Abstract
The 5.2-kb ColJs plasmid of a colicinogenic strain of Shigella sonnei (colicin type 7) was isolated and sequenced. pColJs was partly homologous to pColE1 and to pesticin-encoding plasmid pPCP1, mainly in the rep, mob, and cer regions. A 1.2-kb unique region of pColJs showed significantly different G+C content (34%) compared to the rest of pColJs (53%). Within the unique region, seven open reading frames (ORFs) were identified. ORF94 was shown to code for colicin Js activity (cja), a 94-amino-acid polypeptide (molecular mass, 10.4 kDa); ORF129 (cji) was shown to code for the 129-amino-acid colicin Js immunity protein (molecular mass, 14.3 kDa); and ORF65 was shown to be involved in colicin Js release by producer bacteria (cjl) coding for a 65-amino-acid polypeptide (molecular mass, 7.5 kDa). In contrast to the gene order in other colicin operons, the cjl gene was found upstream from cja. Moreover, the promoter upstream from cjl was similar to promoters described upstream from several colicin activity genes. The cji gene was found to be located downstream from cja with a transcription polarity opposite to that of the cjl and cja genes. The cja, cji, and cjl genes were not similar to other known colicin genes. Colicin Js was purified as an inactive fusion protein with an N-terminal histidine tag. Activity of the purified fusion form of colicin Js was restored after cleavage of the amino acids fused to the colicin Js N terminus.
Colicins are plasmid-encoded toxic exoproteins that are produced by colicinogenic strains of Escherichia coli and some related species of the family Enterobacteriaceae (28, 29). To date, at least 23 colicin types have been described in detail (19, 27, 30, 34). They exert an inhibitory effect on sensitive bacteria of the same family and preferably on strains of the same species. The molecular masses of colicins range between 29,000 and 75,000 Da (7). Colicin polypeptide chains can be divided into separate functional domains, each of which is responsible for one step in the interaction between the colicin and a sensitive bacterium. The central domain of colicins is involved in the attachment of the colicin molecule to a specific outer membrane receptor protein, the N-terminal domain mediates translocation through the cell envelope, and the C-terminal domain exerts the lethal effect (4, 6, 7). At least 12 different outer membrane proteins have been shown to be colicin receptors, two different translocation systems (Ton and Tol) used by colicins have been identified, and six different modes of molecular lethal action of colicins have been described (7, 10, 22, 34). Some molecules initially described as colicins were later reclassified as microcins, e.g., colicin V as microcin V and colicin X as microcin B19. In contrast to these oligopeptide microcins (3), colicins are larger proteins. Moreover, colicins are not posttranslationally modified, are usually inducible by the SOS response, and also differ from microcins by the mode of export from the producer bacteria.
Colicin Js was originally described as a bacteriocin of Shigella sonnei colicinotype 7 (1, 2). In 1987, colicin type 7 was reclassified in accordance with Fredericq's original classification scheme (14) and designated colicin Js. Its particular physicochemical and biological characteristics were published (33). For a number of reasons, colicin Js appeared to be a rather exceptional colicin type: producer bacteria, as well as the indicator strain S. sonnei 17 (colicin type 6), were involved in outbreaks of epidemic diarrhea (12). Colicin Js showed a unique antimicrobial spectrum, being inactive against standard E. coli colicin indicators. Indirect fluorimetry measurements indicated that the mode of action of colicin Js was not analogous to that of either pore-forming or nuclease-type colicins (33). Colicin Js was shown to be active against enteroinvasive E. coli (EIEC) serotypes (17). The sensitivity to Js was 90% associated with the ability of EIEC strains to produce experimental keratoconjunctivitis in rabbits. Strains belonging to EIEC serotypes that were not sensitive to colicin Js were, as a rule, negative in the enteroinvasiveness test (17).
This communication presents a number of new details of the colicin Js system. These include the structure of the colicin Js coding region on plasmid ColJs; identification of genes for colicin activity, immunity, and release; and molecular characterization of the Js polypeptide.
MATERIALS AND METHODS
Media.
Bacterial strains were grown at 37°C in TY medium containing (per liter) 8 g of Bacto Tryptone (Difco Laboratories), 5 g of yeast extract, and 5 g of NaCl (pH 7). For selection and maintenance of plasmids, we added (per liter of liquid medium or 1.5% [wt/vol] TY agar) 25 μg of chloramphenicol, 100 μg of ampicillin, or 25 μg of kanamycin. Isopropyl-β-d-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were used at 0.5 mM and 80 μg ml−1, respectively.
