells might stimulate production of SalA by S. salivarius strains, leading to modulation of the number of S. pyogenes cells. In this way, the number of S. pyogenes cells may be self-limiting, thus promoting maintenance of the population in a carrier state. An alternative scenario is that sensing of SalA by S. pyogenes may influence expression of other genes, including genes related to virulence. In this regard it has recently been demonstrated that a bacteriocin-like peptide (BlpC) in S. pneumoniae induces a set of 16 genes following activation of its cognate two-component sensor-regulator system (6). We are currently investigating the possibility that SalR, like CsrR (17) or Mga (25) proteins, may have a global regulatory function in S. pyogenes and modulate production of virulence factors, such as streptolysin S (28).
In summary, here we describe the genetic locus encoding the polypeptides necessary for autoregulated production of SalA and SalA1 lantibiotics by S. salivarius and S. pyogenes. The SalA and SalA1 peptides act as inhibitors of sensitive streptococcal strains which do not themselves produce these peptides, but they are also sensed by other SalA-producing or SalA1-producing strains and there is concomitant induction of lantibiotic synthesis. It could be envisaged, therefore, that SalA production and SalA1 production have a role in modulating the coexistence of streptococcal populations in vivo, where it is becoming apparent that signaling peptides are likely to have a major influence on oral microbial community structure.
ACKNOWLEDGMENTS
We thank Nancy Ragland for expert technical assistance, K. Ross and K. Dierksen for helpful discussions, R. Burne for advice on electroporation, A. Podbielski for kindly supplying plasmids, J. Ferretti for providing strains, and N. Jakubovics for help with preparation of the manuscript. We gratefully acknowledge the BLAST search facilities at the National Center for Biotechnology Information (National Library of Medicine, Washington, D.C.) and the University of Oklahoma Streptococcal (GAS) Genome Sequencing Project funded by a USPHS/NIH grant to B. A. Roe, S. P. Linn, L. Song, X. Yuan, S. Clifton, R. E. McLaughlin, M. McShan, and J. Ferretti.
This work was supported by grant UO0605 from the Marsden Fund, Royal Society of New Zealand.
REFERENCES
- 1.Bisno A L. Group A streptococcal infections and acute rheumatic fever. N Engl J Med. 1991;325:783–793. doi: 10.1056/NEJM199109123251106. [DOI] [PubMed] [Google Scholar]
- 2.Brurberg M B, Nes I F, Eijsink V G H. Pheromone-induced production of antimicrobial peptides in Lactobacillus. Mol Microbiol. 1997;26:347–360. doi: 10.1046/j.1365-2958.1997.5821951.x. [DOI] [PubMed] [Google Scholar]
- 3.Chan W C, Bycroft B W, Leyland M L, Lian L Y, Roberts G C. A novel post-translational modification of the peptide antibiotic subtilin: isolation and characterization of a natural variant from Bacillus subtilis ATCC 6633. Biochem J. 1993;291:23–27. doi: 10.1042/bj2910023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y Y, Weaver C A, Mendelsohn D R, Burne R A. Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J Bacteriol. 1998;180:5769–5775. doi: 10.1128/jb.180.21.5769-5775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Saizieu A, Gardes C, Flint N, Wagner C, Kamber M, Mitchell T J, Keck W, Amrein K E, Lange R. Microarray-based indentification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J Bacteriol. 2000;182:4696–4703. doi: 10.1128/jb.182.17.4696-4703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vos W M, Jung G, Sahl H-G. Appendix: definitions and nomenclature of lantibiotics. In: Jung G, Sahl H-G, editors. Nisin and novel lantibiotics. Leiden, The Netherlands: Escom Publishers; 1991. pp. 457–464. [Google Scholar]
- 8.de Vos W M, Kuipers O P, van der Meer J R, Siezen R J. Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by gram positive bacteria. Mol Microbiol. 1995;17:427–437. doi: 10.1111/j.1365-2958.1995.mmi_17030427.x. [DOI] [PubMed] [Google Scholar]
