Abstract
WHO-defined SARS-CoV-2 variants of concern (VOC) drive therapeutics and vaccine development. The Omicron VOC is dominating the arena since November 2021, but the number of its sublineages is growing in complexity. Omicron represent a galaxy with a myriad of stars that suddenly rise and expand before collapsing into apparent extinction when a more fit sublineage appears. This has already happened with BA.1, BA.2, and BA.4/5 and is happening with BA.2.75. We review here the current PANGO phylogeny, focusing on sublineages with Spike mutations, and show how frequently xxxxxxxx convergent evolution has occurred in these sublineages. We finally summarize how Omicron evolution has progressively defeated the anti-Spike monoclonal antibodies authorized so far, leaving clinicians to again fall back on COVID19 convalescent plasma from vaccinated donors as the only antibody-based therapy available.
Abbreviations: VOC, variant of concern; VOI, variant of interest; LUM, lineage under monitoring
Keywords: SARS-CoV-2, Spike, Variant of concern, VOC, Omicron, B.1.1.529, BA.1, BA.2, BA.2.12.1, BA.4, BA.5, BA.2.75, BA.4.6
1. Introduction
On November 8, 2021, a novel SARS-CoV-2 lineage named B.1.1.529 in PANGOLIN phylogeny, 21 M in NextStrain, VUI-21NOV-01 in Public Heath England (PHE), and belonging to GISAID clade GR/484 A was reported from 100 cases in South Africa. These strains came primarily from Gauteng, North West and Limpopo regions where it was likely to have been circulating for at least 6 weeks (Yeh and Contreras, 2021). Shortly after the discovery of the Omicron variant in South Africa it was found in numerous countries around the world and it was declared as variant of concern (VOC) Omicron by WHO on November 26, 2021. Omicron initially harbored 32 different Spike mutations ( Table 1). Arising from a third level-branch of the PANGOLIN phylogeny (B.1.1.529) it was immediately clear that, according to nomenclature rules, further Omicron sublineages would have been named with aliases, which should occur every fourth branching. BA was selected as the first alias for Omicron. E.g., at the time of this writing, BA.5 stands for B.1.1.529.5, while BF.1.1 stands for BA.1.1.529.5 or B.1.1.529.5.3.4.1. The nomenclature is growing in complexity, and ambiguities can be expected. For example, it is not immediately clear that BF is a descendant of BA rather than an alias of a different lineage. Furthermore, alphabet exhaustion is driving letter addition to names (e.g. BW.* for sublineages and XBB for recombinants).
Table 1.
Occurrence of selected Spike mutations in Omicron VOC and VOC-LUM, and impact of that mutation on efficacy of mAbs authorized (median fold-reduction in in vitro neutralization activity compared to wild-type SARS-CoV-2). BAM: bamlanivimab; ETE: etesevimab; CAS: casirivimab; IMD: imdevimab; TIX: tixagevimab; CIL: cilgavimab; SOT: sotrovimab; BEB: bebtelovimab; ADI: adintrevimab. REG: regdanvimab. = : < 2-fold reduced susceptibility; ↓: 2–5-fold reduced susceptibility; ↓↓: 5–24.9-fold reduced susceptibility; ↓↓↓: ≥ 25-fold reduced susceptibility. stanford.edu/search-drdb" id= "ce_inter-ref_ir0005">https://covdb.stanford.edu/search-drdb
| Spike mutation | BA.1 | BA.2 | BA.2.10.4 | BA.2.12.1 | BA.2.75 | BA.4/5 | BAM | ETE | CAS | IMD | CIL | TIX | SOT | BEB | ADI | REG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T19I | – | + | – | + | – | – | = | = | = | = | = | = | = | = | = | = |
| L24S/∆25–27 | – | + | – | + | – | – | = | = | = | = | = | = | = | = | = | = |
| W64R | – | – | + | – | – | – | = | = | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| A67V | + | – | – | – | – | – | = | = | = | = | = | = | = | n.a. | = | n.a. |
| ∆69–70 | + | – | – | – | – | + | = | = | = | = | = | = | = | = | = | n.a. |
| T95I | + | – | – | – | – | – | = | = | = | = | = | = | = | = | = | = |
| ∆141–144 | – | – | + | – | – | – | = | = | = | = | = | = | = | = | = | = |
| G142D/∆143–145 | + | + | – | + | – | – | = | = | = | = | = | = | = | = | = | = |
| K147E | – | – | – | – | + | – | = | = | = | = | = | = | = | = | n.a. | = |
| W152R | – | – | – | – | + | – | = | = | = | = | = | = | = | = | n.a. | = |
| F157L | – | – | – | – | + | – | = | = | = | = | = | = | = | = | n.a. | = |
| I210V | – | – | – | – | + | – | = | = | = | = | = | = | = | = | n.a. | = |
| Δ211/L212I | + | – | – | – | – | – | = | = | = | = | = | = | = | = | = | = |
| V213G | – | + | – | + | – | – | = | = | ↓ | = | ↓ | = | = | = | n.a. | = |
| ins214EPE | + | – | – | – | – | – | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| G257S | – | – | – | – | + | – | = | = | = | = | = | = | = | = | n.a. | = |
| G339x | D | D H in BJ.1 |
– | D | H | – | = | = | = | = | = | = | = | = | = | = |
| R346x | -, K in BA.1.1 | - T in BA.2.9.4, BA.2.80 |
– | – | - T in BL.1 (a.k.a. BA.2.75.1.1), BL.2, BA.2.75.2, BM.1.1, BM.2, BM.4.1.1, BR.2, BN.1, and BA.2.75.6 |
- I in BA.5.9 and CE.1 S in BA.4.7 and BF.13 T in BA.4.1.9, BA.4.1.10, BA.4.6, BA.5.2.6, BA.5.2.13, BE.1.2, BE.4.1.BF.7, BF.11 and BQ.1.1 |
= | = | = | = | ↓↓↓ | = | = | = | = | = |
| S371X | L | F | – | F | – | – | = | ↓↓↓ (F) ↓↓ (L) |
↓↓ (F) ↓ (L) |
↓↓↓ (F) ↓↓ (L) |
↓ | ↓↓↓ (F) ↓ (L) |
↓↓ | ↓ | ↓↓↓ (F) ↓↓ (L) |
↓↓↓ |
| S373P | + | + | – | + | – | – | = | = | = | ↓ | = | = | = | = | = | = |
| S375F | + | + | – | + | – | – | = | = | = | = | = | = | = | = | = | = |
| T376A | – | + | – | + | – | – | = | = | = | = | = | = | = | = | = | = |
| D405N | – | + | – | + | – | – | ↓ | ↓↓ | ↓↓↓ | ↓ | = | ↓ | = | = | ↓ | n = |
| R408S | – | + | – | + | – | – | = | = | = | = | = | = | = | = | = | = |
| K417N | + | + | – | + | – | – | = | ↓↓↓ | ↓↓ | = | = | = | = | = | = | = |
| N440K | + | + | – | + | – | – | ↓ | ↓ | ↓ | ↓↓↓ | ↓ | = | ↓ | = | ↓ | = |
| G446S | + | – | + | – | + | – | ↓ | ↓ | ↓ | ↓↓↓ | ↓↓ | ↓ | ↓ | ↓ | ↓ | = |
| L452x | – | – | – | Q | R in BA.2.75.4 | –R | ↓↓↓ | = | = | ↓ | ↓ | = | = | ↓ | ↓ | n.a. |
| N460K | – | – | – | – | + | –, + in BQ.1, BW.1, BU.1, BA.4.6.3 | = | ↓↓↓ | = | = | = | = | = | = | n.a. | n.a. |
| S477N | + | + | – | + | – | – | = | = | = | = | = | = | = | = | = | = |
| T478K | + | + | – | + | – | – | ↓ | = | = | = | ↓ | ↓ | ↓ | = | ↓ | n.a. |
| E484A | + | + | – | + | – | – | ↓↓↓ | ↓ | ↓↓↓ | ↓↓ | ↓ | ↓↓ | = | ↓ | ↓ | n.a. |
| F486X | – | – | – | P | S in BA.2.75.2, BM.1, BM.4.1, BY.1 and BA.2.75.7 I in BR.2, V in CB.1 |
V | ↓↓↓ | ↓↓↓ | ↓↓↓ | = | = | ↓↓↓ | = | = | = . | n.a. |
| Q493R | + | + | - R493Q | + | - R493Q | - R493Q | ↓↓↓ | ↓↓↓ | ↓↓↓ | = | = | ↓↓ | = | = | = | ↓↓↓ |
| S494P | – | + | – | + | – | – | ↓↓ | = | ↓↓ | = | n.a. | n.a. | ↓ | n.a. | ↓ | ↓ |
| G496S | + | – | – | – | – | – | = | = | = | ↓ | = | = | = | = | = | = |
| Q498R | + | + | – | + | – | – | ↓ | ↓↓ | ↓ | ↓ | ↓ | ↓ | = | = | ↓ | = |
| N501Y | + | + | – | + | – | – | = | ↓↓ | = | = | = | = | = | ↓ | ||
| Y505H | + | + | – | + | – | – | = | ↓ | = | ↓ | ↓ | = | = | = | = | = |
| T547K | + | – | – | – | – | – | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| D614G | + | + | + | + | + G614S in BL.1.1 |
+ | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| H655Y | + | + | – | + | – | – | = | ↓ | = | ↓ | ↓ | = | = | = | = | ↓ |
| N679K | + | + | – | + | – | – | = | = | = | = | = | = | = | = | n.a. | = |
| P681H | + | + | – | + | – | – | = | = | = | = | = | = | = | = | = | = |
| N764K | + | + | – | + | – | – | = | = | = | = | = | = | = | = | = | = |
| D796Y | + | + | – | + | – | – | = | = | = | = | = | = | ↓ | ↓ | = | ↓ |
| N856K | + | – | – | – | – | – | = | ↓↓ | = | = | ↓ | = | = | = | = | = |
| Q954H | + | + | – | + | – | – | = | = | = | = | = | = | = | = | = | = |
| N969K | + | + | – | + | – | – | = | ↓↓ | = | = | = | = | = | ↓ | = | = |
| L981F | + | – | – | – | – | – | = | = | = | = | = | = | = | = | = | = |
3. Main Omicron branches
The Omicron VOC members has 5 major sublineages that share a few Spike mutations (K417N, N440K, E484A, and N679K): given that B.1.1.529 had already consumed the three levels of classification allowed by the PANGO nomenclature, the BA alias was introduced for sublineages.
-
•
BA.1 (a.k.a. 21 K in NextStrain) has the hallmark mutations listed in Table 1.