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Colicin Js producer strain S. sonnei type 7 and colicin Js-sensitive S. sonnei 17, colicin type 6, were kindly provided by J. Šmarda, Brno, Czech Republic.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain, plasmid, or cosmid | Genotype and/or phenotype | Source or reference |
|---|---|---|
| Escherichia coli | ||
| K-12 strains | ||
| 5K | hsdR lacZ rpsL ser thi thr | G. Schrempf |
| Row | 58-161 metB1 rpsL λ+ Fdef | P. Fredericq |
| JMXAc | F− Δ(lac pro)XIII ara argE(Am) gyrA rpoB nalA rif su | This laboratory |
| GE2340 | JMXAc supD metB(Ser-Am) | This laboratory |
| GE2341 | JMXAc supE metB(Glu-Am) | This laboratory |
| GE2342 | JMXAc supF metB(Tyr-Am) | This laboratory |
| GE2345 | JMXAc su6(Leu-Am) | This laboratory |
| M15 | F−lac ara gal mtl | Qiagen |
| KK2186 | F− 12 Δlac pro supE thi strA sbcBIS endA nspRA F′ truD36 prcAB lacIqZΔM15 | This laboratory |
| LMG194 | K-12 ΔlacX74 galE thi rpsL ΔphoA (PvuII) Δara-714 leu::Tn10 | Invitrogen |
| BL21 | F−hsdS gal | 36 |
| EIEC strains | ||
| O164 | J. Šmarda | |
| O143 | B. Murray | |
| Shigella sonnei | Colicinotype 7, colicin Js producer | 2 |
| Shigella sonnei 17 | Colicinotype 6; E6im Bres Mres; colicin Js indicator | 2 |
| Plasmids | ||
| pBAD/HisB | ara promoter transcription-translation system with N-terminal His tag | Invitrogen |
| pBluescript SK (+) | Phage T7 gene 10 promoter | Stratagene |
| pCR2.1 | Cloning vector for Taq polymerase-amplified PCR products | Invitrogen |
| pCR2.1-TOPO | Cloning vector for Taq polymerase-amplified PCR products | Invitrogen |
| pPD110 | Phage T7 gene 10 promoter; ori from pSC101 | 11 |
| pQE-30 | Phage T5 promoter transcription-translation system with N-terminal His tag | Qiagen |
| pQE-60 | Phage T5 promoter transcription-translation system with C-terminal His tag | Qiagen |
| pDS1 | pBCSK+ carrying pColU in ClaI restriction site | 32 |
| pDS43 | pBluescript SK (+) with 3.2-kb insert of pColJs carrying cja, cji, and cjl | This work |
| pDS44 | pBluescript SK (+) with 1.4-kb insert of pColJs carrying cji | This work |
| pDS45 | pBluescript SK (+) with 1.7-kb insert of pColJs | This work |
| pDS68 | pCR2.1 with cja and cjl in Js3S4-Js3S7 DNA fragment of pColJs | This work |
| pDS82 | pCR2.1 with cja and cjl in JsUH-Js3S19 DNA fragment of pColJs | This work |
| pDS83 | pCR2.1 with cja in Js3S21-Js3S19 DNA fragment of pColJs | This work |
| pDS85 | pCR2.1 with cja, cji, and cjl in JsUH-JsLH DNA fragment of pColJs | This work |
| pDS87 | pQE-30 with cja (ORF94U-ORF94/93L) | This work |
| pDS96 | pCR2.1 with cja in ORF94U-ORF94/93L DNA fragment of pColJs | This work |
| pDS97 | pCR2.1 with cja and amber stop codon in ORF93 (ORF93Uamb-ORF94/93L) | This work |
| pDS99 | pCR2.1 with cja and opal stop codon in ORF92 (ORF94U-ORF94Lopal) | This work |
| pDS104 | pBluescript SK (+) with 5.2-kb pColJs in ClaI | This work |
| pDS110 | pCR2.1 with cja and amber stop codon in cja (ORF94UAI-ORF94L) | This work |
| pDS112 | pQE-60 with cja (ATG94U-ATG94LHA) | This work |
| pDS117 | pQE-30 with cja preceded by enterokinase cleavage site (ORF94UEK-ATG94LHA) | This work |
| pDS216 | pPD110 with cjl in Js3S4-ORF65L DNA fragment of pColJs | This work |
| pDS257 | pBAD/HisB with JsUH-JsLH fragment pColJs cloned in BglII | This work |
| pDS265 | pCR2.1-TOPO with ORF69, ORF47, and ORF129 (Js3S28-JsLH) | This work |
| pDS270 | pBAD/HisB with cji (ORF129UH-ORF129LH) | This work |
| pDS274 | pCR2.1-TOPO with cji (ORF129UH-ORF129LH) | This work |
Crude colicin preparations.
Cells from the TY cultures of colicinogenic strains (producers of colicins Js and U) induced by mitomycin C (0.5 μg ml−1; Sigma) were harvested, resuspended in distilled water, washed, and sonicated. The sonicates were used as crude colicins.
Determination of sensitivity to colicin Js.
Colicin Js producer bacteria were inoculated onto agar plates with a single stab and subsequently grown for 16 to 48 h at 37°C. After that, plates were either directly overlaid with 100 μl of colicin Js-sensitive bacteria in 3 ml of 0.75% (wt/vol) TY agar or exposed to chloroform vapor for 30 min to lyse the producer bacteria and then overlaid. This was followed by overnight cultivation at 37°C. Bacteria sensitive to colicin Js formed a zone of growth inhibition around the colicin Js producer.
Colicin activity assays.
Colicin Js activity was tested by spotting 10-fold dilutions of colicin-containing crude cell lysates on agar plates seeded by sensitive bacteria; TY agar plates were overlaid with 3 ml of 0.75% (wt/vol) TY agar with 100 μl of an overnight culture of indicator bacteria. Each experiment was performed at least three times, and the data reported are averages of three independent measurements.
Restriction analysis.
Standard methods were used for restriction endonuclease analysis, ligation, and transformation of plasmid DNA (31).
Recombinant DNA methods.
Plasmids were isolated and cells were transformed by using standard techniques. pColJs genes were cloned into the vector pCR2.1 or pCR2.1-TOPO (Invitrogen) after PCR amplification. For each plasmid construct, the PCR primer pairs used for amplification are described in Table 1. PCR primer sequences and their positions on pColJs are shown in Table 2. Cloning in the pQE-30, pQE-60, and pBAD/HisB vectors was done after PCR amplification of insert DNA, restriction digestion of both ends of the amplified DNA, and ligation into the digested plasmid DNA. The primer pairs used for amplification are noted in Table 1, and their characteristics are shown in Table 2. Insert DNA for plasmid pPD110 was prepared by PCR amplification from pColJs by using primers Js3S4 and ORF65L. Amplified DNA was cloned into vector pCR2.1 and then cut out with EcoRI and ligated into pPD110 digested with the same enzyme. Insert DNA was sequenced by using the Taq Dye-deoxy Terminator method and a model 377 DNA sequencing system (Applied Biosystems, Foster City, Calif.).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′–3′)a | Positionb |
|---|---|---|
| ATG94U | GATTCCATGGTTTCTCAAAATGTTTGGGCTCCT | 2586–2615 |
| ATG94LHA | TCAGGGATCCCTATAAGGTAGTGTATACATGAAT | 2869–2847 |
| Js3S4 | TACAGTATATTCTGTGTAAAT | 2335–2355 |
| Js3S7 | CAAAGTTGGTGGTGACAGCAT | 3227–3207 |
| Js3S19 | GGTATAGCTTAGGCAGGATTA | 2967–2947 |
| Js3S21 | CAGGAGATTATATGATTTCTC | 2575–2596 |
| Js3S28 | TAATCCTGCCTAAGCTATACC | 2947–2967 |
| JsUH | GAGGGGATCCATGAAAGAAGAAAAAGTATTGCTTC | 2371–2395 |
| JsLH | GCAGGGATCCTGAAGTTGAAGCCTTATAGCGGC | 3702–3680 |
| ORF65L | CAAAGGATCCGAGAAATCATATAATCTCCTG | 2596–2575 |
| ORF93Uamb | AGGAGGATCCATGATTTCTCAAAATGTTTGGGCTCCTCCTTTCTTTTTTGGTCCTTGGGTAGAAC | 2586–2640 |
| ORF94/93L | TCCTGGATCCGGTATAGCTTAGGCAGGATTA | 2907–2887 |
| ORF94L | TTATGGATCCTTAGCTCTTATTATAAGGTAG | 2848–2879 |
| ORF94Lopal | TTATGGATCCTTCAGCTCTTATTATAAGGTAGTGTATACATGAATAATCTGCCCTGACCCA | 2880–2830 |
| ORF94U | AGGAGGATCCATGATTTCTCAAAATGTTTGG | 2586–2609 |
| ORF94UAI | CAGGAGGATCCATGATTTAGCAAAATGTTTGGG | 2586–2610 |
| ORF94UEK | GGAGGATCCGACGATGACGATAAGATGATTTCTCAAAATGTTTGGGCTCC | 2586–2614 |
| ORF129LH | CTAGGATCCTTATTTTGAATAAGCTTTTAT | 2934–2954 |
| ORF129UH | GGAGGATCCATGTTCAAATCATCAGTAAAG | 3323–3303 |
Nucleotides in boldface represent target nucleotides for in vitro mutagenesis, and nucleotides in italics correspond to introduced BamHI and NcoI restriction sites.
