Abstract
Mayaro virus (MAYV) is a mosquito-transmitted alphavirus that is being recognized with increasing frequency in South America. As part of on-going surveillance of a school cohort in Haiti, we identified MAYV infections in 5 children across a 7-month time span, at two different school campuses. All had a history of fever, and three had headaches; none complained of arthralgias. On analysis of whole genome sequence data, three strains were genotype D, and two were genotype L; phylogenetic and molecular clock analysis was consistent with at least 3 independent introductions of the virus into Haiti, with ongoing transmission of a common genotype D strain in a single school. Our data highlight the clear potential for spread of the virus in the northern Caribbean and North America.
Mayaro virus (MAYV; genus Alphavirus, family Togaviridae) is a single-stranded positive RNA virus. First isolated in Trinidad in 1954 (Anderson et al., 1957), it is one of the viruses that comprise the Semliki Forest virus complex (Acosta-Ampudia et al., 2018). Members of this group are reported to cause illness characterized by fever, arthralgias, and skin manifestations. While illness is generally mild, sequelae have been reported, including severe persistent arthralgias (Acosta-Ampudia et al., 2018; Halsey et al., 2013). The primary vectors include mosquitoes within the genus Haemagogus, although Aedes spp may also be competent vectors (Acosta-Ampudia et al., 2018). While originally isolated in Trinidad (Anderson et al., 1957), the majority of MAYV cases have been reported from the Amazon basin region, including Brazil, Peru, and Ecuador (Acosta-Ampudia et al., 2018; Halsey et al., 2013; Blohm et al., 2019; Mavian et al., 2017; Pan American Health Organization/World Health Organization, 2019; Auguste et al., 2015).
Methods
Since May of 2014, our research group has maintained surveillance for viral causes of undifferentiated febrile illness in a cohort of approximately 1,250 school children in the four campuses of the Christianville Foundation school system in the Gressier/Leogane region of Haiti (Figure 1) (Ball et al., 2018; Lednicky et al., 2016). A central school clinic provides free medical care to all children attending these schools. All children who present to the clinic with a history of fever, without clear localizing symptoms, are invited to participate in the study, which includes collection of a blood sample. Written informed consent is obtained from parents of all participants. The study protocol has been approved by the University of Florida Institutional Review Board (IRB) and the Haitian National IRB.
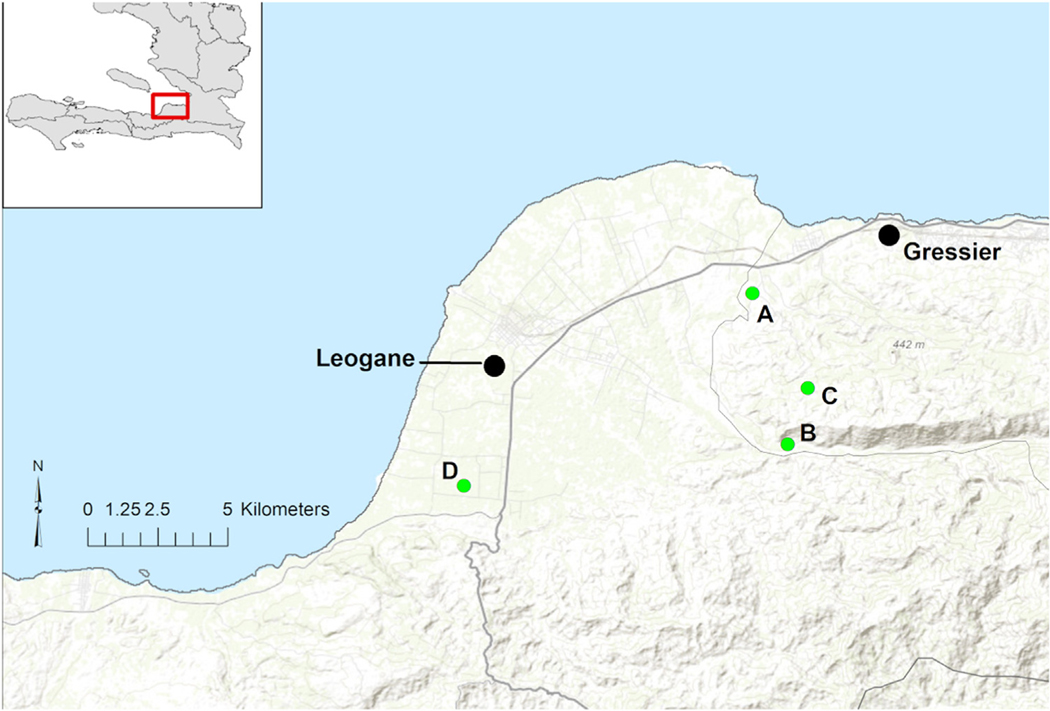
Figure 1.
