Abstract
Background
Inflammatory bowel disease (IBD) is a gastrointestinal chronic disease with an unpredictable disease course. Computational methods such as machine learning (ML) have the potential to stratify IBD patients for the provision of individualized care. The use of ML methods for IBD was surveyed, with an additional focus on how the field has changed over time.
Methods
On May 6, 2021, a systematic review was conducted through a search of MEDLINE and Embase databases, with the search structure (“machine learning” OR “artificial intelligence”) AND (“Crohn* Disease” OR “Ulcerative Colitis” OR “Inflammatory Bowel Disease”). Exclusion criteria included studies not written in English, no human patient data, publication before 2001, studies that were not peer reviewed, nonautoimmune disease comorbidity research, and record types that were not primary research.
Results
Seventy-eight (of 409) records met the inclusion criteria. Random forest methods were most prevalent, and there was an increase in neural networks, mainly applied to imaging data sets. The main applications of ML to clinical tasks were diagnosis (18 of 78), disease course (22 of 78), and disease severity (16 of 78). The median sample size was 263. Clinical and microbiome-related data sets were most popular. Five percent of studies used an external data set after training and testing for additional model validation.
Discussion
Availability of longitudinal and deep phenotyping data could lead to better modeling. Machine learning pipelines that consider imbalanced data and that feature selection only on training data will generate more generalizable models. Machine learning models are increasingly being applied to more complex clinical tasks for specific phenotypes, indicating progress towards personalized medicine for IBD.
Keywords: artificial intelligence, machine learning, inflammatory bowel disease
Introduction
Inflammatory bowel disease (IBD) is an umbrella term for a set of chronic diseases, of which there are 2 main subtypes: Crohn’s disease (CD) and ulcerative colitis (UC). The global prevalence of IBD increased to 84.3 per 100 000 by 2017, and with it comes a greater burden to patients and health services.1 Due to a number of factors contributing to its etiology, IBD disease course is highly variable. Patients can experience a mild disease or a severe, refractory disease requiring many interventions. A patient’s disease course is often unclear at diagnosis.
There has been a relative explosion in the use of artificial intelligence and machine learning (ML) techniques for complex diseases, after the success of these algorithms in fields like oncology.2 Unlike traditional statistical techniques, ML infers patterns from data, allowing model application to unseen cases. Key concepts for this field are included in Box 1, and a further breakdown of ML terms, metrics, and methods are detailed elsewhere.3,4 For IBD, ML has the potential to improve patient care at every stage of their disease course through prediction modeling: from a quick subtype diagnosis so appropriate treatment can be identified, to assessing disease activity and identifying those patients more likely to develop complications and require surgery. For clinicians, this potential is exciting but comes with many questions about which ML methods may be successful. Here, practical guidelines are provided to guide interpretation of current and future research in this field (Appendix 1). Although this systematic review centers around applications to IBD, these are general guidelines for ML interpretation.
Key Messages.
What is already known?
Machine learning has been applied with success to cancer diagnostics, and now these methods are increasingly being applied to complex conditions, such as inflammatory bowel disease.
What is new here?
In the past 2.5 years, the number of articles published in this field has increased by 68%; this has been accompanied by a shift in machine learning applications from diagnostics to prognostics, and the use of more complex methods such as neural networks.
How can this study help patient care?
Two main requirements were identified for translation of machine learning models into the clinic: generalizable models generated from robust pipelines, and the collection of deep and specific patient phenotype data.
Box 1. – Key Concepts.
Artificial Intelligence: methods that enable computers to mimic human intelligence.
Machine Learning: methods that infer patterns from data to perform a specific task, usually classification or regression.
Deep Learning: neural network–based approaches that enable machines to train themselves to perform tasks.
Supervised Learning: the ML model learns patterns in data and associates this information with an already present label. The model can then apply this learning to new data and predict these labels.
Unsupervised Learning: the ML model identifies patterns and clusters the data in a way that explains the data structure (not according to labels).
Feature Selection: a collection of methods that reduce the dimensionality of a data set, such that ML is performed on a subset of the most informative variables for the task.
Cross Validation: a method that can reduce the overfitting of ML models, meaning the results will generalize well to new data. During training the data are split into k folds, and the ML model trained on k-1 folds. The model performance is tested on the final fold, and the process is repeated so each fold becomes the test fold exactly once.
In a previous, broader systematic review of artificial intelligence and ML applications to autoimmune diseases,3 a number of popular methods and applications were identified, and the research assessed guided some recommendations for the field. In addition, other systematic reviews have been published commenting on this area, including Tontini et al’s review of artificial intelligence for gastrointestinal endoscopy5 and Nguyen et al’s study on machine learning for diagnosis and prognosis in IBD.6 The aim of this systematic review was to assess common data types, applications, and methods in the field of ML for IBD and to evaluate changes in the field over the past few years. The broad scope of this review allows for the assessment of trends and the recording of the full range of ML applications to IBD.
Methods
Literature Search
An electronic literature search was performed using 2 databases available through OvidSP: MEDLINE(R) and Epub Ahead of Print, In-Process, In-Data-Review & Other Non-Indexed Citations and Daily 1946, and Embase 1974. The literature search was completed in May 2021. Search terms were combined using Boolean operators as follows: (“machine learning” OR “artificial intelligence”) AND (“Crohn* Disease” OR “Ulcerative Colitis” OR “Inflammatory Bowel Disease”). Any record in which these search terms were identified in the title, abstract, and/or subject headings would appear in the list of records (last search May 6, 2021).
Inclusion and Exclusion Criteria
This systematic review sought to expand and better characterize a subsection of a previous review of ML in autoimmune disease; therefore the same criteria was employed. Studies that applied ML to IBD, or a subtype of IBD, were included. Studies that used ML for analysis of complications that arise from IBD were also included. Studies that were not written in English, were published prior to 2001, that did not use real human patient data, were not peer reviewed, or were not original research articles were also excluded. Therefore, the following publication types (as labeled by OvidSP) were not assessed during screening: conference abstracts, conference review, editorial, erratum, journal article comment, journal article review, letter, letter comment, note, and review. The abstract of each study was assessed by 2 reviewers independently for inclusion in the systematic review. The full text was assessed when a decision on inclusion could not be made based on the abstract, and a consensus was reached by the 2 reviewers. The following data items were collected for each study that met the criteria: the task ML was applied to, the type of ML (supervised or unsupervised), all ML algorithms trialed by the researchers, the best performing ML algorithm, sample size, clinical population (IBD, UC, or CD), data type, the best results achieved, whether a training and testing split was used, whether other cross-validation was used, whether the model was applied to independent test data, and the year of publication. This systematic review conforms to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) standards.7
Graphical Representations of Data
Articles were graphically summarized in sunburst or pie chart diagrams using a custom R script utilizing the Plotly library.8,9 The R scripts can be downloaded from Github (github.com/isstafford/review_ml_ibd_2021). Briefly, all articles were classified according to the machine-learning approach used (method), the type of information being analyzed (data type), and the outcome which is being predicted (task). These categories and the sorting of studies into categories were agreed upon by all authors. The R script counts unique titles under each level of the 3-class hierarchy, and the results are displayed in a sunburst diagram. The innermost ring represents the highest level of the hierarchy, whereas the outermost ring represents the individual articles. Since some articles discuss multiple methods, tasks, or outcomes, they may be represented more than once on the diagram.
