Abstract
A possible role for posttranslational modifications in regulating the activity of ATP-binding cassette (ABC) transporters has not been well established. In this study, the drug efflux ABC transporter gene KlPDR5 was isolated from the budding yeast Kluyveromyces lactis, and it was found that the encoded KlPdr5 drug pump is posttranslationally regulated by the type 2A-related Ser/Thr protein phosphatase, Sit4p. The KlPdr5 transporter is a protein of 1,525 amino acids sharing 63.8% sequence identity with its Saccharomyces cerevisiae counterpart, ScPdr5p. Overexpression of the KlPDR5 gene confers resistance to oligomycin, antimycin, econazole, and ketoconazole, whereas cells with a disrupted allele of KlPDR5 are hypersensitive to the drugs and have a decreased capacity to carry out efflux of the anionic fluorescent dye rhodamine 123. It was found that a chromosomal disruption of KlPDR5 abolishes the drug-resistant phenotype associated with sit4 mutations and that a synergistic hyperresistance to the drugs can be created by overexpressing KlPDR5 in sit4 mutants. These data strongly indicate that the multidrug-resistant phenotype of sit4 mutants is mediated by negatively modulating the activity of KlPdr5p. As the transcriptional level of KlPDR5 and the steady-state level of KlPdr5p are not significantly affected by mutations in SIT4, the regulation by Sit4p appears to be a posttranslational process.
Multidrug resistance (MDR) is a ubiquitous biological phenomenon that occurs in living organisms to evade chemotherapy or to resist naturally occurring toxic chemical compounds (for reviews, see references 10, 23, 29, and 50). One common type of MDR is caused by an increased activity of specific transporters on the plasma membrane that carry chemotherapeutic drugs out of cells and subsequently lower intracellular drug concentration. These drug pumps belong to a superfamily of evolutionarily conserved proteins known as ATP-binding cassette (ABC) transporters and are characterized by a similar molecular architecture involving one or two ATP-binding sites and 6 to 17 predicted transmembrane domains (for recent reviews, see references 3, 5, and 35). Drugs are actively extruded across the plasma membrane at the expense of ATP hydrolysis (reviewed in reference 47). Among the well-studied drug transporters are the human P glycoprotein and the MDR-associated proteins (MRPs) that render tumor cells highly resistant to anticancer drugs (10, 23, 29, 50). In recent years, attention has also been turned to the ABC transporters that confer drug resistance in pathogenic microorganisms such as Candida albicans (42, 45, 59), Plasmodium falciparum (43), and infectious bacteria (41). The ABC transporter-mediated drug efflux appears to be an evolutionarily conserved mechanism, as exemplified by the Lactococcus lactis drug pump, LmrA, which is able to confer MDR on human cells (55).
The unicellular eukaryote Saccharomyces cerevisiae has been extensively studied in recent years as a model system for understanding the molecular mechanisms underlying the development of MDR and the structural and functional aspects of ABC proteins. The S. cerevisiae genome contains genes for as many as 31 distinct ABC proteins that can be phylogenetically classified into at least six subfamilies (3, 14). Among these proteins are the members of the PDR and MRP subfamilies, classified as the functional orthologues of the human MDR and MRP systems, respectively. Well-studied transporters include Pdr5p, Pdr12p, Snq2p, Ycf1p, and Yor1p, which are involved in the transport of antibiotics, antifungal drugs, and other toxic chemical compounds.
To understand how cells develop MDR in response to chemotherapy, efforts have been directed toward elucidating the regulation of the yeast ABC transporter genes. It has been found that S. cerevisiae has a highly complex regulatory network that modulates expression of the ABC genes. At least three major transcriptional activators of the Cys6 zinc finger type have been genetically identified in S. cerevisiae (2, 12, 16, 18, 51). The Pdr1 and Pdr3 proteins control the transcriptional levels of PDR5, SNQ2, YOR1, PDR10, and PDR15 by direct binding to DNA in the promoter region of the target genes (13, 17, 26, 33, 39, 40), whereas the expression of SNQ2 involves a third transcriptional activator, Yrr1p (12). Recent studies have also revealed additional proteins, such as the yeast homologues of the stress-dependent transcriptional factor AP1 and the heat shock protein Hsp70p, that are implicated in the transcriptional activation of ABC transporter genes (25, 27, 32, 56, 57).
Much less is known about whether posttranscriptional regulations play any role in controlling activity of ABC drug pumps in yeast. Recent investigations have shown that the S. cerevisiae Pdr5, Snq2, and Yor1 transporters are subject to phosphorylation in vitro and that phosphorylation of Pdr5p appears to involve the two genes encoding casein kinase I (15). Another S. cerevisiae ABC protein, Pdr12p, has been reported to be under a negative posttranscriptional control by the Cmk1 Ca2+-calmodulin-dependent protein kinase (30). In an earlier study, a putative phosphorylation site in the Ycf1 transporter was proposed, as mutations in this phosphorylation motif render the protein nonfunctional in the detoxification of cadmium (53). Regulation of ABC transporters by protein phosphorylation is thus emerging as an important alternative mechanism for modulating a cell's capacity to resist drugs and toxic compounds.
In respect to the above possible mechanism, we have recently isolated from the budding yeast Kluyveromyces lactis the SIT4 gene, encoding a Ser/Thr protein phosphatase. SIT4 has been found to have a broad role in regulating MDR (9). It negatively regulates the resistance of cells to oligomycin, antimycin, ketoconazole, and econazole and positively modulates tolerance to paromomycin, sorbic acid, and 4-nitroquinoline-N-oxide. An explanation for these observations is that the Sit4 protein phosphatase (Sit4p) controls the activity of the ABC transporters specific for these drugs. As a first step towards an understanding of the role of protein phosphorylation in regulating MDR, this study describes the isolation and characterization of the S. cerevisiae PDR5 homologue of K. lactis, KlPDR5, that is responsible for the efflux of oligomycin, antimycin, and the antifungal drugs ketoconazole and econazole. Unequivocal genetic evidence that the MDR phenotype of sit4 mutants is mediated via an activation of the KlPdr5 transporter by Sit4p is provided.
MATERIALS AND METHODS
Strains and media.
Table 1 lists the yeast strains used in this study. The K. lactis strain CK432/8 was derived from CK254/1 by selecting for Ura− on 5-fluoroorotic acid medium. Complete medium (GYP) contains 0.5% Bacto yeast extract, 1% Bacto Peptone, and 2% glucose. Glycerol medium (GlyYP) contains 2% glycerol in place of glucose. Glucose minimal medium (GMM) contains 0.17% Difco yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, and 2% glucose. GMM is supplemented at 25 μg/ml for bases and 50 μg/ml for amino acids. Resistance to antimycin A and oligomycin was tested on GlyYP plates. Resistance to ketoconazole and econazole was tested on GYP. Antimycin A, oligomycin, econazole, and rhodamine 123 were all purchased from Sigma and dissolved in ethanol. The stock solution for ketoconazole (ICN Pharmaceuticals) was prepared with dimethyl sulfoxide.
TABLE 1.
Genotypes and sources of yeast strains
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| K. lactis | ||
| PM6-7A | MATaadeT-600 uraA1 | 6 |
| CK254/1 | Same as PM6-7A, but sit4Δ::URA3 | 9 |
| CK373/1 | Same as PM6-7A, but pdr5::kan | This study |
| CK413 | Same as PM6-7A, but sit4Δ::URA3 pdr5::kan | This study |
| CK432/8 | Same as CK254/1, but sit4Δ::ura3 | This study |
| CW2-8B | MATα ade1 lysA1 uraA1 sit4-1 | 9 |
| S. cerevisiae | ||
| CY4029 | MATaura3-1 leu2-3,112 his3-11,15 trp1-1 ade2-1 can1-100, SSD1-v1 | 38 |
| CY3938 | Same as CY4029, but sit4Δ::HIS3 | 38 |
Manipulation of K. lactis.
