Abstract
Background
Myxopapillary ependymoma (MPE) is a heterogeneous disease regarding histopathology and outcome. The underlying molecular biology is poorly understood, and markers that reliably predict the patients’ clinical course are unknown.
Methods
We assembled a cohort of 185 tumors classified as MPE based on DNA methylation. Methylation patterns, copy number profiles, and MGMT promoter methylation were analyzed for all tumors, 106 tumors were evaluated histomorphologically, and RNA sequencing was performed for 37 cases. Based on methylation profiling, we defined two subtypes MPE-A and MPE-B, and explored associations with epidemiological, clinical, pathological, and molecular characteristics of these tumors.
Results
MPE-A occurred at a median age of 27 years and were enriched with tumors demonstrating papillary morphology and MGMT promoter hypermethylation. Half of these tumors could not be totally resected, and 85% relapsed within 10 years. Copy number alterations were more common in MPE-A. RNA sequencing revealed an enrichment for extracellular matrix and immune system-related signatures in MPE-A. MPE-B occurred at a median age of 45 years and included many tumors with a histological diagnosis of WHO grade II and tanycytic morphology. Patients within this subtype had a significantly better outcome with a relapse rate of 33% in 10 years (P = 3.4e-06).
Conclusions
We unraveled the morphological and clinical heterogeneity of MPE by identifying two molecularly distinct subtypes. These subtypes significantly differed in progression-free survival and will likely need different protocols for surveillance and treatment.
Keywords: DNA methylation, histology, myxopapillary ependymoma, outcome, RNA sequencing
Key Points.
Myxopapillary ependymomas can be split into two biologically distinct groups (MPE-A and MPE-B).
MPE-A occur at younger age and at lower parts of the spinal cord.
MPE-A have a shorter PFS and need closer surveillance.
Importance of the Study.
Myxopapillary ependymoma (MPE) is a clinically heterogenous disease. While many patients show a benign course with excellent long-term outcome, relapses occur in around half of the patients and reliable prognostic factors are missing. To understand the underlying biology and to establish prognostic factors, we analyzed 185 tumors classified as MPE based on DNA methylation. The methylation profiles were related to clinicopathological and RNA sequencing data and allowed to define two distinct subtypes MPE-A and MPE-B. MPE-A occurred in younger patients and were enriched with tumors demonstrating papillary morphology and MGMT promoter hypermethylation. MPE-B occurred in older patients and included many tumors with a histological diagnosis of WHO grade II and tanycytic morphology. The outcome was significantly worse in MPE-A (10-year relapse rate 85% vs. 33%, P = 3.4e-06). The identified subtypes explain the disparate clinical behavior of MPE and may be useful for patient stratification and planning of future clinical trials.
Myxopapillary ependymomas (MPE) are tumors of glial origin almost exclusively located in the lower part of the spinal cord.1 They account for up to 20% of ependymal tumors, and the reported incidence is around 1.00 per million person-years.2,3 MPE occur in all age groups with peak incidences in early adulthood and the middle ages.2 The incidence is increased in men compared to women with a ratio of approximately 1.4.2,4
MPE is a histopathologically defined type of ependymoma in the 2021 WHO classification, characterized by a radial arrangement of tumor cells around hyalinized fibrovascular cores in a papillary fashion, with accumulations of myxoid material, and with low mitotic activity.1 The histopathology is heterogeneous, comprising predominantly papillary, myxoid, or tanycytic patterns.
Molecular analyses of ependymomas based on global DNA methylation led to the definition of 10 types with distinct anatomical localizations, molecular alterations, and clinical characteristics, including MPE.5–7 Recent studies on smaller series comparing morphologically and molecularly diagnosed ependymal tumors showed that almost all histopathologically defined MPE belonged to the molecular MPE subgroup.8,9 However, up to half of the tumors with a molecular type of MPE were morphologically diagnosed as WHO grade II spinal ependymoma.8 As the clinical utility of a molecular classification of MPE is still unclear, the WHO 2021 classification remains solely based on morphological criteria. Further, the clinical relevance of the histopathological patterns is unknown and therefore also not included in the current classification.1
MPE has been regarded as a benign WHO grade I tumor. However, a 10-year progression-free survival (PFS) of only 60-80% as well as a distinct proneness towards metastatic behavior led to reassignment as grade 2 tumor in the WHO 2021 brain tumor classification.1,4,10–12
Surgical resection is the gold-standard treatment, and gross total resection (GTR) was consistently associated with a favorable prognosis.4,11,13 The role of adjuvant radiotherapy is less clear. It was strongly and independently associated with an improved PFS in a study including 183 patients,4 but others report no significant positive effect of adjuvant radiotherapy14 or high PFS rates in a cohort not treated with radiotherapy.11 Although there are currently no established predictive factors for radiotherapy, younger patients seem to profit more from adjuvant irradiation.13 There are no established chemotherapeutic regimens neither in pediatric nor in adult patients.15,16
Knowledge about prognostic and predictive factors in MPE is limited. A sacral tumor location was significantly associated with an unfavorable prognosis in a French study with 101 patients.11 However, well-established molecular markers of prognostic or predictive value are missing. DNA methylation and gene expression-based analyses were limited to small cohorts of less than 30 MPE patients and could not establish further molecular subtypes and predictive or prognostic factors.5,8,17 Further, the relation between histology and methylation profile remains unclear.1 As the clinical course of MPE is heterogeneous and as it is currently poorly understood why some patients have a high risk for tumor recurrence, whereas others have an excellent long-term outcome, a better understanding of the underlying biological mechanisms and the establishment of prognostic or predictive factors is needed.
Towards those ends, we characterized 185 tumors classified as MPE based on DNA methylation. The methylation patterns were related to transcriptomic data as well as clinical and histopathological features to establish a more precise tumor classification and optimize prognostic and predictive patient stratification.
