Abstract
Nonhost resistance (NHR) is a plant immune response that prevents many microorganisms in the plant’s environment from pathogenicity against the plant. Since successful pathogens have adapted to overcome the immune systems of their host, the durable nature of NHR has potential in the management of plant disease. At present, there is genetic and molecular evidence that the underlying molecular mechanisms of NHR are similar to the plant immune responses that occur in host plants following infection by adapted pathogens. We consider that the molecular basis of NHR is multilayered, conferred by physicochemical barriers and defense responses that are induced following molecular recognition events. Moreover, the relative contribution of each component may depend on evolutionary distances between host and nonhost plants of given pathogen species. This mini-review has focused on the current knowledge of plant NHR, especially the recognition of non-adapted pathogens by nonhost plants at the cellular level. Recent gains in understanding the roles of plasma membrane-localized pattern-recognition receptors (PRRs) and the cytoplasmic nucleotide-binding leucine-rich repeat receptors (NLRs) associated with these processes, as well as the genes involved, are summarized. Finally, we provide a theoretical perspective on the durability of receptor-mediated NHR and its practical potential as an innovative strategy for crop protection against pathogens.
Keywords: effector, NLR, nonhost resistance, PAMP, Pattern Recognition Receptor, plant defense
What is nonhost resistance?
In their natural environment, plants coexist with numerous microorganisms. Most relationships between plants and microbes are benign or even mutually beneficial. For any species of plant, only a few of the microbes it encounters are pathogenic to it [1]. Plants have evolved a multilayered defense system to fend off invading pathogens, and only microbe species or strains that have adapted to successfully bypass or incapacitate those defense systems are able to infect plants. This defense of plants against most microbes is considered nonhost resistance (NHR).
NHR can be simply defined as a plant immune response against non-adapted pathogens. The relationship between pathogen and host has been referred to as a co-evolutionary arms race, in which the adapted pathogen must continuously evolve to overcome host defense systems, which are, in turn, also evolving. In this scenario, it is clear that host resistance is not necessarily durable [2]. This specialization of pathogenicity and host defense is inefficient as the machinery by which plants provide resistance to a wide range of potential pathogens. Thus, lack of adaptation is assumed to be an essential concept of the durable nature of NHR [3].
Molecular components of NHR
Recently, numerous studies on the interactions between nonhost plants and non-adapted pathogens have been performed by inoculating various nonhost plants with non-adapted pathogens. The cytologic, genetic, and molecule-based evidence has led to the suggestion that nonhost plant responses to non-adapted pathogens and host response to pathogens share pre- and post-invasion components [4,5].
Physicochemical barriers
In many studies, non-adapted pathogens failed to penetrate the plant epidermis. Apparently, the pathogens could not overcome the physicochemical barriers on the plant surfaces. Consequently, there was no macroscopic evidence of infection [6]. Failure to invade host tissue was in some instances due to the composition of the cell wall or the waxy cuticle on the epidermis.
Evidence for the importance of physicochemical barriers comes from genetic studies. The irg1/palm1 mutant of the legume Medicago truncatula does not synthesize epicuticular wax, and could be colonized by non-adapted rust pathogens [7]. The P450 CYP96B22 gene of barley is believed to be related to the synthesis of epicuticular wax, and silencing the gene enabled isolates of the non-adapted fungal pathogen Magnaporthe oryzae to infect nonhost barley [8]. It is also interesting to note that the germination rates of spores of the fungus Melampsora larici-populina in cuticle extracts derived from host and nonhost plants were markedly reduced in nonhost extracts compared with host extracts [9]. From these results, we suggest that the composition of plant cuticles not only functions as a physical barrier to non-adapted pathogens but also acts as an inductive signal for initiating infection by adapted pathogens.