Numbers indicate position on pColJs with respect to its unique ClaI restriction site (accession no. AF282884).
Protein analysis procedures.
Separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed as previously described (32). Western blot analysis was performed after semidry transfer of proteins from the electrophoretic gel to nitrocellulose membranes and detected with a 1:1,000-diluted mouse anti-His primary monoclonal antibody (Qiagen). Membranes were then treated with a 1:4,000-diluted rabbit anti-mouse secondary antibody conjugated to horseradish peroxidase (Rockland Immunochemicals). Proteins were visualized with a chemiluminescence detection kit (ECL; Amersham Pharmacia Biotech).
Purification of colicin Js.
Colicin Js was purified as an inactive fusion protein with an N-terminal histidine tag and an enterokinase recognition site (MRGSH6GFD4K-). E. coli M15(pDS117) bacteria were grown in 50 ml of TY medium to an optical density of 0.3, subsequently induced with 1 mM IPTG, and cultivated for an additional 3 h. Cells were harvested and resuspended in 5 ml of buffer B (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris · Cl [pH 8.0]; Qiagen). Cells were subsequently stirred for 60 min at room temperature, until the solution became translucent. Lysates were centrifuged at 10,000 × g for 30 min at room temperature to pellet the cell debris. Supernatants were mixed with 0.5 ml of the 50% Ni-nitrilotriacetic acid agarose and incubated for 60 min at room temperature. Lysate-resin was then loaded onto an empty column (Qiagen) and washed twice with 4 ml of buffer C (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris · Cl [pH 6.3]; Qiagen). To isolate the fraction containing maximum colicin Js and minimum contaminating proteins, His-tagged colicin Js was eluted four times with 0.5 ml of buffer D (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris · Cl [pH 5.9]; Qiagen) and four times with 0.5 ml of buffer E (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris · Cl [pH 4.5]; Qiagen). Purified colicin Js was dialyzed overnight at 4°C against 10 mM Tris · Cl [pH 7.4] to remove urea. The dialyzed colicin Js (50 μl) was digested with 5 μl of enterokinase (light chain; 0.4 U μl−1; New England Biolabs) for 8 h at room temperature. The activity of purified and enterokinase-treated colicin Js was measured by colicin activity assay.
Computer-assisted sequence analysis.
Computer-assisted sequence analysis was performed by using programs in the Genetics Computer Group (Madison, Wis.) software package. ProtParam at ExPASy was used for theoretical polypeptide molecular weight and isoelectric point calculations. For prediction of protein localization and the signal peptide sequence, transmembrane prediction, and protein motif identification, ExPASy programs were used.
Nucleotide sequence accession number.
The nucleotide sequence reported in this study was deposited in the GenBank database under accession no. AF282884.
RESULTS
Plasmid pColJs.
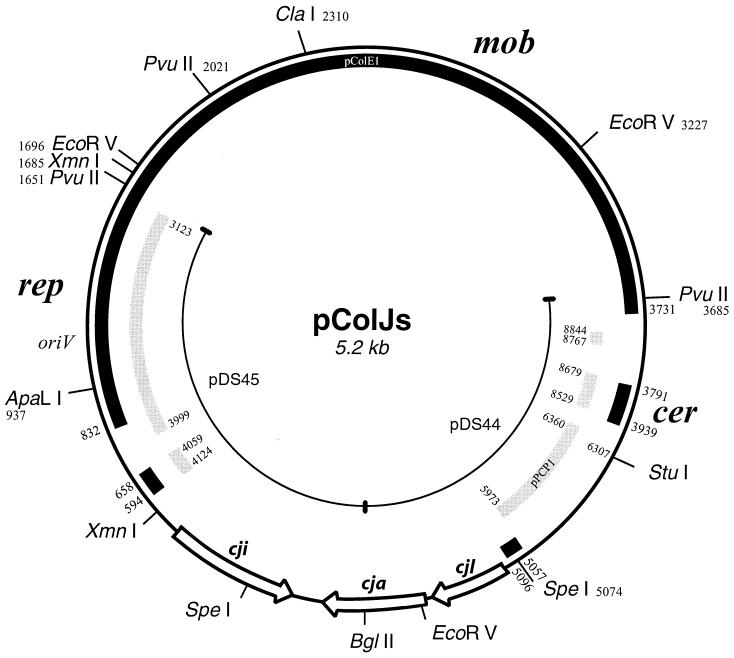
Plasmid DNA was prepared from the original producer of colicin Js, S. sonnei, colicin type 7. To isolate the colicin Js coding region of pColJs, the product of restriction digestion with PvuII was ligated into pBSSK(+) digested with EcoRV. A colicin Js-producing clone was isolated with a 3.2-kb DNA insert (pDS43). The DNA insert was further subcloned by using the BglII restriction site of the insert and the XbaI (pDS44, 1.4-kb insert) or HindIII (pDS45, 1.7-kb insert) site of the pBluescript multiple cloning site. Neither pDS44 nor pDS45 conferred colicin Js activity, localizing the BglII restriction site to the colicin Js coding determinant. Plasmid pDS45 coded for colicin Js immunity. The whole pColJs plasmid was cloned in pBSSK(+) by using a unique ClaI restriction site of pColJs, resulting in pDS104. The insert of pDS43 and the rest of pColJs were sequenced by using synthetic oligonucleotides. The resulting restriction map of pColJs is shown in Fig. 1. The pColJs (5,210 bp) map was numbered from its unique ClaI restriction site. A substantial part of pColJs was homologous to pColE1, mainly in the mob, rep, and cer regions of pColE1 (9). The 2.9-kb region of pColJs, which was similar to the region between bp 832 and 3731 of pColE1, showed 94.7% identity; shorter regions of homology showed similar degrees of identity (3791 to 3939, 89.3%; 5057 to 5096, 87.5%; 594 to 658, 95.4%). Moreover, homology to pesticin-encoding plasmid pPCP1 (18) was also observed in five distinct regions of pPCP1 (3123 to 3999, 93.4%; 4059 to 4124, 100%; 5973 to 6360, 95.9%; 8529 to 8629, 86.8%; 8767 to 8844, 85.0%). The rep region of pColJs was homologous to both pColE1 and pPCP1, while the region downstream of mob and cer was similar only to pPCP1 (Fig. 1). The complete sequence of pColJs was deposited in the GenBank database under accession number AF282884.
