Abstract
In Pseudomonas putida DOT-T1E multidrug efflux pumps of the resistance-nodulation-division family make a major contribution to solvent resistance. Two pumps have been identified: TtgABC, expressed constitutively, and TtgDEF, induced by aromatic hydrocarbons. A double mutant lacking both efflux pumps was able to survive a sudden toluene shock if and only if preinduced with small amounts of toluene supplied via the gas phase. In this article we report the identification and characterization in this strain of a third efflux pump, named TtgGHI. The ttgGHI genes form an operon that is expressed constitutively at high levels from a single promoter. In the presence of toluene the operon is expressed at an even higher level from two promoters, the constitutive one and a previously unreported one that is inducible and that partially overlaps the constitutive promoter. By site-directed mutagenesis we constructed a single ttgH mutant which was shown to be unable to survive sudden 0.3% (vol/vol) toluene shocks regardless of the preculture conditions. The mutation was transferred to single and double mutants to construct mutant strains in which two or all three pumps are knocked out. Survival analysis of induced and noninduced cells revealed that the TtgABC and TtgGHI pumps extruded toluene, styrene, m-xylene, ethylbenzene, and propylbenzene, whereas the TtgDEF pump removed only toluene and styrene. The triple mutant was hypersensitive to toluene, as shown by its inability to grow with toluene supplied via the vapor phase.
Organic solvents with a partition coefficient in an octanol-water mixture (log Pow) between 1.5 and 3 are extremely toxic to microorganisms (34). Organic solvents cause irreversible lesions to cell membranes (loss of ions, metabolites, lipids, and proteins, dissipation of the pH gradient and electrical potential, etc.) followed by cell lysis and death (5, 36, 37).
Several Pseudomonas putida strains have been isolated as able to grow in the presence of high concentrations of toxic organic solvents, such as toluene, styrene, and p-xylene (4, 10, 11, 31, 40). Tolerance to organic solvents in these P. putida strains is mainly achieved by a series of efflux pumps that actively remove the organic solvent from the cell membranes (10–14, 19, 25, 32, 33, 36). These efflux pumps, which belong to the resistance-nodulation-division (RND) family, are formed by a translocase made of an inner membrane transporter and a periplasmic fusion protein anchored in the inner membrane plus an outer membrane protein that spans the periplasm and forms a tunnel. It has been proposed that this tunnel is contacted by the cytoplasmic membrane pump, presumably assisted by the inner membrane-periplasmic fusion protein to facilitate the direct removal of drugs across the two membranes and the intervening periplasmic space (17). Evidence for this mode of operation has been obtained with antibiotic efflux pumps in Escherichia coli (17, 46) and Pseudomonas aeruginosa (43, 44).
We previously identified two efflux pumps that expel toluene in the solvent-tolerant P. putida DOT-T1E strain (25, 33). The TtgABC pump is synthesized at a high basal level in cells growing in the absence of toluene and to a significant but lower level in cells growing with toluene (6). The TtgABC pump extrudes solvents and several antibiotics (33). The other pump, called TtgDEF, is highly homologous to TtgABC (70% similarity at the protein level) and is able to expel toluene but not antibiotics (25). The ttgDEF genes are expressed in response to aromatic hydrocarbons in the culture medium, and the genes are linked to the chromosomal tod genes (24, 25), which encode the enzymes of the so-called toluene dioxygenase pathway and which allow the strain to grow with toluene as the sole C source.
In the wild-type P. putida DOT-T1E strain, toluene tolerance is influenced by growth conditions. Preexposure of cells to low concentrations of this aromatic hydrocarbon led to survival of almost 100% of the cells after a sudden toluene shock; in contrast, only a fraction (10−4) of cells that had not been preexposed survived (33). The TtgABC pump seemed to contribute to the innate tolerance of P. putida DOT-T1E to solvents, and both the TtgABC and TtgDEF pumps contributed to solvent tolerance when cells were preinduced (25). P. putida strain DOT-T1E-82 is a derivative of the wild type with knockouts in the ttgB and ttgD genes. Inactivation of the TtgABC and TtgDEF pumps in this strain still allowed the survival of 10−5 cells if they were preexposed to low concentrations of toluene, although none (<10−8) survived without induction. This result suggests the presence of at least one undiscovered pump.
We now report the identification of a third toluene efflux pump in P. putida DOT-T1E and show that inactivation of this pump in the double mutant described above produced a strain that was not only unable to stand sudden toluene shock, regardless of the growth conditions, but was also unable to grow in the presence of toluene even when it was supplied via the gas phase.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture medium.