Transformation and genetic manipulation of K. lactis were carried out essentially as previously described (7, 8). Genomic DNA was extracted from protoplasts obtained by Zymolyase treatment (49). The K. lactis-S. cerevisiae-Escherichia coli shuttle vector pCXJ15 (X. J. Chen, unpublished data) was used for the expression of KlPDR5 and its derivatives in K. lactis and S. cerevisiae, as this plasmid can be replicated in both yeasts in multiple copies. For low-level expression in K. lactis, the centromeric vector pCXJ18 was used (8).
Isolation of KlPDR5.
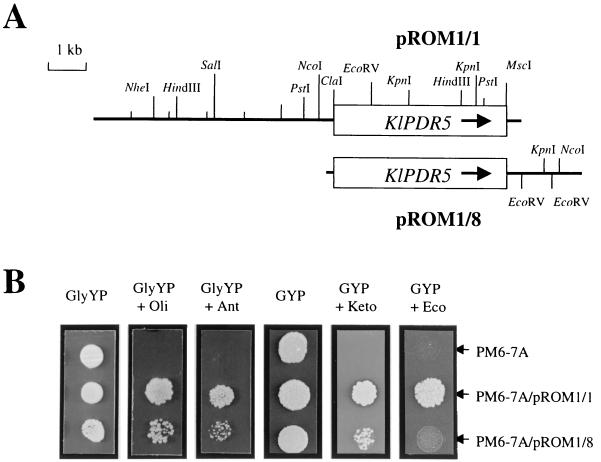
The K. lactis wild-type strain PM6-7A was transformed with a K. lactis partial Sau3AI genomic library based on the K. lactis-E. coli shuttle vector KEp6 (58). KEp6 contains the S. cerevisiae URA3 gene, which upon transformation complements the uraA1 defect of K. lactis. Approximately 30,000 Ura+ transformants were scored and replica plated onto GlyYP supplemented with 0.5 μg of oligomycin/ml. After incubation at 28°C for 3 days, 16 Olir colonies were obtained. Six of these clones showed a cosegregation of the Ura+ and the Olir phenotypes after growth in nonselective medium. The plasmids were rescued, amplified in E. coli, and reintroduced into PM6-7A by transformation to confirm the Olir phenotype. Restriction enzyme analysis showed that the six clones represent two categories of overlapping plasmids. Physical maps were established from two of the plasmids, pROM1/1 and pROM1/8, which share an overlapping insert DNA of about 5.0 kb (Fig. 1A).
FIG. 1.
Physical map of the K. lactis chromosomal region containing the KlPDR5 gene and the MDR phenotype conferred by KlPDR5 overexpression. (A) Restriction map of the overlapping genomic clones pROM1/1 and pROM1/8. Open box, KlPDR5 open reading frame; arrow, direction of transcription. (B) Growth and MDR phenotype of K. lactis cells overexpressing KlPDR5. The strain PM6-7A (wild type) was transformed with pROM1/1 and pROM1/8. The Ura+ transformants were diluted to 5 × 104 cells/ml, and 10-μl aliquots were applied to GlyYP, GlyYP supplemented with oligomycin (Oli; 0.5 μg/ml) and antimycin (Ant; 0.2 μM), GYP, and GYP supplemented with ketoconazole (Keto; 4.0 μg/ml) and econazole (Eco; 0.2 μg/ml). The plates were incubated at 28°C for 4 days before being photographed.
In a parallel experiment, the sit4-1 mutant CW2-8B was transformed by the genomic library DNA. Approximately 30,000 Ura+ transformants were scored and tested for Olir on GlyYP supplemented with oligomycin at 4.0 μg/ml. This selection procedure was based on the assumption that an overexpression of a putative oligomycin efflux pump in the absence of a functional SIT4 gene could produce cells with a hyper-active drug efflux capacity. These cells would be resistant to an oligomycin concentration of 4.0 μg/ml, on which sit4 mutants alone cannot grow. In this way, 14 genomic clones conferring Olir were identified. Restriction analysis revealed that all the clones are identical to pROM1/1.
Chromosomal disruption of KlPDR5.
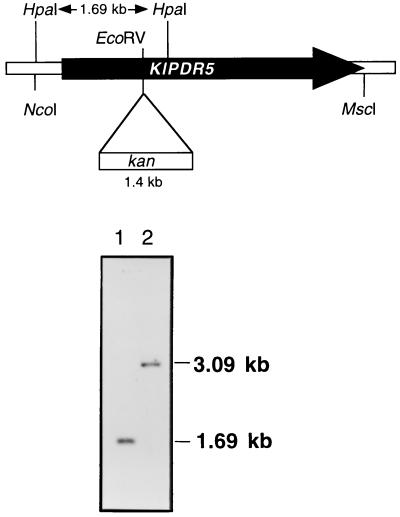
The 4.8-kb PstI fragment containing the promoter and the majority of the coding sequence of KlPDR5 was isolated from pROM1/1 and subcloned into pTZ19U to produce pTZ19-MDR1/2. pTZ19-MDR1/2 was then linearized by digestion with EcoRV, which cuts at codon 344 of KlPDR5. A 1.4-kb BglII-XhoI fragment containing the kan expression module (24) was inserted after filling in with Klenow polymerase. The resulting plasmid, pKlPDR5::kan/3, was digested with HpaI to release the 3.09-kb Klpdr5::kan cassette (Fig. 2), which was subsequently used to transform K. lactis strains by selecting for G418-resistant colonies. Among 24 transformants scored, only 2 had a stable G418r phenotype after growth in GYP in the absence of the antibiotic. Correct replacement of the chromosomal KlPDR5 locus was confirmed by digestion of genomic DNA with HpaI and hybridization to a 32P-labeled probe.
FIG. 2.
Chromosomal disruption of KlPDR5. (Top) Strategy used for disrupting KlPDR5 by the insertion of the kan gene into the EcoRV site located in the coding region of the gene. (Bottom) Southern blot analysis confirming KlPDR5 disruption. Total DNA was extracted from the disruptant CK373/1 (lane 2) and its parental strain PM6-7A (lane 1), digested with HpaI, transferred to a nylon membrane, and hybridized with the 32P-labeled 1.69-kb HpaI fragment containing the KlPDR5 sequence. The hybridization signals are indicated as bands of 1.69 kb in the control and 3.09 kb in the disruptant as a result of the insertion of the 1.4-kb kan gene.
Flow cytometry of K. lactis cells.
K. lactis strains were grown in GMM to an A600 of about 1.0. To 0.5 ml of cell culture, 5 μl of rhodamine 123 at 0.5 mg/ml was added, and dye loading was allowed for 60 min at 30°C. Rhodamine 123 efflux was stopped by a 1:10 dilution of the cells in ice-cold water. Intracellular rhodamine fluorescence was analyzed with a Becton Dickinson FACScan flow cytometer using WinMDI software.
Transcriptional analysis of KlPDR5.
Total RNA was extracted according to the method described by Schmitt and coworkers (46). RNA was fractionated by electrophoresis in 1.2% agarose-formaldehyde gels, transferred to nylon membranes, and hybridized at high stringency to a 32P-labeled 0.6-kb EcoRI fragment from the KlPDR5 coding sequence. As an internal control for sample loading, the S. cerevisiae ACT1 gene was used as a probe to detect K. lactis ACT1 mRNA. Measurement of band intensity was carried out by using the PhosphorImager analyzer (Molecular Dynamics).