-
•
BA.2 (a.k.a. 21 L in NextStrain, VUI-22JAN-01 in PHE, or colloquially as “Omicron 2”) was first reported in Gauteng on Nov 17, 2021 and carries almost all the spike RBD mutations first noted in Omicron, and both furin cleavage adjacent mutations as well as the NSP6 deletion seen in other VOCs. As shown in Table 1, BA.2 has 7 additional Spike mutations not found in BA.1 and 4 lost compared to BA.1.
-
•
BA.3 harbors R408S and has been a relatively minor sublineage.
-
•
BA.4 (a.k.a. 22 A in NextStrain, V-22APR-03 first and VOC-22APR-03 later in PHE) shares all mutations/deletions with the BA.2 lineage except the following: NSP4: L438F reverted to WT (wild type); S: 69/70 deletion, L452R, F486V, Q493 (WT); ORF 6: D61 (WT); ORF 7b: L11F; N: P151S. The Spike 69/70 deletion produces an undetectable S-gene target (S-gene target failure) in the TaqPath assay. The earliest BA.4 sample in GISAID was from South Africa with a sample collection date of 10 January 2022. BA.4/BA.5 shows the weakest receptor-binding activity due to F486V and R493Q reversion (Cao et al., 2022c). The growth advantages for BA.4 and BA.5 of 0.08 (95% CI: 0.07–0.09) and 0.12 (95% CI: 0.09–0.15) per day respectively over BA.2 in South Africa (Tegally et al. (2022)).
-
•
BA.5 (a.k.a. 22B in NextStrain, V-22APR-04 first and VOC-22APR-04 later in PHE) shares the same mutations/deletions as BA.4 except the following: M: D3N; ORF7b: L11 (WT); N: P151 (WT); synonymous SNPs: A27038G, and C27889T.
4. VOC lineages under monitoring
Among the currently circulating BA.2 and BA.4/5 sublineages, several ones deserve a separate discussion, having been classified by WHO as VOC lineages under monitoring (VOC-LUM: https://www.who.int/activities/tracking-SARS-CoV-2-variants) or listed at https://cov-spectrum.org/collections/18 because of their relative growth advantage:
-
•second-generation BA.2 appeared after BA.4/5 had become dominant over BA.2:
-
oBA.2.12.1 (a.k.a. 22 C in NextStrain and V-22MAY-01 in PHE) has been a prevalent lineage in USA in Spring 2022, and carries L452Q and S704F
-
oOther lineages first reported from India:
-
▪BA.2.10 descendants. BA.2.10.4 (nicknamed “Pisces”) harbors W64R, Δ 141–144, Δ243-244, G446S, F486P, R493Q and S494P, and P1143L. It was first detected in India in August 2022, showing 10% daily growth advantage over other BA.2. It additionally harbors and is resistant to all mAbs except bebtelovimab (Sheward et al., 2022b). BJ.1 (nicknamed "Argus") harbors R346T, G446S, and F490V : it has recombined with BM.1.1.1 (nicknamed " Mimas") to generate XBB.1 (nicknamed "Gryphon).
-
▪BA.2.75 (nicknamed “Centaurus”) was detected in India in May 2022. It carries K147E, W152R, F157L, I210V, G257S, D339H (mutated from G339D), G446S (which causes escape from cilgavimab and imdevimab), N460K, R493Q (reversion). R493Q and N460K cause BA.2.75 to exhibit higher ACE2-binding affinity than BA.2 and BA.4/BA.5 (Cao et al., 2022b; Saito et al., 2022) and the longest evolutionary distance of the S gene (Sugano et al., 2022). BA.2.75 is neutralized by vaccine-elicited and BA.1/2 breakthrough infection-induced sera at the same levels of BA.2, but at lower level from convalescent sera than BA.2 and BA.5. Nirmatrelvir, molnupiravir and ritonavir retain efficacy. Fusogenicity, growth efficiency in human alveolar epithelial cells, and intrinsic pathogenicity in hamsters of BA.2.75 were comparable to those of BA.5 but were greater than those of BA.2 (Saito et al., 2022). Of concern, the replicative ability of BA.2.75 in hamster lungs was higher than that of BA.2 and BA.5, causing focal viral pneumonia (Uraki et al., 2022). Nine sublineages have developed so far. BA.2.75.1 has acquired R346T to generate BL.1 and BL.2. BA.2.75.2 (nicknamed “Alcyon”, short for “ Alcyoneus”, which includes R346T, F486S and D1199N) is the most immune escaping (5-times reduced neutralization by vaccinated donors than BA.5 (Sheward et al., 2022b)). BA.2.75.2 has acquired S494P in CA.2 and L452R in CA.1. BA.2.75.3 has acquired F486S in BM.1 and BM.4, and R346T in BM.2: BM.1.1 (nicknamed "Mimas") recombined with BJ.1 to create XBB (nicknamed "Gryphon"), while the R346T-BM.4 serially acquired F486S (BM.4.1), R346T (BM.4.1.1), K444T (CH.1), and L452R (CH.1.1). The L452R-harboring BA.2.75.4 has acquired K444M in BR.1, R346T plus F486I in BR.2 and R346T in BR.3. BA.2.75.5 has acquired R346T plus F490S in BN.1 (nicknamed "Hydra"), which in turns has acbasiliquired S494P in BN.1.1. The R346T-harboring BA.2.75.6 has acquired F486S in BY.1, while the R346T-harboring BA.2.75.9 has acquired F486V in CB.1
-
▪BH.1 (i.e., BA.2.38.3.1, nicknamed “ Almops”) harbors 144del, F446S, and L452Q
-
▪
-
o
-
•
BA.4/5 descendants. BA.4.6 (nicknamed "Aeterna") harbored R346T and later acquired K444N and N460K in BA.4.6.3, while BA.4.7 harbors R346S. BA.5 has instead many more convergent descendants. BA.5.2 acquired R346I in CE.1, R346S in BF.13, R346T in BA.5.2.6, BA.5.2.13, BF.7 (nicknamed "Minotaur"), and BF.11. BA.5.3 acquired R346T in BE.1.2 and BE.4.1 (which in turn acquired K444R in BE.4.1.1), K444T in BE.1.1.1, and both K444N and N460Kin BE.4.2. BE.1.1.1 is of much interest because it acquired in N460K in BQ.1 (nicknamed "Typhon"), which in turn acquired R346T in both BQ.1.1 (nicknamed "Cerberus") and BQ.1.17. BA.5.6 acquired K444T in BA.5.6.2, which in turn acquired N460K in BW.1. The R346T-harboring BA.5.1.20 acquired K444M and N460K in BU.1.
-
•
Omicron sublineages with E340 mutations have been exceedingly rare before massive usage of sotrovimab, which is known to induce this escape mutant (Rockett et al., 2021). E340K has been observed in BA.2.77. BA.1.24, first detected in Spain and harboring the E340K Spike mutation, was previously outcompeted by the BA.2 wave.
5. Co-infections and recombinants
Further complicating the landscape, several Delta-BA.1 (Combes et al., 2022) and BA.1-BA.2 (Gjorgjievska et al., 2022, Vatteroni et al., 2022) co-infections have been reported.
As expected for coronaviruses, multiple recombinants (Focosi and Maggi, 2022) have been detected to date (designed by the PANGOLIN nomenclature with the “X” prefix and progressive alphabet letters, with one more letter added when the alphabet is over), with the left part of the description indicating the Spike donor lineage. The recombinants can be summarized as follows:
-
•DeltaxBA.1: XD (France and Denmark (Zayet et al., 2022)), XF (UK)
-
oDeltaxBA.1.1: XS (USA)
-
o
-
•BA.1xBA.2 (Colson et al., 2022):
-
oXE (a.k.a. V-22APR-02 in PHE), XG (Denmark), XH (Denmark), XJ (Finland), XK (Belgium), XN (UK), XT (South Africa), XU (India), XV (Denmark and Italy), XW (USA, Germany, England), XY (USA), XAA (USA), XAB (Germany), XAC (Canada and USA), XAF (Costa Rica), XAG (Brazil), XAK (Germany), XAL (Germany), XAQ (Canada), XAR (Reunion)
-
oBA.1.1xBA.2: XM (Europe), XP (UK), XQ (UK), XR (UK), and XZ
-
oBA.1.1xBA.2.9: XAM (Panama and USA), XAU (Spain, England, France)
-
o
-
•BA.2xBA.1: XAD (Germany), XAE (USA, Chile), XAH (Slovenia), XAP (USA)
-
oBA.2.3.13xBA.1: XAT (Japan)
-
o
-
•
BA.2xBA.4: BA.2.12.1xBA.4 (England)
-
•BA.2xBA.5: XAV (USA, Scotland)
-
oBA.2xBA.5.1: XAN (Spain, Denmark)
-
oBA.2.5xBA.5: XAZ (France, Germany, Croatia, Denmark and USA)
-
oBA.2.76xBA.5.2: (PANGO issue #896)
-
o
-
•BA.2xDelta:
-
oBA.2xAY.122x: XAW (Russia)
-
oBA.2xAY.45: XAY/C1 (South Africa)
-
oC2
-
o
-
•
BA.5xBA.2: XAS (USA)
-
•
BM.1.1.1xBJ.1: XBB
5.1. Convergent evolution across Omicron sublineages
Each Omicron sublineage has tens of descendent sub-sublineages (see Supplementary Table 1), which often show signs of convergent evolution in the amino acid sequence of the Spike protein (see Fig. 1). A few prominent examples have been selected below, and the reader can find more details on the functional consequences of specific mutations at the references provided. As a primer, it should be considered that the receptor-binding domain (RBD) of Spike consists of amino acid residues from positions 333–527: the specific portion which makes contact with ACE2 is called receptor-binding motif (RBM) and includes amino acids 438–506. In general, mutations that favor increased ACE2 affinity are positively selected, with a few exceptions (e.g., K417T/N) that require compensation by other residues.
-
•
∆HV69-70 was firstseen in VOC Alpha (B.1.1.7), and later detected in lineages B.1.375 (Larsen and Worobey, 2021, Moreno et al., 2021) and B.1.346 reported from USA (Larsen and Worobey, 2021), B.1.1.298, B.1.177 (EU1), B.1.160 (EU2), and B.1.258∆ (Brejová et al., 2021). Among Omicron sublineages, it is a hallmark of both BA.1 and BA.4/5. ΔH69/ΔV70 diminishes protrusion of the 69–76 loop, increasing Spike-mediated infectivity by 2 folds. Interestingly for screening purposes, the deletion causes false negativity in the Spike target (so called S-dropout variant or S-gene target failures (SGTF)) of a 3-target TaqPath® RT-PCR COVID19 assay (Thermo Fischer Scientific) (Bal et al., 2020, Gravagnuolo et al., 2021, Washington et al., 2020). The deletion can also be detected as a positive signal using a pair of molecular beacons paired with loop mediated isothermal amplification (LAMP) (Sherrill-Mix et al., 2021).