The bacterial strains and plasmids used in this study are shown in Table 1. Bacterial strains were routinely grown in liquid Luria-Bertani (LB) medium (35). E. coli cultures were incubated at 37°C, while P. putida cultures were incubated at 30°C. Cultures were shaken on an orbital platform operating at 200 strokes per min. When P. putida cultures were supplied with toluene via the gas phase we used culture flasks with a central vessel in which 0.15 ml of toluene was introduced to avoid direct contact with the liquid medium. The flasks were closed tightly with a Teflon screw top. Under these culture conditions the concentration of toluene in the liquid medium was about 2 mM.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| P. putida strains | ||
| DOT-T1E | Rifr | 33 |
| DOT-T1E-1 | Rifr TelrttgD::kilAB | 25 |
| DOT-T1E-18 | Rifr KmrttgB::′phoA-Km | 43 |
| DOT-T1E-82 | Rifr Kmr TelrttgD::kilAB ttgB::′phoA-Km | 25 |
| DOT-T1E-PS28 | Rifr SmrttgH::ΩSm | This study |
| DOT-T1E-PS30 | Rifr Telr SmrttgD::kilAB ttgH::ΩSm | This study |
| DOT-T1E-PS32 | Rifr Kmr SmrttgB::′phoA-Km ttgH::ΩSm | This study |
| DOT-T1E-PS34 | Rifr Kmr Smr TelrttgB::′phoA-Km ttgD::kilAB ttgH::ΩSm | This study |
| Other strains | ||
| E. coli DH5αF′ | recA1 | Stratagene Inc. |
| Plasmids | ||
| pUC18 | Apr, cloning vector | 45 |
| pHP45ΩSm | Apr Smr | 30 |
| pGG1 | pUC18 bearing a 7.7-kb BamHI insert with the ttgGHI genes | This study |
| pGG2 | ttgGH′::Smr::′ttgHI | This study |
Rifr, Telr, Kmr, Smr, Apr, resistance to RIF, potassium tellurite, KAN, STR, and AMP, respectively.
Plasmid pUC18 was used for cloning experiments (45) and E. coli DH5αF′ was used as a recipient for recombinant plasmids. The relevant characteristics of plasmids constructed in this study are described below. Potassium tellurite was used at a concentration of 15 μg/ml for P. putida and 5 μg/ml for E. coli DH5αF′. The antibiotics used were ampicillin (AMP) (100 μg/ml), kanamycin (KAN) (50 μg/ml), streptomycin (STR) (300 μg/ml), piperacillin (PIP) (100 μg/ml), and rifampin (RIF) (20 μg/ml).
DNA techniques.
Preparation of chromosomal DNA, digestion with restriction enzymes, electrophoresis, and Southern blotting were carried out using standard methods (3, 35). Hybridizations were done using a digoxigenin (DIG) DNA labeling and detection kit (Boehringer Mannheim) in accordance with the manufacturer's instructions. For plasmid isolation a Qiagen kit was used. Plasmid DNA was sequenced on both strands with universal, reverse, or specifically designed primers using an automatic DNA sequencer (ABI-PRISM 310; Applied Biosystems, Inc.).
Primer extension analysis.
P. putida DOT-T1E was grown overnight in LB medium. Cells were then diluted 100-fold in fresh medium, and aliquots were incubated in the absence or presence of toluene supplied via the gas phase until the culture reached a turbidity of about 1.0 at 660 nm. Cells (30 ml) were pelleted and processed for RNA isolation according to the method of Marqués et al. (23). About 20 μg of RNA/ml was treated with RNase-free DNase I and RNase inhibitor (Boehringer Mannheim) to ensure the complete removal of DNA and to maintain the integrity of mRNA. The primer used for extension was complementary to the ttgG gene. This primer was labeled at its 5′ end with [γ-32P]ATP and T4 polynucleotide kinase as described before (3). About 105 cpm of the labeled primer was hybridized to 20 μg of total RNA, and extension was carried out as described previously (3, 23). Electrophoresis of cDNA products was done in a urea-polyacrylamide sequencing gel to separate the reaction products, and dry gels were exposed to X-ray films and visualized. The relative intensity of the bands was quantitated using the Molecular Imagen System GS 525 (Bio-Rad) with the multianalyst program (3, 23).
Cloning of the pump genes and construction of the mutant strains.
To check if a gene homologous to the P. putida srpB gene was present in the genome of P. putida DOT-T1E, a PCR with primers designed based on the srpB gene sequence was performed in 25 μl using 1 μg of DOT-T1E chromosomal DNA/ml as a template. The PCR conditions were 94°C for 1 min, 60°C for 1 min, and 72°C for 30 s for 30 cycles. This yielded a 565-bp fragment that was labeled with DIG by standard procedures and used to hybridize a DOT-T1E genomic library previously generated in our laboratory in pUC18 (33). A positive clone, named pGG1, was obtained with a BamHI insert of 7,674 bp. This clone was digested with EcoRV (which cuts once in the plasmid within the ttgH gene) and ligated to a 2.2-kb SmaI fragment from pHP45Ω-Sm containing the STR resistance cassette to yield pGG2 (Apr, Smr).
About 600 ng of the suicide pGG2 plasmid was electroporated into P. putida DOT-T1E cells prepared as previously described (6, 7), and transformants that integrated the pGG2 plasmid into the host chromosome via homologous recombination were selected on LB solid medium with RIF, STR, and PIP (successful integration was confirmed by a Southern blot). A random merodiploid clone was grown overnight in LB medium to allow for a second recombination event in which the wild-type gene was replaced by the mutant allele. For this selection, colonies were plated again on LB medium with RIF and STR. Among these colonies, those that did not grow in the presence of PIP were selected as putative resolved clones. These mutant clones were checked again by Southern blot, and one of the clones which exhibited the correct replacement (not shown) was kept for further studies and was called P. putida strain DOT-T1E-PS28.
