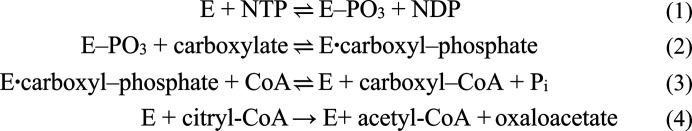The mutation of a glutamate residue that is conserved among ATP-citrate lyases (ACLYs) and succinyl-CoA synthetases allowed capture of the catalytic intermediate succinyl-phosphate in a high-resolution crystal structure of succinyl-CoA synthetase. This structure clarifies the enzymatic mechanism of ACLY and supports the conclusion that the amino-terminal portion of ACLY catalyzes the formation of citryl-CoA, while the carboxy-terminal portion catalyzes its cleavage into oxaloacetate and acetyl-CoA.
Keywords: succinyl-CoA synthetase, succinyl-phosphate, cryogenic electron microscopy, catalytic intermediates, ATP-citrate lyase
Abstract
Succinyl-CoA synthetase (SCS) catalyzes a three-step reaction in the citric acid cycle with succinyl-phosphate proposed as a catalytic intermediate. However, there are no structural data to show the binding of succinyl-phosphate to SCS. Recently, the catalytic mechanism underlying acetyl-CoA production by ATP-citrate lyase (ACLY) has been debated. The enzyme belongs to the family of acyl-CoA synthetases (nucleoside diphosphate-forming) for which SCS is the prototype. It was postulated that the amino-terminal portion catalyzes the full reaction and the carboxy-terminal portion plays only an allosteric role. This interpretation was based on the partial loss of the catalytic activity of ACLY when Glu599 was mutated to Gln or Ala, and on the interpretation that the phospho-citryl-CoA intermediate was trapped in the 2.85 Å resolution structure from cryogenic electron microscopy (cryo-EM). To better resolve the structure of the intermediate bound to the E599Q mutant, the equivalent mutation, E105αQ, was made in human GTP-specific SCS. The structure of the E105αQ mutant shows succinyl-phosphate bound to the enzyme at 1.58 Å resolution when the mutant, after phosphorylation in solution by Mg2+-ATP, was crystallized in the presence of magnesium ions, succinate and desulfo-CoA. The E105αQ mutant is still active but has a specific activity that is 120-fold less than that of the wild-type enzyme, with apparent Michaelis constants for succinate and CoA that are 50-fold and 11-fold higher, respectively. Based on this high-resolution structure, the cryo-EM maps of the E599Q ACLY complex reported previously should have revealed the binding of citryl-phosphate and CoA and not phospho-citryl-CoA.
1. Introduction
In humans, ATP-citrate lyase (ACLY) links carbohydrate and lipid metabolism since it is the primary source of acetyl-CoA in the cytosol. The enzyme is a target for the design of drugs to treat obesity and cancer, both of which rely on the synthesis of lipids, either to store in fat cells or to form the membranes of fast-proliferating cells. The proposed catalytic mechanism of ACLY, which is important for the design of compounds targeting the enzyme, was based on studies of succinyl-CoA synthetase (SCS), the prototype for this family of ADP- or GDP-forming acyl-CoA synthetases (Sánchez et al., 2000 ▸). While human ACLY is a homotetramer with 1101 amino-acid residues in each subunit, human GTP-specific SCS is a heterodimer, with the α- and β-subunits possessing 306 and 395 amino-acid residues, respectively. The first structures of ACLY supported the view that the N-terminal portion of the enzyme, which shows similarity to SCS, catalyzes the first three steps of the reaction cycle, while the C-terminal portion, which shows similarity to citrate synthase, catalyzes the cleavage of citryl-CoA (Fig. 1 ▸; Sun et al., 2010 ▸; Nguyen et al., 2019 ▸; Verschueren et al., 2019 ▸; Wei et al., 2019 ▸).
Figure 1.
Proposed mechanism for succinyl-CoA synthetase and ATP-citrate lyase. The first three steps are catalyzed by both enzymes. The carboxylate is succinate in succinyl-CoA synthetase and citrate in ATP-citrate lyase. The fourth step is catalyzed by ATP-citrate lyase.
Controversy arose when the N-terminal portion of ACLY was proposed to catalyze all four steps, with the C-terminal portion serving only an allosteric role (Wei et al., 2020a ▸; Verstraete et al., 2021 ▸; Wei & Marmorstein, 2021 ▸). This interpretation was based on the partial loss of catalytic activity of ACLY when Glu599, a conserved residue in the N-terminal portion of ACLY, was mutated to Gln or Ala, and on the interpretation that the phospho-citryl-CoA intermediate was trapped when the E599Q mutant of ACLY was mixed with ATP, citrate and CoA. However, the 2.85 Å resolution map from cryogenic electron microscopy (cryo-EM) did not clearly indicate the thioester bond between citrate and CoA (Wei et al., 2020b ▸). A higher resolution structure is needed to determine whether the cryo-EM map shows phospho-citryl-CoA or citryl-phosphate and CoA and to better understand the role of Glu599.