Location of school campuses. Location of the school campuses within the Christianville Foundation School System in the Gressier/Leogane region of Haiti (labeled as A, B, C, and D). National Route 2, the main highway through the area is identified with a bolded line and smaller roads appear lighter (gray). Base map data from ESRI Online.
Methods for viral identification and phylogenetic analysis have been previously described (Blohm et al., 2019; Mavian et al., 2017; Ball et al., 2018; Lednicky et al., 2016). In brief: plasma was separated and screened by rtRT-PCR for dengue, chikungunya, and zika viruses, with virus isolation then attempted in cell culture. In the cases reported here, an alphavirus was suspected based on characteristic cytopathic effects in vero-cell culture (Blohm et al., 2019; Lednicky et al., 2016). This was followed by screening with a duplex RT-PCR test for alphavirus and flavivirus vRNAs, which yielded an ~434 base pair PCR amplicon consistent with alphavirus vRNA. Subsequent sequencing of cDNA obtained from the duplex RT-PCR test provided further documentation that the virus present was MAYV. The MAYV genomes were Sanger-sequenced using a gene-walking approach with overlapping primers (Blohm et al., 2019; Ball et al., 2018; Lednicky et al., 2016), and the virus sequence deposited in GenBank (Accession #KY985361; KX496990; MK837006; MK837007; MN138459). A multiple sequence alignment using the non-recombinant portion of the genome was obtained as previously described (Mavian et al., 2017). Phylogenetic signal in the multiple sequence alignment was quantified by likelihood mapping analysis (Strimmer and von Haeseler, 1997), using the program TREE-PUZZLE (Schmidt et al., 2002). Temporal signal for the reliable calibration of a molecular clock was, then, assessed by tip-to-root divergence vs. time plots, using the program TempEst (Rambaut et al., 2016) and a maximum likelihood tree, with HKY + G estimated genetic distances, inferred with IQ-TREE (Nguyen et al., 2015). Time-scaled phylogeny was inferred using the Bayesian phylogenetic framework with BEAST v.1.8.4 (Drummond et al., 2012; Drummond and Rambaut, 2007). The best-fit model determined by marginal likelihood (Xie et al., 2011) was HKY substitution model, empirical base frequencies, gamma distribution of site-specific rate heterogeneity, strict molecular clock and constant size demographic prior.
Results and comment
MAYV was identified in plasma samples from five patients. Two of the five cases have been previously reported (Ball et al., 2018; Lednicky et al., 2016), albeit as single cases, with the existence of two other strains noted in another paper (Blohm et al., 2019); the current manuscript combines data for all strains, and provides a unified phylogenetic and molecular clock analysis. In one case, the patient was found to be infected with both MAYV and DENV1 (Lednicky et al., 2016); a second case involved a dual infection with MAYV and CHIKV (Ball et al., 2018). Cases occurred between May, 2014, and February, 2015. Four case patients were male. Age ranged from 4 to 7 years. While all patients reported a history of fever, only three were febrile at the time of their clinic visit, with one child having a temperature of 39 degrees C. Three complained of headache; none noted arthralgias. All recovered without sequelae. While arthralgias have been linked with MAYV infection (Acosta-Ampudia et al., 2018; Halsey et al., 2013; Auguste et al., 2015), lack of arthralgias clearly does not exclude the diagnosis.