FIG. 1.
Restriction map of pColJs. Dark areas represent DNA sequences homologous to pColE1. Grey areas show sequences similar to pesticin-encoding plasmid pPCP1. The region of pColJs not homologous to either pColE1 or pPCP1 codes for colicin Js activity. Numbers outside the pColJs circle correspond to the nucleotide sequence of pColE1, and numbers inside correspond to that of pPCP1. The positions of the rep, mob, and cer regions and cloned DNA fragments in pDS44 and pDS45 are indicated. pColJs is numbered starting from its unique ClaI restriction site.
A 1.2-kb region of pColJs flanking the unique BglII restriction site showed no significant similarity to other colicinogenic plasmids or sequences. This region between bp 2343 and 3508 of pColJs showed significantly lower G+C content (33.6%) compared to the G+C content of the rest of pColJs (52.9%). Within the 1.2-kb unique region, seven potential open reading frames (ORFs) were identified. Each ORF was named according to the number of amino acids it encodes. The orientations, lengths, and names of the ORFs are shown in Fig. 2.
FIG. 2.
(A) Schematic drawing of the 1.2-kb unique (nonhomologous) region of pColJs. ORFs are designated according to the number of amino acids encoded. Cloned DNA regions in pDS68, pDS82, pDS83, pDS85, and pDS274 are indicated. pDS83 has the smallest insert conferring colicin Js activity, pDS274 has the smallest insert conferring colicin Js immunity, and pDS85 codes for colicin Js activity, immunity, and release. The functions of ORF92, ORF93, ORF69, and ORF47 are unknown. (B) Nucleotide sequence of the promoter upstream from the cjl gene. The putative promoter regions (−10 and −35), SOS boxes, ribosome binding site sequences (S.D.), and transcriptional polarity (arrow) of cjl are indicated. Numbers correspond to positions in pColJs.
Subcloning of PCR-amplified DNA segments of the 1.2-kb unique region of pColJs into the pCR2.1 or pCR2.1-TOPO cloning vector placed the colicin Js activity gene within a DNA fragment with ORF94, ORF93, and ORF92 (plasmid pDS83, Fig. 2). pDS82 with ORF65, ORF94, ORF93, and ORF92 codes for colicin Js activity, as well as for colicin Js release. In contrast to sensitive bacteria with pDS83, the colicin Js-sensitive strains with pDS82 were unstable and in overnight culture, colicin Js-resistant colonies appeared. The same phenotype was observed for pDS68, which has ORF69 in addition. pDS85 with the complete 1.2-kb unique region of pColJs coded for colicin Js activity, immunity, and release. To identify the ORF responsible for colicin Js synthesis, stop codons in all three ORFs of pDS83 were independently introduced and colicin Js activity in amber suppressor and nonsuppressor strains was measured. E. coli strain JMXAc was used as a nonsuppressing strain; strains GE2340, GE2341, GE2342, and GE2345 were used for suppression of amber stop codon mutations, while the opal stop codon in ORF92 was not suppressed. Only the introduction of a stop codon into ORF94 resulted in the complete loss of colicin Js activity, which was partially restored in a suppressor strain. Stop codons in ORF93 and ORF92 did not interfere with colicin Js synthesis. Thus, ORF94 codes for colicin Js, a 94-amino-acid polypeptide (molecular mass, 10.4 kDa; pI 6.70) with no sequence similarity to other known colicins. ORF94 was named cja (for colicin Js activity).
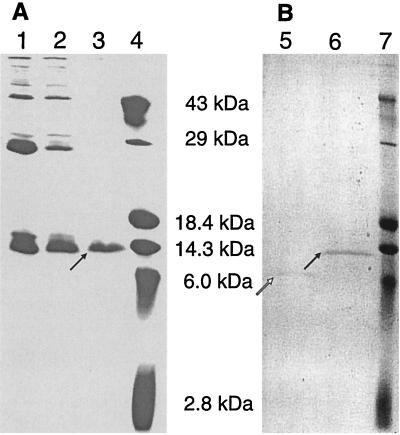
Western blot detection with a primary antibody recognizing the penta-His sequence of the Cja protein fused to the N-terminal His tag expressed from pQE-30 is shown in Fig. 3. His-tagged colicin Js showed a molecular mass of approximately 14 kDa, which is slightly more than the predicted 11.8 kDa. The colicin Js polypeptide was predicted to be a cytoplasmic protein (psort), but one possible transmembrane region between amino acids 30 and 50 (tmpred) was predicted. No signal peptide sequence (signalp) or prokaryotic protein motifs were identified (prosite).
FIG. 3.
