Abstract
During the essential processes of DNA replication and transcription, RNA-DNA hybrid intermediates are formed that pose significant risks to genome integrity when left unresolved. To manage RNA-DNA hybrids, all cells rely on RNase H family enzymes that specifically cleave the RNA portion of the many different types of hybrids that form in vivo. Recent experimental advances have provided new insight into how RNA-DNA hybrids form and the consequences to genome integrity that ensue when persistent hybrids remain unresolved. Here we review the types of RNA-DNA hybrids, including R-loops, RNA primers, and ribonucleotide misincorporations, that form during DNA replication and transcription and discuss how each type of hybrid can contribute to genome instability in bacteria. Further, we discuss how bacterial RNase HI, HII, HIII and bacterial FEN enzymes contribute to genome maintenance through the resolution of hybrids.
Keywords: RNA-DNA hybrids, RNase HI, RNase HII, RNase HIII, mutation rate, genome instability
Introduction
In all cells, faithful inheritance of genetic information relies on RNA-DNA hybrid formation during the essential processes of DNA replication and transcription (24; 42; 46; 91; 111). Essential RNA-DNA hybrids occur predominately as RNA primers, which prime DNA polymerase activity during replication, and as R-loops, which form when the nascent RNA molecule base pairs with the complementary DNA strand during transcription (46; 91; 111). Evidence suggests that R-loops can also be converted into RNA polymers with a covalent RNA-DNA junction known as “R-tracks”(43). RNA-DNA hybrids also form when replicative DNA polymerases make single sugar errors (65; 117), incorporating an rNMP in place of the cognate dNMP, or when lower fidelity DNA polymerases incorporate ribopatches, short polymers of 4 or more rNMPs, during synthesis (75). Despite the necessity and pervasiveness of RNA-DNA hybrid formation, persistent hybrids compromise genome integrity, such that all organisms must be able to resolve the variety of hybrids to ensure proper replication and inheritance of genetic information.
To manage the myriad of hybrids that occur in vivo, all cells contain several different types of enzymes that help mitigate formation of persistent hybrids (15; 30; 91; 108; 111) (Table 1). In all three domains of life, as well as numerous viruses and mobile genetic elements, ribonuclease H (hybrid) enzymes specifically incise RNA within RNA-DNA hybrids contributing to their removal (35). RNase H enzymes are traditionally classified into types 1 and 2, where type 1 refers to RNase HI and type 2 includes RNase HII and RNase HIII (72). The presence of RNase HI is pervasive across biology, where these enzymes resolve hybrids containing four or more consecutive ribonucleotides, such as RNA primers in Okazaki fragments, ribopatches, R-tracks and R-loops (99) (see Table 2). The RNase HI domain is also prevalent in the retroviral integrase superfamily (66), which includes diverse enzymes important for many nucleic acid processes, including replication of HIV-1 (3; 44; 67).
Table 1.
Bacterial proteins involved in RNA-DNA hybrid formation or resolution.
| Protein | Hybrid type | Function |
|---|---|---|
| RNases | ||
| RNase HI | R-loops, Okazaki fragments, ribopatches | Hybrid resolution (30). |
| RNase HII | Single ribonucleotide errors, ribopatches, Okazaki fragments | Hybrid resolution (30). |
| RNase HIII | R-loops, Okazaki fragments, ribopatches | Hybrid resolution (30). |
| RNase E | R-loop | Reduces R-loop formation (104; 105). |
| Exonucleases | ||
| Exo I | R-loops | Hybrid removal in rnhA− strains (41). |
| FEN | Okazaki fragments, possible ribopatches and single ribonucleotide errors | Hybrid removal (85). |
| Recombination | ||
| RecA | R-loops | Hybrid formation-promotes base pairing between mRNA and the template strand (29). |
| RecBCD (Exo V) | R-loops | Hybrid removal in rnhA− strains (41). |
| Topoisomerases | ||
| DNA Gyrase | R-loops | Promotes R-loop formation through inducing negative supercoiling (16). |
| Topoisomerase I | R-loops | Reduces formation of R-loops (49). |
| Topoisomerase III | R-loops | Reduces formation of R-loops (5). |
| Topoisomerase IV | R-loops | Overexpression reduces R-loop formation (5). |
| Transcription and translation | ||
| RNA polymerase | R-loops | R-loop formation during transcription and backtracked states (18). |
| DksA | R-loops | Conflict resolution during starvation (101) and contribution to cSDR initiation (64). |
| GreA | R-loops | Reduces RNAP backtracking and R-loop formation (18). |
| GreB | R-loops | Reduces RNAP backtracking and R-loop formation (18). |
| Mfd | R-loop | Increases R-loop formation and mutagenesis (81). |
| NusG | R-loops | Limits R-loop formation by regulating Rho-dependent termination (50). |
| Rho | R-loops | Limits R-loop formation (83). |
| Ribosomes | R-loops | R-loop resolution by reducing RNAP backtracking (18). |
| DNA replication | ||
| DNA polymerase I (A-family) | Single ribonucleotide errors, R-loops, and Okazaki fragments | Insertion of ribonucleotide errors during replication, replication from R-loops during cSDR, removal of RNA from Okazaki fragments (15; 91). |
| C-family DNA polymerases | R-loops and single ribonucleotide errors | Insertion of ribonucleotide errors (117) and removal of R-loops by strand displacement synthesis (119). |
| Y-family DNA polymerases | Single ribonucleotide errors and ribopatches | Insertion of ribonucleotide errors (75; 108). |
| Primase (DnaG) | Okazaki fragments | RNA primers during Okazaki fragment synthesis (87). |
| SSB | R-loops | Targets RNase HI for R-loop resolution (114). |
| Helicases | ||
| DnaB | R-loops | R-loop resolution by unwinding hybrids (94). |
| DinG | R-loops | R-loops resolution by unwinding hybrids (4). |
| PcrA (Gram-positive bacteria) | R-loops | R-loops resolution by unwinding hybrids (102). |
| RecG | R-loops | R-loops resolution by unwinding hybrids (29). |
| Rep | R-loops | R-loops resolution by unwinding hybrids (4). |
| UvrD | R-loops | R-loops resolution by unwinding hybrids (4). |
Table 2.
Bacterial RNase H enzymes and substrates cleaved
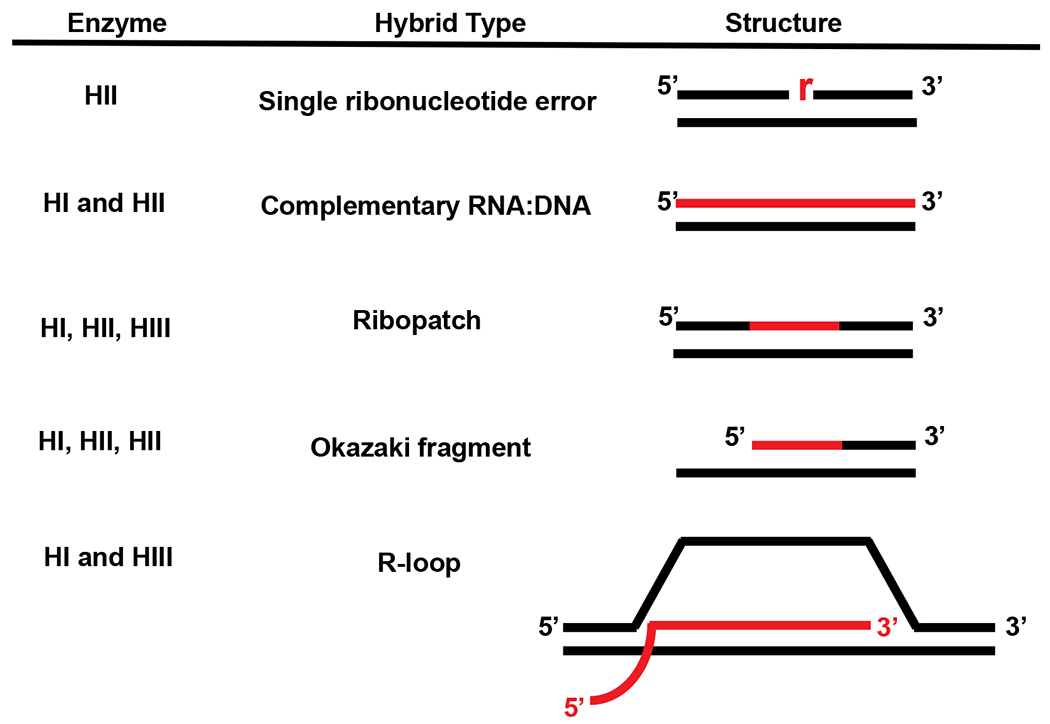
|
Adapted from Reference (91).
The red lines indicate RNA and the black lines denote DNA. It should be noted that RNase HI and RNase HIII have been described to cleave single ribonucleotides in vitro under specific conditions (53; 100). RNase HII is expected to recognize and cleave at single rNMPs in most organisms under physiological conditions.