Material and Methods
Patient Samples
The study cohort consisted of 112 unpublished and 73 previously published cases of MPE. DNA methylation profiling was performed for all cases, and samples with the highest score for the class “ependymoma, myxopapillary” according to the Heidelberg brain tumor classifier v11b6 were included.7 Raw methylation data and clinical data were obtained from Gene Expression Omnibus (GEO, RRID:SCR_005012) and the corresponding authors for the published cases (Supplementary Table S1).5,7,8 For the remaining cases, clinical samples and data were collected after consent was obtained from each patient according to protocols approved by the institutional review boards of the participating institutions (Supplementary Table S1). For the comparison of MPE with molecular types of ependymoma (Figure 1A), raw methylation data were obtained from Pajtler et al. (n = 462) and Raffeld et al. (n = 8).5,6 Further reference cases of spinal gliomas (Supplementary Figure 1, n = 34) were obtained from the archive of the Institute of Neuropathology of the University Medical Center Hamburg-Eppendorf.
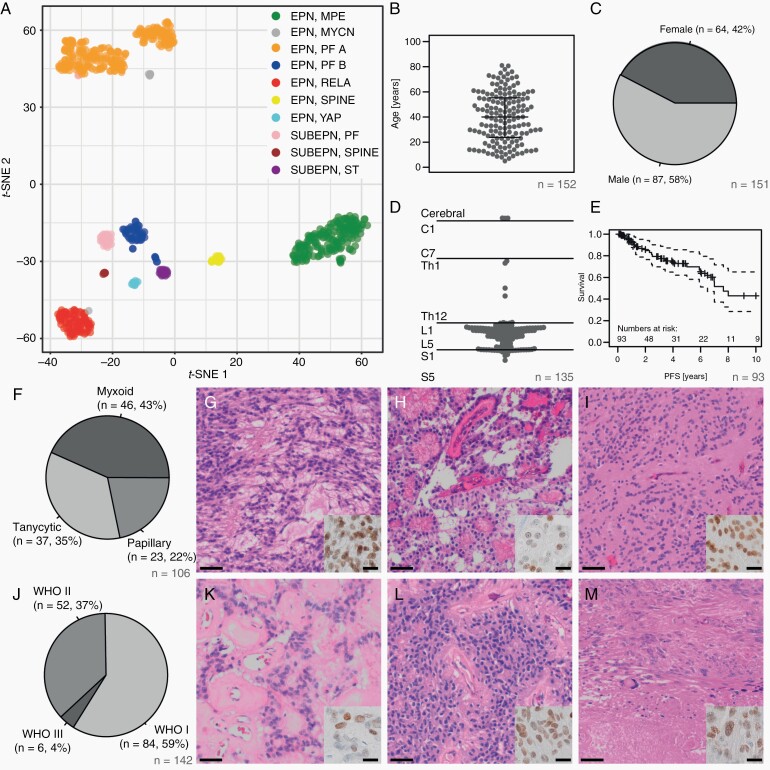
Fig. 1.
Clinical and histopathological features of myxopapillary ependymoma. (A) T-SNE analysis of the 10 molecular ependymoma types based on methylation profiling. (B) Age distribution of the MPE samples (beeswarm plot). Lines indicate the median, upper and lower quartile. (C) Sex distribution in the MPE cohort. (D) Schematic representation of the MPE tumor localization along the neural axis. (E) Progression-free survival in the MPE cohort. (F-I) Distribution of histological MPE subtypes with typical patterns: myxoid (G), papillary (H), and tanycytic/solid (I) (scale bar = 50 µm). Inset: immunohistochemical staining for HOXB13 (scale bar = 10 µm). (J–M) Distribution of WHO grades in the study cohort with examples of WHO grade I (K), II (L), and III (M) tumors (scale bar = 50 µm). Inset: immunohistochemical staining for HOXB13 (scale bar = 10 µm).
Histology and Immunohistochemistry
Tumors were histomorphologically reassessed by at least two neuropathologists based on H&E-stained tumor sections and categorized by their predominant patterns as being either papillary, tanycytic, or myxoid. Immunohistochemistry was performed using antibodies against HOXB13 (Santa Cruz Biotechnology, sc-28333, RRID:AB_627744, 1:100 dilution) according to standard procedures (Roche, Ventana Benchmark systems).
DNA Methylation Profiling
DNA was isolated using the ReliaPrep™ FFPE gDNA Miniprep System (Promega) according to the manufacturer’s instructions. About 100-500 ng DNA was used for bisulfite conversion by the EZ DNA Methylation Kit (Zymo Research). The DNA Clean & Concentrator-5 (Zymo Research) and the Infinium HD FFPE DNA Restore Kit (Illumina) were employed to clean and restore the converted DNA. Finally, Infinium BeadChip arrays (Illumina) were used to quantify the methylation status on an iScan (Illumina).
RNA Sequencing
RNA was isolated using the Maxwell® RSC RNA FFPE Kit (Promega) and integrity was analyzed with the RNA 6000 Nano Chip on a 2100 Bioanalyzer (Agilent Technologies). Ribosomal RNA was depleted with the RiboCop rRNA Depletion Kit (Lexogen) followed by RNA sequencing library generation using the CORALL Total RNA-Seq Library Prep Kit (Lexogen). Concentrations were measured with a Qubit 2.0 Fluorometer (Thermo Fisher Scientific) and fragment lengths distribution was analyzed with the DNA High Sensitivity Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies). Samples were normalized to 2 nM and pooled at equimolar concentrations. The library pool was sequenced on the NextSeq500 (Illumina) with 1 × 75 bp. Reads were processed using fastp (v.0.21.0) to remove sequences of low quality and artificial origin and aligned to the human reference assembly (GRCh38.103) using STAR (RRID:SCR_004463, v2.7.8a) in two-pass mode.