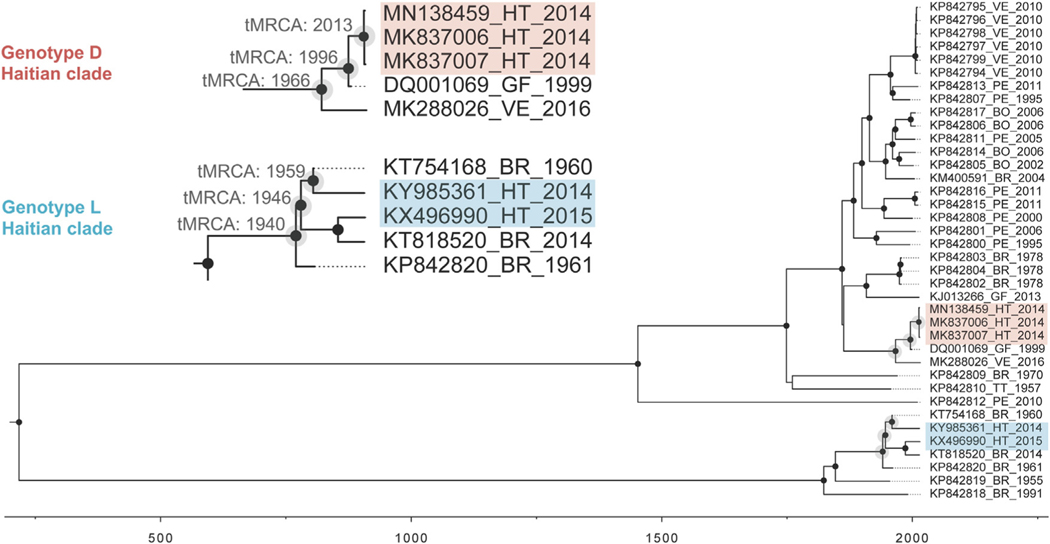
The aligned, non-recombinant full genome sequences displayed negligible phylogenetic noise (<10% in the likelihood mapping analysis) and robust temporal signal (r2> 0.8 in the TempEst analysis), assuring the reliability of both phylogeny inference and molecular clock calibration. On phylogenetic analysis (Figure 2), three strains were in MAYV genotype D (“widely Dispersed”). All three case patients came from a single school (School C, Figure 1), with illnesses occurring in June, October, and November of 2014. Molecular clock analysis suggested that these closely related Haitian strains diverged from a common ancestor in 2013 (95% high posterior density confidence interval 2009–2014). This Haitian strain group, in turn, diverged in 1996 (95%CI 1993–1999) from a strain group that includes a 1999 strain from French Guiana, with the entire strain group diverging in 1966 (95%CI 1952–1979) from a group that includes a recently identified Venezuelan strain (Blohm et al., 2019). The remaining two Haitian strains (both from case patients in School A, Figure 1) were in Genotype L (prior to this, “Limited” to sites in northcentral Brazil). However, these strains are more closely related to other Brazilian strains than they are to each other, with divergence of the respective clades dating back to 1946 (95%CI 1936–1955). These findings are consistent with at least three independent introductions of MAYV into Haiti, with the serogroup D introduction leading to recurrent infections in a single school across a five-month time period.
Figure 2.
Temporal reconstruction of the history of MAYV. Maximum Clade Credibility time-scaled phylogenetic maximum clade credibility tree inferred using strict clock and constant demographic priors implemented in BEAST v1.8.4. Circles at nodes represent branches supported by posterior probability >0.90.
As recently noted in an Epidemiological Alert from the Pan American Health Organization, there have been increasing reports of infections due to MAYV in Peru and, most recently, Ecuador (Pan American Health Organization/World Health Organization, 2019). Given that MAYV is reported to cause severe symptoms, including persistent arthralgias, in a subset of patients, the increasing number of reported cases and movement of the virus from its “home base” in the Amazon basin to other parts of South American and into the northern Caribbean is of clear concern (Acosta-Ampudia et al., 2018; Blohm et al., 2019; Mavian et al., 2017). Our findings, indicating circulation of at least 3 distinct clades of the virus within a student population in a relatively limited geographic area, substantiate this concern. In this study, we identified MAYV because we were using research diagnostic approaches, including tissue culture, to monitor what appears to be ongoing spread of the virus. In keeping with recent recommendations from PAHO (Pan American Health Organization/World Health Organization, 2019), there is a clear need for more easily accessible diagnostic tools for MAYV infection; coupled with this, there is also a need for ongoing surveillance for the virus, to monitor incidence and spread of MAYV into new areas.