Graphical summaries of sample sizes and uses of ML method types over time were generated using ggplot2 in R.10 For the sample size graphical summary, the ML method was counted as used if it was recorded as a method in the research article, even if the ML method did not generate the optimal model. Machine learning methods were sorted into type groups (eg, ridge regression and logistic regression were both included under “regression”). Multiple methods from the same type group within the same article were counted once to avoid skewing the data. In cases where articles investigated multiple classification problems with different sample sizes, each classification problem counted as a separate entry. All ML method groups with sufficient data for a boxplot (n ≥ 5) were included in the visualization. The same ML method groups were plotted for the use of ML types over time.
Results
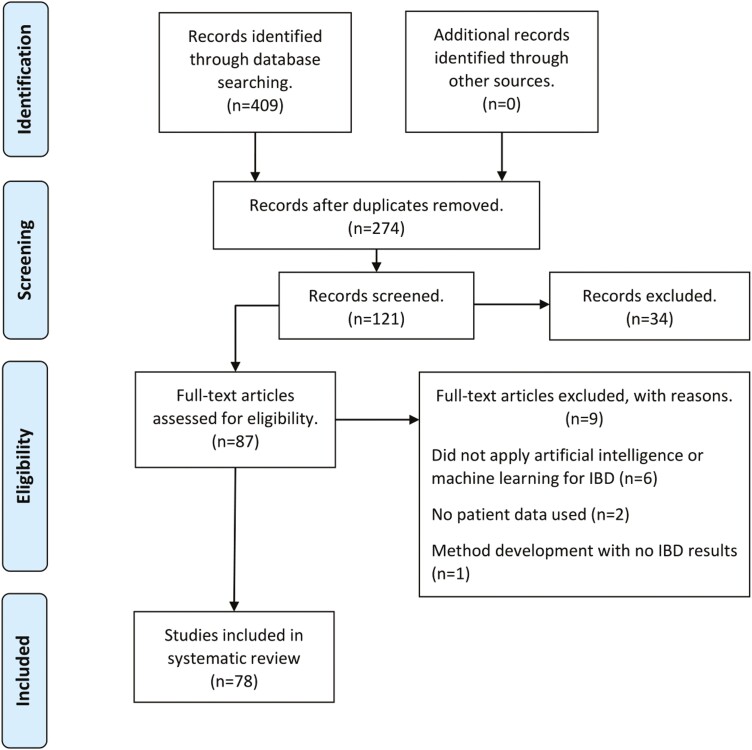
Initially, 409 records were identified, and 135 records were subsequently removed as duplicates. When the study criteria regarding original research articles, year of publication, and language were applied, 153 entries were removed. Of the remaining 121 screened articles, 33 were excluded after assessing the abstract against the inclusion and exclusion criteria, and a further 9 were excluded after a full-text read (Figure 1). A technical analysis of the ML applications in these studies is outside the scope of this review. Here, summary statistics are provided regarding popular methods, applications, and data; summary statistics are also provided regarding the sample sizes, cross-validation, and trends in ML usage in recent years. The chosen ML models and data types used for each type of task are detailed in Table 1.
Figure 1.
Flowchart documenting number of records found and reviewed at each stage.
Table 1.
Summary of ML Models Chosen as Most Optimal for the Clinical Task, and the Types of Data Used (ML models and data types sorted alphabetically).
| Task | No. Studies | Chosen ML Models | Data Types Used |
|---|---|---|---|
| Disease Course | 22 | Bayes Network, Boosting, Decision Tree, Hierarchical Clustering, Neural Network, Partial Least Squares Discriminant Analysis, Random Forest, Regression, Support Vector Machine | Clinical, Gene Expression, Genetic, Imaging, Metabolomic, Metatranscriptomic, Microbiome |
| Diagnosis | 18 | Boosting, Hierarchical Clustering, Neural Network, Random Forest, Regression, Support Vector Machine | Gene Expression, Genetic, Imaging, Metabolomic, Microbiome |
| Disease Severity | 16 | Bayes Network, Boosting, Decision Tree, Hierarchical Clustering, Intelligent Monitoring, Neural Network, Regression, Support Vector Machine | Clinical, Gene Expression, Genetic, Imaging, Protein Biomarkers |
| Disease Subtype | 8 | Boosting, Hierarchical Clustering, Random Forest, Similarity Network Fusion Clustering, Support Vector Machine | Clinical, Gene Expression, Metabolomic, Microbiome |
| Treatment Response | 7 | Neural Network, Random Forest | Clinical, Gene Expression, Microbiome |
| Risk of Disease | 6 | Ensemble Model, Random Forest, Regression | Clinical, Gene Expression, Genetic |
| Patient Clustering | 4 | Gaussian Mixture Model, Hierarchical Clustering, Latent Dirichlet Allocation, Neural Network | Immunoassay, Metagenomic, Online Posts, Questionnaire |
| Medication Adherence | 1 | Support Vector Machine | Clinical |
| Metabolite Abundance | 1 | Sparse Neural Encoder-Decoder Network | Metabolomic, Microbiome |
| Identification of Patients | 1 | Natural Language Processing | Clinical |
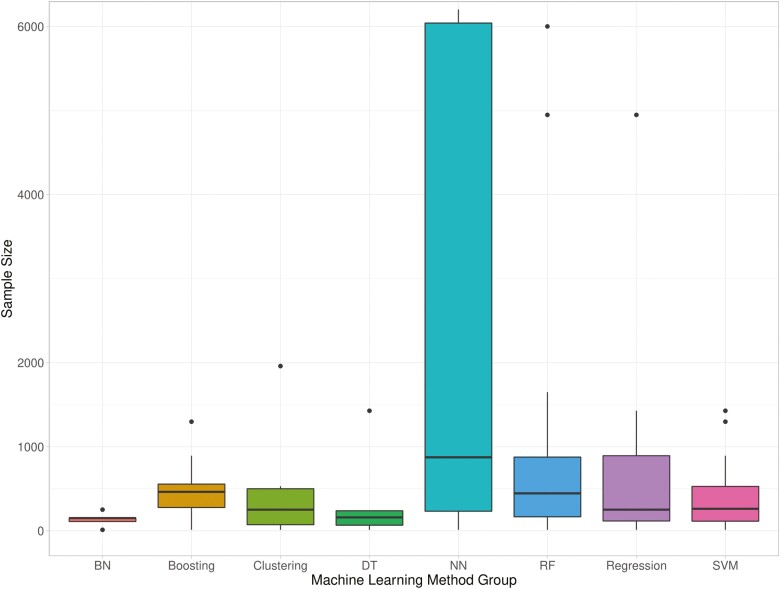
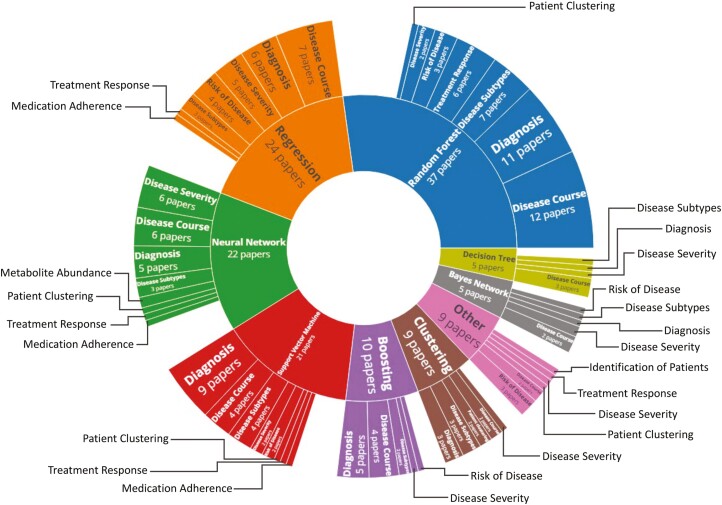
Of 78 studies included in the systematic review, the majority used supervised ML, with 4 articles employing unsupervised methods,11–14 and 5 utilizing both supervised and unsupervised ML15–19 for varied clinical applications. Many articles trialed different ML methods before selecting the optimal one, and some researchers implemented ML for multiple IBD applications. Three main clinical application areas were identified: diagnosis (23%),15,20–36 disease course (28%),15,30,37–56 and disease severity (21%).19,57–71 Diagnosis classification tasks involved differentiating IBD patients (or one subtype) from controls. Studies of disease course examined relapse, remission, and surgery ML classifiers. Disease severity studies sought to predict patients’ IBD activity or those who may develop complications. The most prevalent method implemented was random forest (47%), with regressions, neural networks, and support vector machines also used regularly (31%, 28%, and 27%, respectively. Percentages here sum to over 100%, as multiple methods