Data Processing and Statistics
Data processing was performed in R. Methylation data of MPE (n = 185) were processed using minfi and noob normalization. CpG sites associated with SNPs, sex chromosomes, cross-reactive sites, and sites that were not on the 450k or the EPIC BeadChip were excluded as described previously.7 Differentially methylated sites between EPIC and 450k BeadChips (absolute difference of mean beta > 0.2, two-sided t-test P-value < .05 after Bonferroni correction) were excluded to avoid batch effects (n = 1711), resulting in a dataset of 426 702 sites. The 10 000 most variable sites were subsequently used. The k-means algorithm was used to define methylation-based clusters and the optimal number of clusters was computed with the silhouette method (factoextra). Consensus clustering was performed with ConsensusClusterPlus (RRID:SCR_016954) using default parameters. T-SNE plots were generated with Rtsne (RRID:SCR_016342), initial_dims=20, and perplexity=30. ComplexHeatmap (RRID:SCR_017270) was used for heatmap generation with the previously defined k-means clusters, average linkage, and Pearson correlation. Conumee and GISTIC2 (RRID:SCR_000151) were used for the segmentation of arm-level methylation-based copy number profiles using default parameters (Supplementary Table S2). Methylation-based estimates of tumor-infiltrating lymphocytes (TILs) were computed with DIMEImmune.18 Global methylation of a tumor sample was defined as the mean of all beta values. MGMT promoter methylation status was assessed from methylation data as described previously.7,19 The data used for t-SNE analysis of all ependymomas (Figure 1A) were processed analogously and the plot was generated using the 10 000 most variable sites, initial dims = 50, and perplexity = 30.
For RNAseq analysis (n = 37), data normalization was performed using DESeq220 (RRID:SCR_000154). Sex-associated genes were removed (n = 44) to avoid batch effects. The data were matched to HUGO symbols (n = 18 219). The variance stabilization transformation (vst) as implemented in DESeq2 was used to normalize data for the unsupervised analysis. ComplexHeatmap (RRID:SCR_017270) was used for heatmap generation using the 2000 most variable genes and k-means clustering for the samples with average linkage and Pearson correlation. The optimal number of clusters (k = 2) was determined with the silhouette method (factoextra). Highly correlating gene clusters (R > 0.3, defined by cutting the row dendrogram with cutree) of at least 200 genes were annotated and subjected to enrichment analysis using Fisher’s exact test (one-sided) with the Reactome (RRID:SCR_003485) pathways from fgsea (RRID:SCR_020938). Differential expression between subtypes was analyzed with DESeq2. Genes were considered differentially expressed if the Benjamini-Hochberg corrected P-value (P-BH) was less than 0.05 and the absolute log2-foldchange was greater than 1. Gene set enrichment analysis was performed with fgsea and the Reactome database. Pathways with P-BH for enrichment < .05 were considered significantly enriched (n = 24). Redundant pathways were removed using collapsePathways (n = 8). Signatures of immune cell populations and functional immune signatures were computed as described previously.21,22 Metaprogram expression of distinct tumor cell subpopulations was computed by averaging the marker genes defined by Gojo et al.23 Fusions in RNAseq data were analyzed with Arriba24 using default parameters and GRCh38.103.
Differences between clinical and biological characteristics of MPE subtypes were assessed with the Wilcoxon test for ordinal/metric variables and Fisher’s exact test for categorical variables (two-sided). Survival analyses were performed with the Kaplan-Meier method, the coxph function (multivariate), and the log-rank test (univariate) from survival (RRID:SCR_021137). P-values < .05 were considered statistically significant.
Data Availability
The methylation data are deposited at the GEO repository GSE184900. Processed RNAseq data are available in Supplementary Table S3.
Results
Study Dataset
We retrospectively assembled a cohort of 185 MPE from over 15 institutions (Supplementary Table S1). The inclusion criterion was methylation-based classification with the highest score for MPE using the Heidelberg brain tumor classifier.7 Of the classified samples, 175 had a clear match with a score > 0.9, and 10 samples had lower scores (Supplementary Table S1). In a t-SNE analysis including the 10 currently defined types of ependymoma, all MPE grouped together and clearly separated from the remaining samples (Figure 1A). A grouping of the 10 cases with lower scores was not detectable. Histology was available for n = 106/185 cases (57.3%, Supplementary Table S1).
MPE is a Clinically and Histopathologically Heterogeneous Type of Ependymoma
MPE showed a homogenous age distribution in patients between 5 and 81 years (median 40.1 years, Figure 1B, Supplementary Table S1). Males were more frequently afflicted (58%, Figure 1C). Most tumors occurred in the lower part of the spinal cord, and there were few thoracic and cerebral cases. The most common locations were around the conus medullaris and the cauda equina (Figure 1D). PFS was relatively low in our cohort with a 5- and 10-year PFS of 69.8% (95% CI: 58.3–83.7%) and 43% (95% CI: 28.4–65.0%), respectively (Figure 1E). There were three main histological features with myxoid, tanycytic, and papillary histology as predominant growth pattern in 43%, 35%, and 22% of the cases with available material, respectively (Figure 1F–I). WHO grading according to the 2016 WHO classification was available for 142 tumors. Of these, 59%, 37%, and 4% were graded as WHO I, II, and III, respectively (Figure 1J–M). HOXB13, which has been reported to be expressed in spinal ependymal tumors,25 showed a specific nuclear expression in 75/77 analyzed MPE (97.4%), whereas all other analyzed spinal gliomas were negative (insets in Figure 1G–I and K–M, Supplementary Figure S1).