The presence of RNase HII is also widespread in biology, where these enzymes specifically recognize the RNA-DNA covalent junction, cleaving on the 5' side of the ribonucleotide (73; 74; 99). Such activity allows for RNase HII to recognize single ribonucleotide errors in otherwise double-stranded DNA, RNA primers in Okazaki fragments, ribopatches, but not R-loops, which lack the RNA-DNA covalent junction necessary for activity (73; 74; 90). Most bacteria that contain RNase HII also have either RNase HI or RNase HIII (37; 72; 99), while many archaeal organisms have only RNase HII (37). RNase HIII is typically classified as a type 2 enzyme based on amino acid and structural conservation with RNase HII, however it hydrolyzes the same types of hybrids as RNase HI and does not have activity on single ribonucleotide errors (71; 85), with very rare exceptions (53). RNase HIII is not widespread and is only present in a subset of bacterial and archaeal organisms that predominately lack a chromosomally encoded RNase HI (37; 99).
The structures and enzymology of RNases H enzymes have been studied extensively for decades in diverse systems including viruses, bacteria, and mammalian cells and are extensively reviewed elsewhere (3; 7; 30; 34). Over the last fifteen years, many studies have contributed to our understanding of the contribution of bacterial RNase H enzymes to genome integrity. Recent technical advancements have allowed for genome-wide mapping of RNA-DNA hybrids (8; 45; 83), replication fork fate (45; 69), and measurement of sugar misincorporation errors by replicative DNA polymerases (90; 117), stimulating new advancements in our understanding of how RNA-DNA hybrids impact genome integrity. The goal of this Review is to discuss how different RNA-DNA hybrids affect genome stability, the enzymes involved in the resolution of different types of RNA-DNA hybrids, and the consequences that occur to cells when RNase H activity is compromised in bacteria. For readers interested in how RNA-DNA hybrids are formed or resolved in eukaryotic cells, please see the following excellent reviews (79; 82; 111; 112; 120).
Genome instability caused by replication-transcription conflicts
R-loops form when nascently transcribed RNA base pairs with the complementary DNA strand, forming a stable RNA-DNA hybrid. Due in part to their stability, “collisions” can occur between R-loops and the replisome (45; 69) that cause replication fork stalling (13; 23; 45; 69; 109) and genomic rearrangements in both E. coli and mammalian cells (26). In E. coli, R-loops are resolved by RNase HI, encoded by the rnhA gene (33), and loss of RNase HI activity is associated with several phenotypes, including constitutive SOS induction (41), which could be a result of increased replication-transcription conflicts in vivo. Bacterial genomes are typically organized such that highly expressed essential genes are encoded on the leading strand, so that progression of RNA polymerase (RNAP) and the replisome are oriented co-directionally (86; 92) (Figure 1). Conversely, in genes encoded on the lagging strand, RNAP and the replisome encounter each other in the head-on direction (86; 92). Experiments in different organisms using various loci have shown that when highly expressed genes are oriented head-on to DNA replication encounters between RNAP and the replisome result in replication fork movement that is slowed, stalled or arrested, (23; 45; 62; 69; 97; 109) which can lead to double-strand break formation (12). Replication fork progression can also be slowed or stopped in the presence of R-loops (45; 69; 109). Therefore, abundant information demonstrates that conflicts between the replisome and R-loops (or RNAP) have severe consequences to genome integrity. What remains unclear is the types of mutations that result from replication-transcription conflicts that occur in the head-on orientation [for recent perspectives (46; 92)].
Figure 1.
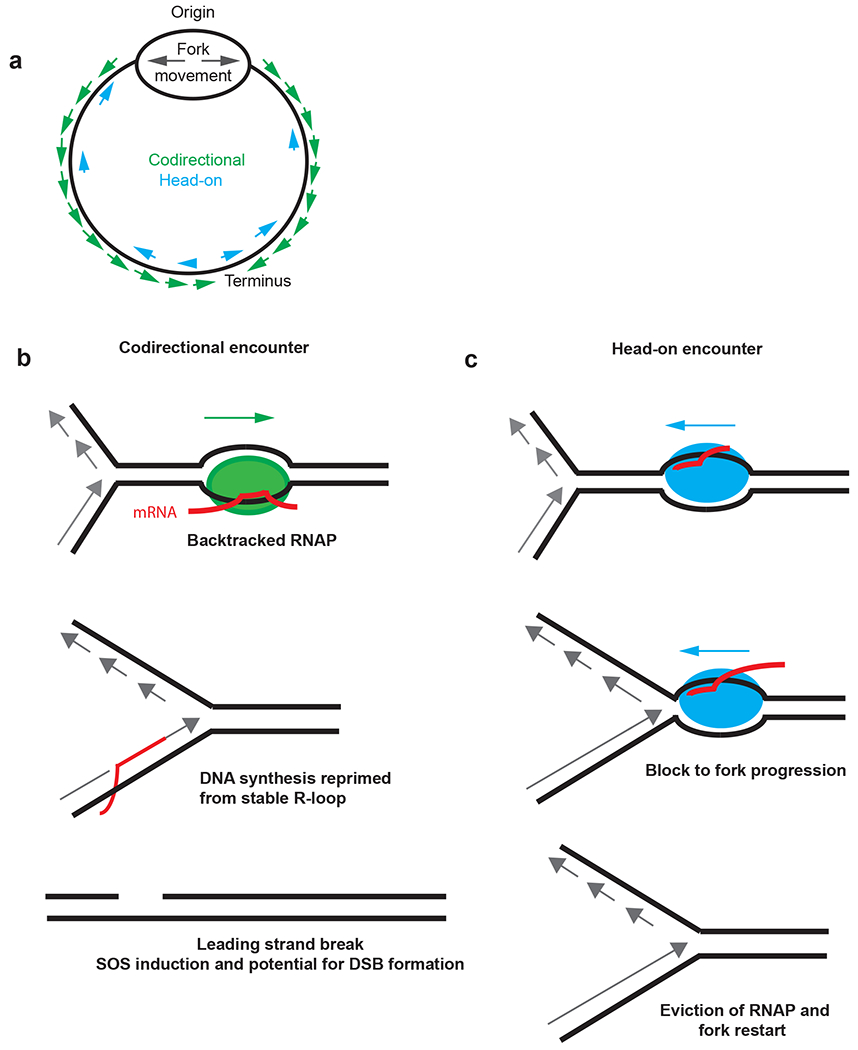
Genome instability caused by codirectional and head-on encounters. (a) Schematic of a circular chromosome with genes oriented codirectional or head-on to replication fork movement (b) Codirectional encounter between backtracked RNAP and a replication fork. DNA synthesis by the replisome can be reprimed by mRNA or primase-initiated RNA synthesis following the encounter with RNAP. The replisome and RNAP collision creates a gap on the leading strand behind the repriming event (28, 81). Following excision of the repriming RNA, a gap remains in the leading strand that could be converted to a double-strand break during the next round of DNA replication when the gap is encountered by the replisome. (c) RNAP moving head-on toward a replication fork. Head-on encounters between RNAP and the replisome can block fork progression. Although several outcomes can occur, the fork can be restored by evicting RNAP and enabling primosome-dependent fork restart (47,61a). Abbreviation: RNAP, RNA polymerase. Part of this figure is adapted from Reference 18.
Head-on conflicts with R-loops
One model suggests that head-on encounters result in a modest increase in mutagenesis, which is supported by evidence from engineered reporters, using selection for reversion of auxotrophic markers to measure mutagenesis (61; 77). In these experiments, the modest increase in mutation rate (~2.5 to 3-fold) is dependent on head-on orientation and highly induced gene expression with reporters that select for specific changes in the coding sequence (45; 61; 77). Other evidence using reporters that sample a much wider range of mutations suggested that deleterious mutations, including insertions and deletions (indels), are the main mutations arising from head-on conflict (88; 118). Base-pair substitutions (BPSs) were enriched in the promoter region, with no enrichment for BPSs observed in coding sequences demonstrating signature mutations for head-on replication-transcription conflicts are detrimental to gene function (88; 118).
In contrast to reporter-based experiments, use of mutation accumulation lines, which examine genes in their native contexts, suggests that any modest increase in mutagenesis observed in gene coding sequences oriented head-on is dependent upon sequence context as opposed to gene orientation or gene expression (21; 48; 52; 56; 89; 98). A recent study interrogating ~30,000 BPSs in E. coli found no increase in point mutations in genes oriented in the head-on direction relative to co-directional genes (21). Collectively, studies by multiple groups in different organisms find that the major driver of mutagenesis is explained by neighboring sequence context which causes DNA polymerases to become more error-prone (19; 20; 22; 48; 52; 56; 58; 89; 92; 98), with no detectable contribution from replication-transcription conflicts in head-on oriented genes (21; 56).