were trialed by one study in many instances). Other tree-based methods were used by 22% of studies (13% boosting with trees, 9% decision trees). Clinical data (41%) and data related to the microbiome (23%) were the most commonly used in ML modeling. The median sample size, not including external validation data sets that were additional to usual training and testing data, was 263 (range, 12-7 400 000). A breakdown of sample sizes per ML method used can be viewed in Figure 2. Validation data sets in addition to the expected training and testing sets were used in 5% of studies.12,32,41,72 Another 7 studies trained their models with cross-validation on one data set and tested their method on an external, independent data set.23,29,45,59,61,66,73 Crohn’s disease data (only) was used in 27 studies,12,17,19,26–29,32,37–39,41,44,47–50,58,60,63,65,74–79 and UC data (only) was used in 15 studies,25,40,42,46,52,55,59,61,62,64,68–70,80,81 with the remainder (n = 36) using a mix of CD and UC data, or IBD data as one class.11,13–16,18,20–24,30,31,33–36,43,45,51,53,54,56,57,66,67,71–73,82–88 Half of the research using UC-only data focused on predicting disease activity with endoscopy data, whereas the aims of ML classifications on CD data were varied. A breakdown of the method and classification task can be found in Figure 3, which can be customized here (isstafford.github.io/review_ml_ibd_2021/). More details regarding each study included in this review are found in Supplementary Table 1.
Figure 2.
Sample sizes used for each group of machine learning methods. Abbreviations: BN, Bayes Network; DT, decision tree; NN, neural network; RF, random forest; SVM, support vector machine. Note that 10 outlier entries (sample sizes 20 368 to 7 400 000) have been excluded from the visualization.
Figure 3.
Sunburst of machine learning methods and the classification tasks used in conjunction with them.
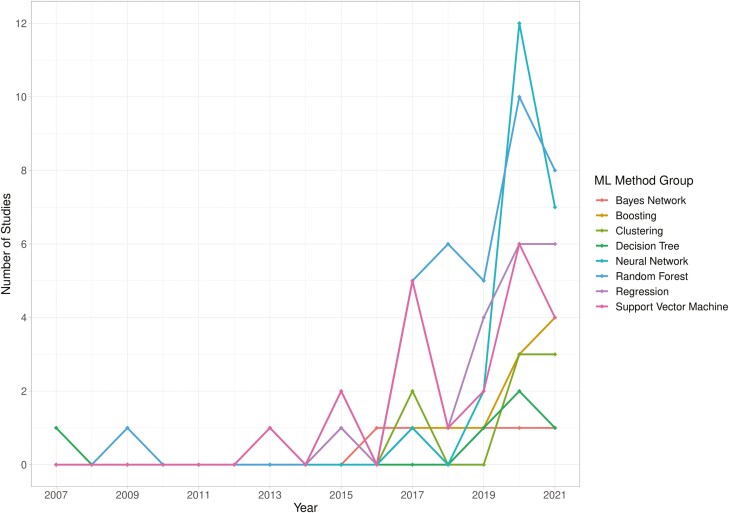
The literature search assessed in the previous systematic review was completed on the December 18, 2018; therefore, comparisons were made between studies published before and during 2018 and those published from 2019 to the literature search date. Fifty-three articles have been published from 2019 to May 2021. If a publication is published online and subsequently printed in a different year, the first publication date is used. Since the end of 2018, there has been a rapid expansion in the use of neural networks (a deep learning method) for IBD, with 21 studies trialing this method on their data from 2019 onwards, compared with 1 study prior to this. This increase coincides with more imaging data sets (4% 2007-2018, 18% 2019-2021), specifically colonoscopy data; the majority of neural networks were applied to this data type. Support vector machine, random forest, and regression-based methods were popular during both time periods (year on year breakdown of ML method group use, Figure 4). More studies utilized 2 data types in 2019-2021 (8% vs 17%), almost always combining clinical data with another data type. The median sample size of studies has not increased in recent years (N = 273 2007-2018, N = 257.5 2019-2021). Diagnosis has continued to be a popular ML application, but prior to 2019, investigating treatment response was more popular (24% vs 1.8%); and exploring classification tasks connected to disease course is now the most popular application (12% vs 35.8%).
Figure 4.
Implementation of machine learning methods over time; incomplete data for 2021.
Discussion
The increased use of ML methods for IBD demonstrates the wider interest in artificial intelligence for health care. Due to the heterogeneity of ML model workflows, data types and reported metrics, it was not possible to ascertain any superior approaches. It is possible that some studies may have been excluded from the review, as Medical Subject Headings (MeSH) were not utilized in the search strategy. However, when the ML subject heading was expanded, the only algorithm specified as a search term was “support vector machine,” which could have biased the search strategy towards only identifying additional articles that used this classifier. An additional limitation was the search of only 2 databases, as the systematic search focused on capturing models with clinical application. An assessment of the risk of bias was not performed, as there is no clear equivalent of PROBAST (Prediction model Risk Of Bias ASsessment Tool) to assess ML modeling. The construction of a tool that could assess potential ML pipeline bias would be beneficial for the transition of models into clinical settings. Minimizing bias in modeling and creating generalizable models go hand in hand.