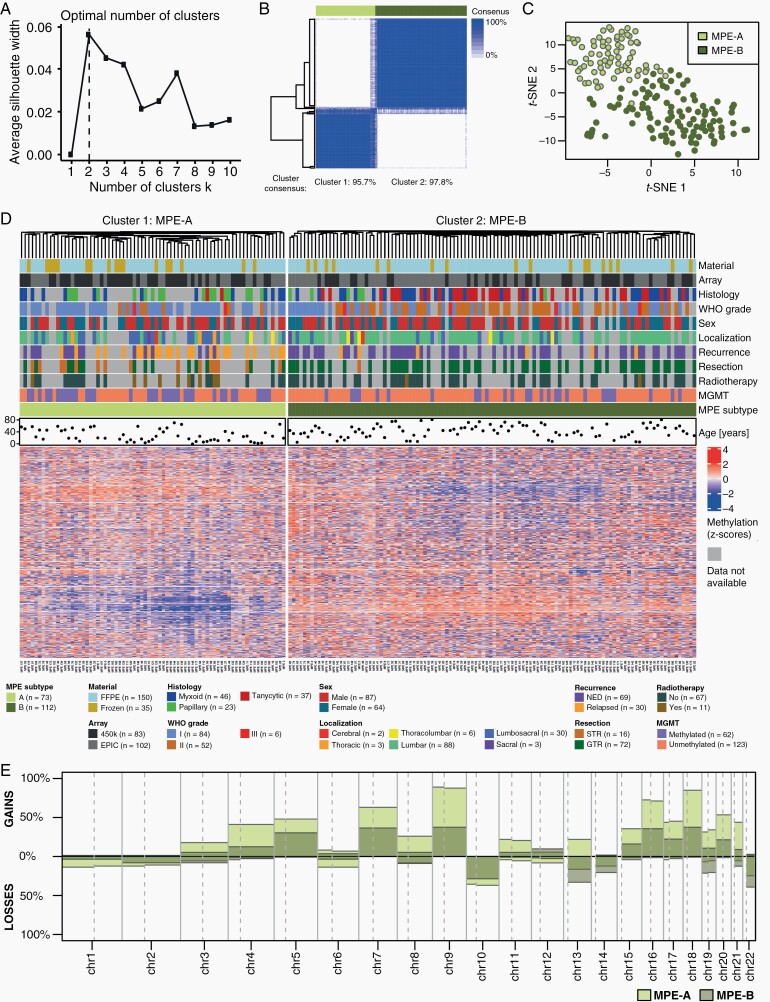
DNA Methylation Profiling Defines Two Molecular Subtypes
Methylation profiling was further evaluated for all 185 MPE attempting to define tumor subtypes and to relate them to clinicopathological variables. K-means clustering was used to define methylation-based subtypes, and the silhouette method determined an optimal number of two clusters (Figure 2A). Consensus clustering showed a consensus of the defined clusters of over 95% (Figure 2B). In the following, we define these clusters as tumor subtypes MPE-A and MPE-B. In a t-SNE analysis, MPE-A and MPE-B formed two rather distinct groups, although there was no clear separation of the two subtypes (Figure 2C). The heatmap of methylation data and clinicopathological covariates showed different methylation patterns in MPE-A and MPE-B (Figure 2D). No visible batch effects associated with differential material types (Frozen/FFPE) and the used methylation array (450k/EPIC) were detected. Histology, WHO grade, tumor localization, resection outcome, adjuvant radiotherapy, the MGMT promoter status, and age are depicted case by case in Figure 2D and will also be discussed in detail later in this manuscript.
Fig. 2.
Methylation analysis of myxopapillary ependymoma. (A) Determination of the optimal number of methylation-based k-means clusters using the silhouette method. (B) Consensus clustering for the k-means algorithm. The column sidebar shows the k-means cluster for k = 2 with 10 000 CpG sites optimal in (A). (C) T-SNE analysis of methylation data. The colors represent the k-means cluster for k = 2 with 10 000, CpG sites. (D) Cluster analysis with heatmap of methylation data and clinicopathological variables of MPE (n = 185). (E) Overview of chromosome arm-wise copy number alterations in methylation-based subtypes of MPE.
MPE-A Have More Copy Number Alterations Than MPE-B
Arm-wise copy number alterations were computed based on methylation data. The overall pattern was similar in MPE-A and MPE-B, but alterations were more common in MPE-A (Figure 2E, Supplementary Table S2). MPE-A had a median number of 17 arm-wise alterations, whereas MPE-B only had a median of 6 alterations (P = 3.41e-10). Gains were more common than losses. The most frequent gains were on chromosomes 9, 16, and 18 affecting over 70% of MPE-A and 35–40% of MPE-B. The most frequent loss affecting chromosome 22q was more common in MPE-B (39%) than in MPE-A (25%). Further, frequent losses included chromosome 10 (approx. 36% in MPE-A, 29% in MPE-B) and chromosome 13q (16% in MPE-A, 33% in MPE-B).
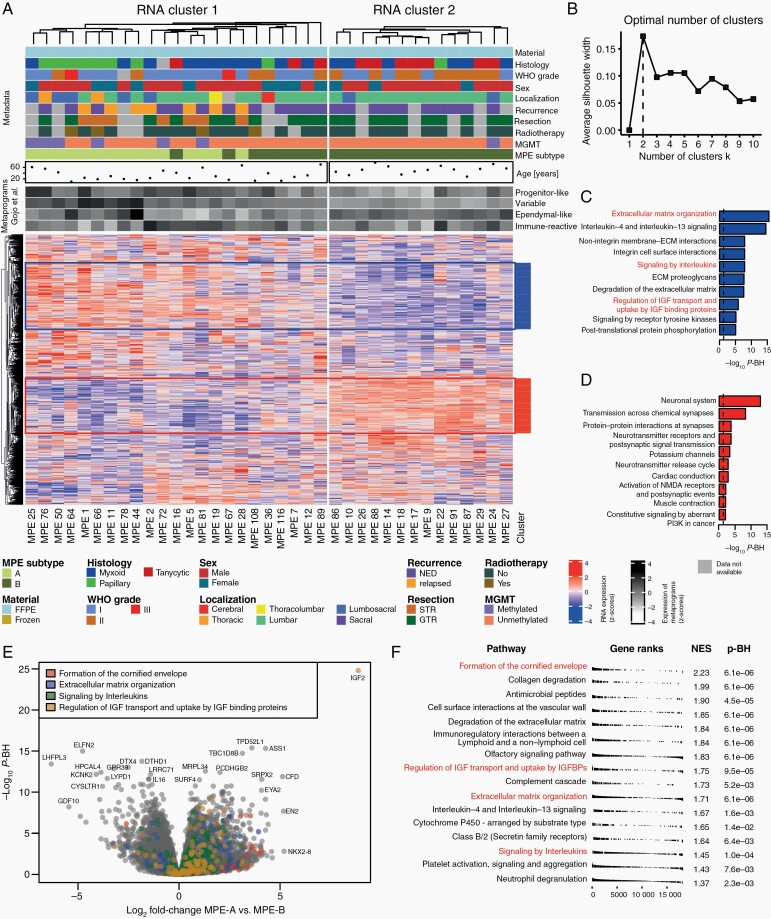
RNA Sequencing Reveals Differentially Expressed Genes in MPE Subtypes
RNA sequencing was performed for a subset of 37 tumors with a sufficient amount of high-quality RNA available (Supplementary Table S3). Heatmap analysis was performed using the k-means algorithm (Figure 3A), and the optimal number of clusters was determined by the silhouette method (Figure 3B). As for methylation profiling, there was an optimal number of two clusters. There was a reasonable agreement between RNA-based clusters and methylation-defined subtypes. RNA cluster 2 was solely composed of MPE-B tumors, and RNA cluster 1 contained all MPE-A tumors, accounting for 65% of the samples in this cluster. All recurrent tumors belonged to RNA cluster 1, whereas most tumors with tanycytic histology and WHO grade II belonged to RNA cluster 2. Gene set enrichment analysis was performed on two large and highly correlating gene clusters (Figure 3A,C,D). The most significantly enriched categories for the annotated clusters were “Neuronal System” (P-BH = 1.1e-13) and “Extracellular matrix organization” (P-BH = 3.3e-16). Differential expression analysis and enrichment analysis of differentially expressed genes between MPE-A and B (Supplementary Tables 4 and 5) identified 1,936 significantly differentially expressed genes (Figure 3E,F). The significantly enriched gene sets included “Formation of the cornified envelope” (P-BH = 6.1e-6), “Regulation of insulin-like growth factor (IGF) transport and uptake of IGF binding