Co-directional conflicts with R-loops
There is also evidence that formation of R-loops in co-directionally oriented genes contributes to genome instability (18; 60). While in vitro work examining collisions shows that R-loops do not present a block to fork progression in the head-on orientation, R-loops can affect fork progression in the co-directional orientation (6). Prior work in B. subtilis has shown that highly expressed co-directionally oriented genes require primosome-dependent reloading of the replisome, which also suggests that co-directional encounters are deleterious to fork progression (60). Using an engineered plasmid-based system in E. coli, the contribution of gene orientation to double-strand break (DSB) formation was tested (18). In this system, RNAP stalls and backtracks (or backslides), which causes a more stable association of the R-loop due to increased base pairing between the newly transcribed RNA and the template DNA (18). Replisome encounters with a backtracked RNAP result in DSBs at the site of the backtracked RNAP (18). Elongation factors GreA, GreB and actively translating ribosomes were found to reduce DSB formation at backtracked RNAPs by acting as anti-backtracking factors (18; 68). DSBs were detected for co-directional conflicts between backtracked RNAP and the replisome (Figure 1), but not for the plasmid reporter in the head-on orientation (18). Importantly, expression of RNase HI reduced DSB formation, supporting the conclusion that breaks in co-directional genes are caused by R-loop formation associated with backtracked RNAP. Therefore, the simplest model is that when RNAP backtracks, it creates a longer R-loop that forms a more stable association with DNA (18). When the replisome encounters the stable R-loop in the co-directional orientation the replisome “skips” over RNAP with synthesis continuing through either re-priming by DnaG (primase) or when the mRNA serves to re-prime synthesis ahead of RNAP (6; 80). This leaves gaps in the leading strand (6) with DSB formation occurring when successive rounds of replication encounter the gap (Figure 1). These studies provide mechanistic insight into how DNA breaks occur when the replisome encounters a backtracked RNAP in the co-directional orientation (18).
DksA and R-loop removal
In addition to RNAP associated proteins GreA and GreB that affect backtracked RNAP, another RNAP associated protein DksA contributes to replisome bypass of encountered R-loops. Under the stress condition of amino acid starvation the transcription elongation factor DksA is critical for the replisome to complete genome replication (101). Starvation induces a rapid arrest of replication fork progression in E. coli in the absence of dksA, leading to recruitment of RecA and activation of the SOS response (101). Since the effect of DksA on maintaining forks during starvation depends on active transcription, this work suggests that DksA helps the replisome navigate conflicts with sites of active transcription by reducing pausing of RNAP during elongation (101). Therefore, in addition to RNase H enzymes, transcription associated factors including GreA, GreB (18) and DksA contribute to maintaining genome integrity (101). A common theme is that RNAP pausing, or backtracking, can increase the barrier to replisome progression (18; 101). One possibility is that the barrier is enhanced by stable R-loop formation or a stalled and stable RNAP associated with an R-loop (6). Some recent evidence indicates that the transcription coupled repair factor Mfd (59) actually helps stabilize R-loop formation leading to an increase in mutation rate (81). Therefore, factors associated with RNAP contribute significantly to R-loop mediated genome instability, with RNase H enzymes helping to mitigate the effects.
R-loop formation at GC-rich DNA repeats contributes to genome instability
R-loops also contribute to genome instability through the contraction of DNA repeats. The base pairing of GC-rich DNA repeats in the template strand with nascently transcribed RNA creates stable R-loops due to the stability of rGC base pairs (51). In E. coli, R-loop formation is stabilized in GC-rich CTG•CAG repeat tracks. To test whether R-loops at these regions affected genome stability, researchers generated two IPTG inducible plasmids with 98 CTG•CAG repeats on either the leading or the lagging strand and transformed wild-type and RNase HI mutant cells with the plasmids (51). This work found that while the repeats contracted in RNase HI mutant cells for both orientations, repeats on the lagging strand had a much more dramatic contraction, with only 20% of the full length repeat tracts remaining by the final growth period. This study also showed that the contraction of the repeat tracts was dependent on transcription from the IPTG-inducible promoter, suggesting that repeat instability in E. coli is transcription-dependent and R-loop mediated.
Replication initiation from R-loops: Constitutive stable DNA replication
Replication in bacteria is normally initiated at the single origin of replication, oriC, by DnaA (31; 32). While each round of initiation at oriC requires newly synthesized proteins, mutations in the E. coli rnhA gene, encoding RNase HI, allow for replication initiation in the absence of new protein synthesis (27). Further, initiation in RNase HI-defective cells can occur in the absence of DnaA and oriC, demonstrating a non-canonical mechanism of replication initiation. In RNase HI-deficient cells initiation occurs from R-loops that accumulate at sites termed oriK, with oriK sites enriched in the replication terminus (14; 57). This oriC-independent replication initiation is referred to as constitutive stable DNA replication (cSDR) because it occurs in the absence of new protein synthesis (40; 57). cSDR initiates from R-loops with the assistance of a suite of enzymes, including RecA, the primosome, DNA polymerase I and DNA polymerase III holoenzyme (40; 57) (Figure 2a).
Figure 2.
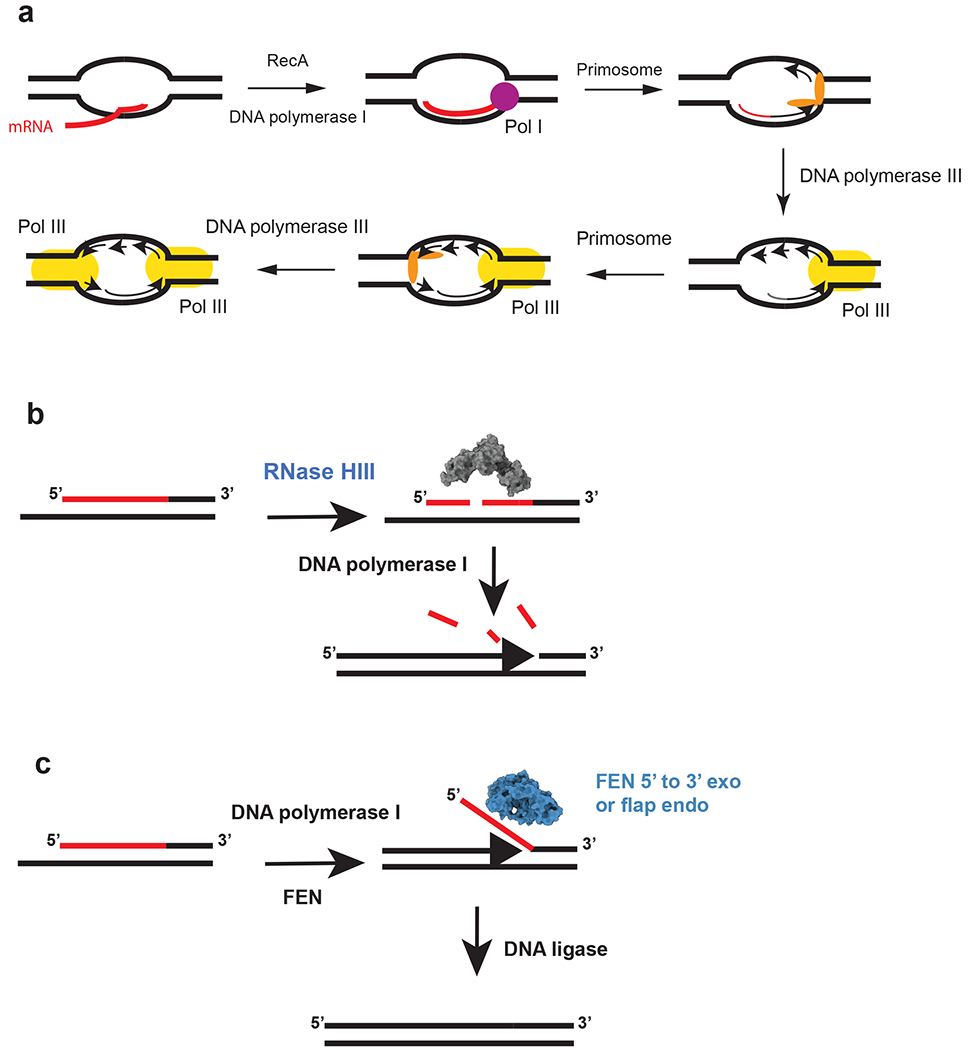
Initiation of replication from R-loops and a model for Okazaki fragment processing in RNase HIII–containing bacteria. The red lines indicate RNA, and the black lines denote DNA. (a) Model for cSDR. The nascent transcript is paired with DNA by RecA in negative supercoiled DNA behind RNAP. The transcript primes synthesis by DNA polymerase I followed by primosome assembly and loading of DNA polymerase III. The second fork is activated following primosome assembly and loading of DNA polymerase III. This figure is based on models from the following References 15 and 41 (b) RNase HIII incises the RNA portion of the Okazaki fragment. The incisions allow for DNA polymerase I to efficiently remove the RNA during DNA synthesis from the 3′-OH on the adjacent fragment (86). (c) Bacteria that have a stand-alone FEN can cleave the flap resulting from DNA polymerase I strand displacement synthesis. For bacteria that contain RNase HIII and FEN, it is expected that both function during Okazaki fragment maturation (1, 86). The model is based on the following reference (86). The space filling models for RNase HIII and FEN were generated using the B. subtilis protein sequences modeled with I-TASSER (116). Abbreviation: FEN, flap endonuclease/5′-to-3′ exonuclease.