HA tagging of KlPDR5.
A BglII site was created by PCR immediately upstream of the KlPDR5 stop codon. An in-frame fusion was made between the altered KlPDR5 and a stretch of sequence coding for a triple hemagglutinin (HA) tag. The fusion product was then cloned into pCXJ15 and pCXJ18 to produce the expression plasmids pCXJ15-PDR5HA/1 and pCXJ18-PDR5HA/1. To know whether the tagged KlPDR5 is functionally active in vivo, the plasmids were introduced into the Klpdr5 mutant CK373/1 by transformation. Ura+ transformants were found to be resistant to oligomycin and antimycin on GlyYP at 0.5 μg/ml and 0.1 μM, respectively, while the untransformed strain CK373/1 remained hypersensitive to the drugs (see below). Thus, the HA-tagged KlPDR5 gene is functionally active in complementing the chromosomally disrupted Klpdr5 locus.
Preparation of K. lactis cell extracts and Western blot analysis.
K. lactis strains were grown in GMM to an optical density at 600 nm (OD600) of about 1.0. Three OD600 equivalents of cells were harvested and washed with cold water. Cells were lysed by adding 150 μl of 1.85 M NaOH–7.5% β-mercaptoethanol for 10 min on ice. The proteins were then precipitated by adding 150 μl of 50% trichloroacetic acid followed by incubation on ice for 10 min. Precipitates were pelleted by centrifugation in a microcentrifuge for 3 min and resuspended in 50 μl of sample buffer (40 mM Tris-HCl [pH 6.8], 8 M urea, 5% sodium dodecyl sulfate (SDS), 0.1 mM EDTA, 1% β-mercaptoethanol, 0.01% bromophenol blue) and 10 μl of 1 M Tris base. The proteins were dissociated by incubating at 42°C for 15 min before being electrophoresed through a 4-to-20% precast gradient polyacrylamide-SDS gel (Gradipore). After being transfered onto a nylon Immobilon-P membrane (Millipore), the proteins were reacted with antibodies and visualized with the enhanced chemiluminescence detection system (Amersham). The anti-HA monoclonal antibody 12CA5 was purchased from Boehringer Mannheim, and the anti-yeast 3-phosphoglycerate kinase mouse monoclonal antibody 22C5-D8 was from Molecular Probes.
Nucleotide sequence accession number.
The nucleotide sequence of KlPDR5 has been submitted to the GenBank/EBI Data Bank with the accession number AF245358.
RESULTS
Isolation of the K. lactis ABC transporter gene KlPDR5, conferring resistance to oligomycin.
To test the hypothesis that the Sit4 protein phosphatase modulates MDR by regulating the activity of ABC transporters, the isolation of K. lactis ABC transporter genes was attempted. It was expected that the availability of these genes would allow a direct examination for possible functional interactions between Sit4p and the membrane drug pumps. Two different strategies were used to identify a potential oligomycin pump. In the first strategy, a search was made for genes that, when overexpressed, confer an increased resistance to oligomycin in a wild-type strain. PM6-7A was thus transformed with a K. lactis genomic library based on a multicopy vector. The transformants were screened for their ability to grow on GlyYP supplemented with oligomycin at 0.5 μg/ml, a concentration sufficient to inhibit the growth of the host cells. In the second strategy, the sit4-1 mutant CW2-8B was transformed with the multicopy genomic library. Transformants that are hyperresistant to oligomycin were sought. This screen was based on the assumption that if the potential oligomycin pump is negatively controlled by Sit4p, a combination of the high gene dosage for the ABC transporter and a mutation in SIT4 would generate cells with a high drug efflux capacity which therefore could be hyperresistant to oligomycin. An oligomycin concentration of 4 μg/ml was used for screening the hyperresistant colonies from the CW2-8B transformants, as the sit4 host strain cannot grow on GlyYP supplemented with the drug at a concentration above 2 μg/ml.
The two strategies described above identified a single genomic locus that confers resistance to oligomycin when the gene was overexpressed from a multicopy vector. Two types of genomic clones were found, as exemplified by pROM1/1 and pROM1/8 (Fig. 1A) containing inserts of 11.3 and 6.8 kb, respectively. Reintroduction of the two plasmids into PM6-7A clearly showed that the transformants are resistant to oligomycin, in contrast to the host strain (Fig. 1B).
As the Olir-conferring plasmids pROM1/1 and pROM1/8 have an overlapping region of 5.2 kb, it was assumed that a potential ABC gene might reside in this region. The nucleotide sequence of a 5.6-kb fragment from this region was determined. Computer analysis revealed an uninterrupted open reading frame of 1,525 codons with the ability to encode a protein of 172 kDa. Comparison of the deduced protein with sequences in the GenBank databases revealed that it shares 63.8, 62.8, and 59.5% identity with the Pdr5, Pdr15, and Pdr10 drug pumps of S. cerevisiae and 63.0 and 53.8% identity with the CgCdr1 and CaCdr1 proteins from Candida glabrata and C. albicans. By analogy to S. cerevisiae, the gene was designated KlPDR5 and the encoded protein was termed KlPdr5p.
KlPdr5p shares most common molecular characteristics with other yeast ABC transporters. Like ScPdr5p, KlPdr5p is a full-size ABC protein, with each half of the molecule having six putative membrane-spanning domains. In the upstream sequence of the two transmembrane domain clusters is a nucleotide binding site composed of Walker motifs A and B. Between Walker A and B motifs resides a short stretch of sequence, called the ABC signature, that is a hallmark of ABC proteins.
Overexpression of KlPDR5 also confers resistance to antimycin A, ketoconazole, and econazole, whereas Klpdr5 mutants are hypersensitive to the drugs.
KlPDR5 was shown to be a typical multidrug transporter, as overexpression of the gene confers resistance to several other drugs that are structurally and mechanistically unrelated to oligomycin. As shown in Fig. 1B, transformants of the multicopy plasmids pROM1/1 and pROM1/8 are resistant to the mitochondrial inhibitor antimycin and to the antifungal drugs ketoconazole and econazole. Note that the transformants of pROM1/8 show a lower level of resistance to these drugs under the same conditions. This can be explained by the fact that pROM1/8 carries KlPDR5 with a truncated promoter (Fig. 1A), and thus, a lower expression level of the gene is expected. Sequence analysis showed that only 184 bp of sequence upstream of the translation initiation codon is retained in pROM1/8.
The chromosomal KlPDR5 gene was disrupted by the one-step gene replacement procedure (44). A 1.4-kb fragment carrying the G418r gene was inserted into the EcoRV site at codon 344. Correct gene replacement was confirmed by comparing genomic Southern blots of the disruptant CK373/1 and the parental PM6-7A (Fig. 2). Digestion with the restriction enzyme HpaI, followed by hybridization with a 32P-labeled probe, yielded bands of 3.09 kb for CK373/1 and 1.69 kb for PM6-7A, which are the sizes expected for a correct disruption of the chromosomal KlPDR5 locus.