protein” (P-BH = 9.5e-5), “Extracellular matrix organization” (P-BH = 6.1e-6), and “Signaling by Interleukins” (P-BH = 1e-4). Further, we analyzed the association of MPE subtypes with four recently described metaprograms associated with distinct tumor cell subpopulations in a single-cell RNA sequencing study by Gojo et al. The primitive neural stem cell-like as well as the variable metaprogram were significantly higher expressed in MPE-A as compared to MPE-B (P = .003 and P = 5.4e-7, respectively). The benign ependymal-like metaprogram was higher expressed in MPE-B, but the results were not statistically significant (Figure 3A, Supplementary Figure S3). Further, we used characteristic signatures of immune cells and immunologic processes to study immunological differences between MPE-A and MPE-B. Overall, all studied immune cell populations and all functional signatures were higher expressed in MPE-A as compared to MPE-B, with 16/19 studied signatures significantly differentially expressed (all P < .05). This indicates a more prominent immune infiltration in MPE-A (Supplementary Figure S4). Finally, analysis of gene fusions detected 90 fusions (0–9 per sample, Supplementary Table S6). However, there was only a single gene with a high confidence fusion (TEKT1- RP11-530N7.3/AC027763.2 in MPE 27), and there were no recurrent fusions in our data.
Fig. 3.
RNA sequencing analysis of myxopapillary ependymoma. (A) Heatmap analysis of RNA sequencing data, clinicopathological variables and expression of metaprograms defined by Gojo et al.23 in 37 MPE. The MPE subtype is based on k-means clustering of methylation data. The optimal number of clusters was determined by the silhouette method (B). Highly correlating gene clusters (R > 0.3, defined by cutting the row dendrogram) with at least 200 genes are annotated and subjected to gene set enrichment analysis (C–D). (E) Volcano plot of differential expression analysis with annotation of four gene sets. (F) Results of gene set enrichment analysis of differentially expressed genes between MPE subtypes.
MPE Subtypes Have Distinct Clinical, Histological, and Biological Features
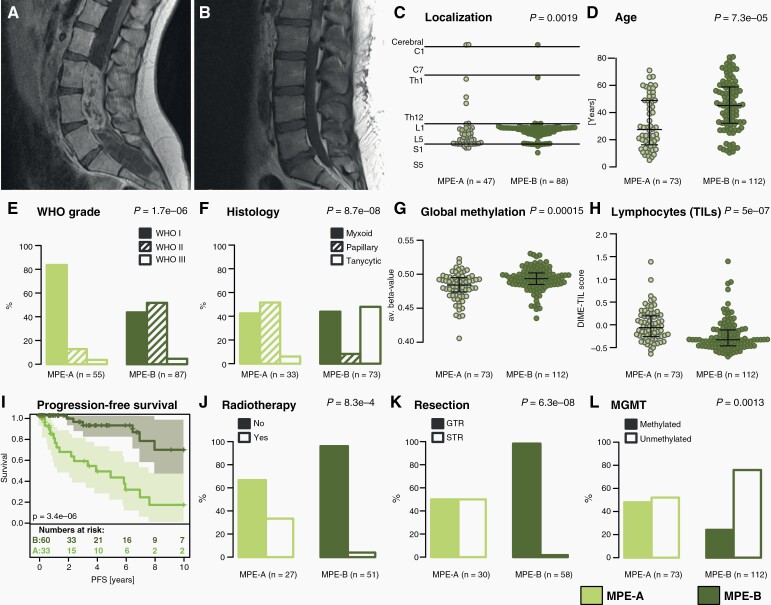
MPE subtypes were correlated with clinicopathological variables to assess their clinical impact. MPE-A and MPE-B had different typical tumor localizations. Most MPE-B tumors were located in the region around the conus medullaris, while MPE-A tumors were located significantly lower in the spinal canal (P = .0019, Figure 4A–C). MPE-A patients were significantly younger than MPE-B patients (median age 27 vs. 45 years, P = 7.3e-5, Figure 4D). The distribution of WHO grades and predominant histological pattern was significantly different between subtypes (P = 1.7e-6 and P = 8.7e-8, respectively, Figure 4E,F). MPE-A was almost exclusively composed of WHO grade I tumors, whereas MPE-B contained most grade II tumors. Papillary and tanycytic tumors typically belonged to MPE-A and MPE-B, respectively, whereas predominantly myxoid tumors appeared in both subtypes. Global DNA methylation levels showed weak, but significant differences between subtypes (P = .00015, Figure 4G). As gene expression profiling indicated an enrichment of immune system-associated signatures in MPE-A, we used DIMEimmune18 to compare estimates for tumor-infiltrating lymphocytes (TIL) between MPE-A and B, showing more estimated TILs in MPE-A (P = 5e-7, Figure 4H). Furthermore, 10-year PFS was strikingly better for MPE-B than for MPE-A patients (relapse rate 33% vs. 85%, P = 3.4e-6, Figure 4I), although the molecular subtype did not turn out to be an independent prognostic marker after a multivariate survival analysis including histology, localization, and resection status (Supplementary Figure 2). MPE-A patients were more often treated with adjuvant radiotherapy (P = 8.3e-4, Figure 4J). Almost all MPE-B (57/58) tumors could be gross totally resected compared to only half of MPE-A tumors (15/30, P = 6.3e-8, Figure 4K). Finally, we assessed the MGMT promoter methylation status, which was hypermethylated in 48% of MPE-A and 24% of MPE-B (P = 0.0013, Figure 4L).