Plant secondary metabolites with antimicrobial activity may also be determinants of plant host status. Phytophthora infestans is the oomycetes pathogen that causes late blight of potato and tomato but is not a pathogen of pepper and tobacco. The P. infestans was more susceptible to the sesquiterpene phytoalexin capsidiol produced by pepper and tobacco compared with the Phytophthora capsici, a species adapted to be pathogenic to tobacco and pepper [10,11]. Similarly, avenacin is an antimicrobial agent present in the roots of many oats. Gaeumannomyces graminis pv. avenae, a pathogen of oats, produces avenacinase, an enzyme that can degrade avenacin [12]. G. graminis pv. tritici, in contrast, cannot infect oats that accumulate avenacin [12]. Glucosinolate compounds are suggested to be a component of NHR in Brassicaceae. Arabidopsis plants deficient in PEN2, a glycosyl hydrolase, can be infected by the grass pathogen Blumeria graminis f. sp. hordei, a non-adapted pathogen of Arabidopsis plants [13]. Also in Arabidopsis, mutants that were deficient in a tryptophan-derived antimicrobial compound were colonized by the non-adapted pathogen P. infestans [14,15].
We conclude, from the studies discussed above, that physicochemical barriers contribute to NHR. Especially, while adapted pathogens are able to suppress or bypass these barriers of host plants, non-adapted pathogens cannot overcome them.
Receptor-mediated recognition
In addition to physicochemical barriers, plants have evolved surveillance systems composed of plasma membrane-localized pattern-recognition receptors (PRRs) and cytoplasmic nucleotide-binding leucine-rich repeat receptors (NLRs) that recognize nonself-derived molecules [16]. PRRs generally recognize pathogen-associated molecular patterns (PAMPs), such as bacterial flagellin and fungal chitin, and induce PAMP-triggered immunity (PTI). NLRs directly or indirectly recognize pathogen effectors, which are secreted into plant cells by pathogens to modulate the plant immune system and induce effector-triggered immunity (ETI). Both defense responses lead to oxidative burst, transcriptional reprogramming, and biosynthesis of defense-related compounds through intracellular signaling to limit invading pathogens [17]. While the repertoire of downstream signaling is broadly shared in both processes, responses in ETI are known to be more prolonged and robust [18] and often culminate in highly localized hypersensitive response (HR) cell death, which restricts pathogen growth in invading tissues.
PTI
PAMPs are generally (but not always) conserved in microbial pathogens. Thus, plants can recognize the PAMPs of non-adapted pathogens. For example, when Arabidopsis was inoculated with the non-adapted Pseudomonas syringae pv. phaseolicola, PTI-like transcriptional changes similar to flg22 treatment were induced [19]. Genetic analysis of this response revealed that the flg22 receptor FLS2 is involved in this type of NHR [20]. In addition, studies with loss-of-function mutants have been interpreted to support the involvement of multiple PTI-related components, specifically nonhost 1 (NHO1) [21], PEN2 [22,23], BRI1-associated receptor kinase (BAK1) [24], and the PRR receptor HvLEMK1 [25], in NHR. More examples and references have been well documented in earlier reviews [26].