-
•R346 mutations were exceedingly rare before Omicron, but are now hallmarks of various Omicron sublineages (e.g.:
-
oR346I in BA.5.9 and CE.1
-
oR346K (previously seen in VOC Mu/B.1.621) in BA.1.1
-
oR346S in BA.4.7 and BF.13
-
oR346T in BA.1.23, BA.2.9.4, BL.1, BL.2 BA.2.75.2, BM.1.1, BM.2, BM.4.1.1, BR.2, BN.1, BP.1, BA.2.75.6, BA.2.75.9, BA.2.80, BA.2.82, BA.4.1.8, BA.4.6, BF.7, BF.11 and BJ.
BA.4.6, BA.4.7, and BA.5.9 display higher humoral immunity evasion capability than BA.4/BA.5, causing 1.5–1.9-fold decrease in NT50 of the plasma from BA.1 and BA.2 breakthrough-infection convalescents compared to BA.4/BA.5. Importantly, plasma from BA.5 breakthrough-infection convalescents also exhibits significant neutralization activity decrease against BA.4.6, BA.4.7, and BA.5.9 than BA.4/BA.5, showing on average 2.4–2.6-fold decrease in NT50. R346S causes resistance to class 3 antibodies: bebtelovimab remains potent, while Evusheld™ is completely escaped by these subvariants (Jian et al., 2022).
-
o
-
•
K417T occurs in BA.2.18, BA.2.40.1 and BA.3.1. K417N previously occurred in VOC Beta and Delta “plus” (AY.1 and AY.2 sublineages) and in VOI Mu, while K417T occurred in VOC Gamma. Both mutations eliminate a hydrogen bond in the interaction with ACE2 thus reducing affinity (Villoutreix et al., 2021). Despite the loss in the binding affinity (1.48 kcal/mol, i.e. 6.4-fold drop (Wang et al., 2021b)) between RBD and ACE2 (Laffeber et al., 2021), the K417N/T mutations abolish a buried interfacial salt-bridge between RBD, and escapes neutralization by mAbs etesevimab (Luan and Huynh, 2021, Starr et al., 2021b, Wang et al., 2021b). However, these mutations have only a modest effect on binding by polyclonal antibodies in a few convalescent samples studied (Greaney et al., 2020). K417T leads to resistance to etesevimab (Luan and Huynh, 2021, Starr et al., 2021b, Wang et al., 2021b) and casirivimab (Hoffmann et al., 2021a), while K417R leads to resistance to the REGN-COV2 cocktail (Copin et al., 2021).
-
•K444 mutations were also exceedingly rare in the pre-Omicron era, while they occur in many Omicron sublineages as:
-
oK444M in BR.1.
-
oK444N in BA.2.38.1, BA.2.38.2 and BA.4.6.3
-
oK444T in BE.1.1.1, BA.5.6.2.1
-
oK444R in BF.16, BA.2.3.20 (nicknamed " Basilisk"). BA.5.2.18, and BE.4.1.1.
-
o
-
•L452 mutations have probably been the best example of convergent evolution so far, having been previously detected in multiple VOC (Focosi et al., 2022b).
-
oL452R does not have a major impact in ACE2 affinity when tested in the context of recombinant monomeric RBD but presents enhanced binding within the context of full-length membrane-anchored Spike (Gong et al., 2021). L452R was found in A.21, A.2.4, A.2.5, B.1.1.10, B.1.1.130, B.1.232 (a.k.a. CAL.20 A (Tchesnokova et al., 2021)), B.1.427/B.1.429 (a.k.a. CAL 0.20 C) (Zhang et al., 2021), B.1.617.1 and B.1.617.2 Delta VOCs (Cherian et al., 2021), B.1.362 +L452R, C.16 and C.36. L452R also causes evasion from HLA-A24-restricted CTL response (Motozono et al., 2021). Among Omicron sublineages, L452R is a universal hallmark of BA.4 and BA.5 and occurs in BA.2.11, BA.2.35, BA.2.75.4, BA.2.77, and CA.1
-
oL452Q increases ACE2 binding by 3-folds and in vitro infectivity by 2-folds (Tada et al., 2021b). Of interest, the C.37 VOI and a single B.1.74 strain harbors the L452Q mutation (Tchesnokova et al., 2021), . Among Omicron sublineages L452Q occurs in BA.2.12.1, BH.1, BA.2.75.7,
-
oL452M occurs in BA.2.9.1. BA.2.13, BA.2.56, BA.2.74, BP.1 and XBC.1
The effective reproduction numbers of these L452R/M/Q-bearing BA.2-related Omicron variants are greater than the original BA.2. Neutralization experiments revealed that the immunity induced by BA.1 and BA.2 infections is less effective against BA.4/5. Cell culture experiments showed that BA.2.12.1 and BA.4/5 replicate more efficiently in human alveolar epithelial cells than BA.2, and particularly, BA.4/5 is more fusogenic than BA.2. Furthermore, experimental infection in hamsters indicated that BA.4/5 was more pathogenic than BA.2 (Kimura et al., 2022). L452 mutations facilitate escape from some antibodies directed to the so-called Class 2 and Class 3 regions of the receptor-binding domain (RBD) (Wang et al., 2022a) ( Table 2).
-
o
-
•
E484 mutations were very common before Omicron, in the form of E484K (nicknamed "Eeek", found in VOC Beta and Gamma, several sublineages of VOC Alpha, B.1.1.33(E484K), VOI Eta and Zeta, B.1.220 from New York (Lesho et al., 2021), B.1.243.1 from Arizona (Skidmore et al., 2021), R.1 from Japan and USA (Hirotsu and Omata, 2021), and B.1.1.318 and B.1.621 lineages from UK) or E484Q (VOCs B.1.617.1 and B.1.617.3 (Cherian et al., 2021)). Both Omicron BA.1 and BA.2 instead show E484A, while BA.4/5 have wild-type E484.
-
•F486 mutations were also rare before Omicron, while nowadays occur across many Omicron sublineages:
-
oF486P in BA.2.10.4
-
oF486V in BA.4/5 (further changed to F486I in BR.2) , driving immune escape (Cao et al., 2022c, Khan et al., 2022, Tegally et al., 2022, Wang et al., 2022a).
-
oF486S in BA.2.75.2, BM.1 (a.k.a. BA.2.75.3.1), BM.4.1, BY.1, and BA.2.75.7
-
oF486L had been previously found in Dutch and US minks (Cai and Cai, 2021, Eckstrand et al., 2021, Lu et al., 2021).
-
o
-
•
R493Q occurs in BA.2.75, BA.2.77, BA.2.10.4 and BA.4/5. It is called a reversion, because prior to Omicron all previous VOCs had Q493, then BA.1 and BA.2 came with Q493R. Q493 mutations were exceedingly rare in other SARS-CoV-2 isolates, suggesting selective pressure rather than spontaneous convergent evolution. Q493E was formerly seen only in the Delta VOC sublineage AY.49, and Q493R was associated with resistance to both bamlanivimab and etesevimab mAbs (Starr et al., 2021a; Starr et al., 2021b; Wang et al., 2021a). The Q493R mutation emerged during treatment with this cocktail(Focosi et al., 2021; Guigon et al., 2021; Lohr et al., 2021; Peiffer-Smadja et al., 2021; Pommeret et al., 2021; Truffot et al., 2021; Vellas et al., 2021), while the Q493K mutation has emerged in a single patient after treatment with casirivimab plus imdevimab (Choi et al., 2020; Clark et al., 2021). Amazingly, Q493R was found in mice-adapted SARS-CoV-2 lineage, but infection of mice with wild-type SARS-CoV-2 was not possible before Omicron. In this regard, VOC Beta (Ravi et al., 2021) and Gamma (Montagutelli et al., 2021) could establish infection in wild-type mice, but with a milder infection than in human ACE2 transgenic mice (Tarres-Freixas et al., 2021). Omicron, because of its ability to infect mice (Bentley et al., 2021, Liu et al., 2022, Rissmann et al., 2022, Suryawanshi et al., 2022), was hypothesized to emerge from partially fit jump species(Wei et al., 2021), and reversions after widespread circulation in humans could support such hypothesis. The F486V mutation found in BA.4/5 facilitates escape from certain Class 1 and Class 2 antibodies to the RBD but compromises the Spike affinity for the cellular receptor ACE2, but the R493Q reversion mutation restores receptor affinity, and consequently, the fitness of BA.4/5 (Wang et al., 2022a).
Further examples of convergent evolution that are worth mentioning include:
-
•
W64 occurs as W64R in BA.2.9.3, BA.2.10.4, BA.2.61 and BA.2.71, while W64L occurs in BA.2.14 and BA.2.15.
-
•
I68T occurs in BA.2.10.3 and BA.2.36.
-
•
G181 mutates as G181E in BA.4.5 and as G181A in BF.6.
-
•
L212S occurs in BA.2.50 and in BA.2.72. Δ211/L212I was a defining mutation of BA.1
-
•
Y248 mutates as Y248H in BA.2.47, Y248S in BA.2.54, and as Y248N in BA.2.76
-
•
G339 mutations occurs as G339D in BA.1 and BA.2, and as G339H in BA.2.75
-
•
K356T is found in BA.2.75.5, BA.2.77 and BA.2.80
-
•
S371Y is found in both BA.2.43 and BA.2.44.
-
•
N440K was found in BA.1 and BA.2, and previously in B.1.1.420 (Garcia et al., 2021).
-
•
G446 mutations occur as G446V in BF.3.1 and as G446S in BA.2.75, BA.2.10.4 and BJ.1
-
•
Y449 mutates as Y449H in BA.2.45 and Y449N in BA.2.53
-
•
N450D occurs in BA.5.5.1 and BF.14
-
•
F490 mutations occur as F490V in BJ.1, F490S in BM.1.1.1, BN.1 and BN.2.1
-
•
S494P occurs in several Alpha VOC sublineages and in BA.2.10.4, BN.1.1 and CA.2 Omicron sublineages. It causes resistance to bamlanivimab (Chen et al., 2021).
-
•
T547I occurs in BA.5.2.4 and BF.3 (an alias for BA.5.2.4.3)
-
•
Q628K occurs in both BA.1.15.2 and BD.1 (an alias for BA.1.17.2.1)
-
•
S704 mutations occur as S704L in both BA.1.9 and BA.2.12, including BA.2.12.1
-
•
V1264L occurs in BG.2 (an alias for BA.12.2.1.2) and BA.2.58. It was previously found in AY.4.2.2 and sometimes in VOI kappa.