Higher resolution structural data are available for GTP-specific SCS (GTPSCS) in complex with the substrates GTP/GDP (Fraser et al., 2006 ▸), CoA (Huang et al., 2015 ▸) and Mg2+-succinate (Huang & Fraser, 2016 ▸) and the inhibitor tartryl-CoA (Huang & Fraser, 2020 ▸), and with different conformations of the phosphohistidine loop (Huang & Fraser, 2021 ▸), providing insights into the first three steps of the catalytic mechanism (Fig. 1 ▸). Glu599 of ACLY is conserved not only in ACLYs but also in SCSs, and corresponds to Glu105α in the heterodimeric human GTPSCS (Fig. 2 ▸). While structures of the N-terminal portion of human ACLY have been determined at resolutions of up to 1.70 Å, this protein was in an open conformation even in solution, preventing transfer of the phosphoryl group from the active-site histidine to citrate or its analogues (Hu et al., 2017 ▸). Thus, SCS is the better system to study the impact of the mutation on catalysis both in solution and in crystals. For this reason, the E105Qα mutant of human GTPSCS was obtained, studied using kinetic analyses and phosphorylated for co-crystallization with and without Mg2+-succinate, CoA and desulfo-CoA. One of the resulting structures showed the catalytic intermediate succinyl-phosphate bound at the active site.
Figure 2.
Sequence alignment of parts of ATP-citrate lyase (ACLY) and succinyl-CoA synthetase (SCS) showing the conservation of the glutamate residue Glu599 of ACLY. Crystal structures have been determined of the enzymes shown. While human ACLY is a homotetramer, ACLY from Chlorobium limicola is a heterooctamer and the equivalent residue is in the A subunit. Human GTPSCS is a heterodimer, while E. coli and Thermus aquaticus SCS are heterotetramers; however, the similar residue is in the α-subunit in all cases.
2. Methods
2.1. Site-directed mutagenesis to obtain E105αQ GTPSCS
The two primers used to generate the human E105αQ GTPSCS mutant are given in Table 1 ▸. 30 ng plasmid, 125 ng forward primer, 5 µl 5× Q5 buffer (NEB), 0.66 U Q5 polymerase (NEB) and 0.5 µl 10 mM dNTP were brought to 25 µl with distilled water in one reaction tube; the second tube had the same composition but contained the reverse primer. Elongation was performed for 11 cycles before the contents of the two tubes were combined. Another 0.66 U of Q5 polymerase was added and elongation was continued for another 17 cycles. The mutated genes were sequenced at the University Core DNA Services, University of Calgary.
Table 1. Oligonucleotides for mutagenesis.
| Forward primer | GTTGTGTGTATCACTCAAGGAATTCCCCAGCAGGAC |
| Reverse primer | GTCCTGCTGGGGAATTCCTTGAGTGATACACACAAC |
2.2. Protein production and purification
The pET-42b(+) expression plasmid containing the human E105αQ GTPSCS genes was transformed into Escherichia coli strain BL21(DE3). The protocols for protein production, cell lysis and protein purification have been described previously (Huang & Fraser, 2020 ▸). During protein purification, pooled fractions from HiTrap SP HP column chromatography (GE Healthcare) were mixed with 5 mM GTP and 5 mM MgCl2 and left to phosphorylate overnight prior to gel filtration with a Superdex 200 prep-grade column (GE Healthcare) using running buffer consisting of 25 mM potassium phosphate pH 7.5, 150 mM KCl, 5 mM 2-mercaptoethanol (2ME). Fractions with high absorbance at 280 nm were pooled, and the purity of the protein was verified by SDS–PAGE. E105αQ GTPSCS was concentrated using Amicon Ultra-15 Centrifugal Filter Units with a nominal cutoff of 30 kDa (Millipore Sigma) and exchanged into buffer consisting of 10 mM Tris–HCl pH 8.0, 5 mM 2ME. The concentration of the protein was determined from the absorbance at 280 nm using the calculated value of 0.255 for 1 mg ml−1 (Gasteiger et al., 2005 ▸). 20 µl aliquots of concentrated protein solution were flash-frozen in thin-walled PCR tubes using liquid nitrogen and stored at −80°C for future use.
2.3. Crystallization and X-ray diffraction
Human E105αQ GTPSCS was first crystallized with Mg2+-succinate and CoA at 21°C. Microseeding was used to crystallize the complex with Mg2+-succinyl phosphate and desulfo-CoA by diluting seeds 1:102, 1:103, 1:104 and 1:105. The crystallization conditions are given in Table 2 ▸. To obtain the best diffraction, rounds of optimization were performed to achieve single, rod-shaped crystals.