Funding source
The study was funded, in part, by a grant from N.I.H/NIAID to JGM (NIH R01 AI126357–01S1). Funders had no role in study design, data collection or analysis, or preparation of the manuscript.
Footnotes
Ethical statement
Written informed consent was obtained from parents of all participants. The study protocol was approved by the University of Florida Institutional Review Board (IRB) and the Haitian National IRB.
Conflict of interest
No authors declare any conflict of interest relevant to this publication.
References
- Acosta-Ampudia Y, Monsalve DM, Rodríguez Y, Pacheco Y, Anaya JM, Ramírez-Santana C. Mayaro: an emerging viral threat?. Emerg Microbes Infect 2018;7 (1):163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CR, Downs WG, Wattley GH, Ahin NW, Reese AA. Mayaro virus: a new human disease agent. II. Isolation from blood of patients in Trinidad, B.W.I. Am J Trop Med Hyg 1957;6:1012–6. [DOI] [PubMed] [Google Scholar]
- Auguste AJ, Liria J, Forrester NL, Giambalvo D, Moncada M, Long KC, et al. Evolutionary and ecological characterization of Mayaro virus strains isolated during an outbreak, Venezuela, 2010. Emerg Infect Dis 2015;21:1742–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball JD, Elbadry MA, Telisma T, White SK, Chavannes S, Anilis MG, et al. Clinical and epidemiological patterns of Chikungunya virus infection and coincident arboviral disease in a school cohort in Haiti, 2014/2015. Clin Infect Dis 2018; (August), doi: 10.1093/cid/ciy582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blohm GM, Márquez-Colmenarez MC, Lednicky JA, Bonny TS, Mavian C, DelgadoNoguera L, et al. Isolation of Mayaro virus from a Venezuelan patient with febrile illness, arthralgias, and rash; further evidence of regional strain circulation and possible long-term endemicity. Am J Trop Med Hyg 2019; [Accepted for publication]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 2007;7(November):214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 2012;29:1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsey ES, Siles C, Guevara C, Vilcarromero S, Johnston EJ, Ramal C. Mayaro virus infection, Amazon Basin region, Peru, 2010–2013. Emerg Infect Dis 2013;19:1839–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky JA, Beau De Rochars VM, El Badry MA, Loeb JC, Telisma T, Chavannes S, et al. Mayaro virus in a child with acute febrile illness, Haiti, 2015. Emerg Infect Dis 2016;22:2000–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavian C, Rife BD, Dollar JJ, Cella E, Ciccozzi M, Prosperi MC, et al. Emergence of recombinant Mayaro virus strains from the Amazon basin. Sci Rep 2017;7:8718, doi: 10.1038/s41598-017-07152-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol 2015;32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan American Health Organization/World Health Organization. Epidemiological alert: Mayaro fever. 1 May 2019. Washington, D.C: PAHO/WHO; 2019. https://www.paho.org/hq/index.php?option=com_docman&view=download&category_slug=mayaro-fever-2323&alias=48374-1-may-2019-mayaro-fever-epidemiological-alert&Itemid=270&lang=en. [Google Scholar]
- Rambaut A, Lam TT, Carvalho LM, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst. Virus Evol 2016;2:vew007, doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 2002;18:502–4. [DOI] [PubMed] [Google Scholar]
- Strimmer K, von Haeseler A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc Natl Acad Sci USA 1997;94:6815–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Lewis PO, Fan Y, Kuo L, Chen MH. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst Biol 2011;60 (March (2)):150–60. [DOI] [PMC free article] [PubMed] [Google Scholar]