Fig. 1.
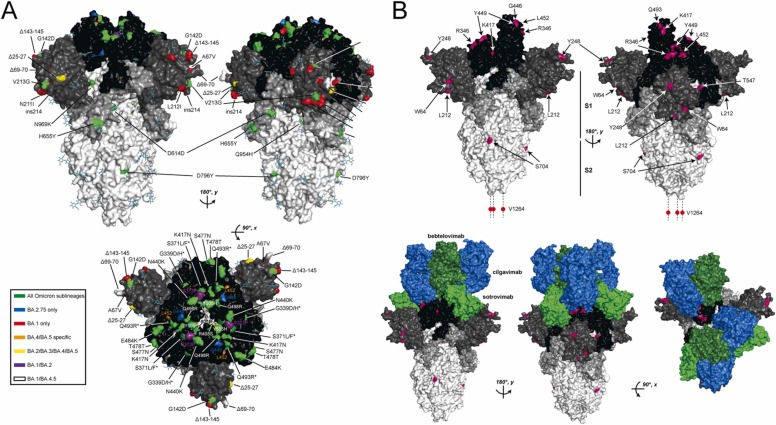
SARS-CoV-2 S protein mutations found in Omicron variant sublineages. A) Positions on the S protein where amino acid mutations are found in different Omicron sublineages are mapped to the spike trimer structure (PBD 6VXX). The S trimer is rotated 180 degrees along the y-axis to show both faces of the long axis and 90 degrees about the x-axis to display mutations on the receptor binding domains. Positions with amino acid mutations found in all Omicron sublineages are colored green. Positions that are BA.1 and BA.2.75 strain-specific are colored blue and red, respectively, and mutations found only in BA.4/BA.5, BA.2/BA.3/BA.4/BA.5, BA.1/BA.2 and BA.1/BA.2 are colored orange, yellow, purple and white, respectively. Positions where amino acid substitutions are different across different sublineages are indicated with an asterisk. B) Instances of convergent evolution in the Omicron sublineages are highlighted in magenta on the homotrimeric S protein structure (PBD 7C2L). The domain organization of S is highlighted with RBD, NTD, S1, and S2 subunits colored black, dark grey, light grey, and white, respectively. The homotrimer is rotated 180 degrees about the y-axis to show both faces. In the top panel, the sites of convergent evolution on the S homotrimer alone are indicated. In the bottom panel, the interfaces with clinically important mAbs from 3 different classes (Focosi et al., 2022c) (cilgavimab, bebtelovimab, and sotrovimab; colored blue, dark green and light green, respectively) are displayed to highlight the importance of the positions of convergent evolution in the Omicron S protein.
Table 2.
Efficacy of anti-Spike monoclonal antibodies against Omicron sublineages. PHE: Public Health England. VOC: Variant of concern. LUM: lineage under monitoring. = <1-fold reduced susceptibility; ↓: 1–5-fold reduced susceptibility; ↓↓: – 5–24.9-fold reduced susceptibility; ↓↓↓: – ≥ 25-fold reduced susceptibility.
Cao et al. demonstrated that such convergent mutations partly evade convalescent plasma, including those from BA.5 breakthrough infection, and totally evade authorized mAbs, including Evusheld™ and bebtelovimab. BA.2.75.2, BQ.1.1 and XBB are the most immune evasive strains tested so far. BA.2 and especially BA.5 breakthrough infection caused significant reductions of nAb epitope diversity and increased proportion of non-neutralizing mAbs, which in turn concentrated humoral immune pressure and promoted the convergent RBD evolution (Cao et al., 2022a).
5.2. Escape to anti-Spike monoclonal antibodies
The most concerning implication of Omicron evolution so far has been the progressive escape to vaccine-elicited immunity and anti-Spike monoclonal antibody (mAb) therapies (Table 1, Table 2), including representatives (Focosi et al., 2022c). Models have been developed to predict polyclonal antibody binding to Omicron sublineages remaining after mutating one or more sites in the SARS-CoV-2 RBD, based on deep mutational scanning of RBD targeting antibodies (e.g.: https://jbloomlab.github.io/SARS2_RBD_Ab_escape_maps/escape-calc/ (Greaney et al., 2022)), but to date most interest focuses on mAb efficacy.
Eli Lilly’s bamlanivimab and etesevimab never worked against any Omicron VOC or VOC-LUM, as well as Roche’s casirivimab and imdevimab (Ronapreve™). Sotrovimab worked against BA.1, but lost efficacy against BA.2 and BA.4/5. Similarly, the tixagevimab component of the Evusheld™ mAb cocktail has not ever worked against any Omicron VOC or VOC-LUM: the remaining efficacy being based on the cilgavimab component only. Cilgavimab did not work against BA.1, recovered efficacy against BA.2, but there are in vitro evidences of 30-fold reduced efficacy against BA.4/5, and totally lost efficacy against the above-mentioned R346X harboring sublineages (see Table 1, Table 2). Despite attempts at increasing the therapeutic dose (AstraZeneca, 2022, FDA, 2022), to date there is no clearcut evidence of clinical benefit from Evusheld™ at the time of BA.4/5.
No authorized mAb other than bebtelovimab has convincing in vitro activity against BA.4/5 and BA.2.75.2 (Sheward et al., 2022b), and bebtelovimab has anyway reduced activity against BA.2.75 (Yamasoba et al., 2022a): being administered as a monotherapy and based on previous experiences with other mAb monotherapies (Huygens et al., 2022, Rockett et al., 2021), the likelihood of treatment-emergence immune escape is around 10% (Focosi et al., 2022c). Lack of actions by regulatory authorities at withdrawing authorization for mAbs that were no longer effective or troubles in medical education at the time of a pandemic have often driven inappropriate prescription of mAbs (Anderson et al., 2022, Focosi and Tuccori, 2022), with detrimental consequences on both healthcare budgets and public health.
Most importantly, Omicron has severely hit the pipeline of mAbs that were under advanced stages of development, such as amubarvimab and romlusevimab, as well as adintrevimab. So far, among the advanced candidates, only P5C3 and P2G3 retain efficacy against BA.4/5.
6. Perspective
The progressive emergence of the Omicron sublineages has now defeated most, if not all, authorized mAb therapies. Whereas the epitopes recognized by these mAbs were obliterated by Spike protein substitutions, those same protein changes created new epitopes that could in theory be targeted by new mAbs. Furthermore, it may still be possible to find some conserved protein regions that could be targeted in new mAb development. Hence, new mAb therapies remain feasible but the problems with developing new mAb therapeutics are time and economics. Developing new mAb therapies takes at least several months and given the history of variant generation it is unlikely that the virus will be antigenically stable during that time. Even if a new mAb maintains activity during development time the experience detailed above means that the period of therapeutic usefulness will be short, which greatly reduces the incentives of pharmaceutical companies to invest in new reagents. The creation of mAb cocktails could prolong the useful life of a mAb product, but combining immunoglobulins increases costs and the experience with cocktails has shown that they are also fully vulnerable to new variants. So, while the future of mAb therapies against SARS-CoV-2 is uncertain there is one antibody-based therapy that keeps up with the variants – COVID-19 convalescent plasma (CCP). In the first year of the pandemic CCP was deployed in the USA where over half a million patients were treated and there was a strong inverse correlation between population usage and mortality, suggesting that it averted about 100,000 deaths (Casadevall et al., 2021). However, several randomized controlled trials showed no effect in mortality, an effect that could be explained by their design, which in most instances involved late use with units of insufficient antibody titer (Focosi et al., 2022a). This led to the abandonment of CCP but more recent work showing efficacy when used early (Sullivan et al., 2021): but the loss of mAbs therapies, the finding that CCP from donors who had both COVID-19 and vaccination has very high titers that neutralized all VOCs (Sullivan et al., 2022), and the need for antibody-based therapies in immunocompromised patients have reinvigorated interest in CCP. In contrast to mAbs, CCP is a polyclonal preparation including antibodies with many specificities and representing all isotypes, making it much less vulnerable to defeat by any single variant. As the pandemic evolves the immunocompromised are emerging as a population at great risk in whom SARS-CoV-2 infection can become chronic providing an opportunity for evolution of further variants. CCP appears to be effective in such patients and is currently undergoing a renaissance in use as it remains the only antibody-based therapy capable to neutralize all variants.
7. Conclusions
Omicron sublineages have represented a major turning point in the pandemic (Dhawan et al., 2022). While small molecule antivirals have preserved efficacy, mAbs have been defeated by immune escape, hitting manufacturer confidence with development plans. In the field of passive immunotherapies, CCP is likely to remain a pillar for immunocompromised patients that get infected with novel Omicron sublineages. Additionally, the design of upcoming Omicron-based vaccines (either monovalent or bivalent) will be a hard exercise, given the myriad of diverse sublineages that are circulating at this time and the rapidity with which the landscape is evolving. Ongoing genomic surveillance will be required in the coming years to promptly interecept emerging sublineages with growth advantages and dissect their immune escape abilities, in order to predict waves and update vaccines.
Author contributions
D.F. wrote the firstdraft and curated Tables, S.M. designed Figure, A.C. and S.M. revised themanuscript.
Conflict of Interest
We declare we have no conflict of interest related to this manuscript.
Acknowledgements
D.F. is grateful to Federico Gueli for help with recently designated Omicron sublineages.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.drup.2022.100882.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.drup.2022.100882.
Appendix A. Supplementary material
Supplementary material
.
Appendix B. Supplementary material
Supplementary material
.
References
- A. Aggarwal, A. Ospina Stella, G. Walker, A. Akerman, V. Milogiannakis, A.C. Hoppe, V. Mathivanan, C. Fichter, S. McAllery, S. Amatayakul-Chantler, N. Roth, G. Coppola, M.L. Munier, D.R. Darley, D.S. Khoury, C.S.P. Foster, Y. Lu, P. Schofield, J. Jackson, J. Henry, O. Mazigi, H.-M. Jaeck, D. Langles, D. Cromer, M.P. Davenport, D. Christ, G. Matthews, W. Rawlinson, A.D. Kelleher and S.G. Turville, , 2021. SARS-CoV-2 Omicron: reduction of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern, (2021), p. 2021.2012.2014.21267772.