Plasmid pGG2 was also electroporated into P. putida DOT-T1E-18 and DOT-T1E-1 cells to generate double mutants that lacked functional TtgABC and TtgGHI pumps (strain DOT-T1E-PS32) and functional TtgDEF and TtgGHI pumps (strain DOT-T1E-PS30), respectively. Finally, pGG2 was also electroporated into strain DOT-T1E-82 to yield DOT-T1E-PS34, which lacked all three toluene efflux pumps.
MIC assays.
The assays were done in LB medium by the microtiter broth dilution method (2). Microtiter plates were inoculated with 105 CFU/ml and incubated for 20 h at 30°C.
Survival in response to toluene shocks.
Cells were grown in 30 ml of LB medium with or without toluene in the gas phase overnight. On the following day the cultures were diluted 1:100 and grown under the same conditions until the culture reached a turbidity of about 0.8 at 660 nm. These cultures contained about 108 CFU/ml. The cultures were divided into two halves; to one we added 0.3% (vol/vol) toluene, and the other was kept as a control. The number of viable cells was determined before toluene was added and 10, 30, and 60 min later.
RT-PCR.
P. putida DOT-T1E cells (5 ml) growing exponentially on LB medium were harvested by centrifugation (5,000 × g for 10 min). RNA from P. putida DOT-T1E was isolated with the RNeasy Total RNA kit. About 100 μg of total RNA was treated with RNase-free DNase I (50 U) in the presence of 3 U of an RNase inhibitor (Boehringer Mannheim) to avoid DNA contamination and RNA degradation. Reverse transcriptase (RT)-PCR was performed with 1 μg of RNA/ml in a 20-μl final volume using the Titan OneTube RT-PCR system according to the manufacturer's instructions (Boehringer Mannheim). The annealing temperature used for the PCR was 60°C, and the cycling conditions were as follows: 94°C for 30 s, 60°C for 30 s, and 68°C for 1 min. Positive and negative controls were included in all assays. The primers used to test contiguity in the mRNA of the ttgG and ttgH genes and the ttgH and ttgI genes are available upon request.
Computer analysis.
Open reading frames (ORFs) in DNA sequences were predicted with various programs included in the DNA Strider 1.1 package. Sequences were compared with the BlastX programs (1) available from the National Institute for Biotechnology Information server. Protein sequences were aligned with the multiple sequence alignment Clustal W program (38).
Nucleotide sequence accession number.
The nucleotide sequences of the efflux system genes and adjacent DNA described here have been submitted to the GenBank/EMBL data bank under accession number AF299253.
RESULTS AND DISCUSSION
Cloning of the ttgGHI genes of P. putida DOT-T1E.
SrpABC is an inducible efflux pump involved in solvent tolerance described in P. putida S12 (12, 13, 41). Sequence comparison of the SrpABC proteins with those of the TtgABC and TtgDEF pumps identified in DOT-T1E as involved in toluene tolerance revealed 54 to 70% similarity. Because our previous data pointed towards the involvement of a third inducible pump in solvent tolerance in P. putida DOT-T1E, we tested whether the srpABC genes or their homologues were present in P. putida DOT-T1E. To this end we designed two oligonucleotides based on the srpB gene sequence of P. putida S12 and used them in a PCR with P. putida DOT-T1E chromosomal DNA as the template (see Materials and Methods). A 565-bp fragment was amplified, sequenced, and compared with the srpB sequence. The amplified DNA fragment sequence differed by only one nucleotide among the 565 bp.
After labeling this DNA fragment with DIG and hybridizing it against a BamHI chromosomal DOT-T1E library (33), we rescued a clone bearing a 7.7-kb DOT-T1E chromosomal fragment in plasmid pGG1. P. putida DNA was sequenced on both strands, and sequence analysis revealed the presence of two putative ORFs and an incomplete third one that were highly homologous to the srpABC genes (overall identity was 98%). DNA sequence analysis revealed that the two complete ORFs and the incomplete one could potentially form an operon because of the overlap of the stop codon of the second gene and the ATG of the third one and the 17-bp separation between the first ORF and the second one. The corresponding genes were called ttgGHI (toluene tolerance genes) to follow DOT-T1E nomenclature and because the corresponding gene products were found to be involved in tolerance to toluene (see below). The ttgGH genes encode the putative translocase consisting of a periplasmic lipoprotein anchored to the inner membrane (the ttgG gene product) and an inner membrane pump of the RND family (the TtgH protein). The ttgI gene encodes an outer membrane protein that may extend into the periplasmic space to form a channel (17). Because of the high conservation of the sequences between the genes in strains DOT-T1E and S12, the complete ttgI gene could be rescued after PCR amplification with an internal primer and a primer based on the 3′ end of srpC. The BamHI recognition site was located 286 bp from the ATG start codon of ttgI.