The process of cSDR priming of replication from R-loops can impact genome stability in a couple of ways (14). First, replisome recruitment to oriK sites results in a portion of the replication forks proceeding in a different direction compared to that of oriC-initiated replication forks. As most highly expressed essential genes are encoded co-directionally with oriC-dependent replisome progression, the oriK-dependent switch in replication fork direction increases replisome-RNAP conflicts. As discussed above, replication-transcription conflicts can result in increased genome instability.
Replication initiated at R-loops has also been shown to contribute to genome instability through stress-induced mutagenesis (SIM) in non-replicating E. coli cells (113). The formation of double-stranded breaks (DSBs), discussed previously in the context of co-directional collisions between RNAP and the replisome, is a major contributor to genome instability (18). While most mechanisms for the generation of DSBs are hypothesized to occur in actively replicating cells, work in starved E. coli cells found that R-loops contribute to the formation of DSBs by priming replication fork progression that subsequently collapses at independently generated ssDNA knicks, ultimately resulting in formation of a double-stranded end (DSE) (113). In attempt to repair the DSEs, induction of SIM in starved cells results in genomic alterations through two mechanisms; activation of error-prone DNA polymerases, which results in point mutations, and gene amplification, which can result in genome rearrangements (113). This mechanism may not be limited to non-replicating cells and could underly a portion of the SOS induction that is observed during replication from R-loops in proliferating bacterial cells during cSDR when nicks or gaps in the template strand are encountered.
For more information on the genetic requirements for cSDR please see the following excellent reviews (15; 40).
SSB protein cooperates with RNase HI to remove R-loops
As the replisome separates double-stranded DNA during replication, ssDNA is generated and bound by single-stranded DNA binding protein (SSB) to stabilize and prevent reannealing to dsDNA (93). In addition to functioning during normal DNA replication, SSB has been shown to serve as a “hub” for the recruitment of factors critical for DNA repair (10; 47), including RNase HI. Recent work also shows that SSB stimulates RNase HI activity in vitro (78). The interaction between SSB and RNase HI is formed through docking of the C-terminal end of SSB into a binding pocket on RNase HI suggesting that SSB is critical for recruitment of RNase HI to substrates in vivo (78; 114).
RNase HI fused to YPet shows an SSB-mediated localization to replication forks when compared to β-clamp as a proxy for replisome position (114). Interaction between RNase HI and SSB is mediated by K60 in RNase HI (78). Mutation of K60E prevented interaction between RNase HI and SSB, abolishing recruitment of RNase HI into foci while still maintaining normal levels of RNase HI nuclease activity (78). Wolack and co-workers found that localization of RNase HI to the replisome is independent of its function in Okazaki fragment processing or removal of R-loops that could allow for oriC and DnaA-independent DNA replication initiation through cSDR (114). However, it was shown that the rnhAK60E had a growth phenotype when combined with disruption of the DNA helicase Rep (114). Rep is known to move ahead of the replication fork and aid in replication through highly transcribed regions (4; 27). The increase in SOS induction of rnhAK60E rep- cells, along with synergistic effects of RNAP variants known to increase or decrease R-loop formation (39), shows that RNase HI interaction with SSB is important for removing R-loops encountered by the replisome during DNA replication (114). It will be important to learn if the SSB interaction with RNase HI contributes to reducing orientation specific conflicts or if there are specific loci in E. coli that are more prone to R-loop formation that impede replisome progression.
RNase HI functions in R-loop removal and Okazaki fragment maturation
As described above, RNase HI contributes to genome stability through the removal of R-loops reducing replication-transcription conflicts and by preventing persistent R-loops from priming replication outside of oriC (15; 46). RNase HI is also involved in RNA primer removal during Okazaki fragment maturation, in addition to pol I and, in a subset of bacteria, a flap endonuclease/5' to 3' exonuclease (FEN) (discussed in greater detail below) (1; 38). The prevailing model is that RNase HI cleaves the RNA portion of the Okazaki fragment followed by either strand displacement synthesis by pol I or concomitant degradation of the RNA moiety using its flap processing N-terminal domain (54; 70). E. coli cells that are pol I deficient contain short fragments with RNA at the 5' end (70). The appearance of Okazaki fragments with 5' linked RNA increases in cells that are compromised for both RNase HI and pol I activity, with 10 to 30 times more intact RNA primer (36). Although RNase HI has an important role in Okazaki fragment maturation, the strong phenotypes of rnhA mutants stem from R-loop removal (114), making it challenging to separate the role of RNase HI in RNA removal on the lagging strand from R-loop removal elsewhere on the chromosome.
Another limitation to the study of the effects of RNase HI (or RNase HIII) deficiencies on hybrid removal lies in the current approaches to detect genome-wide RNA-DNA hybrids. Recent evidence shows that the antibody S9.6, which is typically used for precipitating RNA-DNA hybrids in a the genome-wide RNA-DNA hybrid detection approach, DNA/RNA immunoprecipitation followed by sequencing (DRIP-seq), has a strong affinity for double-stranded RNA (95). When interpreting DRIP-seq, the procedure should include data showing that the enrichment is RNase HI-sensitive to ensure that the signal is coming from an RNA-DNA hybrid along with other technical considerations (8). Thus, an important area moving forward will be to demonstrate where RNA-DNA hybrids form in vivo through development of new approaches or by including procedures that require pulldowns are specific for RNA-DNA hybrids. Future experiments in bacteria will need to determine where RNA-DNA hybrids form and discriminate between RNA-DNA hybrids that are from true R-loops and RNA primers used during lagging strand replication to understand the importance of RNase H enzymes to removal of each hybrid in vivo.
Contributions of bacterial FEN to hybrid resolution and genome integrity
During lagging strand synthesis the RNA used to prime Okazaki fragment synthesis must be replaced with DNA (42). In bacteria, this is accomplished through the combined actions of pol I and RNase HI (or RNase HIII) (54; 70; 85). Pol I catalyzes strand-displacement synthesis, displacing the RNA primer and creating a bifurcated structure with a single-stranded 5' overhang, referred to as a flap (54). For complete maturation of the Okazaki fragment, this flap must be removed, and the remaining nick sealed by DNA ligase (54). The resolution of flapped structures is primarily carried out by a class of structure-specific proteins called FENs (1; 103), with involvement from RNase HI or HIII in cleaving RNA primers (25). Substrate preference similarities between FENs and RNase Hs has caused confusion, as the FEN in T4 bacteriophage was initially identified as an RNase H (85).
In bacteria, FENs can be found as part of the N-terminal domain of pol I (1) or as a stand-alone protein in addition to pol I (1; 103). While they are found in all domains of life (55), FENs do not share a well-conserved sequence (1). Rather, this family has essential metal-coordinating residues and a conserved structure (103). For Mycobacterium FenA three Mn2+ ions are coordinated that are important for catalysis (103). The primary substrate associated with FEN activity is the 5' flap described earlier, however, FENs have also been shown to interact with nucleic acid structures that are single-stranded, double-stranded, have a 5' overhang, a 3' overhang, or are nicked (1; 85). Recently, in vitro assays using bacterial FENs have indicated a preference for RNA-DNA hybrids over DNA-only substrates (28; 85) suggesting that bacterial FENs are important for hybrid resolution (Figure 2). The mechanism by which FENs cleave their substrate is less clear (115), but recent studies, including crystallography of the T5 bacteriophage FEN bound to substrate, support a mechanism by which FEN binds to the substrate and the single-stranded flap is subsequently threaded through the enzyme prior to cleavage (2). Importantly, at least one FEN is required for bacterial organisms to be viable, highlighting the importance of such proteins to hybrid removal during replication (25). Therefore, active, stand-alone FENs clearly contribute to genome integrity and some show preference for RNA-DNA hybrid removal. The role of these independent FENs and how they work in conjunction with pol I remains unclear. It is also unclear if bacterial FENs are primarily responsible for Okazaki fragment maturation (85), contribute to repair, (76) or both.
Contributions of RNase HIII to genome integrity
As mentioned earlier, RNase HIII is usually classified as a type 2 RNase H and encoded by the rnhC gene (37; 99), but it is sometimes placed in its own group as a type 3 enzyme (30). Some RNase HIII enzymes have been shown to cleave at single ribonucleotides when complexed with Mn2+, although under most circumstances this activity occurs under conditions that are not physiological (53; 84). Therefore, in most instances, RNase HIII behaves like RNase HI, cleaving RNA primers during Okazaki fragment maturation, ribopatches, and R-loops. RNase HIII is also characterized as having a TATA-binding protein-like domain which aids in substrate recognition (9; 63; 71). Unlike RNase HI and RNase HII, RNase HIII is rather limited in its phylogenetic distribution. A comparative study found that RNase HIII was present in 17% of bacterial genomes and 4% of archaeal genomes analyzed (37). For comparison, RNase HII was present in 94% of bacteria and 100% of archaeal genomes analyzed (37). Bioinformatic studies have suggested that RNase HIII and HI are mutually exclusive because active enzymes from each of these classes were not found to coincide (37; 72). Recently, a clear exception to this rule was described in the wild B. subtilis strain NCIB 3610, which encodes an active RNase HI (RnhP) on a large naturally occurring plasmid (69). This study opens the possibility that RNase HI and RNase HIII may be more coincident than previously appreciated if one of the genes is carried on a plasmid or mobile genetic element allowing for loss, transfer, and reacquisition when needed (69).