- Aggarwal A., Akerman A., Milogiannakis V., Silva M.R., Walker G., Kidinger A., Angelovich T., Waring E., Amatayakul-Chantler S., Roth N., Coppola G., Yeang M., Jean T., Foster C., Hoppe A.C., Ling Munier C.M., Darley D., Churchill M., Starck D., Christ D., Matthews G., Rawlinson W., Kelleher A.D., Turville S. SARS-CoV-2 Omicron BA.5: Evolving tropism and evasion of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern. EBioMedicine. 2022 doi: 10.1016/j.ebiom.2022.104270. p. 2022.2007.2007.22277128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson T.S., O’Donoghue A., Mechanic O., Dechen T., Stevens J. Administration of Anti–SARS-CoV-2 monoclonal antibodies after us food and drug administration deauthorization. JAMA Netw. Open. 2022;5 doi: 10.1001/jamanetworkopen.2022.28997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AstraZeneca, 2022. Updated EVUSHELD Emergency Use Authorization (EUA) Dosage Recommendations for Patients Who Received an Initial Dose of 150 mg tixagevimab and 150 mg cilgavimab. Accessed online at 〈https://www.fda.gov/media/156617/download〉 on 17 August 2022.
- A. Bal, G. Destras, A. Gaymard, H. Regue, Q. Semanas, C. d'Aubarde, G. Billaud, F. Laurent, C. Gonzales, M. Valette, M. Bouscambert, B. Lina, F. Morfin, L. Josset, 2020. Two-step strategy for the identification of SARS-CoV-2 variants co-occurring with spike deletion H69-V70, Lyon, France, August to December 2020, (2020), p. 2020.2011.2010.20228528.
- Boschi C., Colson P., Bancod A., Moal V., La B. Scola, Omicron Variant Escapes Therapeutic Monoclonal Antibodies (mAbs) Including Recently Released Evusheld®, Contrary to 8 Prior Main Variant of Concern (VOC) Clin. Infect. Dis.: Off. Publ. Infect. Dis. Soc. Am. 2022;75:e534–e535. doi: 10.1093/cid/ciac143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- B. Brejová, V. Hodorová, K. Boršová, V. Čabanová, L. Reizigová, E. Paul, P. Čekan, B. Klempa, J. Nosek, T. Vinař, 2021. B.1.258∆, a SARS-CoV-2 variant with ∆H69/∆V70 in the Spike protein circulating in the Czech Republic and Slovakia. (2021). [DOI] [PMC free article] [PubMed]
- Bruel T., Hadjadj J., Maes P., Planas D., Seve A., Staropoli I., Guivel-Benhassine F., Porrot F., Bolland W.-H., Nguyen Y., Casadevall M., Charre C., Péré H., Veyer D., Prot M., Baidaliuk A., Cuypers L., Planchais C., Mouquet H., Baele G., Mouthon L., Hocqueloux L., Simon-Loriere E., André E., Terrier B., Prazuck T., Schwartz O. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat. Med. 2022 doi: 10.1038/s41591-022-01792-5. [DOI] [PubMed] [Google Scholar]
- Cao Y., Jian F., Wang J., Yu Y., Song W., Yisimayi A., Wang J., An R., Zhang N., Wang Y., Wang P., Zhao L., Sun H., Yu L., Yang S., Niu X., Xiao T., Gu Q., Shao F., Hao X., Xu Y., Jin R., Wang Y., Xie X.S. Impr. SARS-CoV-2 Humor Immun. induces Converg. Omicron RBD Evol. 2022 p. 2022.2009.2015.507787. [Google Scholar]
- Cao Y.R., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., Wang J., Wang Y., Niu X., Yang S., Liang H., Sun H., Li T., Yu Y., Cui Q., Liu S., Yang X., Du S., Zhang Z., Hao X., Shao F., Jin R., Wang X., Xiao J., Wang Y., Xie X.S. Omicron escapes the majority of SARS-CoV-2 neutralizing antibodies of diverse epitopes. Nature. 2021;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.R., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., Wang P., Zhang Z., Liu P., An R., Hao X., Wang Y., Wang J., Feng R., Sun H., Zhao L., Zhang W., Zhao D., Zheng J., Yu L., Li C., Zhang N., Wang R., Niu X., Yang S., Song X., Zheng L., Li Z., Gu Q., Shao F., Huang W., Jin R., Shen Z., Wang Y., Wang X., Xiao J., Xie X.S. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022 doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Dragotakes Q., Johnson P.W., Senefeld J.W., Klassen S.S., Wright S.R., Joyner M.J., Paneth N., Carter R. Convalescent plasma use in the United States was inversely correlated with COVID-19 mortality: did convalescent plasma hesitancy cost lives? eLife. 2021;4 doi: 10.7554/eLife.69866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Gao K., Wang R., Wei G.-W. Revealing the threat of emerging SARS-CoV-2 mutations to antibody therapies. bioRxiv. 2021 doi: 10.1016/j.jmb.2021.167155. p. 2021.2004.2012.439473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., Das S., Agarwal A., Singh S., Abraham P., Panda S., Mande S., Swarup R., Bhargava B., Bhushan R. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. bioRxiv. 2021 doi: 10.3390/microorganisms9071542. p. 2021.2004.2022.440932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.-H., Boucau J., Bowman K., Adhikari U.D., Winkler M.L., Mueller A.A., Hsu T.Y.-T., Desjardins M., Baden L.R., Chan B.T., Walker B.D., Lichterfeld M., Brigl M., Kwon D.S., Kanjilal S., Richardson E.T., Jonsson A.H., Alter G., Barczak A.K., Hanage W.P., Yu X.G., Gaiha G.D., Seaman M.S., Cernadas M., Li J.Z. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.A., Clark L.E., Pan J., Coscia A., McKay L.G.A., Shankar S., Johnson R.I., Brusic V., Choudhary M.C., Regan J., Li J.Z., Griffiths A., Abraham J. SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell. 2021;184:2605–2617. doi: 10.1016/j.cell.2021.03.027. e2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Delerce J., Marion-Paris E., Lagier J.C., Levasseur A., Fournier P.E., La Scola B., Raoult D. A 21L/BA.2–21 K/BA.1 "MixOmicron" SARS-CoV-2 hybrid undetected by qPCR that screen for variant in routine diagnosis. Infect., Genet. Evol.: J. Mol. Epidemiol. Evolut. Genet. Infect. Dis. 2022;105 doi: 10.1016/j.meegid.2022.105360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combes P., Bisseux M., Bal A., Marin P., Latour J., Archimbaud C., Brebion A., Chabrolles H., Regagnon C., Lafolie J., Destras G., Simon B., Izopet J., Laurence J., Henquell C., Mirand A. Evidence of co-infections during Delta and Omicron SARS-CoV-2 variants co-circulation through prospective screening and sequencing. Clin. Microbiol. Infect.: Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2022 doi: 10.1016/j.cmi.2022.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copin R., Baum A., Wloga E., Pascal K.E., Giordano S., Fulton B.O., Zhou A., Negron N., Lanza K., Chan N., Coppola A., Chiu J., Ni M., Atwal G.S., Romero Hernandez A., Saotome K., Zhou Y., Franklin M.C., Hooper A.T., McCarthy S., Hamon S., Hamilton J.D., Staples H.M., Alfson K., Carrion R., Ali S., Norton T., Somersan-Karakaya S., Sivapalasingam S., Herman G.A., Weinreich D.M., Lipsich L., Stahl N., Murphy A.J., Yancopoulos G.D., Kyratsous C.A. The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies. Cell. 2021;184:3949–3961. doi: 10.1016/j.cell.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan M., Saied A.A., Mitra S., Alhumaydhi F.A., Emran T.B., Wilairatana P. Omicron variant (B.1.1.529) and its sublineages: What do we know so far amid the emergence of recombinant variants of SARS-CoV-2? Biomed. Pharmacother. 2022;154 doi: 10.1016/j.biopha.2022.113522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E. Andreano, I. Paciello, G. Pierleoni, G. Maccari, G. Antonelli, V. Abbiento, P. Pileri, L. Benincasa, G. Giglioli, G. Piccini, C. De Santi, C. Sala, D. Medini, E. Montomoli, P. Maes and R. Rappuoli, mRNA vaccines and hybrid immunity use different B cell germlines to neutralize Omicron BA.4 and BA.5, (2022), p. 2022.2008.2004.502828.
- E.G. Bentley, A. Kirby, P. Sharma, A. Kipar, D.F. Mega, C. Bramwell, R. Penrice-Randal, T. Prince, J.C. Brown, J. Zhou, G.R. Screaton, W.S. Barclay, A. Owen, J.A. Hiscox and J.P. Stewart, 2021. SARS-CoV-2 Omicron-B.1.1.529 Variant leads to less severe disease than Pango B and Delta variants strains in a mouse model of severe COVID-19, (2021), p. 2021.2012.2026.474085.
- Eckstrand C., Baldwin T., Torchetti M.K., Killian M.L., Rood K.A., Clayton M., Lott J.A., Wolking R.M., Bradway D.S., Baszler T. An outbreak of SARS-CoV-2 with high mortality in mink (Neovison vison) on multiple Utah farms, PLoS Pathog. 2021 doi: 10.1371/journal.ppat.1009952. p. 2021.2006.2009.447754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA, 2022. FDA authorizes revisions to Evusheld dosing. Accessed online on April 29, 2022 at 〈https://www.fda.gov/drugs/drug-safety-and-availability/fda-authorizes-revisions-evusheld-dosing〉, (2022).