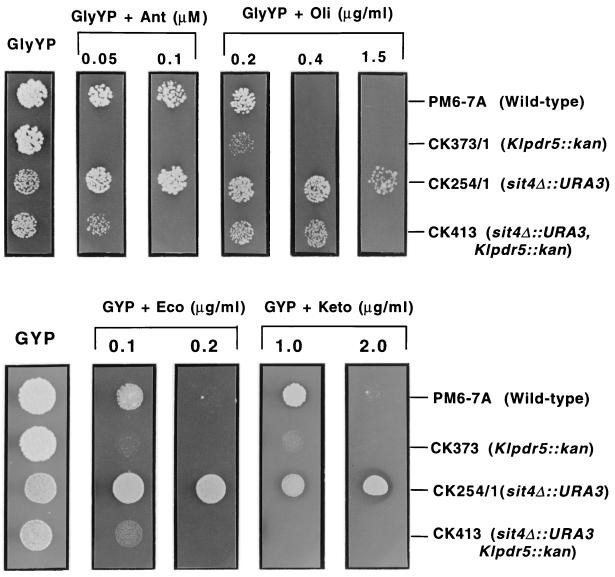
The KlPDR5 disruption mutant, CK373/1, does not display any detectable growth defect on GYP. However, Klpdr5 mutants were found to be hypersensitive to different drugs. Growth of CK373/1 is clearly inhibited by econazole and ketoconazole (Fig. 3, bottom) at concentrations of 0.1 and 1.0 μg/ml, respectively, while the same concentrations do not inhibit the growth of the isogenic wild-type strain PM6-7A. Likewise, CK373/1 is hypersensitive to antimycin at 0.05 μM, in contrast to PM6-7A (Fig. 3, top). However, the Klpdr5 mutant is only slightly more sensitive to oligomycin than PM6-7A, as judged from the colony size on oligomycin at 0.2 μg/ml. It is possible that in K. lactis, KlPDR5 is not the only ABC transporter involved in the efflux of oligomycin. An ABC transporter similar to Yor1p of S. cerevisiae that could contribute to the detoxification of oligomycin might also be present in K. lactis (11, 34).
FIG. 3.
Growth and drug sensitivity of K. lactis strains showing the drug-hypersensitive phenotypes of KlPDR5-disruption strains and the genetic interaction between KlPDR5 and SIT4. Cells were grown in liquid GYP medium and diluted to 5 × 104/ml, and 10-μl aliquots were applied to GlyYP or GYP supplemented with antimycin (Ant), oligomycin (Oli), econazole (Eco), and ketoconazole (Keto). Photographs were taken after incubation of plates at 28°C for 4 days.
Mutations in KlPDR5 increase accumulation of rhodamine 123.
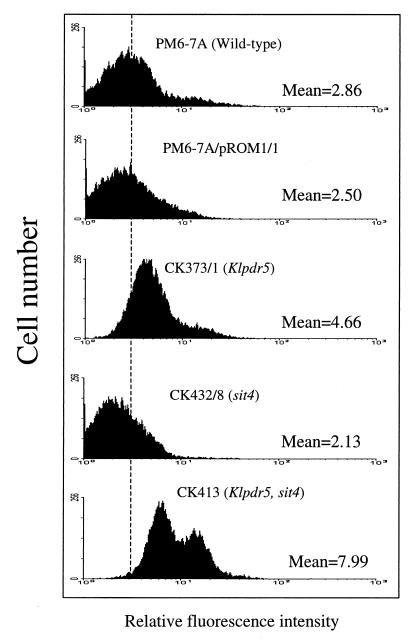
To demonstrate directly that the transport of drugs or toxic compounds is affected in Klpdr5 mutants, the accumulation of the anionic fluorescent dye rhodamine 123 was measured by flow cytometry (Fig. 4). It was found that the Klpdr5 cells have a significantly higher accumulation of the dye, with a 63% increase in mean intracellular fluorescence intensity compared with the wild-type cells. In contrast, the dye accumulation is decreased in cells overexpressing KlPDR5. It is relevant that this decrease seems to be rather marginal compared with the magnitude of the effects of KlPDR5 overexpression on the resistance to oligomycin, antimycin, and the antifungal drugs in the plate assays (see above). This discrepancy could be explained either by a low affinity of KlPdr5p for rhodamine 123 compared with the other drugs or by a difference in the copy number of the KlPDR5-bearing plasmid in the two different assays. It would be expected that a selection for cells carrying a higher copy number of the plasmid is applied under the growth conditions in the plate assay and such a selection does not occur in the rhodamine 123 efflux experiment.
FIG. 4.
Flow cytometry analysis showing the steady-state fluorescence intensities of K. lactis cells. All the strains used are isogenic and include the wild-type control PM6-7A, PM6-7A transformed with the multicopy KlPDR5-expressing plasmid pROM1/1 the Klpdr5 mutant CK373/1, the sit4 mutant CK432/8, and the sit4 Klpdr5 double mutant CK413. The rhodamine 123 accumulation in steady-state cells was measured after dye loading for 60 min at 30°C. The mean fluorescence intensity for each strain is shown.
The MDR phenotype of sit4 mutants is dependent on the function of KlPDR5.
Previous studies have shown that mutations in the Ser/Thr protein phosphatase gene SIT4 render cells resistant to oligomycin, antimycin, econazole, and ketoconazole (9). As described above, the KlPDR5 gene is required for the detoxification of the four drugs. It was therefore suggested that the MDR phenotype of sit4 mutants is caused by an increased activity of KlPdr5p that is under the negative control of Sit4p. If this is the case, it can be expected that the MDR phenotype of sit4 mutants would be dependent on KlPDR5 and disruption of KlPDR5 would abolish drug resistance of sit4 cells.
Comparison of the sit4 mutant CK254/1, which can resist antimycin and oligomycin at 0.1 μM and 1.5 μg/ml, respectively, with the sit4 pdr5 double mutant CK413 showed that the latter is more sensitive to the drugs (Fig. 3, top). Likewise, when the cells were examined for resistance to econazole and ketoconazole (Fig. 3, bottom), CK413 was hypersensitive, in contrast to the drug-resistant strain CK254/1 (sit4). CK413 (sit4 pdr5) exhibits a drug tolerance level comparable to that of CK373/1 (Klpdr5), indicating that KlPDR5 is the only transporter for econazole and ketoconazole activated by the sit4 mutation. However, in cases with antimycin and oligomycin, as CK413 displays a higher drug tolerance than the Klpdr5 single mutant CK373/1, sit4 mutation might activate a distinct mechanism that contributes, to some extent, to drug tolerance.
The genetic interaction between SIT4 and KlPDR5 can also be observed when the rhodamine 123 efflux capacity of the mutants is measured. As shown in Fig. 4, the sit4 mutant CK432/8 has a significantly lower level of dye accumulation inside the cells, with a mean fluorescence intensity of 2.13, compared with 2.86 in the wild-type strain PM6-7A. However, a drastic increase in the intracellular accumulation of rhodamine 123 in the sit4 pdr5 double mutant CK413 was noticed, with a mean fluorescence intensity as high as 7.99, indicating that the strong rhodamine 123 efflux in the sit4 mutants is dependent on KlPDR5. The reason for the biphasic nature of CK413 in dye efflux is unknown.
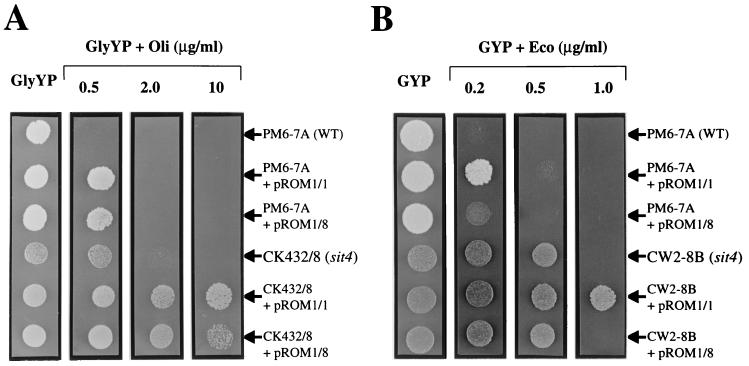
A synergistic hyperresistance can be created by overexpressing KlPDR5 in sit4 mutants.