Fig. 4.
Clinicopathological and radiological features of MPE subtypes. (A–B) Exemplary MRI images of MPE subtype A and B, respectively (T1+CA). (C) Tumor localization of MPE subtypes along the neural axis. (D) Age distribution of MPE subtypes. (E–F) Distribution of WHO grades (2016) and main histological patterns in MPE subtypes. (G–H) Estimation of global DNA methylation level and tumor-infiltrating lymphocytes (TILs) in MPE subtypes. (I) Kaplan-Meier plot comparing PFS between MPE subtypes. (J–L) Distribution of patients having received adjuvant radiotherapy, postoperative resection status, and MGMT promoter methylation in MPE subtypes. Lines in beeswarm diagrams indicate the median, upper, and lower quartile. P-values assess differences between MPE subtypes.
Discussion
Based on a cohort of 185 MPE, we defined two subtypes A and B. The classification was based on clustering of global DNA methylation data, which is now widely available and increasingly used in routine diagnostics of brain tumors worldwide, allowing for a direct practical implementation. The defined subtypes were not only apparent on the methylation level, but also showed strong associations with RNA sequencing data and clinicopathological features underlining their relevance.
DNA methylation-based classification showed a clear separation of MPE from all other ependymoma types, and this separation was well reflected by the expression of HOXB13, which we found to be a robust biomarker of this tumor type. Previous studies had indicated its high expression on an mRNA level,5,25 but validations on larger series using antibodies that allow a fast, robust, and cheap protein detection were missing. In our hands, this antibody turned out to be extremely useful for the detection of epigenetically defined MPE in daily routine diagnostics. Of note, cauda equina paragangliomas also express HOXB13, but the distinction from MPE can usually be performed based on histomorphology.26 Within the group of MPE, histology is well known to be heterogeneous. We also confirmed that tumors with a rather classic morphology lacking myxoid or papillary features may fall into the methylation group of MPE. On the other hand, we realized that none of the cases with a typical myxopapillary histology analyzed in the frame of this study and beyond turned out to cluster or match with other reference groups regarding global DNA methylation profiling.
The methylation-based separation of MPE-A and B was evident, but less clear than the separation between well-established ependymoma types. Although consensus clustering results support the robustness of the defined MPE subtypes, the clustering pattern in the heatmap and the t-SNE analysis indicate that there is partial overlap. In particular, tumors with predominantly myxoid histology can less clearly be assigned to a subtype (Figure 2). We refrained from defining more subtypes as this did not increase the robustness of the clusters, and as it is likely to impede the application in a clinical setting. MPE subtypes were strongly associated with clinicopathological variables. MPE-A tumors occurred in younger patients, were typically located in the (lumbo-)sacral region, and associated with papillary histology. Over 80% were WHO grade I, and most were histologically diagnosed as MPE according to the 2016 WHO classification. Gene expression profiling revealed functional associations with IGF signaling, interleukin signaling, and the extracellular matrix. Interestingly, a recently identified metaprogram from a single-cell RNA sequencing study by Gojo et al., which was characteristic of progenitor-like neural stem cells and associated with worse outcome, was significantly overexpressed in MPE-A. This indicates that MPE-A might contain less differentiated tumor cells and is in line with our survival results showing a significantly worse outcome in MPE-A. A DNA methylation-based estimation of TILs and the analysis of immunological gene signatures indicated more immune infiltration in MPE-A. MPE-B occurred in older patients and tumors were typically located in the upper lumbar region and associated with tanycytic histology. This group comprised many cases that were histologically diagnosed as ependymoma WHO grade II. Together, these findings may even point to a different temporal and/or spatial cellular origin of MPE-A and MPE-B.
A limitation of this study is the comparably low number of tumors, for which RNA sequencing was feasible. This limits functional conclusions and underlines the need for prospective collection of tumor samples. Nonetheless, unsupervised analysis of 37 cases underlines the existence of 2 subtypes with a good concordance to the methylation-based subtypes.
The clinical course of MPE is heterogeneous. Still, it was considered a benign tumor (WHO grade I) until recently, but has been graded as CNS WHO 2 in the 2021 classification of brain tumors due to the high number of recurrences.1 The aggressive behavior of the tumors histologically diagnosed as MPE is also visible in our data, as grade I tumors (WHO 2016) showed a trend towards shorter survival than grade II tumors. The presented subtypes were of high prognostic relevance and might therefore be used for a more precise grading and therapeutic stratification of MPE in the future. Indeed, for MPE-A, GTR failed in approximately 50% of cases and the 10-year PFS of 15% was poor. In MPE-B, GTR was achieved for 57/58 tumors with available resection status and the 10-year PFS of 67% was strikingly better. Although the resection status was not available for all relapsed tumors, we found that relapse occurred in only 3/50 MPE-B, but in 4/13 MPE-A, where GTR was achieved. In multivariate survival analyses, the subtype was not an independent predictor for PFS, most likely because of insufficient sample size. Due to the high correlation between resection status, tumor recurrence, and subtype, the precise relevance of subtotal tumor resection (STR) and intrinsic biological features underlying MPE-A recurrence remains unclear. However, the different growth patterns and tumor localizations appear likely to be causal for lower resection rates. Also, the higher rate of tumor recurrence in totally resected MPE-A as compared to MPE-B indicates that subtype-specific biological features might be causal for the observed clinical heterogeneity.