- Focosi D., Maggi F. Recombination in coronaviruses, with a focus on SARS-CoV-2. Viruses. 2022;14:1239. doi: 10.3390/v14061239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focosi D., Tuccori M. Prescription of anti-spike monoclonal antibodies in COVID-19 patients with resistant SARS-CoV-2 variants in Italy. Pathog. (Basel, Switz. ) 2022;11 doi: 10.3390/pathogens11080823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focosi D., Novazzi F., Genoni A., Dentali F., Dalla Gasperina D., Baj A., Maggi F. Emergence of SARS-CoV-2 Spike Escape Mutation Q493R After Treatment for COVID-19. Emerg. Infect. Dis. 2021;27 doi: 10.3201/eid2710.211538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focosi D., Franchini M., Pirofski L.A., Burnouf T., Paneth N., Joyner M.J., Casadevall A. COVID-19 convalescent plasma and clinical trials: understanding conflicting outcomes. Clin. Microbiol. Rev. 2022 doi: 10.1128/cmr.00200-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focosi D., Maggi F., McConnell S., Casadevall A. Spike mutations in SARS-CoV-2 AY Sublineages of Delta variant of concern: implications for the future of the pandemic. Future Microbiol. 2022;17:219–221. doi: 10.2217/fmb-2021-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focosi D., McConnell S., Casadevall A., Cappello E., Valdiserra G., Tuccori M. Monoclonal antibody therapies against SARS-CoV-2. Lancet Infect. Dis. 2022;22:00311–00315. doi: 10.1016/S1473-3099(22)00311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V., Vig V., Peillard L., Ramdani A., Mohamed S., Halfon P. First description of two immune escape indian B.1.1.420 and B.1.617.1 SARS-CoV2 variants in France. bioRxiv. 2021 p. 2021.2005.2012.443357. [Google Scholar]
- Gjorgjievska M., Mehandziska S., Stajkovska A., Pecioska-Dokuzovska S., Dimovska A., Durmish I., Ismail S., Pavlovska T., Stojchevska A., Amedi H., Andonova J., Nikolovska M., Velickovikj S., Mitrev Z., Kungulovski I., Kungulovski G. Case report: omicron BA.2 subvariant of SARS-CoV-2 outcompetes BA.1 in two co-infection cases. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.892682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S.Y., Chatterjee D., Richard J., Prevost J., Tauzin A., Gasser R., Bo Y., Vezina D., Goyette G., Gendron-Lepage G., Medjahed H., Roger M., Cote M., Finzi A. Contribution of single mutations to selected SARS-CoV-2 emerging variants spike antigenicity. Virology. 2021 doi: 10.1016/j.virol.2021.09.001. p. 2021.2008.2004.455140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravagnuolo A.M., Faqih L., Cronshaw C., Wynn J., Burglin L., Klapper P., Wigglesworth M. Epidemiol. Investig. N. SARS-CoV-2 Var. Concern 202012/01 Engl. 2021 p. 2021.2001.2014.21249386. [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., Dingens A.S., Nargi R.S., Sutton R.E., Suryadevara N., Rothlauf P.W., Liu Z., Whelan S.P.J., Carnahan R.H., Crowe J.E., Jr., Bloom J.D. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Bloom J.D. An antibody-escape estimator for mutations to the SARS-CoV-2 receptor-binding domain. Virus Evol. 2022;8 doi: 10.1093/ve/veac021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Schommers P., Lehmann C., Kurth F., Sander L.E., Klein F. mRNA Boost. Immun. elicits potent neutralizing Serum Act. SARS-CoV-2 Omicron Var. 2021 doi: 10.1038/s41591-021-01676-0. p. 2021.2012.2014.21267769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Sander L.E., Kurth F., Klein F. Neutralization sensitivity of the SARS-CoV-2 Omicron BA.2.75 sublineage. Lancet Infect. Dis. 2022 doi: 10.1016/S1473-3099(22)00580-1. p. 2022.2008.2004.502609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigon A., Faure E., Lemaire C., Chopin M., Tinez C., Assaf A., Lazrek M., Hober D., Bocket L., Engelmann I., Kazali E., Alidjinou E.K. Emergence of Q493R mutation in SARS-CoV-2 spike protein during bamlanivimab/etesevimab treatment and resistance to viral clearance. J. Infect. 2021;S0163–4453:00435–00437. doi: 10.1016/j.jinf.2021.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H.Y., Cai A. SARS-CoV-2 spike protein gene variants with N501T and G142D mutation dominated infections in minks in the US. J. Vet. Diagn. Investig. 2021 doi: 10.1177/10406387211023481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Omata M. Household transmission of SARS-CoV-2 R.1 lineage with spike E484K mutation in Japan. medRxiv. 2021 p. 2021.2003.2016.21253248. [Google Scholar]
- Hoffmann M., Arora P., Gross R., Seidel A., Hoernich B., Hahn A., Krueger N., Graichen L., Hofmann-Winkler H., Kempf A., Winkler M.S., Schulz S., Jaeck H.-M., Jahrsdoerfer B., Schrezenmeier H., Mueller M., Kleger A., Muench J., Poehlmann S. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Krüger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.-S., Winkler M.S., Lier M., Dopfer-Jablonka A., Jäck H.-M., Behrens G., Pöhlmann S. The Omicron variant is highly resistant against antibody-mediated neutralization - implications for control of the COVID-19 pandemic. Cell. 2021 doi: 10.1016/j.cell.2021.12.032. p. 2021.2012.2012.472286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huygens S., Oude Munnink B., Gharbharan A., Koopmans M., Rijnders B. High incidence of sotrovimab resistance and viral persistence after treatment of immunocompromised patients infected with the SARS-CoV-2 Omicron variant. medRxiv. 2022 doi: 10.1093/cid/ciac601. p. 2022.2004.2006.22273503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura N., Hoshino A., Higuchi Y., Taminishi S., Inaba T., Matoba S. SARS-CoV-2 Omicron variant escapes neutralization by vaccinated and convalescent sera and therapeutic monoclonal antibodies. medRxiv. 2021 p. 2021.2012.2013.21267761. [Google Scholar]
- Iketani S., Liu L., Guo Y., Liu L., Huang Y., Wang M., Luo Y., Yu J., Yin M.T., Sobieszczyk M.E., Huang Y., Wang H.H., Sheng Z., Ho D.D. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian F., Yu Y., Song W., Yisimayi A., Yu L., Gao Y., Zhang N., Wang Y., Shao F., Hao X., Xu Y., Jin R., Wang Y., Xie X.S., Cao Y. Furth. Humor Immun. Evas. Emerg. SARS-CoV-2 Ba. 4 Ba. 5 subvariants. 2022 doi: 10.1016/S1473-3099(22)00642-9. p. 2022.2008.2009.503384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan K., Karim F., Ganga Y., Bernstein M., Jule Z., Reedoy K., Cele S., Lustig G., Amoako D., Wolter N., Samsunder N., Sivro A., San J.E., Giandhari J., Tegally H., Pillay S., Naidoo Y., Mazibuko M., Miya Y., Ngcobo N., Manickchund N., Magula N., Abdool Karim Q., von Gottberg A., Abdool Karim S., Hanekom W., Gosnell B., Team C.-K., Lessells R., de Oliveira T., Moosa Y., Sigal A. Omicron sub-Linea Ba. 4/Ba. 5 Escape Ba. 1 Infect. elicited neutralizing Immun. 2022 p. 2022.2004.2029.22274477. [Google Scholar]
- Kimura, I., Yamasoba, D. , Tamura, T. , Nao, N., Oda, Y., Mitoma, S., Ito, J., Nasser, H., Zahradnik, J., Uriu, K., Fujita, S., Kosugi, Y., Wang, L., Tsuda, M., Kishimoto, M., Ito, H., Suzuki, R., Shimizu, R., Begum, M.M., Yoshimatsu, K., Sasaki, J., Sasaki-Tabata, K., Yamamoto, Y., Nagamoto, T., Kanamune, J., Kobiyama, K., Asakura, H., Nagashima, H. M., Sadamasu, K. , Yoshimura, K., Kuramochi, J., Schreiber, G., Ishii, K.J., Hashiguchi, T., The Genotype to Phenotype Japan Consortium, Ikeda, T., Saito, A., Fukuhara, T., Tanaka, S., Matsuno, K., and Sato, K., 2022. Virological characteristics of the novel SARS-CoV-2 Omicron variants including BA.2.12.1, BA.4 and BA. 5, (2022), p. 2022.2005.2026.493539.
- Laffeber C., de Koning K., Kanaar R., Lebbink J. Exp. Evid. Enhanc. Recept. Bind. rapidly Spread SARS-CoV-2 Var. 2021 p. 2021.2002.2022.432357. [Google Scholar]
- Larsen B., Worobey M. Identif. a Nov. SARS-CoV-2 Spike 69-70 deletion Linea Circ. U. S. 2021 [Google Scholar]
- Lesho E., Corey B., Lebreton F., Ong A., Swierczewski B., Bennett J., Walsh E., McGann P. Émerg. E484K Mutat. SARS-CoV-2 Linea B. 1. 1. 220 Upstate N. Y. 2021 p. 2021.2003.2011.21253231. [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F.-W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., Yu J., Chik K.K.-H., Yuen T.T.-T., Yoon C., To K.K.-W., Chen H., Yin M.T., Sobieszczyk M.E., Huang Y., Wang H.H., Sheng Z., Yuen K.-Y., Ho D.D. Striking Antib. Evas. Manif. Omicron Var. SARS-CoV. 2021;2 p. 2021.2012.2014.472719. [Google Scholar]
- Liu S., Selvaraj P., Sangare K., Luan B., Wang T.T. Spike Protein-Indep. Attenuation SARS-CoV-2 Omicron Var. Lab. Mice. 2022 doi: 10.1016/j.celrep.2022.111359. p. 2022.2002.2008.479543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr B., Niemann D., Verheyen J. Bamlanivimab treatment leads to rapid selection of immune escape variant carrying E484K mutation in a B.1.1.7 infected and immunosuppressed patient. Clin. Infect. Dis.: Off. Publ. Infect. Dis. Soc. Am. 2021 doi: 10.1093/cid/ciab392. [DOI] [PubMed] [Google Scholar]