Further support for a genetic interaction between SIT4 and KlPDR5 came from experiments showing that a drug hyperresistant phenotype can be created by overexpressing KlPDR5 in sit4 mutants. As shown in Fig. 5, when CK432/8 (sit4Δ::ura3) and CW2-8B (sit4-1) were transformed with the multicopy plasmids pROM1/1 and pROM1/8, both carrying KlPDR5, the resulting transformants displayed a dramatic increase in resistance to oligomycin and econazole. Expression of KlPDR5 from the plasmids in the wild-type strain PM6-7A or the untransformed sit4 mutant CK432/8 itself cannot tolerate oligomycin at a concentration beyond 2 μg/ml. However, CK432/8 carrying the plasmids can grow in the presence of the drug at a concentration as high as 10 μg/ml. Likewise, the growth of the PM6-7A transformants and the untransformed CW2-8B (sit4-1) is inhibited by econazole at 0.5 μg/ml whereas overexpression of pROM1/1 in CW2-8B results in cells resistant to 1 μg/ml. In both cases, the plasmid pROM1/8, carrying KlPDR5 with a truncated promoter (Fig. 1A), confers a lower level of resistance than pROM1/1, which has a functional promoter. Taken together, these observations provide strong evidence that loss of Sit4 function activates KlPdr5p in detoxifying oligomycin and econazole. Sit4p is therefore a negative regulator of the KlPdr5 transporter.
FIG. 5.
Growth and drug sensitivity test of K. lactis cells showing the synergistic hyperresistant phenotype created by a combination of KlPDR5 overexpression and sit4 mutation. K. lactis cells were grown in liquid minimal medium to stationary phase and tested on GlyYP and GYP supplemented with oligomycin (Oli) and econazole (Eco). The plates were incubated at 30°C for 4 days before being photographed. WT, wild type.
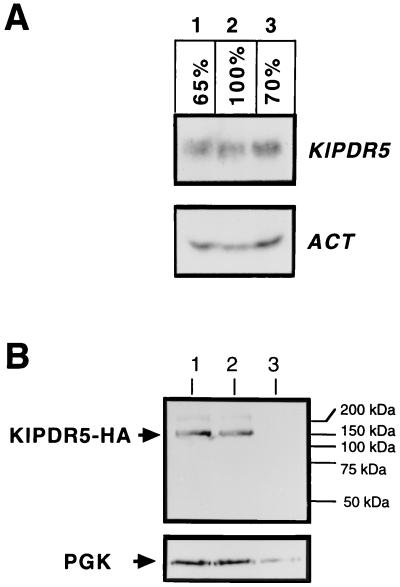
The transcription of KlPDR5 and the steady-state level of the KlPdr5 transporter are not increased in sit4 mutants.
The most common regulatory mechanism for MDR so far described in S. cerevisiae is the transcriptional control of ABC transporter genes. For instance, expression of ABC transporter genes such as PDR5, SNQ2, YOR1, PDR10, and PDR15 is dependent on the Cys6 zinc finger-type transcriptional activators Pdr1p and Pdr3p. As the SIT4 locus has been originally identified by mutations that suppress the transcriptional defect of the S. cerevisiae HIS4 gene (1), it is possible that the Sit4 protein phosphatase has a general role in regulating gene transcription and that in the K. lactis system described above, disruption of SIT4 might lead to transcriptional activation of KlPDR5 and consequently to an MDR phenotype. To examine this possibility, the transcriptional level of KlPDR5 was analyzed by Northern blotting of total RNA extracted from cells with a disruption in SIT4 or overexpressing the SIT4 gene. Relative mRNA levels were quantified by phosphorimaging by using the actin gene as an internal control for sample loading. The KlPDR5 mRNA level is slightly lower in both the sit4 mutant (Fig. 6A, lane 3) and the cells overexpressing SIT4 (lane 1) than in the wild-type strain (lane 2). The results indicate that the disruption or overexpression of SIT4 does not increase the transcriptional level of KlPDR5 and that SIT4 does not execute its regulatory role by affecting the transcription of KlPDR5.
FIG. 6.
Northern and Western blot analysis showing the mRNA abundance of KlPDR5 and the steady-state level of KlPdr5p. (A) Northern blot analysis. K. lactis strains PM6-7A (wild-type, lane 2) and CK254/1 (sit4Δ::URA3, lane 3) were grown in liquid GYP. The KlSIT4-overexpressing strain (lane 1) was PM6-7A transformed with pCXJ3-KlSIT4 (9) and grown in GYP supplemented with G418 at 200 μg/ml to maintain the plasmid. Total RNAs extracted from the strains were electrophoresed on a 1.2% agarose-formaldehyde gels, transferred to nylon membranes, and hybridized at high stringency with the 32P-labeled KlPDR5 and actin probes. The relative abundance of the KlPDR5 mRNA was estimated by PhosphorImager analysis by using the ACT1 mRNA as an internal control for sample loading. (B) Western blot analysis. Protein extracts were prepared from PM6-7A (wild-type, lane 2) and CK432/8 (sit4, lane 1) transformants carrying pCXJ18-KlPDR5HA and separated on a 4-to-20% gradient SDS-polyacrylamide gel before being blotted onto a nylon Immobilon-P membrane (Millipore) and probed with a monoclonal anti-HA antibody. A parallel membrane was probed with the anti-3-phosphoglycerate kinase (PGK) antibody to demonstrate the amounts of proteins loaded on each lane. Cell extracts from untransformed PM6-7A were included as a negative control for the lack of nonspecific cross-reactions of the antibodies with K. lactis proteins.
As the transcriptional level of KlPDR5 is not significantly affected by mutations in SIT4, the possibility was raised that the Sit4 protein phosphatase may influence the steady-state level of KlPdr5p by a mechanism involving the control of protein stability and turnover. To investigate this notion, KlPDR5 was tagged on its C terminus with the HA epitope, and Western blotting was carried out to examine the steady-state level of KlPdr5p in sit4 mutants compared with that in the wild type. The HA-tagged KlPDR5 was found to be functionally active and gave a drug resistance level indistinguishable from that of the wild-type on oligomycin plates (data not shown). The K. lactis centromeric plasmid pCXJ18-KlPDR5HA, carrying the KlPDR5-HA cassette under the control of the native KlPDR5 promoter, was introduced into PM6-7A (wild type) and CK432/8 (sit4Δ::ura3). Protein extracts were prepared from the transformants, followed by Western blotting by using the monoclonal antibody against HA. The experiment revealed that the steady-state level of KlPdr5p in the sit4 mutant (Fig. 6B, lane 1) is comparable to that in the isogenic wild-type strain PM6-7A (lane 2), suggesting that the sit4 mutation does not interfere with the accumulation of KlPdr5p in the cells. Also apparent from Fig. 6B is that there is no difference in the gel mobility of KlPdr5p between the sit4 mutant and the wild type.
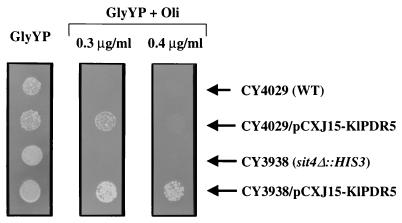
Activation of KlPdr5p by the sit4 mutation in S. cerevisiae.