RNase HIII has received less experimental characterization likely due in part to the more limited distribution of this enzyme in prokaryotic genomes. In B. subtilis, deletion of the rnhC gene results in strong phenotypes including slow growth, sensitivity to a broad range of DNA damage, and growth inhibition on several stressors including hydroxyurea (45; 84; 85). RNase HIII has been suggested to cleave R-loops in vivo (45) due to its activity on complementary RNA-DNA hybrid lacking a covalent RNA-DNA junction (71; 84). Lack of RNase HIII has severe consequences to genome integrity, although the nature of these effects is still unclear. Cells with engineered reporter constructs driven from a strong promoter in the head-on orientation caused strong growth interference in cells lacking RNase HIII, suggesting that RNase HIII is important for the removal of R-loops in the head-on orientation in vivo (45).
Recent work shows that RNase HIII works efficiently with pol I for Okazaki fragment maturation in vitro (85). Since Okazaki fragments have a covalent RNA-DNA junction, RNase HII also cleaves these substrates quite well (85). Therefore, either RNase HII or RNase HIII could contribute to removal of RNA from Okazaki fragments in conjunction with pol I, allowing for functional overlap (85). Using a linear substrate it was shown that RNase HII incision did not stimulate pol I synthesis, whereas RNase HIII incision did stimulate pol I on a model Okazaki fragment (85) (Figure 3). This work suggests that the internal cleavage of the RNA primer aids in pol I removal and resynthesis providing the most efficient pathway for maturation of the lagging strand in vivo (85).
Figure 3. Model for ribonucleotide excision repair.
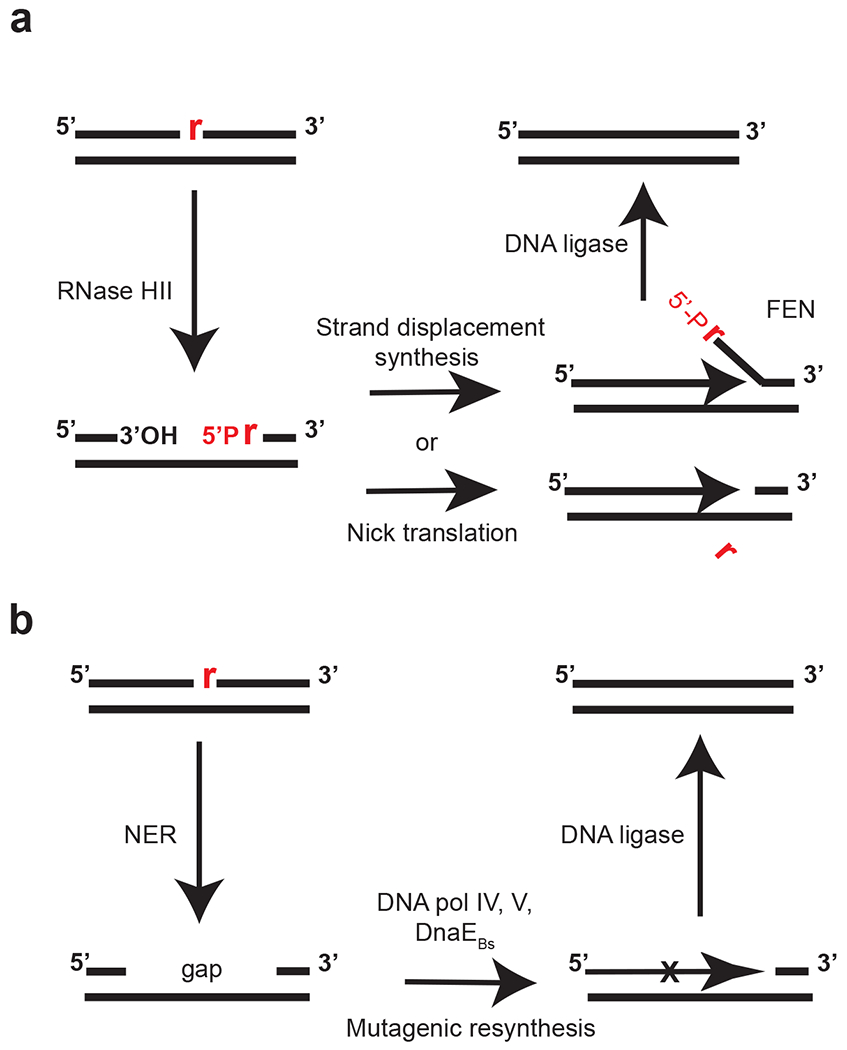
(a) Removal and correction of a single rNMP (red) nested in genomic DNA (black lines). RNase HII incises 5′ to the rNMP. DNA polymerase I synthesizes from the nick, with concomitant 5′-to-3′ exonuclease activity removing the ribonucleotide-containing strand in a process known as nick translation, or by strand displacement synthesis (91, 107). If strand displacement synthesis occurs, DNA polymerase I or a stand-alone FEN cleaves the flap, releasing the fragment (1). (b) In the absence of RNase HII, nucleotide excision repair can recognize single rNMP errors. Nucleotide excision repair action leaves behind a gap allowing mutagenic resynthesis by DNA polymerase IV and DNA polymerase V in Escherichia coli and essential DNA polymerase DnaE in Bacillus subtilis (91, 107, 108). Abbreviation: FEN, flap endonuclease/5′-to-3′ exonuclease.
As mentioned above, the “wild” ancestral strain of B. subtilis NCIB 3610 was shown to contain RNase HIII (rnhC) on the chromosome along with a newly discovered RNase HI gene (rnhP) on a naturally occurring 84 Kbp plasmid (69). This work showed that RnhP is an RNase HI based on substrate preference and sequence similarity to E. coli RNase HI (69). Cells lacking rnhC and rnhP grew poorly and were induced for the SOS response with forks stalling at a 38 Kbp head-on operon (ppsA-E) located near the chromosomal terminus (69). Deletion of the ppsA-E operon in the double rnhC, rnhP deletion alleviated the SOS induction associated with the double RNase H mutant (69). The presence of RnhP in B. subtilis 3610 suggests that this wild strain might either require an additional protein to aid in removal of R-loops or a protein that can cleave R-loops under physiological conditions that do not favor activity from chromosomal encoded RNase HIII (69). Given results with engineered reporters and a single naturally occurring locus in a “wild” strain of B. subtilis, current evidence indicates that an important contribution of RNase HIII is to aid in replication fork progression through long and highly expressed head-on oriented genes. More experiments will be important to determine if there is a true orientation bias.
RNase HII and ribonucleotide excision repair in bacterial genomes
The contribution of R-loops to genome stability has been a major focus of this Review due to the number of ways in which R-loops impact genome integrity in bacterial cells. Another important type of RNA-DNA hybrid comes in the form of single ribonucleotide misincorporation events, which are not resolved by RNase HI or HIII enzymes in vivo (90). During genome replication, replicative DNA polymerases misincorporate ribonucleotides in place of deoxyribonucleotides leading to sugar errors in the chromosome (90; 91; 117). The rate of ribonucleotide errors is a function of ribonucleotide concentrations in the cell (117). For example, rATP out numbers dATP 600 to 1, causing rAMP misincorporations to far exceed that of any other ribonucleotide for E. coli pol III in vitro (117). Once incorporated, the only difference between sugar errors (AMP, CMP and GMP) and their cognate dNMP is an oxygen atom on the 2' carbon of the ribose ring (42). In vitro and in vivo estimates suggest that single ribonucleotide errors account for nearly 2000 errors per round of replication, making ribonucleotide errors the most frequent nucleotide in need of correction (11; 117). Sugar errors are a challenge for the DNA polymerase to detect because the base pairing is correct making it less efficient for replicative enzymes to recognize and proofread sugar errors during synthesis (110). Therefore, once a sugar error is formed, the ribonucleotide will need to be recognized and removed by a process referred to as ribonucleotide excision repair (RER).
In bacteria and eukaryotes RER is initiated by RNase HII (RNase H2 in eukaryotes) (90; 96; 107). RNase HII recognizes the error and cleaves the ribonucleotide 5' to the RNA-DNA junction generating a nick and allowing for entry of a DNA polymerase (Figure 3A). Evidence in B. subtilis and E. coli indicates that pol I is primarily responsible for the resynthesis step in the canonical pathway (90; 106; 107). It is unclear at this point if pol I degrades or cleaves the displaced strand or if a separately encoded FEN homolog could also cleave and release of the ribonucleotide containing strand (Figure 3A). The nick would then be sealed by DNA ligase to complete repair (90; 106; 107).