- 9.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunny G M, Leonard B A B. Cell-cell communication in gram-positive bacteria. Annu Rev Microbiol. 1997;51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 11.Eijsink V G H, Skeie M, Middelhove H, Brurberg M B, Nes I F. Comparative studies of class IIa bacteriocins of lactic acid bacteria. Appl Environ Microbiol. 1998;64:3275–3281. doi: 10.1128/aem.64.9.3275-3281.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelke G, Gutowski-Eckel Z, Hammelmann M, Entian K-D. Biosynthesis of the lantibiotic nisin: genomic organization and membrane localization of the NisB protein. Appl Environ Microbiol. 1992;58:3730–3743. doi: 10.1128/aem.58.11.3730-3743.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelke G, Gutowski-Eckel Z, Kiesau P, Siegers K, Hammelmann M, Entian K-D. Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl Environ Microbiol. 1994;60:814–825. doi: 10.1128/aem.60.3.814-825.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan D, Jessee J, Bloom F R. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
- 15.Havarstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havarstein L S, Diep D B, Nes I F. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates comcomitant with export. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 17.Heath A, Di Rita V J, Barg N L, Engleberg N C. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyogenic extoxin B. Infect Immun. 1999;67:5298–5305. doi: 10.1128/iai.67.10.5298-5305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hynes W L, Ferretti J J, Tagg J R. Cloning of the gene encoding Streptococcin A-FF22, a novel lantibiotic produced by Streptococcus pyogenes, and determination of its nucleotide sequence. Appl Environ Microbiol. 1993;59:1969–1971. doi: 10.1128/aem.59.6.1969-1971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jack R W, Tagg J R. Isolation and partial structure of streptococcin SA-FF22. In: Jung G, Sahl H-G, editors. Nisin and novel lantibiotics. Leiden, The Netherlands: Escom Publishers; 1991. pp. 171–179. [Google Scholar]
- 20.Jung G. Lantibiotics: a survey. In: Jung G, Sahl H-G, editors. Nisin and novel lantibiotics. Leiden, The Netherlands: Escom Publishers; 1991. pp. 1–34. [Google Scholar]
- 21.Kleerebezem M, de Vos W M, Kuipers O P. The lantibiotics nisin and subtilin act as extracellular regulators of their own biosynthesis. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: American Society for Microbiology; 1999. pp. 159–174. [Google Scholar]
- 22.Klein C, Kaletta C, Entian K-D. Biosynthesis of the lantibiotic subtilin is regulated by a histidine kinase/response regulator system. Appl Environ Microbiol. 1993;59:296–303. doi: 10.1128/aem.59.1.296-303.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuipers O P, Beerthuyzen M M, de Reuyter P G G A, Luesink E J, de Vos W M. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem. 1995;270:27299–27304. doi: 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- 24.Louw M E, Reid S J, James D M, Watson T G. Cloning and sequencing the degS-degU operon from an alkalophilic Bacillus brevis. Appl Microbiol Biotechnol. 1994;42:78–84. doi: 10.1007/BF00170228. [DOI] [PubMed] [Google Scholar]
- 25.McIver K, Thurman A S, Scott J R. Regulation of mga transcription in the group A streptococcus: specific binding of Mga within its own promoter and evidence for a negative regulator. J Bacteriol. 1999;181:5373–5383. doi: 10.1128/jb.181.17.5373-5383.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLaughlin R E, Ferretti J J, Hynes W L. Nucleotide sequence of the streptococcin A-FF22 lantibiotic regulon: model for the production of the lantibiotic SA-FF22 by strain of Streptococcus pyogenes. FEMS Microbiol Lett. 1999;175:171–177. doi: 10.1111/j.1574-6968.1999.tb13616.x. [DOI] [PubMed] [Google Scholar]
- 27.Meyer C, Bierbaum G, Heidrich C, Reis M, Suling J, Iglesias-Wind M I, Kempter C, Molitor E, Sahl H-G. Nucleotide sequence of the lantibiotic Pep5 biosynthetic gene cluster and functional analysis of PepP and PepC. Evidence for a role of PepC in thiether formation. Eur J Biochem. 1995;232:478–489. doi: 10.1111/j.1432-1033.1995.tb20834.x. [DOI] [PubMed] [Google Scholar]
- 28.Nizet V, Beall B, Bast D J, Datta V, Kilburn L, Low D E, de Azavedo J C S. Genetic locus for streptolysin S production by group A streptococcus. Infect Immun. 2000;68:4245–4254. doi: 10.1128/iai.68.7.4245-4254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Podbielski A, Spellberg B, Woschnik M, Pohl B, Lüticken R. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS) Gene. 1996;177:137–147. doi: 10.1016/0378-1119(96)84178-3. [DOI] [PubMed] [Google Scholar]