To determine whether the SIT4-mediated regulation of MDR is evolutionarily conserved, we introduced KlPDR5 into S. cerevisiae to see whether the activity of KlPdr5p is affected by the state of the SIT4 gene. As shown in Fig. 7, although the S. cerevisiae sit4 mutant CY3839 does not show an increased tolerance to oligomycin compared with the isogenic wild-type strain CY4029, overexpression of KlPDR5 from the multicopy plasmid pCXJ15-KlPDR5 clearly confers resistance to oligomycin at concentrations up to 0.3 μg/ml in CY4029. The resistance is further increased to 0.4 μg/ml in the sit4 mutant CY3938, carrying pCXJ15-KlPDR5. The synergistic oligomycin resistance suggests that, like in K. lactis, the activity of KlPdr5p is activated by disruption of SIT4 in S. cerevisiae, although the magnitude of activation in this heterologous system seems to be much lower than that observed in K. lactis.
FIG. 7.
Deletion of SIT4 in S. cerevisiae enhances oligomycin resistance promoted by overexpression of KlPDR5. The strains were grown in liquid minimal medium, diluted in water, and spotted onto GlyYP supplemented with oligomycin. The plates were incubated at 30°C for 5 days before being photographed. WT, wild type.
DISCUSSION
A possible role of protein phosphatases in regulating MDR has not been well established. In previous work it was indicated that the Sit4 protein phosphatase of K. lactis plays a critical role in controlling drug transport (9). This finding makes K. lactis an attractive model system for investigating the novel regulatory mechanism of MDR for several reasons. First, Sit4p is a nonessential protein in K. lactis, so the effect of sit4 mutations on drug transport can be readily monitored. In S. cerevisiae, it has been shown that SIT4 is a cell cycle gene and sit4 mutations cannot be tolerated in many laboratory strains (52). Second, Sit4p in K. lactis seems to be a key regulator of MDR, as a number of drug transport systems are simultaneously affected by sit4 mutations. Third, Sit4p appears to control MDR through distinct regulatory pathways. It has been shown that the transport of oligomycin, antimycin, ketoconazole, and econazole is negatively regulated by Sit4p, whereas the efflux of sorbic acid and 4-nitroquinoline-N-oxide is under the positive control of the protein phosphatase. Finally, sit4 mutants exhibit strong genetic phenotypes in response to various drugs on a simple plate assay, which would enormously facilitate the analysis of genetic interactions between different components of the regulatory network.
In this study the first drug efflux ABC transporter gene, KlPDR5, was isolated from K. lactis and its functional interaction with the Sit4 protein phosphatase was examined. The deduced KlPdr5 protein displays 63.8% sequence identity to the Pdr5 transporter of S. cerevisiae. Structurally, the two proteins have a similar molecular architecture with the same organization of their functional domains, which include the two nucleotide binding sites, two ABC signatures, and 12 putative transmembrane regions. Functionally, KlPdr5p and ScPdr5p do not display totally overlapping substrate specificities. KlPdr5p transports the antifungal drugs ketoconazole and econazole, the mitochondrial inhibitors oligomycin and antimycin, and the fluorescent dye rhodamine 123. In S. cerevisiae, although ScPdr5p mediates the efflux of ketoconazole, econazole, and rhodamine 123 (20, 36, 45), oligomycin is extruded mainly by Yor1p (34). In addition, ScPdr5p does not seem to be involved in the detoxification of antimycin (Chen, unpublished). It has been reported that transmembrane domain 10 of Pdr5, at the predicted positions from 1355 to 1375, is an important determinant of substrate specificity (20, 21). Mutations at residues S1360 and T1364 alter both substrate specificity and inhibitor susceptibility. It is interesting that these two amino acids, which correspond to S1350 and T1354 in KlPdr5p, are conserved in the latter protein.
One of the most important objectives of this work was to provide direct evidence for genetic interaction between Sit4p and a drug efflux ABC transporter. With the availability of the KlPDR5 gene, it has been possible to demonstrate that KlPDR5 is epistatic to SIT4 for the transport of oligomycin, antimycin, and the antifungal drugs econazole and ketoconazole. This notion has been supported by several lines of evidence. It has been shown that disruption of KlPDR5 completely abolished the resistance of sit4 mutants to econazole, ketoconazole, and to lesser extent the mitochondrial inhibitors oligomycin and antimycin. The drug-resistant phenotype of sit4 mutants is therefore dependent on the function of KlPdr5p. Consistent with these observations, it has been shown that upon disruption of KlPDR5, the sit4 mutants have a reduced capacity to extrude rhodamine 123 and displayed a strong intracellular accumulation of the dye. Finally, overexpression of KlPDR5 in the sit4 background was found to be correlated with hyperresistance of the cells to oligomycin and econazole. All these observations strongly indicate that the drug efflux activity of KlPdr5p is negatively regulated by Sit4p. The Sit4p-responsive regulatory pathway seems to be conserved in S. cerevisiae, as the KlPdr5 transporter can also be activated to some extent by sit4 mutations in baker's yeast.
Phosphorylation of ABC transporters in S. cerevisiae has been described by several laboratories. The S. cerevisiae Ycf1 transporter seems to require a protein kinase A-type phosphorylation, as the S908A substitution in the PKA motif renders the proteins nonfunctional in the detoxification of cadmium (53). More recently, the S. cerevisiae Pdr12 transporter has been reported to be phosphorylated by the Cmk1 Ca2+-calmodulin-dependent protein kinase that exerts a negative posttranscriptional regulation over the drug efflux activity of the transporter (30). Although a phosphorylated form of Pdr12p was detected, the phosphorylation was independent of Cmk1p. The mechanism of regulation by Cmk1p therefore remains unknown. In a series of in vitro experiments, Decottignies and coworkers (15) showed that the S. cerevisiae Pdr5, Snq2, and Yor1 transporters are also subject to phosphorylation. In the case of Pdr5p, the in vitro phosphorylation involves the YCK1 and YCK2 genes, which encode the two isoforms of casein kinase I. Under semipermissive conditions for a yck1 yck2 double mutant, the steady-state level of Pdr5p is slightly decreased. This has been interpreted by a possible increased instability of Pdr5p. One serine residue, Ser420, has been identified in Pdr5p as a phosphorylation site for casein kinase I.
For K. lactis, it has yet to be determined how KlPdr5p is regulated by the Sit4 protein phosphatase. This study has shown that the transcriptional level of KlPDR5 is not increased compared with that in the wild type. Furthermore, the steady-state level of KlPdr5p is also unchanged in cells disrupted in SIT4. In considering the possible regulatory mechanisms, one can speculate that the activity of KlPdr5p can be modulated by controlling the membrane targeting, the intracellular distribution, and the ATP-hydrolyzing activity of the protein. Alternatively, it is possible that sit4 mutations could alter the membrane environment that in turn may affect the activity of the drug transporter. Mechanistically, Sit4p may directly act on KlPdr5p by dephosphorylating a Ser/Thr residue or indirectly alter KlPdr5p via a cascade of protein-protein interactions. A role of Sit4 in signal transduction has been well established in S. cerevisiae (4, 19, 31).
Mutation in SIT4 does not alter the gel mobility of KlPdr5p under the conditions employed in the present study. It is possible that the protein has a complex phosphorylation pattern which involves an interplay of several protein kinases and phosphatases. The effect of sit4 mutations on the gel mobility of the protein could be masked by fortuitous phosphorylation events that do not have any functional implications. In fact, scanning of KlPdr5p by the NetPhos software (version 2.0) identifies 56 serine and 17 threonine residues having a high probability of being phosphorylated. The demonstration of a specific phosphorylation state correlated with mutations in SIT4 and the identification of functionally meaningful phosphorylation sites could be an extremely challenging task.