Table 2. Crystallization conditions.
| Complex | Human E105αQ GTPSCS complexed with Mg2+-succinyl phosphate and desulfo-CoA | Human E105αQ GTPSCS complexed with Mg2+-succinate and CoA | Phosphorylated human E105αQ GTPSCS complexed with CoA |
|---|---|---|---|
| PDB code | 7msr | 7mss | 7mst |
| Method | Hanging-drop vapour diffusion | Hanging-drop vapour diffusion | Hanging-drop vapour diffusion |
| Plate type | Hampton Research VDX | Hampton Research VDX | Hampton Research VDX |
| Temperature (K) | 294 | 294 | 294 |
| Composition of protein solution | 10 mM Tris–HCl pH 8.0, 50 mM ammonium succinate pH 7.5, 50 mM MgCl2, 0.76 mM desulfo-CoA, 4.8 mM TCEP, 5 mg ml−1 protein | 10 mM Tris–HCl pH 8.0, 50 mM ammonium succinate pH 7.5, 50 mM MgCl2, 5 mM CoA, 4 mM TCEP, 5 mg ml−1 protein | 10 mM Tris–HCl pH 8.0, 5 mM CoA, 5.5 mM TCEP, 5 mg ml−1 protein |
| Composition of reservoir solution | 20.0–22.5%(w/v) PEG 3350, 100 mM ammonium succinate pH 7.5, 100 mM HEPES pH 7.0 or 100 mM Tris–HCl pH 7.2–7.6 | 18.75–22.5%(w/v) PEG 3350, 100 mM ammonium succinate pH 7.5, 100 mM HEPES pH 7.0 or 100 mM Tris–HCl pH 7.3–7.4 | 17.5–20.0%(w/v) PEG 3350, 160–180 mM sodium thiocyanate, 100 mM Tris–HCl pH 7.5 |
| Volume and ratio of drop | 3 µl, 3:2:1 protein:well solution:seeds | 1 µl, 1:1 protein:well solution | 1 µl, 1:1 protein:well solution |
| Volume of reservoir (ml) | 0.5 | 0.5 | 0.5 |
X-ray diffraction experiments were performed on beamline 17-ID at the Advanced Photon Source (APS). Raster screening was applied to find the best-diffracting portion of each crystal, which was exposed to X-rays for 0.02 s and 0.25° per frame for a total of 720 images. Data were processed using xia2 and DIALS (Winter, 2010 ▸; Winter et al., 2018 ▸; Beilsten-Edmands et al., 2020 ▸). The phase problem was solved by molecular replacement using the structure of Mg2+-succinate bound to pig GTPSCS (PDB entry 5cae; Berman et al., 2000 ▸; Huang & Fraser, 2016 ▸) as a model. Coot (Emsley et al., 2010 ▸) and Phenix (Liebschner et al., 2019 ▸) were used to modify the model and refine the structures. Geometrical restraints for the ligand succinyl-phosphate were generated using the grade web server (Global Phasing Ltd) and subsequently updated to match the values used by the PDB.
2.4. Kinetic assays
Kinetic assays were performed to obtain the Michaelis constants of the wild type and the E105αQ mutant for succinate, CoA and GTP. The assay mixture consisted of 50 mM Tris succinate pH 7.4, 10 mM MgCl2, 0.1 mM CoA, 0.1 mM GTP and 7.4 × 10−9
M wild-type GTPSCS or 3.1 × 10−7
M mutant when the concentrations of the substrates were not varied, and the concentration ranges given in Table 3 ▸ when they were. The assays were conducted at 21°C, measuring the absorbance by the thioester bond of succinyl-CoA at 235 nm (Cha & Parks, 1964 ▸). Michaelis–Menten plots were generated using GraphPad Prism (version 5 for Windows; GraphPad Software, San Diego, California, USA) to determine the
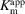 values for succinate, CoA and GTP. Initial velocities were measured in triplicate and are reported with 95% confidence intervals.
values for succinate, CoA and GTP. Initial velocities were measured in triplicate and are reported with 95% confidence intervals.
Table 3. Apparent Michaelis constants in the direction of succinyl-CoA synthesis.
| Substrate concentrations | |||||
|---|---|---|---|---|---|
| Human GTPSCS | Succinate (mM) | CoA (µM) | GTP (µM) | Varied substrate |
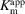 (µM)
(µM) |
| Wild type | 50 | 100 | 1.875–120.000 | GTP | 3.4 ± 0.2 |
| 0.039–5.000 | 100 | 100 | Succinate | 20 × 101 ± 1 × 101 | |
| 50 | 0.664–80.000 | 100 | CoA | 1.6 ± 0.2 | |
| E105αQ | 50 | 100 | 1.875–120.000 | GTP | 7.1 ± 0.6 |
| 1.25–50.00 | 100 | 100 | Succinate | 101 × 102 ± 6 × 102 | |
| 50 | 2.656–85.000 | 100 | CoA | 18 ± 1 | |
3. Results and discussion
To mimic the conditions used to determine the structure of the E599Q mutant of ACLY by cryo-EM, phosphorylated human E105αQ GTPSCS was mixed with magnesium ions, succinate and CoA prior to crystallization. Statistics for the X-ray diffraction data and the model are presented in Tables 4 ▸ and 5 ▸ in the column labelled ‘7mss’. The crystal structure showed that the enzyme was dephosphorylated and Mg2+-succinate, phosphate and CoA were bound in the active site. To prove that the dephosphorylation of the enzyme was due to the presence of Mg2+-succinate along with CoA, crystals were grown with CoA but in the absence of magnesium and succinate. The statistics for these data and the model are labelled ‘7mst’ in Tables 4 ▸ and 5 ▸.