There is a clear dominance of tree-based methods: one or a combination of random forests, decision trees, and tree-based boosting methods were implemented by 55% of studies. This is potentially due to decision trees being highly interpretable, with tree boosting and random forest preventing overfitting of this model type. Random forests are also well known as an ML algorithm that can leverage nonlinear relationships. This popularity is not inherently a drawback; however, a lack of comparison of different ML methods or a lack of reporting of this comparison in studies may make developing ML models for clinical application more challenging.
Overall, there was good reporting of a range of informative metrics in these studies, which was particularly important given many of their data sets had imbalanced classes. In cases where imbalanced data are used, a high accuracy score can mask poor prediction of the minority class. Although some studies sought to correct these imbalances with algorithm weighting82 or oversampling of the minority class,48,64 some researchers did not explicitly address this. Imbalanced data sets may be representative of the patient population, but it is important to consider whether enough samples from both classes were present in ML algorithm training, such that accurate predictions can be made for both the majority and minority class. Another potential ML pipeline issue discovered here was a use of feature selection on the whole data set, rather than just the training set. With this workflow, there is a danger that information regarding features in the test set leaks into the training set. Improvements to ML pipelines can only benefit patients, as more robust workflows allow the identification of the most successful models.
Although an increase in the use of external data sets in the expected workflow of training and testing on one data set and validating on an independent data set was not observed, other interesting approaches were employed, showing researchers bringing data sets together to extract additional information. Some studies used all of their initial data set to train their model with cross-validation and subsequently tested the model on a different external data set.23,29,45,59,61,66,73 Others used a type of cross-validation called LODO, or leave-one-data set-out, allowing researchers to utilize many, smaller data sets.27,84
The range of overall data set sizes used in studies was large. Some of the smaller sample sizes used may not have been appropriate for the chosen ML method, although evaluating whether there was sufficient data to construct a classifier can be challenging; and the required sample size is contingent on the ML task. There is no standard power calculation available for studies using ML. The sample size required depends on the method used, with algorithms such as neural networks requiring more data. This trend was observed in the systematic review: larger data sets were used in conjunction with neural networks. The number of features used for modeling will also affect the required sample size. More features will generally produce a more complex model, so a larger data set is necessary. If the ML model has generalized well from training to testing data (or other independent data), this is a good indicator the data set was sufficient. It is also important to consider how representative the data set is of the patient population. An ML model may perform well in initial training and testing, but if the data set is biased in the demographics or phenotypes represented, then the modeling may translate poorly when implemented in clinical settings.
Although diagnosis (classifying controls and IBD patients) is still a popular application in 2021, it was encouraging to see the highest percentage of articles addressing issues surrounding disease course in recent years. This suggests that more longitudinal and deeper phenotyping data are being collected, allowing a move towards more precise and complex classification tasks. The median size of data sets has not grown in recent years. Although data set size is not an indicator of data quality, it is surprising that even though we are in the era of “big data,” data set sizes are not increasing at a rate they might be expected to. A potential roadblock in garnering larger data sets for more specific classifiers may be linking up these other data types with phenotyping data. A community effort may be necessary to accumulate sufficient data sets for more accurate and generalizable ML models and external validation. Despite the uncontested power of ‘omics data sets in providing a—usually—unbiased representation of a patient profile, detailed clinical information remains fundamental for precise phenotyping and patient stratification. Projects such as UK Biobank89 have progressed this need for data, but phenotyping can be limited. With this data in place, robust pipelines and models that generalize well, the community takes the next step towards personalized medicine for IBD patients. Ways to assess the generalizability of ML models are addressed in the Appendix.
Supplementary Material
Acknowledgments
This study was supported by the Institute for Life Sciences, University of Southampton and the National Institute for Health Research (NIHR) Southampton Biomedical Research Centre. The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care.
Appendix: Application Of AI to IBD in Literature: Points to Consider for Clinicians
Rare indeed is the clinician who possesses the knowledge, experience, and time to master all the intersecting disciplines in health care ML research. Compounding this are the susceptibility of AI in health care to hype cycle effects,90 and a documented deficit in the quality and completeness of reporting in AI in health care articles.91
The interested but busy clinician has aids at their disposal in the form of guidelines and checklists on reporting requirements and quality. Some are tailored for clinicians, others more broadly to peer reviewers, users and authors.91–101 We recommend that clinicians obtain these whenever possible, especially those customized for clinicians91–93,100 for self-paced study and to have on hand as an assessment support tool when reading articles.
However, methodically going through a checklist every time an article is read may still pose a time challenge for the busy clinician, as some lists are quite lengthy. Realistically, it is to be expected that they will often read and assess articles against background knowledge/situational awareness. Herein, we provide a brief exposition on our subjective choice of a subset of critical items for clinicians’ situational awareness in the absence of a checklist. This should also enhance their understanding and use of checklists.
Comparing ML Algorithms
Direct model performance measures include accuracy, recall (sensitivity), precision (positive predictive value), specificity, negative predictive value, area under the curve (AUC), and F1-score. Comparing ML algorithms on these measures across studies is not usually meaningful because of unmatched factors including specific model implementation; database size, type, structure, and quality; reported performance metrics; and specific application. However, studies have compared multiple ML algorithms and curated benchmarking repositories. These repositories document the application of numerous ML algorithms to multiple diverse data sets and provide general insights on performance gradients where consistent performance gradients are observed.102–104 The caveat is these insights may not completely generalize, especially with more modern algorithms such as neural networks. These methods entail many analytical choices that impact ML architecture, performance, and limitations. The algorithms are highly situational and operator-dependent. In fact, it has been argued that a fair machine vs machine comparison would have to eliminate operator interaction, making the question unanswerable.103 Generally, newer ML algorithms can leverage current computational capacity to predict on complex, large, high-dimensional better than older traditional ML algorithms—but not always.102–104
Importantly, choice of an ML model is not only related to direct performance measures. Indirect performance-related ML characteristics are equally important, but perhaps more difficult to compare across different workflows. These include algorithm susceptibility to overfitting, which can lead to difficulties achieving the same results in different data; the transparency of ML model decisions; the time and computational cost of ML algorithms; whether the data set is static (offline learning) or updating over time (online learning); raw data preprocessing (feature engineering); whether the model is robust to outliers; and if the ML algorithm contains statistical assumptions. Even environmental impacts may come to play a role in algorithm selection. Increasing computational complexity correlates with increased energy consumption and greenhouse emissions. One group of investigators estimated that the CO2 emissions of a full training cycle for a neural network emits approximates the lifetime CO2 emissions of 4 cars.105
Machine learning model selection will therefore always entail some trade-offs. A complex algorithm may deliver excellent performance for the task but at high computational cost—and a limited understanding of how this performance was achieved. All ML algorithms have different strengths and weaknesses. Support vector machines, which find a boundary between different classes of points by optimally separating the closest points from the 2 classes, are less susceptible to overfitting and data outliers, but very large and noisy data sets present a challenge for the algorithm to extract a meaningful boundary between 2 classes. In contrast, a properly initialized deep neural network can approximate any complex decision boundary by identifying, exploiting, and revealing complex interactions. The constructed model may be highly accurate but has a propensity to overfit and sensitivity to initialization as the cost. For all ML methods, it is important to be aware of any assumptions contained within the model. This is especially important for more assumption-laden traditional ML. However, if those assumptions are met, the models can return a quick, competitive performance. It is therefore apt to consider whether a statistically significant ML performance translates into clinical impact for a specific application, given the totality of direct and indirect measures.