Whereas MPE-B tumors with GTR have an excellent long-term prognosis, almost all MPE-A tumors with STR recurred, reflecting the need for additional treatment modalities. Radiotherapy was not prognostically relevant in our cohort in univariate analysis (Supplementary Figure 2). This is likely due to a selection bias as 50% of the treated cohort had STR compared to 14% of the untreated patients. Multiple reports demonstrated the benefit of adjuvant radiotherapy in MPE,4,13 especially in younger patients.13 In contrast, another study reported an excellent long-term outcome without radiotherapy in adult patients with GTR and without sacral tumor location.11 These findings suggest that radiotherapy might be needed for MPE-A patients, which are typically younger and have sacral tumor localization. On the other hand, it might not be necessary for completely resected MPE-B tumors.
Our study also showed that MGMT promoter methylation as assessed by DNA methylation profiling19 was present in a substantial number of MPE. As this is known to be associated with increased response to temozolomide (TMZ) in glioblastoma,27 it might also be a potential predictive factor in MPE. The distribution differed between MPE-A and B, where it was present in 48% and 24% of the cases, respectively. Although there is currently no established chemotherapy regimen for MPE, a case report described an excellent response to TMZ in a young patient with MPE and multiple recurrences.28 This points towards TMZ as a possible therapeutic option, in particular for subtotally resected MPE-A tumors with comparably bad prognosis and frequent MGMT promoter methylation.
Supplementary Material
Acknowledgements
We thank Anne Reichstein, Helena Gladkov and Nicole Borgardt (University Medical Center Hamburg-Eppendorf) for excellent technical support. MiB, MM, and AKW are fellows of the Mildred Scheel Cancer Career Center Hamburg/Deutsche Krebshilfe.
Conflict of interest statement. The authors declare no competing interests.
Authorship statement. Conceptualization: MiB, US. Methodology: MiB, KH, US. Formal analysis: MiB, KH, LP, MK, MA, US, AKS. Investigation: MiB, KH, US. Resources: MiB, LS, TM, MMD, CMM, MH, AKW, DC, JH, SF, TER, IT, EL, MS, LE, MaB, AvD, DO, MM, SR, MG, JEN. Data curation: MiB, KH, US. Writing - Original draft: MiB. Writing - Review & Editing: All authors. Visualization: MiB, US. Supervision: US. List of any unpublished papers cited: None.
Contributor Information
Michael Bockmayr, Department of Pediatric Hematology and Oncology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Research Institute Children’s Cancer Center Hamburg, Hamburg, Germany; Mildred Scheel Cancer Career Center HaTriCS4, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Charité—Universitätsmedizin Berlin, Corporate Member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Institute of Pathology, Berlin, Germany.
Kim Harnisch, Institute of Neuropathology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Institute for Neuropathology, University Hospital of Zurich, Zurich, Switzerland.
Lara C Pohl, Department of Pediatric Hematology and Oncology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Research Institute Children’s Cancer Center Hamburg, Hamburg, Germany.
Leonille Schweizer, Charité—Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Department of Neuropathology, Berlin, Germany; German Cancer Consortium (DKTK), Partner Site Berlin, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Theresa Mohme, Department of Neurosurgery, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Meik Körner, Department of Pediatric Hematology and Oncology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Research Institute Children’s Cancer Center Hamburg, Hamburg, Germany.
Malik Alawi, Bioinformatics Core, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Abigail K Suwala, Department of Neuropathology, Institute of Pathology, University of Heidelberg, Heidelberg, Germany; Clinical Cooperation Unit Neuropathology, German Consortium for Translational Cancer Research (DKTK), German Cancer Research Center (DKFZ), Heidelberg, Germany; Department of Neurological Surgery, UCSF, San Francisco, California, USA.
Mario M Dorostkar, Center for Neuropathology, Ludwig-Maximilians-University, Munich, Germany; German Center for Neurodegenerative Diseases, Munich, Germany.
Camelia M Monoranu, Department of Neuropathology, Institute of Pathology, University of Würzburg, Würzburg, Germany.
Martin Hasselblatt, Institute of Neuropathology, University Hospital Münster, Münster, Germany.
Annika K Wefers, Mildred Scheel Cancer Career Center HaTriCS4, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Institute of Neuropathology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
David Capper, Charité—Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Department of Neuropathology, Berlin, Germany; German Cancer Consortium (DKTK), Partner Site Berlin, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Jürgen Hench, Division of Neuropathology, Institute of Medical Genetics and Pathology, University of Basel, Basel, Switzerland.
Stephan Frank, Division of Neuropathology, Institute of Medical Genetics and Pathology, University of Basel, Basel, Switzerland.
Timothy E Richardson, Department of Pathology and Laboratory Medicine, Glenn Biggs Institute for Alzheimer’s & Neurodegenerative Diseases, UT Health San Antonio, San Antonio, Texas, USA.
Ivy Tran, Department of Pathology, NYU Langone Health, New York City, New York, USA.
Elisa Liu, Department of Pathology, NYU Langone Health, New York City, New York, USA.
Matija Snuderl, Department of Pathology, NYU Langone Health, New York City, New York, USA.
Lara Engertsberger, Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, Medical University of Graz, Graz, Austria.
Martin Benesch, Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, Medical University of Graz, Graz, Austria.
Andreas von Deimling, Department of Neuropathology, Institute of Pathology, University of Heidelberg, Heidelberg, Germany.