We prepared total RNA from P. putida DOT-T1E growing on LB medium with toluene in the gas phase, and RT-PCRs were carried out with primers based on ttgG and ttgH on one hand and primers based on ttgH and ttgI on the other. Amplification was obtained in all cases, with the size of the amplified fragment being that predicted from the DNA sequence (not shown). These results thus confirmed the presumed operon structure of the cluster.
Knockout of the ttgH gene in different P. putida DOT-T1E backgrounds resulted in mutants that are extremely sensitive to toluene.
To determine the function of the TtgGHI proteins in solvent tolerance we decided to generate a knockout of the ttgH gene by using the suicide plasmid pGG2, which carries an Ω-Sm cassette (30) within the ttgH gene (see Materials and Methods). The mutation was transferred to the chromosome of the wild-type strain as described above, and a mutant strain was selected and called DOT-T1E-PS28. Mutant DOT-T1E-PS28 grew on LB medium with a generation time similar to that of the wild-type strain (50 ± 2 min), but in contrast to the wild type, it did not grow in LB medium supplemented with 0.3% (vol/vol) toluene (log Pow = 2.5). However, DOT-T1E-PS28 did grow in LB medium supplemented with 0.3% (vol/vol) m-xylene (log Pow = 3.2), ethylbenzene (log Pow = 3.4), propylbenzene (log Pow = 3.6), and heptane (log Pow = 4.5), which suggests an important role for the TtgGHI efflux pump in toluene tolerance.
To further evaluate the role of this pump in toluene tolerance we carried out a solvent shock assay (adding toluene to reach 0.3% [vol/vol]) with cells that were preexposed or not preexposed to low concentrations of toluene supplied via the gas phase. For the wild-type strain survival was about 0.01 to 0.001% when toluene was added to nonpreexposed cells and about 50 to 100% when the cells were preexposed to toluene (31; Table 2). In contrast, P. putida DOT-T1E-PS28, which lacked the TtgGHI pump, did not survive the 0.3% (vol/vol) toluene shock without induction, and only a 10−7 fraction survived when the culture was preinduced. We also tested the effect of a lower toluene concentration (0.075% [vol/vol]) on the survival of induced and noninduced wild-type and DOT-T1E-PS28 cells. We found that while 100% of the wild-type cells survived this shock regardless of the growth conditions, only a fraction of noninduced (10−3) and induced (10−1) cells of the mutant tolerated the 0.075% (vol/vol) toluene shock. These unexpected results suggested that the TtgGHI efflux pump played a role in both innate and inducible tolerance to toluene, in contrast with previous evidence that this third pump would play a role only in induced tolerance.
TABLE 2.
Survival of P. putida DOT-T1E and its mutant derivatives upon sudden solvent shock
| Strain | Nonfunctional Ttg pump(s) | Cell survivala with:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Toluene
|
Propylbenzene
|
Ethylbenzene
|
m-Xylene
|
Styrene
|
|||||||
| NIb | Ib | NI | I | NI | I | NI | I | NI | I | ||
| DOT-T1E | 10−4–10−5 | 0.5–1 | 1 | 1 | 10−2 | 10−2 | 1 | 1 | 10−2 | 1 | |
| DOT-T1E-18 | ABC | <10−8 | 10−3–10−4 | 1 | 1 | 10−2 | 10−2 | 10−3 | 1 | 10−7 | 10−2 |
| DOT-T1E-1 | DEF | 10−4–10−5 | 10−2–10−3 | 1 | 1 | 10−2 | 10−2 | 1 | 1 | 10−2 | 10−2 |
| DOT-T1E-PS28 | GHI | <10−8 | 10−7 | 1 | 1 | 10−2 | 10−2 | 1 | 1 | 10−7 | 10−6 |
| DOT-T1E-82 | ABC, DEF | <10−8 | 10−5 | 1 | 1 | 10−2 | 10−2 | 10−3 | 1 | 10−7 | 10−5 |
| DOT-T1E-PS30 | DEF, GHI | <10−8 | <10−8 | 1 | 1 | 10−2 | 10−2 | 1 | 1 | <10−8 | 10−7 |
| DOT-T1E-PS32 | ABC, GHI | <10−8 | <10−8 | 10−4 | 10−3 | 10−4–10−5 | 10−4 | 10−4–10−5 | 10−4 | <10−8 | <10−8 |
| DOT-T1E-PS34 | ABC, DEF, GHI | <10−8 | NVc | 10−4–10−5 | NV | 10−5 | NV | 10−5 | NV | <10−8 | NV |
Survival refers to the fraction of viable cells 10 min after the addition of 0.3% (vol/vol) solvent to cultures grown in LB medium (noninduced) or in LB medium with toluene supplied through the gas phase (induced).
NI, noninduced cells; I, induced cells.
NV, nonviable cells; the assay was not performed.
We predicted that a combination of the ttgGHI knockout and either the ttgABC knockout or ttgDEF knockout, both of which were characterized previously (25, 33), would lead to higher sensitivity to sudden toluene shock than when the strain lacked the functional TtgABC and TtgDEF pumps. This hypothesis was confirmed when the survival of P. putida strains DOT-T1E-PS30 (which lacked TtgDEF and TtgGHI) and DOT-T1E-PS32 (lacking TtgABC and TtgGHI) was assayed with cells that were preexposed and not preexposed to toluene (Table 2). No viable cells were recovered after a sudden 0.3% (vol/vol) toluene shock regardless of the growth conditions. This contrasts with a survival of a small but significant fraction of double mutant cells lacking both the TtgABC and TtgDEF pumps when preexposed to low concentrations of toluene (Table 2).