Data Set Quality, Construction, and Labelling
Good training, test and validation data sets are foundational. They should have accurate class labels (i.e. true positive and negative instances) that are well defined, and capture the full range of clinical, demographic and practice variables to be generalizable. Clinical expertise is as important as ML expertise to detect unrepresentative data. Training, test and validation sets should originate from independent sources, or the data should be randomly (not manually) split. Care should be taken to prevent data leakage (test data information leaking into training data), causing biased results, poor generalization, and camouflaged overfitting. This can occur in data with multiple samples, especially time series, from individual patients, or with data pre-processing or transformation prior to splitting. For example, normalizing a continuous variable in the prior to splitting data by using its global mean and standard deviation, is leaking some information from the test set into the training set.
Clinician rating schemes for diagnostic labelling (eg, labelling patients in the data set according to IBD disease activity, endoscopic image-based diagnosis) can affect performance, for example assessment by majority vote vs full adjudication on the same reference set can return significantly different error rates.100 Blinded adjudication by an expert panel provided with sufficient information and time is ideal for subjective labels.
Opportunities for poor representativeness abound. An IBD-specific example is generalizing from general/adult-focused IBD data to early-onset IBD, with its unique phenotypes. Dichotomous classification, for example IBD vs healthy controls, is common and does not accurately reflect differential diagnosis in the clinic, pathology lab, radiology reading room, or endoscopy suite, involving multiple diagnostic possibilities, thus generalizing poorly to real-world settings. In endoscopy studies, deficient representation can be caused by many factors such as endoscope brands, endoscopic modality (high vs standard resolution white light, chromoendoscopy), operator skill, number of study sites, inclusion/exclusion criteria (eg, only best archived images, patients with adequate bowel preps). In digital pathology, randomly cropped vs whole slide images presence/absence of standardisation of whole-slide imaging and staining, could return different results.106,107 Distributional shifts occur when characteristics and context of contemporary data, such as evolving clinical phenotypes, diverge from the training data, resulting in an outdated representation. Ongoing acquisition and use of training data representing current disease or practice are required.108,109
Imbalanced Data Sets
Imbalanced data are unavoidable, especially as ML algorithms move towards more complex prediction tasks. It may bias performance towards predicting the majority class at the expense of the minority class and makes overall accuracy an unreliable performance measure, as high accuracy can be achieved by just naïvely guessing the majority class. The severity of this effect is amplified with smaller, poorly separated, and/or suboptimally labeled data sets. Readers should determine if significant imbalance reflects a natural distribution vs an artifact of study execution. It is tempting to just rebalance the data by oversampling the minority class, either with real or synthetic data, or undersampling the minority class. If the imbalance is natural, performance on the unrealistically rebalanced data set may diverge from real-world results, which tend to be biased toward the minority class. Oversampling should occur after training/testing set splits to avoid data leakage. If practical, enlarging the data set without distorting the natural proportions is desirable. Other approaches include prioritizing the importance or more heavily penalizing false positives or false negatives, according to the specific problem. Another approach is finding a feature set in which the classes are more separable. Finally, trying another ML method such as tree-based approach may be indicated.
Articles with imbalanced data should provide a full suite of direct performance metrics, given the untrustworthiness of overall accuracy in this scenario. Examining the confusion matrix is key to assess class-specific performance, and it serves as the basis for calculating other measures such as precision, recall-balanced accuracy, and F1 score. For performance measures that are a function of prevalence such as predictive value, extrapolation from rebalanced data sets to the natural prevalence ratio should be provided.
Supplementary Data
Supplementary data is available at Inflammatory Bowel Diseases online.
Contributor Information
Imogen S Stafford, Human Genetics and Genomic Medicine, University of Southampton, Southampton, UK; Institute for Life Sciences, University Of Southampton, Southampton, UK; NIHR Southampton Biomedical Research, University Hospital Southampton, Southampton, UK.
Mark M Gosink, Pfizer Inc, New York, NY, USA.
Enrico Mossotto, Human Genetics and Genomic Medicine, University of Southampton, Southampton, UK.
Sarah Ennis, Human Genetics and Genomic Medicine, University of Southampton, Southampton, UK.
Manfred Hauben, Pfizer Inc, New York, NY, USA; NYU Langone Health, Department of Medicine, New York, NY, USA.
Funding
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all data in the study and had final responsibility for the decision to submit for publication.
Conflicts of Interest
The authors declare that they have no financial disclosures to make, and there are no conflicts of interest to report.