Denise Obrecht, Department of Pediatric Hematology and Oncology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Martin Mynarek, Department of Pediatric Hematology and Oncology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Mildred Scheel Cancer Career Center HaTriCS4, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Stefan Rutkowski, Department of Pediatric Hematology and Oncology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Markus Glatzel, Institute of Neuropathology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Julia E Neumann, Institute of Neuropathology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Center for Molecular Neurobiology Hamburg, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Ulrich Schüller, Department of Pediatric Hematology and Oncology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Research Institute Children’s Cancer Center Hamburg, Hamburg, Germany; Institute of Neuropathology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Funding
US was supported by the Deutsche Forschungsgemeinschaft (Grant No. SCHU2442/8-1), Gert und Susanna Mayer Stiftung, and Fördergemeinschaft Kinderkrebs-Zentrum Hamburg. The DNA methylation profiling at NYU was in part supported by the Friedberg Charitable Foundation and the Making Headway Foundation (MS).
References
- 1. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bates JE, Choi G, Milano MT. Myxopapillary ependymoma: a SEER analysis of epidemiology and outcomes. J Neurooncol. 2016;129(2):251–258. [DOI] [PubMed] [Google Scholar]
- 3. Truitt G, Gittleman H, Leece R, et al. Partnership for defining the impact of 12 selected rare CNS tumors: a report from the CBTRUS and the NCI-CONNECT. J Neurooncol. 2019;144(1):53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weber DC, Wang Y, Miller R, et al. Long-term outcome of patients with spinal myxopapillary ependymoma: treatment results from the MD Anderson Cancer Center and institutions from the Rare Cancer Network. Neuro Oncol. 2015;17(4):588–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pajtler KW, Witt H, Sill M, et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27(5):728–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Raffeld M, Abdullaev Z, Pack SD, et al. High level MYCN amplification and distinct methylation signature define an aggressive subtype of spinal cord ependymoma. Acta Neuropathol Commun. 2020;8(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697): 469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Witt H, Gramatzki D, Hentschel B, et al. DNA methylation-based classification of ependymomas in adulthood: implications for diagnosis and treatment. Neuro Oncol. 2018;20(12):1616–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Neumann JE, Spohn M, Obrecht D, et al. Molecular characterization of histopathological ependymoma variants. Acta Neuropathol. 2020;139(2):305–318. [DOI] [PubMed] [Google Scholar]
- 10. Vera-Bolanos E, Aldape K, Yuan Y, et al. Clinical course and progression-free survival of adult intracranial and spinal ependymoma patients. Neuro Oncol. 2015;17(3):440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Montero AS, Tran S, Amelot A, et al. Clinical characteristics and long-term surgical outcome of spinal myxopapillary ependymoma: a French cohort of 101 patients. J Neurooncol. 2021;152(3):491–499. [DOI] [PubMed] [Google Scholar]
- 12. Kraetzig T, McLaughlin L, Bilsky MH, Laufer I. Metastases of spinal myxopapillary ependymoma: unique characteristics and clinical management. J Neurosurg Spine. 2018;28(2):201–208. [DOI] [PubMed] [Google Scholar]
- 13. Kukreja S, Ambekar S, Sharma M, Sin AH, Nanda A. Outcome predictors in the management of spinal myxopapillary ependymoma: an integrative survival analysis. World Neurosurg. 2015;83(5):852–859. [DOI] [PubMed] [Google Scholar]
- 14. Kotecha R, Tom MC, Naik M, et al. Analyzing the role of adjuvant or salvage radiotherapy for spinal myxopapillary ependymomas. J Neurosurg Spine. Published online May 1, 2020:1–6. doi: 10.3171/2020.2.SPINE191534. [DOI] [PubMed] [Google Scholar]
- 15. Benesch M, Weber-Mzell D, Gerber NU, et al. Ependymoma of the spinal cord in children and adolescents: a retrospective series from the HIT database. J Neurosurg Pediatr. 2010;6(2):137–144. [DOI] [PubMed] [Google Scholar]
- 16. Rudà R, Reifenberger G, Frappaz D, et al. EANO guidelines for the diagnosis and treatment of ependymal tumors. Neuro Oncol. 2018;20(4):445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ahmad O, Chapman R, Storer LC, et al. Integrative molecular characterization of pediatric spinal ependymoma: the UK Children’s Cancer and Leukaemia Group study. Neurooncol Adv. 2021;3(1):vdab043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Safaei S, Mohme M, Niesen J, Schüller U, Bockmayr M. DIMEimmune: robust estimation of infiltrating lymphocytes in CNS tumors from DNA methylation profiles. Oncoimmunology. 2021;10(1):1932365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bady P, Sciuscio D, Diserens AC, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124(4):547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bockmayr M, Klauschen F, Maire CL, et al. Immunologic profiling of mutational and transcriptional subgroups in pediatric and adult high-grade gliomas. Cancer Immunol Res. 2019;7(9):1401–1411. [DOI] [PubMed] [Google Scholar]
- 22. Doucette T, Rao G, Rao A, et al. Immune heterogeneity of glioblastoma subtypes: extrapolation from the cancer genome atlas. Cancer Immunol Res. 2013;1(2):112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gojo J, Englinger B, Jiang L, et al. Single-cell RNA-Seq reveals cellular hierarchies and impaired developmental trajectories in pediatric ependymoma. Cancer Cell. 2020;38(1):44–59.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uhrig S, Ellermann J, Walther T, et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res. 2021;31(3):448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barton VN, Donson AM, Kleinschmidt-DeMasters BK, et al. Unique molecular characteristics of pediatric myxopapillary ependymoma. Brain Pathol. 2010;20(3):560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bockmayr M, Körner M, Schweizer L, Schüller U. Cauda equina paragangliomas express HOXB13. Neuropathol Appl Neurobiol. 2021;47(6):889–890. [DOI] [PubMed] [Google Scholar]
- 27. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 28. Fujiwara Y, Manabe H, Izumi B, Shima T, Adachi N. Remarkable efficacy of temozolomide for relapsed spinal myxopapillary ependymoma with multiple recurrence and cerebrospinal dissemination: a case report and literature review. Eur Spine J. 2018;27(Suppl 3):421–425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The methylation data are deposited at the GEO repository GSE184900. Processed RNAseq data are available in Supplementary Table S3.