Recent studies have focused on the alternative pathways of RER that occur in the absence of RNase HII (90; 106; 107). For B. subtilis and E. coli the main phenotype for RNase HII deficient cells is a mild increase in mutation rate of about 2.5-fold (90; 117). In B. subtilis use of mutation accumulation lines showed that the increase in mutation rate was almost entirely explained by a specific GC->AT transition that occurred on the lagging strand in a specific sequence context (90; 117). The general model is that in the absence of RNase HII the nucleotide excision repair (NER) system recognizes ribonucleotide errors and removes an 8-10 nucleotide stretch leaving behind a gap (106) (Figure 1B). The resulting gap, as opposed to the nick generated by RNase HII, allows for access by an error-prone DNA polymerase for resynthesis causing an increase in mutagenesis (107). Reconstitution of the reaction using B. subtilis proteins found that essential DNA polymerase DnaE was responsible for mutagenic resynthesis (90). Even though pol I from B. subtilis lacks proofreading activity (17), pol I resynthesis was rather accurate across the sequence context found to undergo the transition in vivo (90). For E. coli, the resynthesis step can become mutagenic when Y-family DNA polymerases gain access to the resulting gap left behind following NER (107). Therefore, the canonical RNase HII-mediated RER pathway in bacteria shows high fidelity. When RNase HII is compromised, or if an overabundance of rNMP errors occurs, then NER can function as a secondary pathway, with the potential to result in mutagenesis (90; 106; 107; 117).
Concluding Statement
The last 10-15 years has seen a growing interest in understanding the contribution of RNase H enzymes to genome stability in bacteria. New experimental approaches for studying RNA-DNA hybrid formation certainly suggest that R-loops and single ribonucleotide incorporations are far more prevalent than previously appreciated and, if left unresolved, can have significant impacts on genome integrity. As discussed, further work will be necessary to resolve the competing models for how gene orientation and expression levels contribute to replication-transcription conflicts resulting in R-loop-dependent mutagenesis. Another important avenue for future study will be to distinguish the contribution of RNase HI, HII and HIII to Okazaki fragment maturation on a genome-wide scale. Current evidence suggests that Okazaki fragments are matured by RNase HI and RNase HIII, however RNase HII/HIII deletions have been reported as either synthetically lethal or causing the accumulation of compensatory mutations (25; 85). Further, deletion of RNase HI and HII in E. coli results in R-loop conversion to “R-tracks” requiring recombinational repair (43). These results suggest that RNase HII is compensating, at least in part, for loss of RNase HI or RNase HIII during Okazaki fragment maturation and if excessive R-loops are converted to ribonucleotide polymers with an RNA-DNA junction (43). Finally, it will be important to continue to establish genome-wide approaches that improve detection of the different RNA-DNA hybrids that form in vivo. Such improvements will allow for studies to differentiate between hybrids participating in R-loop formation and primers required for Okazaki fragment synthesis in bacteria.
Acknowledgements
The authors would like to thank the many labs who have studied RNase H enzymes over the decades since their originally discovery. Due to space limitations the authors apologize for areas and citations that were not included. We thank Jeremy Schroeder for helpful comments on the manuscript and we also thank Justin Randall and Jeremy Schroeder for initiating our studies on RNase H enzymes. This review was funded by National Institutes of Health grant GM131772 to LAS. EKM was funded in part by a Donald R. Shepherd Scholarship and TMN was supported by a pre-doctoral fellowship from the National Science Foundation (#DEG 1256260). The authors have no conflict of interest to declare.
References
- 1.Allen LM, Hodskinson MR, Sayers JR. 2009. Active site substitutions delineate distinct classes of eubacterial flap endonuclease. Biochem J 418:285–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AlMalki FA, Flemming CS, Zhang J, Feng M, Sedelnikova SE, et al. 2016. Direct observation of DNA threading in flap endonuclease complexes. Nat Struct Mol Biol 23:640–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beilhartz GL, Gotte M. 2010. HIV-1 Ribonuclease H: Structure, Catalytic Mechanism and Inhibitors. Viruses 2:900–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boubakri H, de Septenville AL, Viguera E, Michel B. 2010. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J 29:145–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brochu J, Vlachos-Breton E, Sutherland S, Martel M, Drolet M. 2018. Topoisomerases I and III inhibit R-loop formation to prevent unregulated replication in the chromosomal Ter region of Escherichia coli. PLoS Genet 14:e1007668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruning JG, Marians KJ. 2020. Replisome bypass of transcription complexes and R-loops. Nucleic Acids Res 48:10353–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerritelli SM, Crouch RJ. 2009. Ribonuclease H: the enzymes in eukaryotes. FEBS J 276:1494–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chedin F, Hartono SR, Sanz LA, Vanoosthuyse V. 2021. Best practices for the visualization, mapping, and manipulation of R-loops. EMBO J 40:e106394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chon H, Matsumura H, Koga Y, Takano K, Kanaya S. 2006. Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus. J Mol Biol 356:165–78 [DOI] [PubMed] [Google Scholar]
- 10.Costes A, Lecointe F, McGovern S, Quevillon-Cheruel S, Polard P. 2010. The C-terminal domain of the bacterial SSB protein acts as a DNA maintenance hub at active chromosome replication forks. PLoS Genet 6:e1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cronan GE, Kouzminova EA, Kuzminov A. 2019. Near-continuously synthesized leading strands in Escherichia coli are broken by ribonucleotide excision. Proc Natl Acad Sci U S A 116:1251–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Septenville AL, Duigou S, Boubakri H, Michel B. 2012. Replication fork reversal after replication-transcription collision. PLoS Genet 8:e1002622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deshpande AM, Newlon CS. 1996. DNA replication fork pause sites dependent on transcription. Science 272:1030–3 [DOI] [PubMed] [Google Scholar]
- 14.Dimude JU, Stockum A, Midgley-Smith SL, Upton AL, Foster HA, et al. 2015. The Consequences of Replicating in the Wrong Orientation: Bacterial Chromosome Duplication without an Active Replication Origin. mBio 6:e01294–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drolet M, Brochu J. 2019. R-loop-dependent replication and genomic instability in bacteria. DNA Repair (Amst) 84:102693. [DOI] [PubMed] [Google Scholar]
- 16.Drolet M, Phoenix P, Menzel R, Masse E, Liu LF, Crouch RJ. 1995. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci U S A 92:3526–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duigou S, Ehrlich SD, Noirot P, Noirot-Gros MF. 2005. DNA polymerase I acts in translesion synthesis mediated by the Y-polymerases in Bacillus subtilis. Mol Microbiol 57:678–90 [DOI] [PubMed] [Google Scholar]
- 18.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. 2011. Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146:533–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fijalkowska IJ, Jonczyk P, Tkaczyk MM, Bialoskorska M, Schaaper RM. 1998. Unequal fidelity of leading strand and lagging strand DNA replication on the Escherichia coli chromosome. Proc Natl Acad Sci U S A 95:10020–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster PL, Lee H, Popodi E, Townes JP, Tang H. 2015. Determinants of spontaneous mutation in the bacterium Escherichia coli as revealed by whole-genome sequencing. Proc Natl Acad Sci U S A 112:E5990–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster PL, Niccum BA, Lee H. 2021. DNA replication-transcription conflicts do not significantly contribute to spontaneous mutations due to replication errors in Escherichia coli. mBio 12:e02503–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster PL, Niccum BA, Popodi E, Townes JP, Lee H, et al. 2018. Determinants of Base-Pair Substitution Patterns Revealed by Whole-Genome Sequencing of DNA Mismatch Repair Defective Escherichia coli. Genetics 209:1029–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.French S. 1992. Consequences of replication fork movement through transcription units in vivo. Science 258:1362–5 [DOI] [PubMed] [Google Scholar]
- 24.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. 2006. DNA Repair and Mutagenesis: Second Edition. Washington, DC: American Society for Microbiology [Google Scholar]