We next constructed strain DOT-T1E-PS34, which had a knockout in all three efflux pumps. The sensitivity to toluene of this strain was such that growth in LB medium with toluene supplied in the gas phase was not feasible, in contrast with the three double mutants described above, all of which grew as well as the wild type in LB medium with toluene supplied via the gas phase. This strongly suggests that either no other pumps are involved in toluene tolerance or the contribution of other pumps to toluene tolerance is minimal. It should be noted that the lack of these three efflux pumps in P. putida DOT-T1E made the strain more sensitive to toluene than E. coli DH5αF′, which grew on LB medium with toluene supplied via the gas phase.
Contribution of efflux pumps to the extrusion of different solvents.
P. putida DOT-T1E is able to grow in the presence of a number of organic solvents with a log Pow between 3.6 and 2.9, i.e., propylbenzene, m-xylene, ethylbenzene, and styrene. The availability of single, double, and triple mutants for the Ttg pumps allowed us to assess the involvement of these efflux pumps in the extrusion of other solvents. Wild-type and mutant cells were grown in LB medium in the absence (noninduced) and presence (induced) of toluene (except for DOT-T1E-PS34, which is not viable when exposed to toluene) and then challenged with 0.3% (vol/vol) concentrations of different solvents; survival was determined after 10 min (Table 2). We found that noninduced cells of the single mutants lacking the TtgABC or TtgGHI pumps were hypersensitive to styrene, and sensitivity was exacerbated in a double ttgABC ttgGHI mutant (Table 2). These results suggest that these two pumps were involved in styrene efflux. The mutant lacking the TtgDEF pump was more sensitive than the wild type to styrene under induced conditions, suggesting that this pump plays a role in styrene efflux as well. In fact, toluene-induced cells of the single ttgABC and ttgGHI mutants were more tolerant to a sudden styrene shock than noninduced ones. Furthermore, double mutants of TtgDEF with either TtgABC or TtgGHI were more susceptible to the styrene shock than the single mutants, an observation which confirms the role of TtgDEF in styrene efflux.
The only single mutant whose survival was compromised by exposure to m-xylene was DOT-T1E-18, which suggests that TtgABC is involved in m-xylene extrusion. This sensitivity was increased in the double ttgABC ttgGHI mutant, indicating that the TtgGHI pump also contributes to m-xylene extrusion. Because the profile of m-xylene tolerance in induced cells of single and double mutants was almost identical to that of noninduced cells, we suggest that TtgDEF does not contribute significantly to the extrusion of m-xylene.
Induced and noninduced cells of all three double mutants, DOT-T1E-82 (lacking TtgABC and TtgDEF), DOT-T1E-PS32 (lacking TtgABC and TtgGHI), and DOT-T1E-PS30 (lacking TtgDEF and TtgGHI), were as tolerant as the wild type to propylbenzene and ethylbenzene shocks, although the double mutant that lacked the TtgABC and TtgGHI pumps exhibited reduced tolerance towards these two compounds. This suggests a role for TtgABC and TtgGHI in the efflux of ethylbenzene and propylbenzene, whereas the TtgDEF pump does not seem to contribute significantly to the extrusion of these two aromatic hydrocarbons.
The work of Lee et al. (18) suggests an additive effect in the extrusion of drugs by pumps that operate through a common mechanism. This may be the case with the RND solvent extrusion pumps, as tolerance to propylbenzene and ethylbenzene in P. putida DOT-T1E was not compromised in mutants lacking TtgABC or TtgGHI but was compromised in double mutants lacking both these pumps. The additive effect of TtgABC with TtgDEF and of TtgDEF with TtgGHI was seen when we studied tolerance to toluene and styrene in induced cells: these double mutants were less tolerant to styrene and toluene than the corresponding single ones.
The ttgGHI operon is expressed constitutively, but its expression is enhanced by the presence of toluene in the culture medium.