- 30.Ra S R, Qiao M, Immonen T, Pujana I, Saris P E J. Genes responsible for nisin synthesis, regulation and immunity form a regulon of two operons and are induced by nisin in Lactococcus lactis N8. Microbiology. 1996;142:1281–1288. doi: 10.1099/13500872-142-5-1281. [DOI] [PubMed] [Google Scholar]
- 31.Ra S R, Beerthuyzen M M, de Vos W M, Saris P E J, Kuipers O P. Effects of gene disruptions in the nisin gene cluster of Lactococcus lactis on nisin production and producer immunity. Microbiology. 1999;145:1227–1233. doi: 10.1099/13500872-145-5-1227. [DOI] [PubMed] [Google Scholar]
- 32.Rince A, Dufour A, Le Pogam S, Thuault D, Bourgeois C M, Le Pennec J. Cloning, expression, and nucleotide sequence of genes involved in production of lactococcin DR, a bacteriocin from Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1994;60:1652–1657. doi: 10.1128/aem.60.5.1652-1657.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rince A, Dufour A, Uguen P, Le Pennec J, Haras D. Characterization of the lacticin 481 operon: the Lactococcus lactis genes lctF, lctE, and lctG encode a putative ABC transporter involved in bacteriocin immunity. Appl Environ Microbiol. 1997;63:4252–4260. doi: 10.1128/aem.63.11.4252-4260.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross K F, Ronson C W, Tagg J R. Isolation and characterization of the lantibiotic salivaricin A and its structural gene salA from Streptococcus salivarius 20P3. Appl Environ Microbiol. 1993;59:2014–2021. doi: 10.1128/aem.59.7.2014-2021.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salmond G P, Bycroft B W, Stewart G S, Williams P. The bacterial enigma: cracking the code of cell-cell communication. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 36.Siezen R J, Kuipers O P, De Vos W M. Comparison of lantibiotic gene clusters and encoded proteins. Antonie Leeuwenhoek. 1996;69:171–184. doi: 10.1007/BF00399422. [DOI] [PubMed] [Google Scholar]
- 37.Simpson W J, Ragland N L, Ronson C W, Tagg J R. A lantibiotic gene family widely distributed in Streptococcus salivarius and Streptococcus pyogenes. Dev Biol Stand. 1995;85:639–643. . Karger, Basel. [PubMed] [Google Scholar]
- 38.Skaugen M, Nissen-Meyer J, Jung G, Stevanovic S, Sletten K, Abildgaard C I M, Nes I F. In vivo conversion of l-serine to d-alanine in a ribosomally synthesised polypeptide. J Biol Chem. 1994;269:27183–27185. [PubMed] [Google Scholar]
- 39.Smith D J, Anderson J M, King W F, van Houte J, Taubman M A. Oral streptococcal colonization of infants. Oral Microbiol Immunol. 1993;8:1–4. doi: 10.1111/j.1399-302x.1993.tb00535.x. [DOI] [PubMed] [Google Scholar]
- 40.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suvrov A N, Ferretti J J. Physical and genetic map of an M type 1 strain of Streptococcus pyogenes. J Bacteriol. 1996;178:5546–5549. doi: 10.1128/jb.178.18.5546-5549.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tagg J R, Bannister L V. “Fingerprinting” β haemolytic streptococci by their production of and sensitivity to bacteriocin-like inhibitors. J Med Microbiol. 1979;12:379–411. doi: 10.1099/00222615-12-4-397. [DOI] [PubMed] [Google Scholar]
- 43.Upton M, Carter P E, Morgan M, Edwards G F S, Pennington T H. Clonal structure of invasive Streptococcus pyogenes strains in Northern Scotland. Epidemiol Infect. 1995;115:231–241. doi: 10.1017/s0950268800058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van de Kamp M, Horstink L M, van den Hooven H W, Konings R N, Hilbers C W, Frey A, Sahl H-G, Metzger J W, van de Ven F J. Sequence analysis by NMR spectroscopy of the peptide lantibiotic epilancin K7 from Staphylococcus epidermidis. Eur J Biochem. 1995;227:757–771. doi: 10.1111/j.1432-1033.1995.tb20199.x. [DOI] [PubMed] [Google Scholar]
- 45.van der Meer J R, Polman J, Beerthuyzen M M, Siezen R J, Kuipers O P, De Vos W M. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J Bacteriol. 1993;175:2578–2588. doi: 10.1128/jb.175.9.2578-2588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker J E, Sarste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinase and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weinrauch Y, Penchev R, Dubnau E, Smith I, Dubnau D. A Bacillus subtilis regulatory gene product for genetic competence and sporulation resembles sensor protein members of the bacterial two-component signal-transduction systems. Genes Dev. 1990;4:860–872. doi: 10.1101/gad.4.5.860. [DOI] [PubMed] [Google Scholar]