References
- 1. Alatab S, et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5:17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McKinney SM, Sieniek M, Godbole V, et al. . International evaluation of an AI system for breast cancer screening. Nature. 2020;577:89–94. [DOI] [PubMed] [Google Scholar]
- 3. Stafford IS, Kellermann M, Mossotto E, et al. . A systematic review of the applications of artificial intelligence and machine learning in autoimmune diseases. NPJ Digit Med. 2020;3:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fatima M, Pasha M.. Survey of machine learning algorithms for disease diagnostic. J Intell learn Syst Appl. 2017;09:1–16. [Google Scholar]
- 5. Tontini GE, Rimondi A, Vernero M, et al. . Artificial intelligence in gastrointestinal endoscopy for inflammatory bowel disease: a systematic review and new horizons. Therap Adv Gastroenterol. 2021;14:17562848211017730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nguyen NH, Picetti D, Dulai PS, et al. . Machine learning-based prediction models for diagnosis and prognosis in inflammatory bowel diseases: a systematic review. J Crohns Colitis. 2022;16:398–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Plos Med. 2009;6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Team RC. R: A Language and Environment for Statistical Computing. 2019. Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 9. Inc. PT. Collaborative Data Science. Montréal, QC: Plotly Technologies Inc.; 2015. [Google Scholar]
- 10. Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- 11. Liu T, Han L, Tilley M, et al. Distinct clinical phenotypes for Crohn’s disease derived from patient surveys. BMC Gastroenterol. 2021;21:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kieft K, Zhou Z, Anantharaman K.. VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome. 2020;8:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coelho T, Mossotto E, Gao Y, et al. . Immunological profiling of paediatric inflammatory bowel disease using unsupervised machine learning. J Pediatr Gastroenterol Nutr. 2020;70:833–840. [DOI] [PubMed] [Google Scholar]
- 14. Lerrigo R, Coffey JTR, Kravitz JL, et al. The emotional toll of inflammatory bowel disease: using machine learning to analyze online community forum discourse. Crohns Colitis. 2019;360:1. [Google Scholar]
- 15. Clooney AG, Eckenberger J, Laserna-Mendieta E, et al. . Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study. Gut. 2021;70:499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dhaliwal J, Erdman L, Drysdale E, et al. . Accurate classification of pediatric colonic inflammatory bowel disease subtype using a random forest machine learning classifier. J Pediatr Gastroenterol Nutr. 2021;72:262–269. [DOI] [PubMed] [Google Scholar]
- 17. Le V, Quinn TP, Tran T, Venkatesh S.. Deep in the bowel: highly interpretable neural encoder-decoder networks predict gut metabolites from gut microbiome. BMC Genomics. 2020;21:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mossotto E, Ashton JJ, Coelho T, et al. . Classification of paediatric inflammatory bowel disease using machine learning. Sci Rep. 2017;7:2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Niehaus KE, Uhlig HH, Clifton DA.. Phenotypic characterisation of Crohn’s disease severity. Annu Int Conf IEEE Eng Med Biol Soc. 2015;2015:7023–7026. [DOI] [PubMed] [Google Scholar]
- 20. Biernacka KB, Barańska D, Matera K, et al. . The value of magnetic resonance enterography in diagnostic difficulties associated with Crohn’s disease. Pol J Radiol. 2021;86:e143–e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Volkova A, Ruggles KV.. Predictive metagenomic analysis of autoimmune disease identifies robust autoimmunity and disease specific microbial signatures. Front Microbiol. 2021;12:621310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nuzzo A, Saha S, Berg E, et al. . Expanding the drug discovery space with predicted metabolite-target interactions. Commun Biol. 2021;4:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu C, Zhou M, Xie Z, Li M, Zhu X, Zhu H.. LightCUD: a program for diagnosing IBD based on human gut microbiome data. BioData Min. 2021;14:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manandhar I, Alimadadi A, Aryal S, et al. . Gut microbiome-based supervised machine learning for clinical diagnosis of inflammatory bowel diseases. Am J Physiol Gastrointest Liver Physiol. 2021;320:G328–G337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Khorasani HM, Usefi H, Peña-Castillo L.. Detecting ulcerative colitis from colon samples using efficient feature selection and machine learning. Sci Rep. 2020;10:13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Raimondi D, Simm J, Arany A, et al. . An interpretable low-complexity machine learning framework for robust exome-based in-silico diagnosis of Crohn’s disease patients. NAR Genom Bioinform. 2020;2:lqaa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang P, Lai S, Wu S, et al. . Host DNA contents in fecal metagenomics as a biomarker for intestinal diseases and effective treatment. BMC Genomics. 2020;21:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Romagnoni A, Jégou S, Van Steen K, et al. ; International Inflammatory Bowel Disease Genetics Consortium (IIBDGC). Comparative performances of machine learning methods for classifying Crohn Disease patients using genome-wide genotyping data. Sci Rep. 2019;9:10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y, Miller M, Astrakhan Y, et al. Identifying Crohn’s disease signal from variome analysis. Genome Med. 2019;11:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sarrabayrouse G, Elias A, Yáñez F, et al. Fungal and bacterial loads: noninvasive inflammatory bowel disease biomarkers for the clinical setting. mSystems. 2021;6:e01277-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iablokov SN, Klimenko NS, Efimova DA, et al. . Metabolic phenotypes as potential biomarkers for linking gut microbiome with inflammatory bowel diseases. Front Mol Biosci. 2020;7:603740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Douglas GM, Hansen R, Jones CMA, et al. . Multi-omics differentially classify disease state and treatment outcome in pediatric Crohn’s disease. Microbiome. 2018;6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eck A, Zintgraf LM, de Groot EFJ, et al. . Interpretation of microbiota-based diagnostics by explaining individual classifier decisions. BMC Bioinformatics. 2017;18:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hübenthal M, Hemmrich-Stanisak G, Degenhardt F, et al. Sparse modeling reveals miRNA signatures for diagnostics of inflammatory bowel disease. PLoS One. 2015;10:e0140155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cui H, Zhang X.. Alignment-free supervised classification of metagenomes by recursive SVM. BMC Genomics. 2013;14:641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Forbes JD, Chen CY, Knox NC, et al. . A comparative study of the gut microbiota in immune-mediated inflammatory diseases-does a common dysbiosis exist? Microbiome. 2018;6:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Waljee AK, Cohen-Mekelburg S, Liu Y, et al. Assessing clinical disease recurrence using laboratory data in surgically resected patients from the TOPPIC trial. Crohns Colitis. 2020:2. doi: 10.1093/crocol/otaa088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stidham RW, Liu Y, Enchakalody B, et al. . The use of readily available longitudinal data to predict the likelihood of surgery in Crohn disease. Inflamm Bowel Dis. 2021;27:1328–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Udristoiu AL, Stefanescu D, Gruionu G, et al. . Deep learning algorithm for the confirmation of mucosal healing in Crohn’s disease, based on confocal laser endomicroscopy images. J Gastrointestin Liver Dis. 2021;30:59–65. [DOI] [PubMed] [Google Scholar]