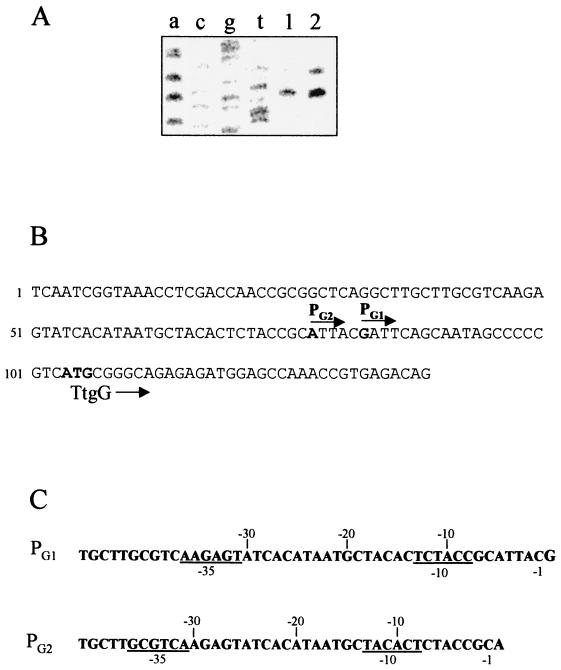
We determined the pattern of expression of the ttgGHI genes in wild-type cells grown on LB medium with and without toluene supplied via the gas phase. Primer extension analysis identified a 213-bp cDNA in cells that were not exposed to toluene, whereas two cDNA bands, of 213 and 218 bp, were found in cells exposed to toluene. It then follows that in the absence of the aromatic hydrocarbon, some basal expression of the ttgGHI operon takes place which increases due to an inducible promoter in cells exposed to toluene. The intensity of the 213-bp cDNA band was three- to fourfold higher in cells grown with toluene than in cells grown in the absence of this aromatic hydrocarbon (Fig. 1A). The intensity of the 218-bp cDNA band was half the intensity of one produced by the 213-bp cDNA in induced cells (Fig. 1A, lane 2). The pattern of expression of the ttgGHI genes explains why the TtgGHI pump is a key element in the innate and inducible efflux of toluene. We observed that the ttgGHI operon was expressed at relatively high levels from a single promoter in noninduced cells and that expression of ttgGHI in induced cells took place at a higher level than in noninduced cells, because in addition to the increase in the levels of expression from the constitutive promoter, expression of this operon also occurred from a second inducible promoter that overlapped the constitutive one. In contrast with our results, Kieboom et al. (13) showed that the srpABC genes were expressed only in response to solvents, with styrene being the best inducer.
FIG. 1.
Pattern of expression of the ttgGHI operon in wild-type cells under different growth conditions. (A) Transcription initiation mapping of the ttgG gene. Lanes 1 and 2, cells grown on LB medium without and with toluene in the gas phase, respectively. The acgt lanes correspond to the sequencing ladder. (B) DNA sequence at the ttgG promoter region. The transcription initiation points are in bold and the arrows indicate the direction of gene transcription. The ATG start codon of the ttgG gene is also in bold. (C) Proposed −10 and −35 boxes (underlined) for PG1 and PG2.
From the size of the cDNA band we inferred that the main transcription initiation points corresponded to bases G83 and A78 in the sequence shown in Fig. 1B for the promoter expressed under both growth conditions and for the inducible promoter, respectively. We analyzed the sequences upstream from the transcription initiation points to define the promoters. The promoter expressed at a certain basal level in cells that were not exposed to toluene was called PG1. Upstream from this point we found an extended −10 region and less conserved bases at the −35 region (Fig. 1C). The inducible promoter, called PG2, also exhibited features of −10 boxes (three matches out of six for a perfect −10 box) but had a poorly conserved −35 region (Fig. 1C), as is also the case in many genes whose transcription is stimulated by a positive regulatory protein. The PG1 and PG2 promoters might overlap by at least 1 bp in their −10 and −35 regions (Fig. 1C), and therefore it is not surprising that toluene increased the expression of both promoters, although the nature of the regulatory protein(s) involved in this process remains to be determined. Recently Wery et al. (41) reported that the srpABC genes can be controlled by the srpS and srpR regulatory genes. Similar genes exist in P. putida DOT-T1E, although at present their role in the regulation of the ttgGHI operon is unknown.
Contribution of solvent efflux pumps to the extrusion of antibiotics.
Several efflux pumps have been described as able to extrude antibiotics as well as solvents. We have previously shown that the TtgABC pump is able to extrude AMP, carbenicillin, nalidixic acid, chloramphenicol, and tetracycline (33; also see Table 3), whereas the TtgDEF pump is specific for organic solvents and does not extrude any of these antibiotics (25).
TABLE 3.
MICs of various antibiotics for P. putida DOT-T1E and its derivatives
| Strain | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|
| Carbenicillin | Nalidixic acid | Chloramphenicol | Tetracycline | AMP | |
| DOT-T1E | 256 | 64 | 32 | 4 | 64 |
| DOT-T1E-18 | 8 | 16 | 8 | 0.25 | 16 |
| DOT-T1E-PS28 | 256 | 64 | 32 | 4 | 64 |
| DOT-T1E-PS32 | 2 | 16 | 8 | 0.05 | 2 |
| DOT-T1E-PS34 | 2 | 16 | 8 | 0.05 | 2 |
We determined the MIC of several antibiotics for DOT-T1E-PS28 (lacking TtgGHI), DOT-T1E-18 (lacking TtgABC), the double mutant DOT-T1E-PS32 (which lacks the TtgABC and TtgGHI pumps), and the triple mutant DOT-T1E-PS34. As shown previously (33), the resistance of DOT-T1E-18 to all antibiotics tested was reduced. The DOT-T1E-PS28 single mutant had the same profile as the wild type (Table 3). However, the DOT-T1E-PS32 double mutant was somewhat more susceptible to these antibiotics than DOT-T1E-18, suggesting that the TtgGHI pump identified in this study plays a role in antibiotic extrusion. Apparently the extrusion of antibiotics by the TtgGHI pump is masked when the TtgABC pump is working in the cell. The triple mutant has the same antibiotic resistance profile as DOT-T1E-PS32. This confirms that TtgDEF is not involved in antibiotic extrusion.