Western blot analysis of expressed cja, cjl, and cji genes cloned in pBAD/HisB or in pQE-30. Proteins were separated by sodium dodecyl sulfate–15% polyacrylamide gel electrophoresis. Genes were expressed after induction with 0.2% arabinose for pBAD-based constructs and with 1 mM IPTG for the pQE-30 vector. Gene products were visualized by using a penta-His-recognizing primary antibody (Qiagen) and a horseradish peroxidase-labeled secondary antibody with chemiluminescence detection. Lanes: 1 and 2, LMG194 BAD/HisB cloning vector uninduced and induced, respectively; 3 and 4, LMG194(pDS257) with cloned cjl uninduced and induced, respectively; 5 and 6, M15(pDS117) with cloned cja uninduced and induced, respectively; 7 and 8, LMG194(pDS270) with cloned cji uninduced and induced, respectively. Expected molecular masses of polypeptides detected: pBAD/HisB, 7.1 kDa; pDS257, 11.5 kDa; pDS117, 11.8 kDa; pDS270, 18.2 kDa.
The promoter region upstream of ORF65 (cjl) was found to be similar to promoter regions upstream of colicin activity genes on, e.g., pColE1 and pColU (9, 32): a highly conserved −35 region (TTGACA), a less-conserved −10 region (TAATTT), and two overlapping SOS boxes (Fig. 2). Since the presence of SOS boxes very similar to that of pColE1 and pColU suggested the possible inducibility of colicin Js synthesis by the SOS response, we measured the inducibility of colicin Js activity after mitomycin C treatment (0.5 μg ml−1). The results (Table 3) show that colicin Js activity increased by 1 order of magnitude in the original producer strain of S. sonnei after induction by mitomycin C in both culture supernatants and cell lysates. Induction of Js activity in E. coli KK2186 with pDS43 was observed only in bacterial supernatants. Induction of colicin U synthesis in E. coli 5K under the same conditions led to increases in colicin U activity in both supernatants and cell lysates of 2 to 3 orders of magnitude (Table 3). The inducibility of colicin Js activity was thus considerably lower than that of colicin U and other colicins.
TABLE 3.
Inducibility of colicin Js synthesis by mitomycin C treatmenta
| Strain | Colicin activity in supernatants
|
Colicin activity in cell lysates
|
||
|---|---|---|---|---|
| Noninduced | Induced | Noninduced | Induced | |
| S. sonnei(pColJs) | 1 (2) | 2 (3) | 2 (3) | 3 (4) |
| E. coli KK2186(pDS43) | 1 (2) | 2 (3) | 2 (3) | 2 (3) |
| E. coli 5K(pDS1) | 0 (1) | 2 (4) | 1 (3) | 3 (5) |
The values indicate the highest colicin dilution that resulted in a clear zone of growth inhibition and the last dilution that resulted in turbid zones (in parentheses) on a lawn of sensitive strain E. coli O164 (colicin Js) or E. coli K-12 5K (colicin U); e.g., 2 = 102. Mitomycin C was used at 0.5 μg ml−1.
Purification of colicin Js.
To demonstrate that the cja gene codes for a polypeptide product with antimicrobial activity identical to that of colicin Js, we purified colicin Js and measured its inhibitory effect on sensitive bacteria. Colicin Js was purified as an inactive fusion protein with an N-terminal histidine tag. The cja gene was cloned into the BamHI restriction site of pQE-30, resulting in pDS87. Because this construct conferred no colicin Js activity, the histidine tag was placed on the C terminus of colicin Js by using the NcoI and BglII sites of pQE-60. The resulting plasmid, pDS112, coded for colicin Js with very low activity. To purify fully active colicin Js, we introduced an enterokinase cleavage site before the start codon of cja and cloned this construct into pQE-30. The resulting plasmid, pDS117, was used for overexpression and purification of colicin Js (Fig. 4). The activity of the purified and renatured fusion form of colicin Js was restored after cleavage of amino acids fused to the colicin Js N terminus (data not shown).
FIG. 4.
(A) Purification of colicin Js with an N-terminal histidine tag by using an Ni column. Lanes: 1, second elution of proteins from the Ni column; 2, third elution; 3, fourth elution; 4, low-molecular-weight protein standard (Gibco). The arrow indicates purified colicin Js. (B) Treatment of colicin Js with enterokinase. Lanes: 5, enterokinase treated colicin Js; 6, untreated, His-tagged colicin Js; 7, low-molecular-weight protein standard (Gibco). The white arrow indicates enterokinase-treated colicin Js, and the black arrow indicates untreated colicin Js. A 15% polyacrylamide gel was stained with Coomassie brilliant blue R250 (Sigma).
Role of ORF65 in colicin Js release.
ORF65 cloned together with the cja gene (in pDS82) in sensitive EIEC strain O143 resulted in an unstable strain giving rise to variants of bacteria resistant to colicin Js. The ORF65 gene was proposed to play a role in colicin Js release and was named cjl (for colicin Js lysis). The cjl gene, located upstream of cja, codes for a 65-amino-acid polypeptide (molecular mass, 7.5 kDa; pI 4.68). Western blot analysis of the cjl gene, expressed from an arabinose-induced pBAD/HisB fusion expression vector, is shown in Fig. 3. The molecular mass (16 kDa) of Cjl fused to 37 N-terminal amino acids derived from pBAD/HisB exceeded the predicted molecular mass of the fusion construct (11.5 kDa). Cjl was predicted to be a cytoplasmic polypeptide (psort). No transmembrane region (tmpred), no signal peptide sequence (signalp), and no prokaryotic protein motifs were identified (prosite).
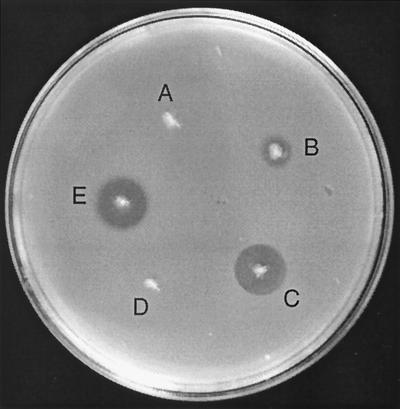
The cjl gene was cloned in the pPD110 plasmid, compatible with ColE1 ori plasmids, resulting in pDS216, and expressed alone or together with the colicin Js activity gene. The results of this experiment are shown in Fig. 5. E. coli BL21 transformed with pDS43 carrying the complete colicin Js coding region showed the same size inhibition zone as BL21 with plasmids pDS96 and pDS216. In contrast, strain BL21 carrying only cja produced a considerably smaller inhibition zone. This suggested that cjl has a role in the increased synthesis or release of colicin Js. To test this, producer bacteria were grown overnight, treated with chloroform vapor for 30 min, and then overlaid with indicator bacteria in top agar. Inhibition zones were the same size (data not shown) for producer strains with or without cjl, indicating that cjl is involved in colicin Js release.