- Lu L., Sikkema R.S., Velkers F.C., Nieuwenhuijse D.F., Fischer E.A.J., Meijer P.A., Bouwmeester-Vincken N., Rietveld A., Wegdam-Blans M.C.A., Tolsma P., Koppelman M., Smit L.A.M., Hakze-van der Honing R.W., van der Poel W.H.M., van der Spek A.N., Spierenburg M.A.H., Molenaar R.J., de Rond J., Augustijn M., Woolhouse M., Stegeman A.J., Lycett S., Oude Munnink B.B., Koppelman M.P.G. Adapt., Spread Transm. SARS-CoV-2 Farm. minks Relat. Hum. Neth. 2021 p. 2021.2007.2013.452160. [Google Scholar]
- Luan B., Huynh T. Insights SARS-CoV-2'S. Mutat. Evading Hum. Antibodies: Sacrif. Surviv. 2021 p. 2021.2002.2006.430088. [Google Scholar]
- Montagutelli X., Prot M., Levillayer L., Baquero Salazar E., Jouvion G., Conquet L., Donati F., Albert M., Gambaro F., Behillil S., Enouf V., Rousset D., Jaubert J., Rey F., van der Werf S., Simon-Loriere E. B1. 351 P. 1 Var. extend SARS-CoV-2 host range mice. 2021 p. 2021.2003.2018.436013. [Google Scholar]
- Moreno G., Braun K., Larsen B., Alpert T., Worobey M., Grubaugh N., Friedrich T., O’Connor D., Fauver J., Brito A. Detect. Non-B. 1. 1. 7 Spike ∆69/70 Seq. (B. 1. 375) U. S. 2021 [Google Scholar]
- Motozono C., Toyoda M., Zahradnik J., Ikeda T., Saito A., Tan T.S., Ngare I., Nasser H., Kimura I., Uriu K., Kosugi Y., Torii S., Yonekawa A., Shimono N., Nagasaki Y., Minami R., Toya T., Sekiya N., Fukuhara T., Matsuura Y., Schreiber G., Nakagawa S., Ueno T., Sato K. Emerg. SARS-CoV-2 Mutant evading Cell. Immun. increasing Viral Infect. 2021 p. 2021.2004.2002.438288. [Google Scholar]
- Ohashi H., Hishiki T., Akazawa D., Kim K.S., Woo J., Shionoya K., Tsuchimoto K., Iwanami S., Moriyama S., Kinoshita H., Yamada S., Kuroda Y., Yamamoto T., Kishida N., Watanabe S., Hasegawa H., Ebihara H., Suzuki T., Maeda K., Fukushi S., Takahashi Y., Iwami S., Watashi K. Differ. Effic. neutralizing antibodies Antivir. Drugs SARS-CoV-2 Omicron subvariants, Ba. 1 Ba. 2. 2022 doi: 10.1016/j.antiviral.2022.105372. p. 2022.2002.2027.482147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer-Smadja N., Bridier-Nahmias A., Ferré V.M., Charpentier C., Garé M., Rioux C., Allemand A., Lavallée P., Ghosn J., Kramer L., Descamps D., Yazdanpanah Y., Visseaux B. Emergence of E484K mutation following bamlanivimab monotherapy among high-risk patients infected with the Alpha Variant of SARS-CoV-2. Viruses. 2021;13 doi: 10.3390/v13081642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Benhassine F.G., Planchais C., Porrot F., Staropoli I., Lemoine F., Pere H., Veyer D., Puech J., Rodary J., Bolland W.H., Buchrieser J., Baele G., Dellicour S., Raymenants J., Gorissen S., Geenen C., Vanmechelen B., Wawina T., Marti J., Cuypers L., Seve A., Hocqueloux L., Prazuck T., Loriere E.S., REY F., Bruel T., Mouquet H., Andre E., Schwartz O. Considerable Escape SARS-CoV-2 Var. Omicron Antib. neutralization. 2021 p. 2021.2012.2014.472630. [Google Scholar]
- Pommeret F., Colomba J., Bigenwald C., Laparra A., Bockel S., Bayle A., Michot J.M., Hueso T., Albiges L., Tiberghien P., Marot S., Jary A., Lacombe K., Barlesi F., Griscelli F., Colomba E. Bamlanivimab+ etesevimab therapy induces SARS-CoV-2 immune escape mutations and secondary clinical deterioration in COVID-19 patients with B-cell malignancies. Ann. Oncol. 2021 doi: 10.1016/j.annonc.2021.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi K., Lauri K., Teemu S., Tobias L.F., Sawan Kumar J., Kari A., Seppo M., Tarja S., Kalle S., Tomas S., Anja K., Olli V. Common laboratory mice are susceptible to infection with SARS-CoV2 beta variant. Res. Sq. 2021 [Google Scholar]
- Rissmann M., Noack D., van Riel D., Schmitz K.S., de Vries R.D., van Run P., Lamers M.M., GeurtsvanKessel C., Koopmans M., Fouchier R., Kuiken T., Haagmans B., Rockx B. Pulm. Lesions inoculation SARS-CoV-2 Omicron Ba. 1 (B. 1. 1. 529) Var. Syr. Gold. Hamst. 2022 doi: 10.1080/22221751.2022.2095932. p. 2022.2003.2015.484448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockett R.J., Basile K., Maddocks S., Fong W., Agius J.E., Johnson-Mackinnon J., Arnott A., Chandra S., Gall M., Draper J.L., Martinez E., Sim E.M., Lee C., Ngo C., Ramsperger M., Ginn A.N., Wang Q., Fennell M., Ko D., Lim L., Gilroy N., Sullivan M.V., Chen S.C.-A., Kok J., Dwyer D.E., Sintchenko V.L. Resistance mutations in SARS-CoV-2 Delta variant after sotrovimab use. N. Engl. J. Med. 2021 doi: 10.1056/NEJMc2120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Tamura T., Zahradnik J., Deguchi S., Tabata K., Kimura I., Ito J., Nasser H., Toyoda M., Nagata K., Uriu K., Kosugi Y., Fujita S., Yamasoba D., Shofa M., Begum M.M., Oda Y., Suzuki R., Ito H., Nao N., Wang L., Tsuda M., Yoshimatsu K., Yamamoto Y., Nagamoto T., Asakura H., Nagashima M., Sadamasu K., Yoshimura K., Ueno T., Schreiber G., Takaori-Kondo A., Shirakawa K., Sawa H., Irie T., Takayama K., Matsuno K., Tanaka S., Ikeda T., Fukuhara T., Sato K. Virol. Charact. SARS-CoV-2 Omicron Ba. 2. 75. 2022 doi: 10.1016/j.chom.2022.10.003. p. 2022.2008.2007.503115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrill-Mix S., Van Duyne G.D., Bushman F.D. Mol. Beac. Allow Specif. RT-LAMP Detect. B. 1. 1. 7 Var. SARS-CoV-2. 2021 doi: 10.7171/jbt.21-3203-004. p. 2021.2003.2025.21254356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheward D.J., Kim C., Ehling R.A., Pankow A., Castro Dopico X., Martin D.P., Reddy S.T., Dillner J., Karlsson Hedestam G.B., Albert J., Murrell B. Var. loss Antib. potency SARS-CoV-2 B. 1. 1. 529 (Omicron) 2021 doi: 10.1016/S1473-3099(22)00129-3. p. 2021.2012.2019.473354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheward D.J., Kim C., Fischbach J., Muschiol S., Ehling R.A., Björkström N.K., Karlsson Hedestam G.B., Reddy S.T., Albert J., Peacock T.P., Murrell B. Evas. neutralizing antibodies Omicron Subline Ba. 2. 75. 2022 doi: 10.1016/S1473-3099(22)00524-2. p. 2022.2007.2019.500716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheward D.J., Kim C., Fischbach J., Muschiol S., Ehling R.A., Björkström N.K., Karlsson Hedestam G.B., Reddy S.T., Albert J., Peacock T.P., Murrell B. Omicron Subline Ba. 2. 75. 2 Exhib. extensive Escape neutralising antibodies. 2022 doi: 10.1016/S1473-3099(22)00524-2. p. 2022.2009.2016.508299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skidmore P.T., Kaelin E.A., Holland L.A., Maqsood R., Wu L.I., Mellor N.J., Blain J.M., Harris V., LaBaer J., Murugan V., Lim E.S. Émerg. a SARS-CoV-2 E484K Var. Interest Ariz. 2021 doi: 10.3201/eid2710.211189. p. 2021.2003.2026.21254367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021:eabf9302. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Dingens A.S., Bloom J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cel. Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano A., Takaoka Y., Kataguchi H., Kumaoka M., Ohta M., Kimura S., Araki M., Morinaga Y., Yamamoto Y. SARS-CoV-2 Omicron Ba. 2. 75 Var. may be much more Infect. preexisting Var. 2022 doi: 10.3390/microorganisms10102090. p. 2022.2008.2025.505217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan D., Gebo K., Shoham S., Bloch E., Lau B., Shenoy A., Mosnaim G., Gniadek T., Fukuta Y., Patel B., Heath S., Levine A., Meisenberg B., Spivak E., Anjan S., Huaman M., Blair J., Currier J., Paxton J., Gerber J., Petrini J., Broderick P., Rausch W., Cordisco M., Greenblatt H.J.B., Cluzet V., Cruser D., Oei K., Abinante M., Hammitt L., Sutcliffe C., Forthal D., Zand M., Cachay E., Raval J., Kassaye S., Foster E., Roth M., Marshall C., Yarava A., Lane K., McBee N., Gawad A., Karlen N., Singh A., Ford D., Jabs D., Appel L., Shade D., Ehrhardt S., Baksh S., Laeyendecker O., Pekosz A., Klein S., Casadevall A., Tobian A., Hanley D. Early outpatient treatment for Covid-19 with convalescent plasma. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2119657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan D.J., Franchini M., Joyner M.J., Casadevall A., Focosi D. Anal. anti-Omicron neutralizing Antib. titers Differ. Conval. Plasma Sources. 2022 doi: 10.1038/s41467-022-33864-y. p. 2021.2012.2024.21268317v21268314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryawanshi R.K., Chen I.P., Ma T., Syed A.M., Simoneau C.R., Ciling A., Khalid M.M., Sreekumar B., Chen P.-Y., George A.F., Kumar G.R., Montano M., Garcia-Knight M.A., Brazer N., Saldhi P., Sotomayor-Gonzalez A., Servellita V., Gliwa A., Nguyen J., Silva I., Milbes B., Kojima N., Hess V., Shacreaw M., Lopez L., Brobeck M., Turner F., Soveg F.W., Fang X., Maishan M., Matthay M., Morris M.K., Wadford D., Hanson C., Greene W.C., Andino R., Spraggon L., Roan N.R., Chiu C.Y., Doudna J., Ott M. Ltd. Cross-Var. Immun. Infect. SARS-CoV-2 Omicron Var. Vaccin. 2022 p. 2022.2001.2013.22269243. [Google Scholar]
- Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Chivukula V., Herati R., Hubbard S.R., Mulligan M.J., Landau N.R. Increase Resist. SARS-CoV-2 Omicron Var. Neutralization Vaccin. -Elicited Ther. Antibodies. 2021 doi: 10.1016/j.ebiom.2022.103944. p. 2021.2012.2028.474369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Mulligan M.J., Landau N.R. SARS-CoV-2 Lambda variant remains susceptible to neutralization by mRNA vaccine-elicited antibodies and convalescent serum. bioRxiv. 2021 p. 2021.2007.2002.450959. [Google Scholar]