Table 4. Data-collection and processing statistics for complexes of human E105αQ GTPSCS.
Values in parentheses are for the outer shell.
| Ligand(s) | Mg2+-succinyl-phosphate and desulfo-CoA | Mg2+-succinate and CoA | CoA (phosphorylated GTSSCS) |
|---|---|---|---|
| PDB code | 7msr | 7mss | 7mst |
| Diffraction source | 17-ID-B, APS | 17-ID-B, APS | 17-ID-B, APS |
| Wavelength (Å) | 1.00000 | 1.00000 | 1.00000 |
| Temperature (K) | 100 | 100 | 100 |
| Detector | Dectris EIGER2 X 9M | Dectris EIGER2 X 9M | Dectris EIGER2 X 9M |
| Crystal-to-detector distance (mm) | 160.01 | 160.00 | 160.00 |
| Rotation range per image (°) | 0.25 | 0.25 | 0.25 |
| Total rotation range (°) | 180.0 | 162.5 | 180.0 |
| Exposure time per image (s) | 0.02 | 0.02 | 0.02 |
| Space group | P21 | P21 | P21 |
| a, b, c (Å) | 80.45, 81.52, 54.36 | 80.47, 81.09, 54.49 | 87.89, 82.96, 48.85 |
| α, β, γ (°) | 90.0, 103.4, 90.0 | 90.0, 103.4, 90.0 | 90.0, 103.5, 90.0 |
| Mosaicity (°) | 0.105 | 0.137 | 0.228 |
| Resolution range (Å) | 56.43–1.58 (1.61–1.58) | 56.32–1.75 (1.78–1.75) | 59.50–1.61 (1.64–1.61) |
| Total No. of reflections | 320245 (15206) | 215571 (10838) | 302968 (15371) |
| No. of unique reflections | 92537 (4486) | 66677 (3252) | 87400 (4307) |
| Completeness (%) | 99.2 (96.4) | 97.2 (95.2) | 99.3 (97.9) |
| Multiplicity | 3.5 (3.4) | 3.2 (3.3) | 3.5 (3.6) |
| R r.i.m. | 0.071 (2.117) | 0.093 (2.131) | 0.120 (1.524) |
| R p.i.m. | 0.038 (1.131) | 0.051 (1.157) | 0.064 (0.797) |
| CC1/2 | 0.998 (0.382) | 0.997 (0.321) | 0.995 (0.348) |
| 〈I/σ(I)〉 | 9.5 (0.3) | 12.4 (0.5) | 6.3 (0.4) |
| Overall B factor from Wilson plot (Å2) | 23.24 | 24.65 | 17.23 |
Table 5. Structure solution and refinement of complexes of human E105αQ GTPSCS.
Values in parentheses are for the outer shell.
| Ligand(s) | Mg2+-succinyl-phosphate and desulfo-CoA | Mg2+-succinate and CoA | CoA (phosphorylated GTSSCS) |
|---|---|---|---|
| PDB code | 7msr | 7mss | 7mst |
| Resolution range (Å) | 39.13–1.58 (1.60–1.58) | 53.01–1.75 (1.77–1.75) | 46.34–1.61 (1.63–1.61) |
| Completeness (%) | 97.54 (67.00) | 96.80 (92.00) | 99.14 (96.00) |
| σ Cutoff | 1.34 | 1.33 | 1.33 |
| No. of reflections, working set | 90981 (2008) | 66430 (1989) | 87292 (2704) |
| No. of reflections, test set | 6027 (86) | 4846 (81) | 5869 (149) |
| Final R cryst | 0.199 (0.433) | 0.202 (0.416) | 0.178 (0.334) |
| Final R free | 0.221 (0.466) | 0.227 (0.460) | 0.194 (0.343) |
| Coordinate error, maximum-likelihood (Å) | 0.25 | 0.26 | 0.20 |
| No. of non-H atoms | |||
| Protein | 5352 | 5269 | 5320 |
| Ligand | 164 | 155 | 143 |
| Water | 299 | 273 | 445 |
| Total | 5739 | 5628 | 5839 |
| R.m.s. deviations from ideal values | |||