In conclusion, this study has shown that the Sit4p-mediated regulatory mechanism plays a critical role in modulating the activity of the ABC transporter KlPdr5p in K. lactis. Although we do not know whether the expression of KlPDR5 is constitutive or induced by transcriptional activators similar to the Pdr1 and Pdr3 proteins of S. cerevisiae, modulation of drug pump activity by protein kinase and phosphatase provides an intriguing alternative for the regulation of MDR. Phosphorylation has already been known to regulate functions of ABC proteins in higher eukaryotic cells (22, 28, 37, 48, 54). Given that the effect of phosphorylation on the function of KlPdr5p has a strong phenotypic manifestation, the K. lactis system would provide an excellent tool for investigating this fundamentally important regulatory mechanism of ABC transporters.
ACKNOWLEDGMENTS
I thank G.D. Clark-Walker for supporting this work and for critical reading of the manuscript, A. Janssen, L.-J. Ouyang, and H. Liszczynsky for skillful technical support, and S. Grüninger for advice and help with FACS analysis.
REFERENCES
- 1.Arndt K T, Styles C A, Fink G R. A suppressor of a HIS4 transcriptional defect encodes a protein with homology to the catalytic subunit of protein phosphatases. Cell. 1989;56:527–537. doi: 10.1016/0092-8674(89)90576-x. [DOI] [PubMed] [Google Scholar]
- 2.Balzi E, Chen W, Ulaszewski S, Capieaux E, Goffeau A. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J Biol Chem. 1987;262:16871–16879. [PubMed] [Google Scholar]
- 3.Bauer B E, Wolfger H, Kuchler K. Inventory and function of yeast ABC proteins: about sex, stress, pleiotropic drug and heavy metal resistance. Biochim Biophys Acta. 1999;1461:217–236. doi: 10.1016/s0005-2736(99)00160-1. [DOI] [PubMed] [Google Scholar]
- 4.Beck T, Hall M N. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 5.Borst P, Evers R, Kool M, Wijnholds J. The multidrug resistance protein family. Biochim Biophys Acta. 1999;1461:347–357. doi: 10.1016/s0005-2736(99)00167-4. [DOI] [PubMed] [Google Scholar]
- 6.Chen X J, Wesolowski-Louvel M, Fukuhara H. Glucose transport in the yeast Kluyveromyces lactis. II. Transcriptional regulation of the glucose transporter gene RAG1. Mol Gen Genet. 1992;233:97–105. doi: 10.1007/BF00587566. [DOI] [PubMed] [Google Scholar]
- 7.Chen X J, Clark-Walker G D. sir2 mutants of Kluyveromyces lactis are hypersensitive to DNA-targeting drugs. Mol Cell Biol. 1994;14:4501–4508. doi: 10.1128/mcb.14.7.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X J. Low- and high-copy-number shuttle vectors for replication in the budding yeast Kluyveromyces lactis. Gene. 1996;172:131–136. doi: 10.1016/0378-1119(96)00125-4. [DOI] [PubMed] [Google Scholar]
- 9.Chen X J, Bauer B E, Kuchler K, Clark-Walker G D. Positive and negative control of multidrug resistance by the Sit4 protein phosphatase in Kluyveromyces lactis. J Biol Chem. 2000;275:14865–14872. doi: 10.1074/jbc.275.20.14865. [DOI] [PubMed] [Google Scholar]
- 10.Cole S P, Deeley R G. Multidrug resistance mediated by the ATP-binding cassette transporter protein MRP. Bioessays. 1998;20:931–940. doi: 10.1002/(SICI)1521-1878(199811)20:11<931::AID-BIES8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 11.Cui Z, Hirata D, Tsuchiya E, Osada H, Miyakawa T. The multidrug resistance-associated protein (MRP) subfamily (Yrs1/Yor1) of Saccharomyces cerevisiae is important for the tolerance to a broad range of organic anions. J Biol Chem. 1996;271:14712–14716. doi: 10.1074/jbc.271.25.14712. [DOI] [PubMed] [Google Scholar]
- 12.Cui Z, Shiraki T, Hirata D, Miyakawa T. Yeast gene YRR1, which is required for resistance to 4-nitroquinoline N-oxide, mediates transcriptional activation of the multidrug resistance transporter gene SNQ2. Mol Microbiol. 1998;29:1307–1315. doi: 10.1046/j.1365-2958.1998.01027.x. [DOI] [PubMed] [Google Scholar]
- 13.Decottignies A, Lambert L, Catty P, Degand H, Epping E A, Moye-Rowley W S, Balzi E, Goffeau A. Identification and characterization of SNQ2, a new multidrug ATP binding cassette transporter of the yeast plasma membrane. J Biol Chem. 1995;270:18150–18157. doi: 10.1074/jbc.270.30.18150. [DOI] [PubMed] [Google Scholar]
- 14.Decottignies A, Goffeau A. Complete inventory of the yeast ABC proteins. Nat Genet. 1997;15:137–145. doi: 10.1038/ng0297-137. [DOI] [PubMed] [Google Scholar]
- 15.Decottignies A, Owsianik G, Ghislain M. Casein kinase I-dependent phosphorylation and stability of the yeast multidrug transporter Pdr5p. J Biol Chem. 1999;274:37139–37146. doi: 10.1074/jbc.274.52.37139. [DOI] [PubMed] [Google Scholar]
- 16.Delaveau T, Delahodde A, Carvajal E, Subik J, Jacq C. PDR3, a new yeast regulatory gene, is homologous to PDR1 and controls the multidrug resistance phenomenon. Mol Gen Genet. 1994;244:501–511. doi: 10.1007/BF00583901. [DOI] [PubMed] [Google Scholar]
- 17.DeRisi J, van den Hazel B, Marc P, Balzi E, Brown P, Jacq C, Goffeau A. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 2000;470:156–160. doi: 10.1016/s0014-5793(00)01294-1. [DOI] [PubMed] [Google Scholar]
- 18.Dexter D, Moye-Rowley W S, Wu A L, Golin J. Mutations in the yeast PDR3, PDR4, PDR7 and PDR9 pleiotropic (multiple) drug resistance loci affect the transcript level of an ATP binding cassette transporter encoding gene, PDR5. Genetics. 1994;136:505–515. doi: 10.1093/genetics/136.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Como C J, Arndt K T. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 20.Egner R, Rosenthal F E, Kralli A, Sanglard D, Kuchler K. Genetic separation of FK506 susceptibility and drug transport in the yeast Pdr5 ATP-binding cassette multidrug resistance transporter. Mol Biol Cell. 1998;9:523–543. doi: 10.1091/mbc.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egner R, Bauer B E, Kuchler K. The transmembrane domain 10 of the yeast Pdr5p ABC antifungal efflux pump determines both substrate specificity and inhibitor susceptibility. Mol Microbiol. 2000;35:1255–1263. doi: 10.1046/j.1365-2958.2000.01798.x. [DOI] [PubMed] [Google Scholar]
- 22.Gadsby D C, Nairn A C. Control of CFTR channel gating by phosphorylation and nucleotide hydrolysis. Physiol Rev. 1999;79:S77–S107. doi: 10.1152/physrev.1999.79.1.S77. [DOI] [PubMed] [Google Scholar]
- 23.Gottesman M M, Hrycyna C A, Schoenlein P V, Germann U A, Pastan I. Genetic analysis of the multidrug transporter. Annu Rev Genet. 1995;29:607–649. doi: 10.1146/annurev.ge.29.120195.003135. [DOI] [PubMed] [Google Scholar]
- 24.Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hallström T C, Katzmann D J, Torres R J, Sharp W J, Moye-Rowley W S. Regulation of transcription factor Pdr1p function by an Hsp70 protein in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1147–1155. doi: 10.1128/mcb.18.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hallström T C, Moye-Rowley W S. Divergent transcriptional control of multidrug resistance genes in Saccharomyces cerevisiae. J Biol Chem. 1998;273:2098–2104. doi: 10.1074/jbc.273.4.2098. [DOI] [PubMed] [Google Scholar]