- 40. Sakurai T, Nishiyama H, Sakai K, et al. . Mucosal microbiota and gene expression are associated with long-term remission after discontinuation of adalimumab in ulcerative colitis. Sci Rep. 2020;10:19186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kang EA, Jang J, Choi CH, et al. Development of a clinical and genetic prediction model for early intestinal resection in patients with Crohn’s disease: results from the IMPACT study. J Clin Med. 2021;10:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sofo L, Caprino P, Schena CA, et al. . New perspectives in the prediction of postoperative complications for high-risk ulcerative colitis patients: machine learning preliminary approach. Eur Rev Med Pharmacol Sci. 2020;24:12781–12787. [DOI] [PubMed] [Google Scholar]
- 43. Shivaji UN, Bazarova A, Critchlow T, et al. . Clinical outcomes, predictors of prognosis and health economics consequences in IBD patients after discontinuation of the first biological therapy. Therap Adv Gastroenterol. 2020;13:1756284820981216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taylor H, Serrano-Contreras JI, McDonald JAK, et al. . Multiomic features associated with mucosal healing and inflammation in paediatric Crohn’s disease. Aliment Pharmacol Ther. 2020;52:1491–1502. [DOI] [PubMed] [Google Scholar]
- 45. Choi YI, Park SJ, Chung J-W, et al. Development of machine learning model to predict the 5-year risk of starting biologic agents in patients with inflammatory bowel disease (IBD): K-CDM Network Study. J Clin Med. 2020;9:3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ghoshal UC, Rai S, Kulkarni A, Gupta A.. Prediction of outcome of treatment of acute severe ulcerative colitis using principal component analysis and artificial intelligence. JGH Open. 2020;4:889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jones CMA, Connors J, Dunn KA, et al. . Bacterial Taxa and functions are predictive of sustained remission following exclusive enteral nutrition in pediatric Crohn’s disease. Inflamm Bowel Dis. 2020;26:1026–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dong Y, Xu L, Fan Y, et al. . A novel surgical predictive model for Chinese Crohn’s disease patients. Medicine (Baltimore). 2019;98:e17510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Braun T, Di Segni A, BenShoshan M, et al. . Individualized dynamics in the gut microbiota precede Crohn’s disease flares. Am J Gastroenterol. 2019;114:1142–1151. [DOI] [PubMed] [Google Scholar]
- 50. Bottigliengo D, Berchialla P, Lanera C, et al. The role of genetic factors in characterizing extra-intestinal manifestations in Crohn’s disease patients: are bayesian machine learning methods improving outcome predictions? J Clin Med. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Waljee AK, Wallace BI, Cohen-Mekelburg S, et al. . Development and validation of machine learning models in prediction of remission in patients with moderate to severe Crohn disease. JAMA Netw Open. 2019;2(5):e193721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Takenaka K, Ohtsuka K, Fujii T, et al. . Development and validation of a deep neural network for accurate evaluation of endoscopic images from patients with ulcerative colitis. Gastroenterology. 2020;158:2150–2157. [DOI] [PubMed] [Google Scholar]
- 53. Morell Miranda P, Bertolini F, Kadarmideen H.. Investigation of gut microbiome association with inflammatory bowel disease and depression: a machine learning approach version 2; peer review: 2 approved with reservations]. F1000Res. 2019;7:702. [Google Scholar]
- 54. Waljee AK, Lipson R, Wiitala WL, et al. . Predicting hospitalization and outpatient corticosteroid use in inflammatory bowel disease patients using machine learning. Inflamm Bowel Dis. 2017;24:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jain S, Kedia S, Sethi T, et al. . Predictors of long-term outcomes in patients with acute severe colitis: a Northern Indian cohort study. J Gastroenterol Hepatol. 2018;33:615–622. [DOI] [PubMed] [Google Scholar]
- 56. Firouzi F, Rashidi M, Hashemi S, et al. . A decision tree-based approach for determining low bone mineral density in inflammatory bowel disease using WEKA software. Eur J Gastroenterol Hepatol. 2007;19:1075–1081. [DOI] [PubMed] [Google Scholar]
- 57. Dorofeyev AE, Holub SV, Babayeva GH, Аnaniin ОЕ.. Application of intellectual monitoring information technology in determining the severity of the condition of patients with inflammatory bowel diseases. Wiad Lek. 2021;74:481–486. [PubMed] [Google Scholar]
- 58. Ungaro RC, Hu L, Ji J, et al. . Machine learning identifies novel blood protein predictors of penetrating and stricturing complications in newly diagnosed paediatric Crohn’s disease. Aliment Pharmacol Ther. 2021;53:281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gutierrez Becker B, Arcadu F, Thalhammer A, et al. . Training and deploying a deep learning model for endoscopic severity grading in ulcerative colitis using multicenter clinical trial data. Ther Adv Gastrointest Endosc. 2021;14:2631774521990623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li X, Liang D, Meng J, et al. . Development and validation of a novel computed-tomography enterography radiomic approach for characterization of intestinal fibrosis in Crohn’s disease. Gastroenterology. 2021;160:2303–2316.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yao H, Najarian K, Gryak J, et al. . Fully automated endoscopic disease activity assessment in ulcerative colitis. Gastrointest Endosc. 2021;93:728–736.e1. [DOI] [PubMed] [Google Scholar]
- 62. Gottlieb K, Requa J, Karnes W, et al. . Central Reading of Ulcerative Colitis Clinical Trial Videos Using Neural Networks. Gastroenterology. 2021;160:710–719.e2. [DOI] [PubMed] [Google Scholar]
- 63. Wang J, Ortiz C, Fontenot L, et al. High circulating elafin levels are associated with Crohn's disease-associated intestinal strictures. PLoS One. 2020;15:e0231796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Popa IV, Burlacu A, Mihai C, Prelipcean CC.. A machine learning model accurately predicts ulcerative colitis activity at one year in patients treated with anti-tumour necrosis factor α agents. Medicina (Kaunas). 2020;56:628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Reddy BK, Delen D, Agrawal RK.. Predicting and explaining inflammation in Crohn’s disease patients using predictive analytics methods and electronic medical record data. Health Informatics J. 2019;25:1201–1218. [DOI] [PubMed] [Google Scholar]
- 66. Biasci D, Lee JC, Noor NM, et al. . A blood-based prognostic biomarker in IBD. Gut. 2019;68:1386–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mohapatra S, Nayak J, Mishra M, et al. . Wavelet transform and deep convolutional neural network-based smart healthcare system for gastrointestinal disease detection. Interdiscip Sci. 2021;13:212–228. [DOI] [PubMed] [Google Scholar]
- 68. Takenaka K, Ohtsuka K, Fujii T, et al. . Deep neural network accurately predicts prognosis of ulcerative colitis using endoscopic images. Gastroenterology. 2021;160:2175–2177.e3. [DOI] [PubMed] [Google Scholar]
- 69. Bossuyt P, Nakase H, Vermeire S, et al. . Automatic, computer-aided determination of endoscopic and histological inflammation in patients with mild to moderate ulcerative colitis based on red density. Gut. 2020;69:1778–1786. [DOI] [PubMed] [Google Scholar]
- 70. Maeda Y, Kudo SE, Mori Y, et al. . Fully automated diagnostic system with artificial intelligence using endocytoscopy to identify the presence of histologic inflammation associated with ulcerative colitis (with video). Gastrointest Endosc. 2019;89:408–415. [DOI] [PubMed] [Google Scholar]
- 71. Menti E, et al. Bayesian Machine Learning Techniques for revealing complex interactions among genetic and clinical factors in association with extra-intestinal manifestations in IBD patients. AMIA Annu Symp Proc. 2016;2016:884–893. [PMC free article] [PubMed] [Google Scholar]
- 72. Waljee AK, Joyce JC, Wang S, et al. . Algorithms outperform metabolite tests in predicting response of patients with inflammatory bowel disease to thiopurines. Clin Gastroenterol Hepatol. 2010;8:143–150. [DOI] [PubMed] [Google Scholar]