FIG. 5.
Cloning and expression of cjl and the colicin Js gene (cja) in E. coli BL21. Producer bacteria: A, BL21; B, BL21(pDS96); C, BL21(pDS216)(pDS96); D, BL21(pDS216); E, BL21(pDS43). Note the same diameter of the inhibition zone on indicator bacteria for the cloned 3.2-kb pColJs region of pDS43 and for the strain expressing cja and cjl from two different compatible plasmids. EIEC strain O164 was used as an indicator.
Immunity to colicin Js.
To test immunity to colicin Js, we transformed sensitive EIEC strain O143 (a strain with sensitivity to colicin Js lower than that of EIEC strain O164) with plasmids carrying different portions of pColJs (Fig. 2). The results are summarized in Table 4. Plasmids pDS43 and pDS85, containing the whole 1.2-kb unique region of pColJs, coded for immunity to colicin Js. The immunity gene was further localized to a DNA fragment of pDS45 and pDS265 with ORF69, ORF47, and ORF129. EIEC strain O143 with pDS68, harboring cjl, cja, and ORF69, was fully sensitive to Js. The ORF129 gene cloned in pDS274 conferred immunity to colicin Js and was named cji (for colicin Js immunity). The transcription polarity of cji is the opposite of that of cjl and cja, and the promoter region showed no obvious consensus region, suggesting a low level of cji transcription in producer bacteria. The cji gene codes for a 129-amino-acid colicin Js immunity protein (molecular mass, 14.3 kDa). Western blot analysis of the cji gene expressed from an arabinose-controlled pBAD/HisB His tag fusion expression vector is shown in Fig. 3. The molecular mass (18 kDa) matches the predicted 18.3 kDa. Because the arabinose-mediated induction of Cji synthesis was not sufficient for Western blot detection in whole-cell lysates, the fusion protein product was prepurified and concentrated by Ni-nitrilotriacetic acid agarose column chromatography. The colicin Js immunity protein was predicted to be an inner or outer membrane protein (psort), and an uncleaved N-terminal signal-anchor was proposed (signalp; 21). No prokaryotic protein motifs were identified (prosite).
TABLE 4.
Immunity to colicin Js
| EIEC strain | Plasmid (ORF[s] on insert) | Sensitivity to Js |
|---|---|---|
| O143 | 2 (3)a | |
| O143 | pDS43 (3.1 kb; ORF65, ORF94, ORF93, ORF92, ORF69, ORF47, ORF129) | Ib |
| O143 | pDS85 (1.2 kb; ORF65, ORF94, ORF93, ORF92, ORF69, ORF47, ORF129) | Ib |
| O143 | pDS82 (ORF65, ORF94, ORF93, ORF92) | 2 (3)c |
| O143 | pDS83 (ORF94, ORF93, ORF92) | 2 (3) |
| O143 | pDS68 (ORF65, ORF94, ORF93, ORF92, ORF69) | 2 (3)c |
| O143 | pDS265 (ORF69, ORF47, ORF129) | Ib |
| O143 | pDS274 (ORF129) | Ib |
The values indicate the highest colicin dilution that resulted in a clear zone of growth inhibition and the last dilution that resulted in turbid zones (in parentheses) on the lawn of sensitive bacteria; e.g., 3 = 103.
I, immune strain.
Unstable strain.
DISCUSSION
The original producer of colicin Js was first described as a bacteriocin-producing strain of S. sonnei, colicin type 7 (1, 2). Not only colicin Js was identified to be originally produced by Shigella strains: colicin S1 was described as a product of an S. sonnei strain, and colicins S4 and U were first described in S. boydii (13, 16). In contrast to other colicins, even those produced by Shigella strains, colicin Js showed a unique antimicrobial spectrum, being inactive against standard E. coli colicin indicators such as E. coli K-12 5K and K-12 Row. Bacteria sensitive to colicin Js were shown to be EIEC and Shigella strains. Moreover, strains sensitive to colicin Js were shown to be able to invade host enterocytes and to produce experimental keratoconjunctivitis (17).
pColJs is a 5.2-kb plasmid showing striking similarities to the plasmid coding for colicin E1, pColE1. More than 3 kb of pColJs is homologous to pColE1, with an identity of nearly 95%, mainly in regions responsible for maintenance of the pColJs plasmid, in replication and mobilization regions. In addition to this, about 1.5 kb is homologous to pesticin-encoding plasmid pPCP1, with identities ranging from 85 to 100%. About 1.2 kb of pColJs is unrelated to other sequences, coding for colicin Js activity, immunity, and release. This region showed significantly lower G+C content (34%) than pColJs regions similar to pColE1 and pPCP1 (53%). These results suggested involvement of DNA recombination in the evolution of the colicin Js-encoding plasmid. The pColJs sequences for replication and plasmid maintenance are similar to those of other colicin plasmids, while the regions coding for colicin Js activity, immunity, and release are unique. A similar situation was described for other Col plasmids, e.g., Col plasmids coding for colicins 5, 10, K, and U showing similarities to the pColE1 rep, mob, and cer regions with specific DNA coding sequences for particular colicin activity, immunity, and lysis genes. Regions upstream and downstream from a particular colicin operon appear to be recombination sites in the evolution of different Col plasmids (23–25, 32).
In the 1.2-kb unique region of pColJs, seven ORFs were identified, none coding for polypeptides longer than 129 amino acids. The promoter region upstream from cjl was similar the promoter region on Col plasmids upstream from the colicin activity gene: a conserved −35 region, a less-conserved −10 region, and two overlapping SOS boxes. Despite this similarity, cjl does not code for colicin Js activity. Instead, the colicin Js-encoding gene (cja) was found to be located downstream from cjl, coding for a polypeptide of 94 amino acids. This might explain the observed lower rate of SOS inducibility of colicin Js activity after mitomycin C treatment compared to the inducibility of activity of other colicin types, e.g., colicin U. cjl was shown to be involved in colicin Js release from producer bacteria. In other colicin plasmids, including pColE1 and pColU, genes for lytic proteins are located downstream from colicin activity and immunity genes. Immunity to colicin Js was shown to be a function of ORF129 (cji), with transcription polarity that is the opposite of that of cja and cjl.