In P. aeruginosa up to three RND efflux pumps for antibiotics (MexAB-OprM, MexCD-OprJ, and MexEF-OprN) have been described (8, 15, 16, 20, 29). The Mex pumps export a wide range of antibiotics (8, 9, 15–17, 19, 20, 29, 33). These three Mex systems and the AcrAB-TolC multidrug efflux system of E. coli (3, 22, 26, 28, 39) are also able to mediate intrinsic organic solvent tolerance (42). It therefore seems that the RND antibiotic and solvent efflux pumps in E. coli and bacteria of the genus Pseudomonas have a broad substrate specificity and are thus able to extrude compounds of different chemical structures (15, 16, 20, 22, 27–29). It will be interesting to see which determinants are recognized by each pump.
ACKNOWLEDGMENTS
We thank M. Fandila, C. Lorente, and K. Shashok for assistance in the preparation of the manuscript.
This study was supported by a grant from the European Commission, BIO4-CT97-2270.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amsterdam D. Susceptibility testing of antimicrobials in liquid media. In: Lorian V, editor. Antibiotics in laboratory medicine. Baltimore, Md: Williams & Wilkins; 1991. pp. 72–78. [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R F, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y.: Greene Publishing, Associated; 1991. [Google Scholar]
- 4.Cruden D L, Wolfram J H, Rogers R D, Gibson D T. Physiological properties of a Pseudomonas strain which grows with p-xylene in a two-phase (organic-aqueous) medium. Appl Environ Microbiol. 1992;58:2723–2729. doi: 10.1128/aem.58.9.2723-2729.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Smet M M, Kingma J, Witholt B. The effect of toluene on the structure and permeability of the outer and cytoplasmic membranes of Escherichia coli. Biochim Biophys Acta. 1978;506:64–80. doi: 10.1016/0005-2736(78)90435-2. [DOI] [PubMed] [Google Scholar]
- 6.Duque E, Segura A, Mosqueda G, Ramos J L. Global and cognate regulators control the expression of the organic solvent efflux pumps TtgABC and TtgDEF of Pseudomonas putida. Mol Microbiol. 2001;39:1100–1106. doi: 10.1046/j.1365-2958.2001.02310.x. [DOI] [PubMed] [Google Scholar]
- 7.Enderle P J, Farwell M A. Electroporation of freshly plated Escherichia coli and Pseudomonas aeruginosa cells. Biotechniques. 1998;25:954–958. doi: 10.2144/98256bm05. [DOI] [PubMed] [Google Scholar]
- 8.Gotoh N, Tsujimoto H, Poole K, Yamagishi J-I, Oyamada Y, Nishino T. The outer membrane protein OprM of Pseudomonas aeruginosa is encoded by oprK of the mexA-mexB-oprK multidrug resistance operon. Antimicrob Agents Chemother. 1995;39:2567–2569. doi: 10.1128/aac.39.11.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirai K, Suzue S, Irikura T, Iyobe S, Mitsuhashi S. Mutations producing resistance to norfloxacin in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;31:582–586. doi: 10.1128/aac.31.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huertas M J, Duque E, Rosselló-Mora R, Mosqueda G, Godoy P, Christensen B, Molin S, Ramos J L. Tolerance to sudden organic solvent shocks by soil bacteria and characterization of Pseudomonas putida strains isolated from toluene polluted sites. Environ Sci Technol. 2000;34:3395–3400. [Google Scholar]
- 11.Inoue A, Horikoshi K. A Pseudomonas thrives in high concentrations of toluene. Nature. 1989;338:264–266. [Google Scholar]
- 12.Kieboom J, Dennis J J, de Bont J A M, Zylstra G J. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J Biol Chem. 1998;273:85–91. doi: 10.1074/jbc.273.1.85. [DOI] [PubMed] [Google Scholar]
- 13.Kieboom J, Dennis J J, Zylstra G J, de Bont J A M. Active efflux of organic solvents by Pseudomonas putida S12 is induced by solvents. J Bacteriol. 1998;180:6769–6772. doi: 10.1128/jb.180.24.6769-6772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K, Lee S, Lee K, Lim D. Isolation and characterization of toluene-sensitive mutants from toluene-resistant bacterium Pseudomonas putida GM73. J Bacteriol. 1998;180:3692–3696. doi: 10.1128/jb.180.14.3692-3696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köhler T, Kok M, Michea-Hamzehpour M, Plesiat P, Gotoh N, Nishino T, Kocjanici Curty L, Péchère J-C. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:2288–2290. doi: 10.1128/aac.40.10.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Köhler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Péchère J-C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 17.Koronakis V, Sharft A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 18.Lee A, Mao W, Warren M S, Mistry A, Hoshino K, Okumura R, Ishida H, Lomovskaya O. Interplay between efflux pumps may provide either additive or multiplicative effect on drug resistance. J Bacteriol. 2000;182:3142–3150. doi: 10.1128/jb.182.11.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X Z, Zhang L, Poole K. Role of the multidrug efflux systems of Pseudomonas aeruginosa in organic solvent tolerance. J Bacteriol. 1998;180:2987–2991. doi: 10.1128/jb.180.11.2987-2991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X Z, Poole K. Organic solvent-tolerant mutants of Pseudomonas aeruginosa display multiple antibiotic resistance. Can J Microbiol. 1999;45:18–22. doi: 10.1139/cjm-45-1-18. [DOI] [PubMed] [Google Scholar]