- 27.Hallström T C, Moye-Rowley W S. Hyperactive forms of the Pdr1p transcription factor fail to respond to positive regulation by the hsp70 protein Pdr13p. Mol Microbiol. 2000;36:402–413. doi: 10.1046/j.1365-2958.2000.01858.x. [DOI] [PubMed] [Google Scholar]
- 28.Hardy S P, Goodfellow H R, Valverde M A, Gill D R, Sepulveda V, Higgins C F. Protein kinase C-mediated phosphorylation of the human multidrug resistance P-glycoprotein regulates cell volume-activated chloride channels. EMBO J. 1995;14:68–75. doi: 10.1002/j.1460-2075.1995.tb06976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 30.Holyoak C D, Thompson S, Ortiz Calderon C, Hatzixanthis K, Bauer B, Kuchler K, Piper P W, Coote P J. Loss of Cmk1 Ca2+-calmodulin-dependent protein kinase in yeast results in constitutive weak organic acid resistance, associated with a post-transcriptional activation of the Pdr12 ATP-binding cassette transporter. Mol Microbiol. 2000;37:595–605. doi: 10.1046/j.1365-2958.2000.02017.x. [DOI] [PubMed] [Google Scholar]
- 31.Jiang Y, Broach J R. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jungwirth H, Wendler F, Platzer B, Bergler H, Hogenauer G. Diazaborine resistance in yeast involves the efflux pumps Ycf1p and Flr1p and is enhanced by a gain-of-function allele of gene YAP1. Eur J Biochem. 2000;267:4809–4816. doi: 10.1046/j.1432-1327.2000.01537.x. [DOI] [PubMed] [Google Scholar]
- 33.Katzmann D J, Burnett P E, Golin J, Mahe Y, Moye-Rowley W S. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol Cell Biol. 1994;14:4653–4661. doi: 10.1128/mcb.14.7.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katzmann D J, Hallstrom T C, Voet M, Wysock W, Golin J, Volckaert G, Moye-Rowley W S. Expression of an ATP-binding cassette transporter-encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6875–6883. doi: 10.1128/mcb.15.12.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein I, Sarkadi B, Varadi A. An inventory of the human ABC proteins. Biochim Biophys Acta. 1999;1461:237–262. doi: 10.1016/s0005-2736(99)00161-3. [DOI] [PubMed] [Google Scholar]
- 36.Kolaczkowski M, van der Rest M, Cybularz-Kolaczkowska A, Soumillion J P, Konings W N, Goffeau A. Anticancer drugs, ionophoric peptides, and steroids as substrates of the yeast multidrug transporter Pdr5p. J Biol Chem. 1996;271:31543–31548. doi: 10.1074/jbc.271.49.31543. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Salter-Cid L, Vitiello A, Preckel T, Lee J D, Angulo A, Cai Z, Peterson P A, Yang Y. Regulation of transporter associated with antigen processing by phosphorylation. J Biol Chem. 2000;275:24130–24135. doi: 10.1074/jbc.M003617200. [DOI] [PubMed] [Google Scholar]
- 38.Luke M M, Della Seta F, Di Como C J, Sugimoto H, Kobayashi R, Arndt K T. The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol Cell Biol. 1996;16:2744–2755. doi: 10.1128/mcb.16.6.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahé Y, Parle-McDermott A, Nourani A, Delahodde A, Lamprecht A, Kuchler K. The ATP-binding cassette multidrug transporter Snq2 of Saccharomyces cerevisiae: a novel target for the transcription factors Pdr1 and Pdr3. Mol Microbiol. 1996;20:109–117. doi: 10.1111/j.1365-2958.1996.tb02493.x. [DOI] [PubMed] [Google Scholar]
- 40.Meyers S, Schauer W, Balzi E, Wagner M, Goffeau A, Golin J. Interaction of the yeast pleiotropic drug resistance genes PDR1 and PDR5. Curr Genet. 1992;21:431–436. doi: 10.1007/BF00351651. [DOI] [PubMed] [Google Scholar]
- 41.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 42.Prasad R, De Wergifosse P, Goffeau A, Balzi E. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr Genet. 1995;27:320–329. doi: 10.1007/BF00352101. [DOI] [PubMed] [Google Scholar]
- 43.Reed M B, Saliba K J, Caruana S R, Kirk K, Cowman A F. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 44.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 45.Sanglard D, Kuchler K, Ischer F, Pagani J L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Senior A E, al-Shawi M K, Urbatsch I L. The catalytic cycle of P-glycoprotein. FEBS Lett. 1995;377:285–289. doi: 10.1016/0014-5793(95)01345-8. [DOI] [PubMed] [Google Scholar]
- 48.Sheppard D N, Welsh M J. Structure and function of the CFTR chloride channel. Physiol Rev. 1999;79:S23–S45. doi: 10.1152/physrev.1999.79.1.S23. [DOI] [PubMed] [Google Scholar]
- 49.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1983. [Google Scholar]
- 50.Simon S M, Schindler M. Cell biological mechanisms of multidrug resistance in tumors. Proc Natl Acad Sci USA. 1994;91:3497–3504. doi: 10.1073/pnas.91.9.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subik J, Ulaszewski S, Goffeau A. Genetic mapping of nuclear mucidin resistance mutations in Saccharomyces cerevisiae. A new pdr locus on chromosome II. Curr Genet. 1986;10:665–670. doi: 10.1007/BF00410914. [DOI] [PubMed] [Google Scholar]
- 52.Sutton A, Immanuel D, Arndt K T. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szczypka M S, Wemmie J A, Moye-Rowley W S, Thiele D J. A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J Biol Chem. 1994;269:22853–22857. [PubMed] [Google Scholar]
- 54.Vanoye C G, Castro A F, Pourcher T, Reuss L, Altenberg G A. Phosphorylation of P-glycoprotein by PKA and PKC modulates swelling-activated Cl− currents. Am J Physiol. 1999;276:C370–C378. doi: 10.1152/ajpcell.1999.276.2.C370. [DOI] [PubMed] [Google Scholar]
- 55.van Veen H W, Callaghan R, Soceneantu L, Sardini A, Konings W N, Higgins C F. A bacterial antibiotic-resistance gene that complements the human multidrug-resistance P-glycoprotein gene. Nature. 1998;391:291–295. doi: 10.1038/34669. [DOI] [PubMed] [Google Scholar]
- 56.Wemmie J A, Szczypka M S, Thiele D J, Moye-Rowley W S. Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J Biol Chem. 1994;269:32592–32597. [PubMed] [Google Scholar]
- 57.Wendler F, Bergler H, Prutej K, Jungwirth H, Zisser G, Kuchler K, Hogenauer G. Diazaborine resistance in the yeast Saccharomyces cerevisiae reveals a link between YAP1 and the pleiotropic drug resistance genes PDR1 and PDR3. J Biol Chem. 1997;272:27091–27098. doi: 10.1074/jbc.272.43.27091. [DOI] [PubMed] [Google Scholar]
- 58.Wésolowski-Louvel M, Tanguy-Rougeau C, Fukuhara H. A nuclear gene required for the expression of the linear DNA-associated killer system in the yeast Kluyveromyces lactis. Yeast. 1988;4:71–81. doi: 10.1002/yea.320040108. [DOI] [PubMed] [Google Scholar]
- 59.Wirsching S, Michel S, Morschhauser J. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol Microbiol. 2000;36:856–865. doi: 10.1046/j.1365-2958.2000.01899.x. [DOI] [PubMed] [Google Scholar]