- 25.Fukushima S, Itaya M, Kato H, Ogasawara N, Yoshikawa H. 2007. Reassessment of the in vivo functions of DNA polymerase I and RNase H in bacterial cell growth. J Bacteriol 189:8575–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gan W, Guan Z, Liu J, Gui T, Shen K, et al. 2011. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev 25:2041–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guy CP, Atkinson J, Gupta MK, Mahdi AA, Gwynn EJ, et al. 2009. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol Cell 36:654–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernandez-Tamayo R, Oviedo-Bocanegra LM, Fritz G, Graumann PL. 2019. Symmetric activity of DNA polymerases at and recruitment of exonuclease ExoR and of PolA to the Bacillus subtilis replication forks. Nucleic Acids Res 47:8521–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong X, Cadwell GW, Kogoma T. 1995. Escherichia coli RecG and RecA proteins in R-loop formation. EMBO J 14:2385–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyjek M, Figiel M, Nowotny M. 2019. RNases H: Structure and mechanism. DNA Repair (Amst) 84:102672. [DOI] [PubMed] [Google Scholar]
- 31.Kaguni JM. 2006. DnaA: controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol 60:351–75 [DOI] [PubMed] [Google Scholar]
- 32.Kaguni JM. 2011. Replication initiation at the Escherichia coli chromosomal origin. Curr Opin Chem Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanaya S, Crouch RJ. 1983. DNA sequence of the gene coding for Escherichia coli ribonuclease H. J Biol Chem 258:1276–81 [PubMed] [Google Scholar]
- 34.Kanaya S, Ikehara M. 1995. Functions and structures of ribonuclease H enzymes. Subcell Biochem 24:377–422 [DOI] [PubMed] [Google Scholar]
- 35.Keller W, Crouch R. 1972. Degradation of DNA RNA hybrids by ribonuclease H and DNA polymerases of cellular and viral origin. Proc Natl Acad Sci U S A 69:3360–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitani T, Yoda K, Ogawa T, Okazaki T. 1985. Evidence that discontinuous DNA replication in Escherichia coli is primed by approximately 10 to 12 residues of RNA starting with a purine. J Mol Biol 184:45–52 [DOI] [PubMed] [Google Scholar]
- 37.Kochiwa H, Tomita M, Kanai A. 2007. Evolution of ribonuclease H genes in prokaryotes to avoid inheritance of redundant genes. BMC Evol Biol 7:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kogoma T. 1986. RNase H-defective mutants of Escherichia coli. J Bacteriol 166:361–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kogoma T. 1994. Escherichia coli RNA polymerase mutants that enhance or diminish the SOS response constitutively expressed in the absence of RNase HI activity. J Bacteriol 176:1521–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kogoma T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev 61:212–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kogoma T, Hong X, Cadwell GW, Barnard KG, Asai T. 1993. Requirement of homologous recombination functions for viability of the Escherichia coli cell that lacks RNase HI and exonuclease V activities. Biochimie 75:89–99 [DOI] [PubMed] [Google Scholar]
- 42.Kornberg A, Baker TA. 1992. DNA Replication. New York: W. H. Freeman and Company [Google Scholar]
- 43.Kouzminova EA, Kadyrov FF, Kuzminov A. 2017. RNase HII Saves rnhA Mutant Escherichia coli from R-Loop-Associated Chromosomal Fragmentation. J Mol Biol 429:2873–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai L, Yokota H, Hung LW, Kim R, Kim SH. 2000. Crystal structure of archaeal RNase HII: a homologue of human major RNase H. Structure 8:897–904 [DOI] [PubMed] [Google Scholar]
- 45.Lang KS, Hall AN, Merrikh CN, Ragheb M, Tabakh H, et al. 2017. Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis. Cell 170:787–99 e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lang KS, Merrikh H. 2018. The Clash of Macromolecular Titans: Replication-Transcription Conflicts in Bacteria. Annu Rev Microbiol 72:71–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lecointe F, Serena C, Velten M, Costes A, McGovern S, et al. 2007. Anticipating chromosomal replication fork arrest: SSB targets repair DNA helicases to active forks. Embo J 26:4239–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee H, Popodi E, Tang H, Foster PL. 2012. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci U S A 109:E2774–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leela JK, Raghunathan N, Gowrishankar J. 2021. Topoisomerase I Essentiality, DnaA-Independent Chromosomal Replication, and Transcription-Replication Conflict in Escherichia coli. J Bacteriol 203:e0019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leela JK, Syeda AH, Anupama K, Gowrishankar J. 2013. Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc Natl Acad Sci U S A 110:258–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Y, Dent SY, Wilson JH, Wells RD, Napierala M. 2010. R loops stimulate genetic instability of CTG.CAG repeats. Proc Natl Acad Sci U S A 107:692–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Long H, Sung W, Miller SF, Ackerman MS, Doak TG, Lynch M. 2014. Mutation rate, spectrum, topology, and context-dependency in the DNA mismatch repair-deficient Pseudomonas fluorescens ATCC948. Genome Biol Evol 7:262–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu Z, Liang R, Liu X, Hou J, Liu J. 2012. RNase HIII from Chlamydophila pneumoniae can efficiently cleave double-stranded DNA carrying a chimeric ribonucleotide in the presence of manganese. Mol Microbiol 83:1080–93 [DOI] [PubMed] [Google Scholar]
- 54.Lundquist RC, Olivera BM. 1982. Transient generation of displaced single-stranded DNA during nick translation. Cell 31:53–60 [DOI] [PubMed] [Google Scholar]
- 55.Lyamichev V, Brow MA, Dahlberg JE. 1993. Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science 260:778–83 [DOI] [PubMed] [Google Scholar]
- 56.Lynch M, Ackerman MS, Gout JF, Long H, Sung W, et al. 2016. Genetic drift, selection and the evolution of the mutation rate. Nat Rev Genet 17:704–14 [DOI] [PubMed] [Google Scholar]
- 57.Maduike NZ, Tehranchi AK, Wang JD, Kreuzer KN. 2014. Replication of the Escherichia coli chromosome in RNase HI-deficient cells: multiple initiation regions and fork dynamics. Mol Microbiol 91:39–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maslowska KH, Makiela-Dzbenska K, Mo JY, Fijalkowska IJ, Schaaper RM. 2018. High-accuracy lagging-strand DNA replication mediated by DNA polymerase dissociation. Proc Natl Acad Sci U S A 115:4212–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mellon I, Hanawalt PC. 1989. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature (London) 342:95–8 [DOI] [PubMed] [Google Scholar]
- 60.Merrikh H, Machon C, Grainger WH, Grossman AD, Soultanas P. 2010. Co-directional replication-transcription conflicts lead to replication restart. Nature 470:554–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Million-Weaver S, Samadpour AN, Moreno-Habel DA, Nugent P, Brittnacher MJ, et al. 2015. An underlying mechanism for the increased mutagenesis of lagging-strand genes in Bacillus subtilis. Proc Natl Acad Sci U S A 112:E1096–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mirkin EV, Mirkin SM. 2005. Mechanisms of transcription-replication collisions in bacteria. Mol Cell Biol 25:888–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miyashita S, Tadokoro T, Angkawidjaja C, You DJ, Koga Y, et al. 2011. Identification of the substrate binding site in the N-terminal TBP-like domain of RNase H3. FEBS Lett 585:2313–7 [DOI] [PubMed] [Google Scholar]
- 64.Myka KK, Kusters K, Washburn R, Gottesman ME. 2019. DksA-RNA polymerase interactions support new origin formation and DNA repair in Escherichia coli. Mol Microbiol 111:1382–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, et al. 2010. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proceedings of the National Academy of Sciences of the United States of America 107:4949–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nowotny M. 2009. Retroviral integrase superfamily: the structural perspective. EMBO Rep 10:144–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. 2007. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol Cell 28:264–76 [DOI] [PubMed] [Google Scholar]
- 68.Nudler E. 2009. RNA polymerase active center: the molecular engine of transcription. Annu Rev Biochem 78:335–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nye TM, McLean EK, Burrage AM, Dennison DD, Kearns DB, Simmons LA. 2021. RnhP is a plasmid-borne RNase HI that contributes to genome maintenance in the ancestral strain Bacillus subtilis NCIB 3610. Mol Microbiol 115:99–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ogawa T, Okazaki T. 1984. Function of RNase H in DNA replication revealed by RNase H defective mutants of Escherichia coli. Mol Gen Genet 193:231–7 [DOI] [PubMed] [Google Scholar]
- 71.Ohtani N, Haruki M, Morikawa M, Crouch RJ, Itaya M, Kanaya S. 1999. Identification of the genes encoding Mn2+-dependent RNase HII and Mg2+-dependent RNase HIII from Bacillus subtilis: classification of RNases H into three families. Biochemistry 38:605–18 [DOI] [PubMed] [Google Scholar]
- 72.Ohtani N, Haruki M, Morikawa M, Kanaya S. 1999. Molecular diversities of RNases H. J Biosci Bioeng 88:12–9 [DOI] [PubMed] [Google Scholar]
- 73.Ohtani N, Tomita M, Itaya M. 2008. Junction ribonuclease activity specified in RNases HII/2. FEBS J 275:5444–55 [DOI] [PubMed] [Google Scholar]
- 74.Ohtani N, Tomita M, Itaya M. 2008. Junction ribonuclease: a ribonuclease HII orthologue from Thermus thermophilus HB8 prefers the RNA-DNA junction to the RNA/DNA heteroduplex. Biochem J 412:517–26 [DOI] [PubMed] [Google Scholar]