| Bond lengths (Å) | 0.003 | 0.004 | 0.004 |
| Angles (°) | 0.59 | 0.71 | 0.69 |
| Average temperature factors (Å2) | |||
| Protein | 36.7 | 38.4 | 27.4 |
| Ligand | 36.1 | 33.2 | 27.5 |
| Water | 40.9 | 38.9 | 33.2 |
| Ramachandran plot (%) | |||
| Favoured regions | 97.84 | 98.28 | 98.26 |
| Allowed | 2.01 | 1.58 | 1.45 |
| Outliers | 0.14 | 0.14 | 0.29 |
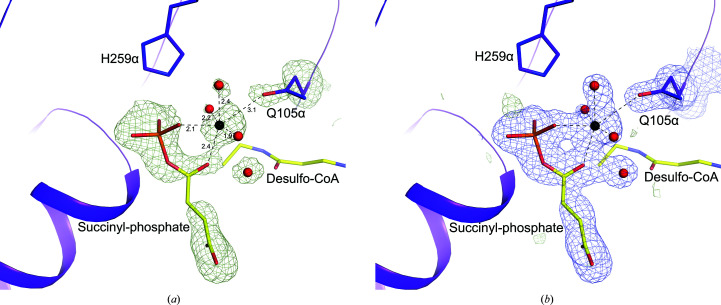
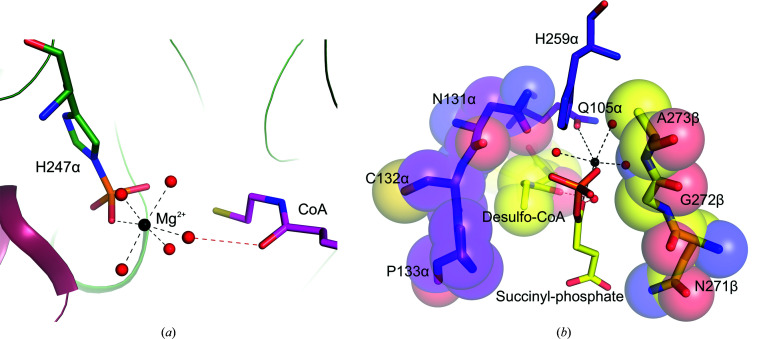
To determine the crystal structure of the catalytic intermediate, CoA was replaced by desulfo-CoA in the crystallization experiment. We had been unsuccessful in trapping the intermediate in wild-type SCS using desulfo-CoA (PDB entry 6xru; Huang & Fraser, 2021 ▸), but were successful in trapping the intermediate in the mutant. The crystal structure of E105αQ GTPSCS with desulfo-CoA shows the intermediate Mg2+-succinyl-phosphate bound to the enzyme. At the resolution obtained, electron density for water molecules and the magnesium ion is clearly shown in the active site (Fig. 3 ▸). Two O atoms of the succinyl-phosphate coordinate the magnesium ion and three additional coordination sites are occupied by water molecules, but the site opposite the O atom of phosphate is occupied by the side-chain carbonyl of the glutamine that substitutes for glutamate in the mutant. In the complex of dephosphorylated E105αQ GTPSCS with phosphate, Mg2+-succinate and CoA (Fig. 4 ▸), this glutamine residue is turned away from the active site and a water molecule completes the octahedral coordination of the magnesium ion. The orientation of the side chain of the glutamine residue is different from Glu105α, the carboxylate of which interacts with two of the water molecules coordinating the magnesium ion (PDB etnry 5cae; Huang & Fraser, 2016 ▸; Fig. 4 ▸).
Figure 3.
Binding of succinyl-phosphate to E105αQ GTPSCS. The maps show the binding of succinyl-phosphate, a magnesium ion, water molecules and the side chain of Gln105α. Succinyl-phosphate and part of desulfo-CoA are shown as sticks with yellow C atoms. The magnesium ion and water molecules are shown as black and red spheres, with ionic interactions represented by dashed lines. Portions of the α-subunit are represented by a purple ribbon diagram with the side chain of Gln105α and the active-site histidine, His259α, shown as sticks with purple C atoms. Other atoms are coloured according to type. (a) The omit map is contoured in light green at 3σ. Distances for the ionic interactions are given in Å. (b) The F o − F c map is contoured in light green at 3σ and the 2F o − F c map is contoured in blue at 1σ. This figure and Figs. 4 ▸, 6 ▸ and 7 ▸ were drawn using PyMOL (version 2.4.0; Schrödinger).
Figure 4.