- 73. Han L, Maciejewski M, Brockel C, et al. . A probabilistic pathway score (PROPS) for classification with applications to inflammatory bowel disease. Bioinformatics. 2018;34:985–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang L, Fan R, Zhang C, et al. . Applying machine learning models to predict medication nonadherence in Crohn’s disease maintenance therapy. Patient Prefer Adherence. 2020;14:917–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Taylor KM, Hanscombe KB, Prescott NJ, et al. . Genetic and inflammatory biomarkers classify small intestine inflammation in asymptomatic first-degree relatives of patients with Crohn’s disease. Clin Gastroenterol Hepatol. 2020;18:908–916.e13. [DOI] [PubMed] [Google Scholar]
- 76. Pal LR, Kundu K, Yin Y, Moult J.. CAGI4 Crohn’s exome challenge: Marker SNP vs exome variant models for assigning risk of Crohn disease. Hum Mutat. 2017;38:1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Doherty MK, et al. Fecal microbiota signatures are associated with response to ustekinumab therapy among Crohn’s disease patients. mBio. 2018;9. doi: 10.1128/mBio.02120-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Daneshjou R, Wang Y, Bromberg Y, et al. . Working toward precision medicine: Predicting phenotypes from exomes in the Critical Assessment of Genome Interpretation (CAGI) challenges. Hum Mutat. 2017;38:1182–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Giollo M, Jones DT, Carraro M, et al. . Crohn disease risk prediction-Best practices and pitfalls with exome data. Hum Mutat. 2017;38:1193–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kang T, Ding W, Zhang L, et al. . A biological network-based regularized artificial neural network model for robust phenotype prediction from gene expression data. BMC Bioinformatics. 2017;18:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Waljee AK, Liu B, Sauder K, et al. . Predicting corticosteroid-free endoscopic remission with vedolizumab in ulcerative colitis. Aliment Pharmacol Ther. 2018;47:763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tong Y, Lu K, Yang Y, et al. . Can natural language processing help differentiate inflammatory intestinal diseases in China? Models applying random forest and convolutional neural network approaches. BMC Med Inform Decis Mak. 2020;20:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. McDonnell M, Harris RJ, Borca F, et al. High incidence of glucocorticoid-induced hyperglycaemia in inflammatory bowel disease: metabolic and clinical predictors identified by machine learning. BMJ Open Gastroenterol. 2020;7(1):e000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jiang P, Wu S, Luo Q, Zhao XM, Chen WH.. Metagenomic analysis of common intestinal diseases reveals relationships among microbial signatures and powers multidisease diagnostic models. mSystems. 2021;6:e00112–e00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Waljee AK, Sauder K, Patel A, et al. . Machine learning algorithms for objective remission and clinical outcomes with thiopurines. J Crohns Colitis. 2017;11:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Isakov O, Dotan I, Ben-Shachar S.. Machine learning-based gene prioritization identifies novel candidate risk genes for inflammatory bowel disease. Inflamm Bowel Dis. 2017;23:1516–1523. [DOI] [PubMed] [Google Scholar]
- 87. Wei Z, Wang W, Bradfield J, et al. ; International IBD Genetics Consortium. Large sample size, wide variant spectrum, and advanced machine-learning technique boost risk prediction for inflammatory bowel disease. Am J Hum Genet. 2013;92:1008–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yu S, Chakrabortty A, Liao KP, et al. . Surrogate-assisted feature extraction for high-throughput phenotyping. J Am Med Inform Assoc. 2017;24:e143–e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sudlow C, Gallacher J, Allen N, et al. . UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hauben M, Reynolds R, Caubel P.. Deconstructing the pharmacovigilance hype cycle. Clin Ther. 2018;40:1981–1990.e3. [DOI] [PubMed] [Google Scholar]
- 91. Cabitza F, Campagner A.. The need to separate the wheat from the chaff in medical informatics: Introducing a comprehensive checklist for the (self)-assessment of medical AI studies. Int J Med Inform. 2021;153:104510. [DOI] [PubMed] [Google Scholar]
- 92. Scott I, Carter S, Coiera E.. Clinician checklist for assessing suitability of machine learning applications in healthcare. BMJ Health Care Inform. 2021;28:e100251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Olczak J, Pavlopoulos J, Prijs J, et al. . Presenting artificial intelligence, deep learning, and machine learning studies to clinicians and healthcare stakeholders: an introductory reference with a guideline and a Clinical AI Research (CAIR) checklist proposal. Acta Orthop. 2021;92:513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Norgeot B, Quer G, Beaulieu-Jones BK, et al. . Minimum information about clinical artificial intelligence modeling: the MI-CLAIM checklist. Nat Med. 2020;26:1320–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Collins GS, Reitsma JB, Altman DG, Moons KG.. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Bmj. 2015;350:g7594. [DOI] [PubMed] [Google Scholar]
- 96. Collins GS, Moons KGM.. Reporting of artificial intelligence prediction models. Lancet. 2019;393:1577–1579. [DOI] [PubMed] [Google Scholar]
- 97. Luo W, Phung D, Tran T, et al. . Guidelines for developing and reporting machine learning predictive models in biomedical research: a multidisciplinary view. J Med Internet Res. 2016;18:e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ibrahim H, Liu X, Rivera SC, et al. . Reporting guidelines for clinical trials of artificial intelligence interventions: the SPIRIT-AI and CONSORT-AI guidelines. Trials. 2021;22:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hernandez-Boussard T, Bozkurt S, Ioannidis JPA, Shah NH.. MINIMAR (MINimum Information for Medical AI Reporting): developing reporting standards for artificial intelligence in health care. J Am Med Inform Assoc. 2020;27:2011–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Liu Y, Chen PC, Krause J, Peng L.. How to read articles that use machine learning: users’ guides to the medical literature. JAMA. 2019;322:1806–1816. [DOI] [PubMed] [Google Scholar]
- 101. Mongan J, Moy L, Kahn CE Jr. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): a guide for authors and reviewers. Radiol Artif Intell. 2020;2:e200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Finch H, Schneider MK.. Classification accuracy of neural networks vs. discriminant analysis, logistic regression, and classification and regression trees. Methodology. 2007;3:47–57. [Google Scholar]
- 103. Maroco J, Silva D, Rodrigues A, et al. . Data mining methods in the prediction of Dementia: a real-data comparison of the accuracy, sensitivity and specificity of linear discriminant analysis, logistic regression, neural networks, support vector machines, classification trees and random forests. BMC Res Notes. 2011;4:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Olson RS, La Cava W, Orzechowski P, et al. . PMLB: a large benchmark suite for machine learning evaluation and comparison. Biodata Min. 2017;10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Strubell E, Ganesh A, McCallum A.. Energy and policy considerations for deep learning in NLP. arXiv. preprint arXiv:190602243. 2019. [Google Scholar]
- 106. Eelbode T, Sinonquel P, Maes F, Bisschops R.. Pitfalls in training and validation of deep learning systems. Best Pract Res Clin Gastroenterol. 2020;52-53:101712. [DOI] [PubMed] [Google Scholar]
- 107. Pannala R, Krishnan K, Melson J, et al. . Artificial intelligence in gastrointestinal endoscopy. Videogie. 2020;5:598–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Challen R, Denny J, Pitt M, et al. . Artificial intelligence, bias and clinical safety. BMJ Qual Saf. 2019;28:231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yu KH, Kohane IS.. Framing the challenges of artificial intelligence in medicine. BMJ Qual Saf. 2019;28:238–241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.