- 21.Ma D, Cook D N, Alberti M, Hearst J E, Nikaido H. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 1994;2:489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 22.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 23.Marqués S, Ramos J L, Timmis K N. Analysis of the mRNA structure of the Pseudomonas putida TOL meta fission pathway operon around the transcription initiation point, the xylTE and the xylFJ region. Biochim Biophys Acta. 1993;1216:227–237. doi: 10.1016/0167-4781(93)90149-8. [DOI] [PubMed] [Google Scholar]
- 24.Mosqueda G, Ramos-González M I, Ramos J L. Toluene metabolism by the solvent-tolerant Pseudomonas putida DOT-T1 strain, and its role in solvent impermeabilization. Gene. 1999;232:69–76. doi: 10.1016/s0378-1119(99)00113-4. [DOI] [PubMed] [Google Scholar]
- 25.Mosqueda G, Ramos J L. A set of genes encoding a second toluene efflux system in Pseudomonas putida DOT-T1E is linked to the tod genes for toluene metabolism. J Bacteriol. 2000;182:937–943. doi: 10.1128/jb.182.4.937-943.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura H. Genetic determination of resistance to acriflavin, phenyl alcohol, and sodium dodecyl sulfate in Escherichia coli. J Bacteriol. 1968;96:987–996. doi: 10.1128/jb.96.4.987-996.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poole K, Heinrichs D, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol Microbiol. 1993;10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 29.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X-Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 30.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 31.Ramos J L, Duque E, Huertas M J, Haidour A. Isolation and expansion of the catabolic potential of a Pseudomonas putida strain able to grow in the presence of high concentrations of aromatic hydrocarbons. J Bacteriol. 1995;177:3911–3916. doi: 10.1128/jb.177.14.3911-3916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramos J L, Duque E, Rodríguez-Herva J J, Godoy P, Haïdour A, Reyes F, Fernández-Barrero A. Mechanisms for solvent tolerance in bacteria. J Biol Chem. 1997;272:3887–3890. doi: 10.1074/jbc.272.7.3887. [DOI] [PubMed] [Google Scholar]
- 33.Ramos J L, Duque E, Godoy P, Segura A. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J Bacteriol. 1998;180:3323–3329. doi: 10.1128/jb.180.13.3323-3329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rekker R F, de Kort H M. The hydrophobic fragmental constant and extension to a 1,000 data point set. Eur J Med Chem. 1979;14:479–488. [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Segura A, Duque E, Mosqueda G, Ramos J L, Junker F. Multiple responses of Gram-negative bacteria to organic solvents. Environ Microbiol. 1999;1:191–198. doi: 10.1046/j.1462-2920.1999.00033.x. [DOI] [PubMed] [Google Scholar]
- 37.Sikkema J, de Bont J A M, Poolman B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev. 1995;59:201–222. doi: 10.1128/mr.59.2.201-222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsukagoshi N, Aono R. Entry and release of solvents by Escherichia coli in an organic-aqueous two-liquid phase system and substrate specificity of the AcrAB-TolC solvent-extruding pump. J Bacteriol. 2000;182:4803–4810. doi: 10.1128/jb.182.17.4803-4810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber F J, Isken S, de Bont J A M. Cis/trans isomerization of fatty acids as a defense mechanism of Pseudomonas putida strains to toxic concentrations of toluene. Microbiology. 1994;140:2013–2017. doi: 10.1099/13500872-140-8-2013. [DOI] [PubMed] [Google Scholar]
- 41.Wery J, Hidayat B, Kieboom J, de Bont J A M. An insertion sequence prepares Pseudomonas putida S12 for severe solvent stress. J Biol Chem. 2001;276:5700–5706. doi: 10.1074/jbc.M007687200. [DOI] [PubMed] [Google Scholar]
- 42.White D G, Goldman J D, Demple B, Levy S B. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong K K Y, Brinkman F S L, Benz R S, Hancock R E W. Evaluation of a structural model of Pseudomonas aeruginosa outer membrane protein OprM, an efflux component involved in intrinsic antibiotic resistance. J Bacteriol. 2001;183:367–374. doi: 10.1128/JB.183.1.367-374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xhian-Zhi L, Poole K. Mutational analysis of the OprM outer membrane component of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol. 2001;183:12–27. doi: 10.1128/JB.183.1.12-27.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 46.Zgurskaya H I, Nikaido H. Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner membrane-associated multidrug efflux pump AcrB from Escherichia coli. J Bacteriol. 2000;182:4264–4267. doi: 10.1128/jb.182.15.4264-4267.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]