Unlike the other known colicins, colicin Js is a polypeptide of 94 amino acids with a molecular mass of 10.4 kDa. This is considerably less than that reported previously (33). Colicin Js is almost three times smaller than the smallest colicin previously described, colicin M, which is composed of 271 amino acid residues with a molecular mass of 29.5 kDa. In this respect, colicin Js resembles microcins more than colicins. In contrast to microcins, no genes were found for colicin Js posttranslational modification or export from producer bacteria. The amino acid composition of the Js polypeptide showed no sequence homology to colicins or other proteins. These data imply that colicin Js represents a novel type of antimicrobial polypeptide not related to colicins or microcins. The colicin Js polypeptide was found to be very sensitive to changes in its amino acid composition. Fusion to a His-tag on either end of the colicin Js polypeptide led to complete or nearly complete loss of antimicrobial activity. A similar activity decrease was observed upon fusion of the N terminus of colicin Js to the N-terminal 28 amino acid residues of the LacZ protein. Hence, both ends of the colicin Js polypeptide appear to be involved in its biological function. Production of colicin Js without its immunity protein was shown to be tolerated in both resistant and colicin Js-sensitive producer strains in the absence of the lysis gene. However, stable maintenance of the plasmid coding for cja and cjl but not cji was observed only for resistant producer strains. These data suggest that colicin Js is active only when it enters the cell from outside and that the synthesized colicin Js is not active in the cytoplasm even when produced without its immunity protein.
The colicin Js immunity gene is located downstream from cja with opposite transcription polarity. This arrangement of genes is typical for colicins attacking the plasma membrane of sensitive bacteria. Moreover, immunity genes with opposite transcription polarity were described for colicin M and pesticin. Colicin M immunity protein protects the producer bacterium against colicin M, which inhibits peptidoglycan synthesis (15), and the pesticin immunity protein prevents pesticin-mediated murein degradation (26). In all cases, the opposite polarity of transcription of the colicin immunity gene is present when the colicin lethal target is not in the cytoplasm. The immunity genes of pore-forming colicins are constitutively transcribed and code for membrane products that protect the producer bacteria against the same colicin type provided outside the cells. Weak promoters of immunity proteins for pore-forming colicins result in low numbers of copies (102 to 103) of protein per cell (35, 38). The sequence upstream from the cji gene revealed no obvious consensus promoter sequence, suggesting a similar low transcription rate. Moreover, based on sequence predictions, Cji appears to be an inner membrane protein.
Colicin Js release from producer bacteria was shown to be encoded by cjl. In this respect, the cjl gene resembles the kil genes of some colicin plasmids coding for semispecific release of colicin molecules from the producer bacteria (5, 8). The expression of the kil (lysis) gene results in lysis of the producer bacteria and is regulated from the same promoter as the colicin structural gene. In all cases, the kil gene is located downstream from the gene coding for colicin activity and immunity genes. The lysis protein of cloacin DF13 has a signal peptide sequence, and the lysis protein was detected in both inner and outer membranes (37). Both signal sequence and lysis proteins contribute to the release of colicin molecules from the producer bacteria (20). In contrast to kil genes, the cjl gene is located as the first gene downstream from the SOS-regulated promoter and the colicin Js activity gene starts 17 nucleotides downstream from the cjl stop codon. Although the function of Cjl appears to be similar to that of Kil proteins, the Cjl polypeptide shows no homology to lysis proteins encoded by Col plasmids and has no predicted signal peptide sequence.
ACKNOWLEDGMENTS
We thank J. Šmarda for S. sonnei producer and indicator strains.
This work was partly supported by a grant from the Grant Agency of the Czech Republic (310/98/0083).
REFERENCES
- 1.Abbott J D, Graham J M. Colicine typing of Shigella sonnei. Mon Bull Minist Health Lab Serv. 1961;20:51–58. [PubMed] [Google Scholar]
- 2.Abbott J D, Shannon R. A new method for typing Shigella sonnei using colicin production as a marker. J Clin Pathol. 1958;11:71–77. doi: 10.1136/jcp.11.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baquero F, Moreno F. The microcins. FEMS Microbiol Lett. 1984;23:117–124. [Google Scholar]
- 4.Baty D, Frenette M, Lloubes R, Geli V, Howard S P, Pattus F, Lazdunski C. Functional domains of colicin A. Mol Microbiol. 1988;2:807–811. doi: 10.1111/j.1365-2958.1988.tb00092.x. [DOI] [PubMed] [Google Scholar]
- 5.Baty D, Lloubes R, Geli V, Lazdunski C, Howard S P. Extracellular release of colicin A is non-specific. EMBO J. 1987;6:2463–2468. doi: 10.1002/j.1460-2075.1987.tb02526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benedetti H, Geli V. Colicin transport, channel formation and inhibition. In: Konings W N, Kaback H R, Lolkema J S, editors. Handbook of biological physics. Vol. 2. Amsterdam, The Netherlands: Elsevier Sciences; 1996. pp. 665–691. [Google Scholar]
- 7.Braun V, Pilsl H, Gross P. Colicins: structures, modes of action, transfer through membranes, and evolution. Arch Microbiol. 1994;161:199–206. doi: 10.1007/BF00248693. [DOI] [PubMed] [Google Scholar]
- 8.Cavard D, Lloubes R, Morlon J, Chartier M, Lazdunski C. Lysis protein encoded by plasmid ColA-CA31. Gene sequence and export. Mol Gen Genet. 1985;199:95–100. doi: 10.1007/BF00327516. [DOI] [PubMed] [Google Scholar]