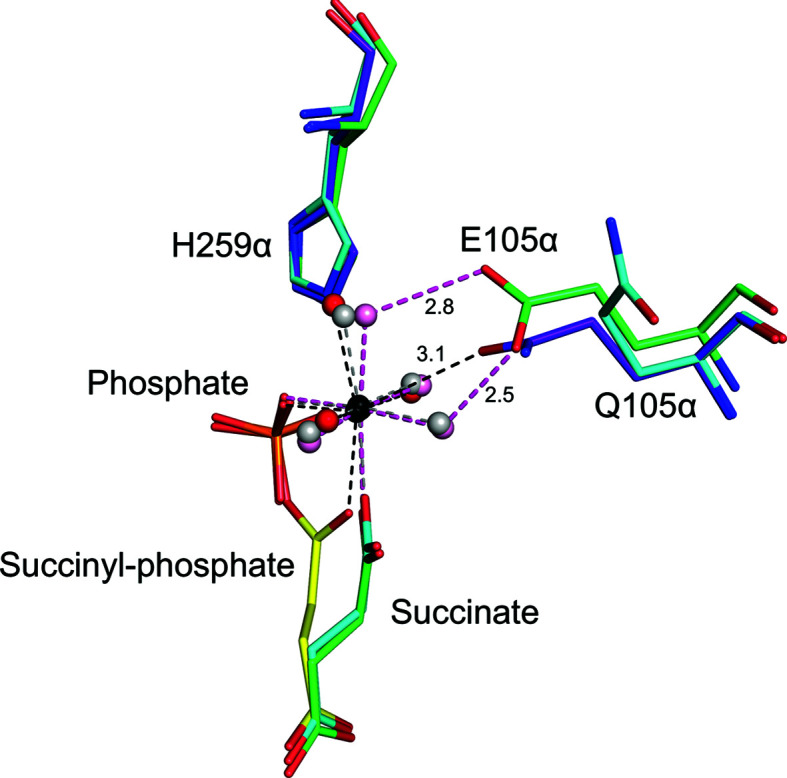
The structure of succinyl-CoA synthetase bound to the succinyl-phosphate intermediate clarifies the catalytic mechanism of ATP-citrate lyase. C atoms are yellow and purple for the succinyl-phosphate complex, green for the wild type with succinate and phosphate, and cyan for the mutant with succinate and phosphate. Water molecules are red for the succinyl-phosphate complex, pink for the wild type and grey for the mutant with succinate and phosphate. Other atoms are coloured according to type. Distances from the side-chain carbonyl O atom of Gln105α to the magnesium ion and from the carboxylate O atom of Glu105α to the water molecules are shown in Å.
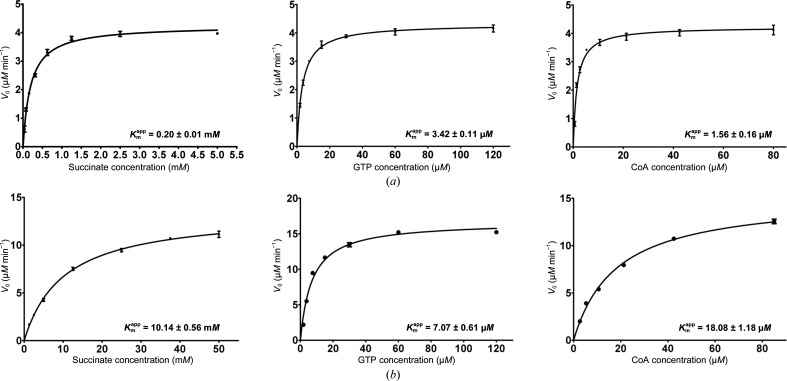
Next, we performed enzymatic and kinetic assays in order to understand why this reaction intermediate could be captured in the crystal structure of mutant GTPSCS. The specific activity of the E105αQ mutant is reduced 120-fold compared with the wild-type enzyme. The apparent K m of the E105αQ mutant for succinate is 50 times higher than that of the wild type and that for CoA is 11 times higher (Fig. 5 ▸ and Table 3 ▸). In contrast, the apparent K m of the mutant for GTP is only twice as high as that of the wild type. We therefore conclude that the mutation impacts the reaction of the phosphorylated enzyme, succinate and CoA at active site I [Joyce et al., 1999 ▸; catalytic steps (2) and (3) in Fig. 1 ▸], but not the phosphorylation by GTP at active site II [step (1)], supporting what is seen in the crystal structure. Specifically, the mutation affects the coordination of the magnesium ion that is essential for binding succinate (Huang & Fraser, 2016 ▸) and succinyl-phosphate. Although the magnesium ion is not required for CoA binding (PDB entry 4xx0; Huang et al., 2015 ▸), the carbonyl of CoA nearest its free thiol can interact with a water molecule that coordinates the magnesium in the structure of phosphorylated SCS from Francisella tularensis even in the absence of succinate (PDB entry 6mgg; Center for Structural Genomics of Infectious Diseases, unpublished work; Fig. 6 ▸ a). In the E105αQ mutant this water molecule is displaced from the coordination shell by the side-chain carbonyl of glutamine, leading to an increase in the apparent K m of the mutant for CoA. Despite the reduction in specific activity, the E105αQ mutant had to be crystallized with desulfo-CoA rather than CoA to capture the reaction intermediate. Over the time course of crystallization, CoA would react with succinyl-phosphate to form succinyl-CoA. Desulfo-CoA is unable to react with succinyl-phosphate, instead working with the protein to protect succinyl-phosphate from hydrolysis by preventing water from approaching the phosphoester (Fig. 6 ▸ b).
Figure 5.
Kinetic analysis of wild-type GTPSCS (a) and the E105αQ mutant (b). Initial velocities are plotted as a function of substrate concentration. The apparent Michaelis constants for succinate, GTP and CoA (
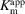 ) calculated using the plots are given. Each point is the mean value of three measurements of the initial velocity at a specific concentration of substrate; the bar represents the standard error of the mean. The plots were generated using GraphPad Prism (version 5 for Windows; GraphPad Software, San Diego, California, USA).