- 75.Ordonez H, Uson ML, Shuman S. 2014. Characterization of three mycobacterial DinB (DNA polymerase IV) paralogs highlights DinB2 as naturally adept at ribonucleotide incorporation. Nucleic Acids Res 42:11056–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Patlan AG, Corona SU, Ayala-Garcia VM, Pedraza-Reyes M. 2018. Non-canonical processing of DNA photodimers with Bacillus subtilis UV-endonuclease YwjD, 5'-->3' exonuclease YpcP and low-fidelity DNA polymerases YqjH and YqjW. DNA Repair (Amst) 70:1–9 [DOI] [PubMed] [Google Scholar]
- 77.Paul S, Million-Weaver S, Chattopadhyay S, Sokurenko E, Merrikh H. 2013. Accelerated gene evolution through replication-transcription conflicts. Nature 495:512–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Petzold C, Marceau AH, Miller KH, Marqusee S, Keck JL. 2015. Interaction with Single-stranded DNA-binding Protein Stimulates Escherichia coli Ribonuclease HI Enzymatic Activity. J Biol Chem 290:14626–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pohl TJ, Zakian VA. 2019. Pif1 family DNA helicases: A helpmate to RNase H? DNA Repair (Amst) 84:102633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pomerantz RT, O’Donnell M. 2008. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature 456:762–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Portman JR, Brouwer GM, Bollins J, Savery NJ, Strick TR. 2021. Cotranscriptional R-loop formation by Mfd involves topological partitioning of DNA. Proc Natl Acad Sci U S A 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Potenski CJ, Epshtein A, Bianco C, Klein HL. 2019. Genome instability consequences of RNase H2 Aicardi-Goutieres syndrome alleles. DNA Repair (Amst) 84:102614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raghunathan N, Kapshikar RM, Leela JK, Mallikarjun J, Bouloc P, Gowrishankar J. 2018. Genome-wide relationship between R-loop formation and antisense transcription in Escherichia coli. Nucleic Acids Res 46:3400–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Randall JR, Hirst WG, Simmons LA. 2017. Substrate specificity for bacterial RNase HII and HIII is influenced by metal availability. J Bacteriol 200:pii: e00401–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Randall JR, Nye TM, Wozniak KJ, Simmons LA. 2019. RNase HIII Is Important for Okazaki Fragment Processing in Bacillus subtilis. J Bacteriol 201: pii: e00686–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rocha EPC. 2004. The replication-related organization of bacterial genomes. Microbiology (Reading) 150:1609–27 [DOI] [PubMed] [Google Scholar]
- 87.Rowen L, Kornberg A. 1978. Primase, the dnaG protein of Escherichia coli. An enzyme which starts DNA chains. J Biol Chem 253:758–64 [PubMed] [Google Scholar]
- 88.Sankar TS, Wastuwidyaningtyas BD, Dong Y, Lewis SA, Wang JD. 2016. The nature of mutations induced by replication-transcription collisions. Nature 535:178–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schroeder JW, Hirst WG, Szewczyk GA, Simmons LA. 2016. The Effect of Local Sequence Context on Mutational Bias of Genes Encoded on the Leading and Lagging Strands. Curr Biol 26:692–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schroeder JW, Randall JR, Hirst WG, O’Donnell ME, Simmons LA. 2017. Mutagenic cost of ribonucleotides in bacterial DNA. Proc Natl Acad Sci U S A 114:11733–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schroeder JW, Randall JR, Matthews LA, Simmons LA. 2015. Ribonucleotides in bacterial DNA. Crit Rev Biochem Mol Biol 50:181–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schroeder JW, Sankar TS, Wang JD, Simmons LA. 2020. The roles of replication-transcription conflict in mutagenesis and evolution of genome organization. PLoS Genet 16:e1008987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. 2008. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol 43:289–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shin JH, Kelman Z. 2006. The replicative helicases of bacteria, archaea, and eukarya can unwind RNA-DNA hybrid substrates. J Biol Chem 281:26914–21 [DOI] [PubMed] [Google Scholar]
- 95.Smolka JA, Sanz LA, Hartono SR, Chedin F. 2021. Recognition of RNA by the S9.6 antibody creates pervasive artifacts when imaging RNA:DNA hybrids. J Cell Biol 220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, et al. 2012. RNase H2-initiated ribonucleotide excision repair. Mol Cell 47:980–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Srivatsan A, Tehranchi A, MacAlpine DM, Wang JD. 2010. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet 6:e1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sung W, Ackerman MS, Gout JF, Miller SF, Williams E, et al. 2015. Asymmetric Context-Dependent Mutation Patterns Revealed through Mutation-Accumulation Experiments. Mol Biol Evol 32:1672–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tadokoro T, Kanaya S. 2009. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J 276:1482–93 [DOI] [PubMed] [Google Scholar]
- 100.Tannous E, Kanaya E, Kanaya S. 2015. Role of RNase H1 in DNA repair: removal of single ribonucleotide misincorporated into DNA in collaboration with RNase H2. Sci Rep 5:9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tehranchi AK, Blankschien MD, Zhang Y, Halliday JA, Srivatsan A, et al. 2010. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell 141:595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Urrutia-Irazabal I, Ault JR, Sobott F, Savery NJ, Dillingham MS. 2021. Analysis of the PcrA-RNA polymerase complex reveals a helicase interaction motif and a role for PcrA/UvrD helicase in the suppression of R-loops. Elife 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Uson ML, Carl A, Goldgur Y, Shuman S. 2018. Crystal structure and mutational analysis of Mycobacterium smegmatis FenA highlight active site amino acids and three metal ions essential for flap endonuclease and 5' exonuclease activities. Nucleic Acids Res 46:4164–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Usongo V, Drolet M. 2014. Roles of type 1A topoisomerases in genome maintenance in Escherichia coli. PLoS Genet 10:e1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Usongo V, Martel M, Balleydier A, Drolet M. 2016. Mutations reducing replication from R-loops suppress the defects of growth, chromosome segregation and DNA supercoiling in cells lacking topoisomerase I and RNase HI activity. DNA Repair (Amst) 40:1–17 [DOI] [PubMed] [Google Scholar]
- 106.Vaisman A, McDonald JP, Huston D, Kuban W, Liu L, et al. 2013. Removal of misincorporated ribonucleotides from prokaryotic genomes: an unexpected role for nucleotide excision repair. PLoS Genet 9:e1003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vaisman A, McDonald JP, Noll S, Huston D, Loeb G, et al. 2014. Investigating the mechanisms of ribonucleotide excision repair in Escherichia coli. Mutat Res Fundam Mol Mech Mutagen 761:21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vaisman A, Woodgate R. 2018. Ribonucleotide discrimination by translesion synthesis DNA polymerases. Crit Rev Biochem Mol Biol 53:382–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang JD, Berkmen MB, Grossman AD. 2007. Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc Natl Acad Sci U S A 104:5608–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Williams JS, Clausen AR, Nick McElhinny SA, Watts BE, Johansson E, Kunkel TA. 2012. Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase varepsilon. DNA repair [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Williams JS, Kunkel TA. 2014. Ribonucleotides in DNA: origins, repair and consequences. DNA Repair (Amst) 19:27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Williams JS, Lujan SA, Zhou ZX, Burkholder AB, Clark AB, et al. 2019. Genome-wide mutagenesis resulting from topoisomerase 1-processing of unrepaired ribonucleotides in DNA. DNA Repair (Amst) 84:102641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wimberly H, Shee C, Thornton PC, Sivaramakrishnan P, Rosenberg SM, Hastings PJ. 2013. R-loops and nicks initiate DNA breakage and genome instability in non-growing Escherichia coli. Nat Commun 4:2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wolak C, Ma HJ, Soubry N, Sandler SJ, Reyes-Lamothe R, Keck JL. 2020. Interaction with single-stranded DNA-binding protein localizes ribonuclease HI to DNA replication forks and facilitates R-loop removal. Mol Microbiol 114:495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu Y, Potapova O, Leschziner AE, Grindley ND, Joyce CM. 2001. Contacts between the 5' nuclease of DNA polymerase I and its DNA substrate. J Biol Chem 276:30167–77 [DOI] [PubMed] [Google Scholar]
- 116.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yao NY, Schroeder JW, Yurieva O, Simmons LA, O’Donnell ME. 2013. Cost of rNTP/dNTP pool imbalance at the replication fork. Proceedings of the National Academy of Sciences of the United States of America 110:12942–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yoshiyama K, Higuchi K, Matsumura H, Maki H. 2001. Directionality of DNA replication fork movement strongly affects the generation of spontaneous mutations in Escherichia coli. J Mol Biol 307:1195–206 [DOI] [PubMed] [Google Scholar]
- 119.Yuan Q, McHenry CS. 2009. Strand displacement by DNA polymerase III occurs through a tau-psi-chi link to single-stranded DNA-binding protein coating the lagging strand template. J Biol Chem 284:31672–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhou ZX, Williams JS, Lujan SA, Kunkel TA. 2021. Ribonucleotide incorporation into DNA during DNA replication and its consequences. Crit Rev Biochem Mol Biol 56:109–24 [DOI] [PMC free article] [PubMed] [Google Scholar]