- 9.Chan P T, Ohmori H, Tomizawa J, Lebowitz J. Nucleotide sequence and gene organization of ColE1 DNA. J Biol Chem. 1985;260:8925–8935. [PubMed] [Google Scholar]
- 10.Cramer W A, Heymann J B, Schendel S L, Deriy B N, Cohen F S, Elkins P A, Stauffacher C V. Structure-function of channel-forming colicins. Annu Rev Biophys Biomol Struct. 1995;24:611–641. doi: 10.1146/annurev.bb.24.060195.003143. [DOI] [PubMed] [Google Scholar]
- 11.Dersch P, Fsihi H, Bremer E. Low-copy-number T7 vectors for selective gene expression and efficient protein overproduction in Escherichia coli. FEMS Microbiol Lett. 1994;123:19–26. doi: 10.1111/j.1574-6968.1994.tb07195.x. [DOI] [PubMed] [Google Scholar]
- 12.Farrant W N, Tomlinson A J H. Some studies on the epidemiology of Sonne dysentery. Changes in colicine type and antibiotic resistance between 1956 and 1965. J Hyg. 1966;64:287–303. doi: 10.1017/s0022172400040572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fredericq P. Actions antibiotiques réciproques chez les Enterobacteriaceae. Rev Belge Pathol Exp Med Exp. 1948;19(Suppl. 4):1–17. [Google Scholar]
- 14.Fredericq P. A note on the classifications of colicins. Zentbl Bakteriol Hyg A. 1965;196:140–142. [Google Scholar]
- 15.Gross P, Braun V. Colicin M is inactivated during import by its immunity protein. Mol Gen Genet. 1996;251:388–396. doi: 10.1007/BF02172531. [DOI] [PubMed] [Google Scholar]
- 16.Horák V. Two new colicins from Shigella. Folia Microbiol. 1990;35:469–470. [Google Scholar]
- 17.Horák V, Sobotková J. Sensitivity to colicin Js, one of important characteristics of Escherichia coli strains belonging to enteroinvasive serovars. Zentbl Bakteriol Hyg A. 1988;269:156–159. doi: 10.1016/s0176-6724(88)80091-9. [DOI] [PubMed] [Google Scholar]
- 18.Hu P, Elliott J, McCready P, Skowronski E, Garnes J, Kobayashi A, Brubaker R R, Garcia E. Structural organization of virulence-associated plasmids of Yersinia pestis. J Bacteriol. 1998;180:5192–5202. doi: 10.1128/jb.180.19.5192-5202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleanthous C, James R, Hemmings A M, Moore G R. Protein antibiotics and their inhibitors. Biochem Soc Trans. 1999;27:63–67. doi: 10.1042/bst0270063. [DOI] [PubMed] [Google Scholar]
- 20.Luirink J, Mol O, Oudega B. Functioning of the pColDF13 encoded BRP. In: James R, Lazdunski C, Pattus F, editors. Bacteriocins, microcins and lantibiotics. Berlin, Germany: Springer-Verlag GmbH; 1992. pp. 307–316. [Google Scholar]
- 21.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Ogawa T, Tomita K, Ueda T, Watanabe K, Uozumi T, Masaki H. A cytotoxic ribonuclease targeting specific transfer RNA anticodons. Science. 1999;283:2097–2100. doi: 10.1126/science.283.5410.2097. [DOI] [PubMed] [Google Scholar]
- 23.Pilsl H, Braun V. Novel colicin 10: assignment of four domains to TonB- and TolC-dependent uptake via the Tsx receptor and to pore formation. Mol Microbiol. 1995;16:57–67. doi: 10.1111/j.1365-2958.1995.tb02391.x. [DOI] [PubMed] [Google Scholar]
- 24.Pilsl H, Braun V. Evidence that the immunity protein inactivates colicin 5 immediately prior to formation of the transmembrane channel. J Bacteriol. 1995;177:6966–6972. doi: 10.1128/jb.177.23.6966-6972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pilsl H, Braun V. Strong function-related homology between the pore-forming colicins K and 5. J Bacteriol. 1995;177:6973–6977. doi: 10.1128/jb.177.23.6973-6977.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pilsl H, Killmann H, Hantke K, Braun V. Periplasmic location of the pesticin immunity protein suggests inactivation of pesticin in the periplasm. J Bacteriol. 1996;178:2431–2435. doi: 10.1128/jb.178.8.2431-2435.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pilsl H, Šmajs D, Braun V. Characterization of colicin S4 and its receptor, OmpW, a minor protein of the Escherichia coli outer membrane. J Bacteriol. 1999;181:3578–3581. doi: 10.1128/jb.181.11.3578-3581.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pugsley A P. The ins and outs of colicins. Part I. Production and translocation across membranes. Microbiol Sci. 1984;1:168–175. [PubMed] [Google Scholar]
- 29.Pugsley A P. The ins and outs of colicins. Part II. Lethal action, immunity and ecological implications. Microbiol Sci. 1984;1:203–205. [PubMed] [Google Scholar]
- 30.Riley M A. Molecular mechanisms of bacteriocin evolution. Annu Rev Genet. 1998;32:255–278. doi: 10.1146/annurev.genet.32.1.255. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Šmajs D, Pilsl H, Braun V. Colicin U, a novel colicin produced by Shigella boydii. J Bacteriol. 1997;179:4919–4928. doi: 10.1128/jb.179.15.4919-4928.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Šmarda J, Petrželová J, Vyskot B. Colicin Js of Shigella sonnei: classification of type colicin “7.”. Zentbl Bakteriol Hyg A. 1987;263:530–540. doi: 10.1016/s0176-6724(87)80196-7. [DOI] [PubMed] [Google Scholar]
- 34.Šmarda J, Šmajs D. Colicins—exocellular lethal proteins of Escherichia coli. Folia Microbiol. 1998;43:563–582. doi: 10.1007/BF02816372. [DOI] [PubMed] [Google Scholar]
- 35.Song H Y, Cramer W A. Membrane topography of ColE1 gene products: the immunity protein. J Bacteriol. 1991;173:2935–2943. doi: 10.1128/jb.173.9.2935-2943.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Studier F W, Moffat B A. Use of bacteriophage T7-RNA-polymerase to direct selective high level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 37.van der Wal F J, Luirink J, Oudega B. Bacteriocin release proteins: mode of action, structure, and biotechnological application. FEMS Microbiol Rev. 1995;17:381–399. doi: 10.1111/j.1574-6976.1995.tb00221.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y L, Cramer W A. Intramembrane helix-helix interaction as the basis of inhibition of the colicin E1 ion channel by its immunity protein. J Biol Chem. 1993;268:1–9. [PubMed] [Google Scholar]