) calculated using the plots are given. Each point is the mean value of three measurements of the initial velocity at a specific concentration of substrate; the bar represents the standard error of the mean. The plots were generated using GraphPad Prism (version 5 for Windows; GraphPad Software, San Diego, California, USA).
Figure 6.
Roles of CoA. (a) CoA interacts with one of the water molecules that is part of the inner coordination shell of the magnesium ion. The carbonyl of CoA nearest its free thiol accepts a hydrogen bond (red dashed line) from this water molecule in the structure of the phosphorylated enzyme (PDB entry 6mgg; Center for Structural Genomics of Infectious Diseases, unpublished work). The water molecules are shown as red spheres, while the magnesium ion is shown as a black sphere and the ionic interactions with the magnesium ion are shown as black dashed lines. C atoms of the α-subunit are shown in green, those of the β-subunit are in pink and those of CoA are in magenta. (b) CoA protects succinyl-phosphate from hydrolysis, as seen in the complex of desulfo-CoA and succinyl-phosphate with E105αQ GTPSCS, where desulfo-CoA and the protein bind on three sides of the phosphoester bond. C atoms of residues of the α-subunit are shown in purple; those of the β-subunit, desulfo-CoA and succinyl-phosphate are shown in yellow. In addition to the stick model, spheres drawn using van der Waals radii surrounding residues Asn131α–Pro133α, Asn271β–Ala273β and desulfo-CoA emphasize how the atoms surround succinyl-phosphate.
The cryo-EM structure of the E599Q ACLY complex reported previously should have revealed the binding of citryl-phosphate and CoA (Fig. 7 ▸), not of phospho-citryl-CoA (Wei et al., 2020a ▸,b ▸). The substantially reduced activity of E599Q ACLY is due to the impact on step (3) of the reaction, the formation of citryl-CoA, not step (4), the cleavage of citryl-CoA to form acetyl-CoA and oxaloacetate. Although the kinetic constants were not measured for E599Q ACLY, 20 nM mutant produced only 25 ± 2% of the product produced by 20 nM ACLY as measured using a coupled-enzyme assay to detect the production of oxaloacetate over 15 min. There is no evidence that Glu599 of ACLY is involved in the cleavage of citryl-CoA to form acetyl-CoA and oxaloacetate. The glutamate residue is conserved in ACLYs and SCSs because it serves the same catalytic role in these enzymes. Slowing down step (3) of the reaction by mutating this glutamate residue to glutamine allowed capture of the citryl-phosphate intermediate even in the presence of CoA during the plunge-freezing technique used in cryo-EM.
Figure 7.
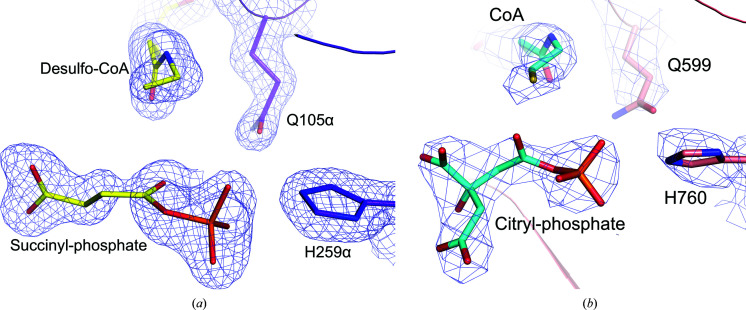
Comparison of the binding of succinyl-phosphate and desulfo-CoA to E105αQ GTPSCS with the binding of citryl-phosphate and CoA to E599Q ACLY. The structures of E105αQ GTPSCS in (a) and E599Q ACLY in (b) are drawn from similar viewpoints. C atoms are shown in yellow and purple for the succinyl-phosphate and desulfo-CoA complex with E105αQ GTPSCS and in cyan and pink for the citryl-phosphate and CoA complex with E599Q ACLY. Other atoms are coloured according to type. The blue maps are contoured at 1σ around succinyl-phosphate, desulfo-CoA, Gln105α and His259α in (a) and at 4σ around citryl-phosphate, CoA, Gln599 and His760 in (b).
4. Conclusions
Our results reveal the binding of succinyl-phosphate to SCS, proving that succinyl-phosphate is a reaction intermediate and showing how it binds to the enzyme. The glutamate residue at position 105α of SCS or 599 of ACLY plays an essential role in catalytic efficiency as it influences the stability of the magnesium complex in active site I. The E105αQ mutant shows decreased specific activity, with increased apparent K m values for succinate and CoA when compared with the wild-type enzyme. The observation of succinyl-phosphate bound to SCS in the presence of desulfo-CoA and of citryl-phosphate bound to ACLY in the presence of CoA shows that the transfer of the phosphoryl group is spontaneous and reveals how the carboxyl-phosphate intermediate is protected from hydrolysis by the protein on two faces of the ester and CoA on another.