- Takashita E., Kinoshita N., Yamayoshi S., Sakai-Tagawa Y., Fujisaki S., Ito M., Iwatsuki-Horimoto K., Halfmann P., Watanabe S., Maeda K., Imai M., Mitsuya H., Ohmagari N., Takeda M., Hasegawa H., Kawaoka Y. Efficacy of antiviral agents against the SARS-CoV-2 Omicron subvariant BA.2. N. Engl. J. Med. 2022;386:1475–1477. doi: 10.1056/NEJMc2201933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashita E., Yamayoshi S., Simon V., van Bakel H., Sordillo E.M., Pekosz A., Fukushi S., Suzuki T., Maeda K., Halfmann P., Sakai-Tagawa Y., Ito M., Watanabe S., Imai M., Hasegawa H., Kawaoka Y. Effic. Antibodies Antivir. Drugs Omicron Ba. 2. 12. 1, Ba. 4, Ba. 5 Subvariants. 2022 doi: 10.1056/NEJMc2207519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarres-Freixas F., Trinite B., Pons-Grifols A., Romero-Durana M., Riveira-Munoz E., Avila-Nieto C., Perez M., Garcia-Vidal E., Perez-Zsolt D., Munoz-Basagoiti J., Raich-Regue D., Izquierdo-Useros N., Blanco I., Noguera-Julian M., Guallar V., Lepore R., Valencia A., Vergara-Alert J., Clotet B., Ballana E., Carrillo J., Segales J., Blanco J. SARS-CoV-2 B. 1. 351 (beta) Var. shows Enhanc. Infect. K18-hACE2 transgenic mice Expand. Trop. wildtype mice Comp. B. 1 Var. 2021 p. 2021.2008.2003.454861. [Google Scholar]
- Tchesnokova V., Kulakesara H., Larson L., Bowers V., Rechkina E., Kisiela D., Sledneva Y., Choudhury D., Maslova I., Deng K., Kutumbaka K., Geng H., Fowler C., Ralston J.D., Greene D., Samadpour M., Sokurenko E.V. Acquis. L452R Mutat. ACE2-Bind. Interface Spike Protein triggers Recent Massiv Expans. SARS-Cov-2 Var. 2021 doi: 10.1128/JCM.00921-21. p. 2021.2002.2022.432189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Moir M., Everatt J., Giovanetti M., Scheepers C., Wilkinson E., Subramoney K., Moyo S., Amoako D.G., Althaus C.L., Anyaneji U.J., Kekana D., Viana R., Giandhari J., Maponga T.G., Maruapula D., Choga W., Mayaphi S.H., Mbhele N., Gaseitsiwe S., Msomi N., Naidoo Y., Pillay S., Sanko Ta, San J.E., Scott L., Singh L., Magini N.A., Smith-Lawrence P., Stevens W.S., Dor G., Tshiabuila D., Wolter N., Preiser W., Treurnicht F.K., Venter M., Davids M., Chiloane G., Mendes A., McIntyre C., O'Toole A., Ruis C., Peacock T.P., Roemer C., Williamson C., Pybus O.G., Bhiman J.N., Glass A.J., Martin D.P., Rambaut A., Gaseitsiwe S., von Gottberg A., Baxter C., Lessells R.J., de Oliveira T. Contin. Émerg. Evol. Omicron South Afr.: N. Ba. 4 Ba. 5 Linea. 2022 p. 2022.2005.2001.22274406. [Google Scholar]
- Touret F., Baronti C., Bouzidi H.S., de Lamballerie X. In vitro evaluation of therapeutic antibodies against a SARS-CoV-2 Omicron B.1.1.529 isolate. Sci. Rep. 2022;12:4683. doi: 10.1038/s41598-022-08559-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truffot A., Andreani J., Le Marechal M., Caporossi A., Epaulard O., Poignard P., Sylvie L. SARS-CoV-2 variants in immunocompromised patient given antibody monotherapy. Emerg. Infect. Dis. 2021;27 doi: 10.3201/eid2710.211509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P., Fenwick C., Raclot C., Genet V., Pantaleo G., Trono D. P2G3 Hum. Monoclon. Antib. neutralizes SARS-CoV-2 Omicron subvariants Incl. Ba. 4 Ba. 5 Bebtelovimab Escape Mutants. 2022 p. 2022.2007.2028.501852. [Google Scholar]
- Uraki R., Iida S., Halfmann P.J., Yamayoshi S., Hirata Y., Iwatsuki-Horimoto K., Kiso M., Ito M., Furusawa Y., Ueki H., Sakai-Tagawa Y., Kuroda M., Maemura T., Kim T., Mine S., Kinoshita-Iwamoto N., Li R., Liu Y., Larson D., Fukushi S., Watanabe S., Maeda K., Wang Z., Ohmagari N., Theiler J., Fischer W., Korber B., Imai M., Suzuki T., Kawaoka Y. Charact. SARS-CoV-2 Omicron Ba. 2. 75 Clin. Isol. 2022 p. 2022.2008.2026.505450. [Google Scholar]
- VanBlargan L.A., Errico J.M., Halfmann P., Zost S.J., Crowe J.E., Purcell L.A., Kawaoka Y., Corti D., Fremont D.H., Diamond M. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022;28:490–495. doi: 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatteroni M.L., Capria A.L., Spezia P.G., Frateschi S., Pistello M. Co-infection with SARS-CoV-2 omicron BA.1 and BA.2 subvariants in a non-vaccinated woman. Lancet Microbe. 2022;3 doi: 10.1016/S2666-5247(22)00119-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellas C., Del Bello A., Alexa D., Steinmeyer Z., Tribaudeau L., Ranger N., Jeanne N., Martin-Blondel G., Delobel P., Kamar N., Izopet J. Influence of neutralizing monoclonal antibodies on the SARS-CoV-2 quasispecies in patients with COVID-19. Clin. Micro Infect. 2021 doi: 10.1016/j.cmi.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villoutreix B., Calvez V., Marcelin A.-G., Khatib A.-M. Silico Investig. N. UK (B. 1. 1. 7) South Afr. (501Y. V2) SARS-CoV-2 Var. a Focus ACE2-Spike RBD Interface. 2021 doi: 10.3390/ijms22041695. p. 2021.2001.2024.427939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Lihong L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M.E., Kyratsous C.A., Shapiro L., Sheng Z., Huang Y., Ho D.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang Q., Guo Y., Iketani S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chan J.Y., Shah J.G., Nguyen N., Meyers K., Yin M.T., Sobieszczyk M.E., Sheng Z., Huang Y., Liu L., Ho D. SARS-CoV-2 Omicron Ba. 2. 12. 1, Ba. 4, Ba. 5 subvariants evolved extend Antib. Evas. 2022 p. 2022.2005.2026.493517. [Google Scholar]
- Wang Q., Iketani S., Li Z., Guo Y., Yeh A.Y., Liu M., Yu J., Sheng Z., Huang Y., Liu L., Ho D.D. Antigen. Charact. SARS-CoV-2 Omicron subvariant BA. 2022;2(75) doi: 10.1016/j.chom.2022.09.002. p. 2022.2007.2031.502235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Li Z., Ho J., Guo Y., Yeh A.Y., Liu M., Wang M., Yu J., Sheng Z., Huang Y., Liu L., Ho D.D. Resist. SARS-CoV-2 Omicron Subvariant Ba. 4. 6 Antib. Neutralization. 2022 p. 2022.2009.2005.506628. [Google Scholar]
- Wang R., Zhang Q., Ge J., Ren W., Zhang R., Lan J., Ju B., Su B., Yu F., Chen P., Liao H., Feng Y., Li X., Shi X., Zhang Z., Zhang F., Ding Q., Zhang T., Wang X., Zhang L. Spike Mutat. SARS-CoV-2 Var. confer Resist. Antib. neutralization. 2021 p. 2021.2003.2009.434497. [Google Scholar]
- Washington N.L., White S., Schiabor Barrett Km, Cirulli E.T., Bolze A., Lu J.T. S gene dropout Patterns SARS-CoV-2 tests Suggest Spread H69del/V70del Mutat. US. 2020 p. 2020.2012.2024.20248814. [Google Scholar]
- Wei C., Shan K.-J., Wang W., Zhang S., Huan Q., Qian W. Evid. a mouse Orig. SARS-CoV-2 Omicron Var. 2021 doi: 10.1016/j.jgg.2021.12.003. p. 2021.2012.2014.472632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm A., Widera M., Grikscheit K., Toptan T., Schenk B., Pallas C., Metzler M., Kohmer N., Hoehl S., Helfritz F.A., Wolf T., Goetsch U., Ciesek S. Limited neutralisation of the SARS-CoV-2 Omicron subvariants BA.1 and BA.2 by convalescent and vaccine serum and monoclonal antibodies. EBioMedicine. 2021;82 doi: 10.1016/j.ebiom.2022.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y. Cao, Y. Yu, W. Song, F. Jian, A. Yisimayi, C. Yue, R. Feng, P. Wang, L. Yu, N. Zhang, J. Wang, T. Xiao, R. An, Y. Wang, J. Wang, L. Liu, S. Yang, X. Niu, Q. Gu, F. Shao, X. Hao, R. Jin, X. Wang, Y. Wang and X.S. Xie, 2022. Neutralizing antibody evasion and receptor binding features of SARS-CoV-2 Omicron BA.2.75, (2022b), p. 2022.2007.2018.500332.
- Yamasoba D., Kimura I., Kosugi Y., Uriu K., Fujita S., Ito J., Sato K. Neutralization Sensit. Omicron Ba. 2. 75 Ther. Monoclon. antibodies. 2022 p. 2022.2007.2014.500041. [Google Scholar]
- Yamasoba D., Kosugi Y., Kimura I., Fujita S., Uriu K., Ito J., et al. Consortium, neutralisation sensitivity of SARS-CoV-2 omicron subvariants to therapeutic monoclonal antibodies. Lancet Infect. Dis. 2022;22:942–943. doi: 10.1016/S1473-3099(22)00365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh T.-Y., Contreras G.P. Tajima D. Test. accurately Forecasts Omicron / COVID-19 Outbreak. 2021 p. 2021.2012.2002.21267185. [Google Scholar]
- Yuan M., Zhu X., He W.-T., Zhou P., Kaku C.I., Capozzola T., Zhu C.Y., Yu X., Liu H., Yu W., Hua Y., Tien H., Peng L., Song G., Cottrell C., Schief W.R., Nemazee D., Walker L.M., Andrabi R., Burton D., Wilson I.A. A broad potent neutralization epitope SARS-Relat. Corona. 2022 doi: 10.1073/pnas.2205784119. p. 2022.2003.2013.484037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayet S., Vuillemenot J.B., Josset L., Gendrin V., Klopfenstein T. Simultaneous co-infection with Omicron (B.1.1.529) and Delta (21A/478K.V1) SARS-CoV-2 variants confirmed by whole genome sequencing. Int. J. Infect. Dis.: IJID: Off. Publ. Int. Soc. Infect. Dis. 2022 doi: 10.1016/j.ijid.2022.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Davis B., Chen S.S., Sincuir Martinez J., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 strain in Southern California, USA. JAMA. 2021 doi: 10.1001/jama.2021.1612. p. 2021.2001.2018.21249786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Tada T., Dcosta B.M., Landau N.R. SARS-CoV-2 Omicron Ba. 2 Var. Evades Neutralization Ther. Monoclon. Antibodies. 2022 p. 2022.2002.2015.480166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material