Supplementary Material
PDB reference: E105αQ mutant, complexed with Mg2+-succinate and CoA, 7mss
PDB reference: E105αQ mutant, phosphorylated, complexed with CoA, 7mst
Acknowledgments
This research used resources at the Industrial Macromolecular Crystallography Association Collaborative Access Team (IMCA-CAT) beamline 17-ID. Use of the IMCA-CAT beamline at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. The Advanced Photon Source is a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Funding Statement
This research was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (grant No. 04185 to MEF).
References
- Beilsten-Edmands, J., Winter, G., Gildea, R., Parkhurst, J., Waterman, D. & Evans, G. (2020). Acta Cryst. D76, 385–399. [DOI] [PMC free article] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Cha, S. & Parks, R. E. Jr (1964). J. Biol. Chem. 239, 1968–1977. [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fraser, M. E., Hayakawa, K., Hume, M. S., Ryan, D. G. & Brownie, E. R. (2006). J. Biol. Chem. 281, 11058–11065. [DOI] [PubMed]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D. & Bairoch, A. (2005). The Proteomics Protocols Handbook, edited by J. M. Walker, pp. 571–607. Totowa: Humana Press.
- Hu, J., Komakula, A. & Fraser, M. E. (2017). Acta Cryst. D73, 660–671. [DOI] [PubMed]
- Huang, J. & Fraser, M. E. (2016). Acta Cryst. D72, 912–921. [DOI] [PubMed]
- Huang, J. & Fraser, M. E. (2020). Acta Cryst. F76, 302–308. [DOI] [PMC free article] [PubMed]
- Huang, J. & Fraser, M. E. (2021). Acta Cryst. D77, 357–368. [DOI] [PMC free article] [PubMed]
- Huang, J., Malhi, M., Deneke, J. & Fraser, M. E. (2015). Acta Cryst. F71, 1067–1071. [DOI] [PMC free article] [PubMed]
- Joyce, M. A., Fraser, M. E., Brownie, E. R., James, M. N. G., Bridger, W. A. & Wolodko, W. T. (1999). Biochemistry, 38, 7273–7283. [DOI] [PubMed]
- Liebschner, D., Afonine, P. V., Baker, M. L., Bunkóczi, G., Chen, V. B., Croll, T. I., Hintze, B., Hung, L.-W., Jain, S., McCoy, A. J., Moriarty, N. W., Oeffner, R. D., Poon, B. K., Prisant, M. G., Read, R. J., Richardson, J. S., Richardson, D. C., Sammito, M. D., Sobolev, O. V., Stockwell, D. H., Terwilliger, T. C., Urzhumtsev, A. G., Videau, L. L., Williams, C. J. & Adams, P. D. (2019). Acta Cryst. D75, 861–877.
- Nguyen, V. H., Singh, N., Medina, A., Usón, I. & Fraser, M. E. (2019). Protein Sci. 28, 1840–1849. [DOI] [PMC free article] [PubMed]
- Sánchez, L. B., Galperin, M. Y. & Müller, M. (2000). J. Biol. Chem. 275, 5794–5803. [DOI] [PubMed]
- Sun, T., Hayakawa, K., Bateman, K. S. & Fraser, M. E. (2010). J. Biol. Chem. 285, 27418–27428. [DOI] [PMC free article] [PubMed]
- Verschueren, K. H. G., Blanchet, C., Felix, J., Dansercoer, A., De Vos, D., Bloch, Y., Van Beeumen, J., Svergun, D., Gutsche, I., Savvides, S. N. & Verstraete, K. (2019). Nature, 568, 571–575. [DOI] [PubMed]
- Verstraete, K., Verschueren, K. H. G., Dansercoer, A. & Savvides, S. N. (2021). Nat. Struct. Mol. Biol. 28, 636–638. [DOI] [PubMed]
- Wei, J., Leit, S., Kuai, J., Therrien, E., Rafi, S., Harwood, H. J., DeLaBarre, B. & Tong, L. (2019). Nature, 568, 566–570. [DOI] [PubMed]
- Wei, X. & Marmorstein, R. (2021). Nat. Struct. Mol. Biol. 28, 639–641. [DOI] [PMC free article] [PubMed]
- Wei, X., Schultz, K., Bazilevsky, G. A., Vogt, A. & Marmorstein, R. (2020a). Nat. Struct. Mol. Biol. 27, 33–41. [DOI] [PMC free article] [PubMed]
- Wei, X., Schultz, K., Bazilevsky, G. A., Vogt, A. & Marmorstein, R. (2020b). Nat. Struct. Mol. Biol. 27, 511–513. [DOI] [PMC free article] [PubMed]
- Winter, G. (2010). J. Appl. Cryst. 43, 186–190.
- Winter, G., Waterman, D. G., Parkhurst, J. M., Brewster, A. S., Gildea, R. J., Gerstel, M., Fuentes-Montero, L., Vollmar, M., Michels-Clark, T., Young, I. D., Sauter, N. K. & Evans, G. (2018). Acta Cryst. D74, 85–97. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: E105αQ mutant, complexed with Mg2+-succinate and CoA, 7mss
PDB reference: E105αQ mutant, phosphorylated, complexed with CoA, 7mst