Not only the PAMPs of non-adapted pathogens are recognized by their nonhost plants, but several lines of evidence are consistent with the contention that non-adapted pathogens are less capable of suppressing the defense mechanisms of nonhost plants than adapted pathogens. One example is the Arabidopsis flg22-induced downstream defense component NHO1. It is suppressed by the adapted bacterium Pseudomonas syringae pv. tomato (Pst), but not by the non-adapted pathovar tabaci [27]. Considering that no less than nine effectors of Pst (HopS1, HopAI1, HopAF1, HopT1-1, HopT1-2, HopAA1-1, HopF2, HopC1, and AvrPto, which are not conserved in P. syringae pv. tabaci except for HopT1-1) are involved in suppressing flg22-induced NHO1 expression [28], effector-mediated suppression would be the crucial strategy of adapted pathogen, Pst, to overcome PTI, while non-adapted pathogens, pv. tabaci, is less-likely to be specialized to suppress PTI of nonhost. Similarly, when the Arabidopsis EF-Tu receptor (EFR), which recognizes the bacterial elongation factor Tu, was transferred into Nicotiana benthamiana and tomato, the transgenic plants acquired broad-spectrum disease resistance. This resistance extended to Ralstonia solanacearum, a bacterial pathogen of Solanaceae plants but not adapted to Arabidopsis [29]. More recently, it was noted that the orthologous PRRs, Rphq2 and Rph22, which are derived from cultivated and wild barley, respectively, modified host status against leaf rust caused by Puccinia hordei. Cultivated barley is resistant to P. hordei-bulbosi, while wild barley is resistant to P. hordei. Barley accessions were generated that were extremely susceptible to both the leaf rust pathogens. The two PRRs were cloned, and each was singly transferred into susceptible accession. Transgenic plants with Rphq2 were more resistant to P. hordei-bulbosi compared with P. hordei, while transgenic plants that received Rph22 exhibited the reverse reaction. Both lines were resistant to the wheat pathogen Puccinia triticina, which is non-adapted to both cultivated and wild barley [30]. These results imply that non-adapted pathogens are less capable of overcoming nonhost-derived PRRs.
Although PRR-mediated PAMP recognition is a highly conserved process in plants, small differences between host and nonhost plants may contribute to durable NHR. Thus, the identification of PRR orthologs from various plant species and further characterization of the differences between hosts and nonhosts PRRs could be a promising strategy for understanding the durable nature of PRR-mediated NHR. In addition, recent studies suggesting PAMPs could be also variable between the species, especially flg22 [31]. Thus, identification of PRR homologs in various plant species could be also prominent strategy for broad-spectrum resistance, as the FLS2XL derived from wild grape (Vitis riparia) recognizes flg22Atum of Agrobacterium tumefaciens which is not recognized by FLS of Arabidopsis [32].
ETI
While PAMPs are relatively conserved in pathogenic microorganisms, effectors have lineage-specific sequence diversity and are prone to mutation [33]. Based on a number of studies, it seems that NLR-mediated effector recognition is also associated with NHR. For example, in several pioneering works, HR-like cell death phenotypes were acquired by inoculating nonhost plants with non-adapted pathogens [34–37]; these phenotypes are hallmarks of ETI. Results from studies with loss-of-function mutants, specifically loss of ETI signaling components such as enhanced disease susceptibility 1 (EDS1), phytoalexin-deficient 4 (PAD4), senescence-associated gene 101 (SAG101) [13], WRKY [38], and SGT1 [39], have also provided evidence that NLRs are components of NHR.
Numerous effectors of non-adapted pathogens elicit HR cell death and defense responses in nonhost plants. For examples, the P. capsici effector PcAvr3a1 triggers HR in nonhost Nicotiana tabacum [37] and multiple RxLR effectors of P. infestans trigger HR-like cell death in nonhost chili pepper [40]. In the latter, it was noted that the HR phenotypes were inherited by Mendelian genetics. The Xanthomonas campestris effector XopQ triggers EDS1-dependent HR in nonhost Nicotiana spp. [41] and multiple Hyaloperonospora arabidopsidis effectors inhibit bacterial growth when transferred into nonhost turnip plants through the bacterial type-three secretion system [42]. Based on these examples, ETI could be a component of NHR, and some instances, recognition of multiple non-adapted pathogen effectors contributes to NHR. This proposal requires a caution, however, as there are examples of single effector deletions, such as HopAS1, HopQ1 of Pseudomonas syringae [43,44], or PWT3 of rice blast [45], enabling host ‘jumps’ in which the pathogens infect previous nonhost plants. The quantitative nature of NHR (a number of genes) might depend on which plant and pathogen species are considered. In other words, it may be context dependent [5,46].
Several plant NLRs that correspond to HR-inducing effectors from non-adapted pathogens have been identified in nonhost plants. Some of these NLRs have been transferred into host species and, indeed, conferred resistance to non-adapted pathogens. Several examples are summarized below (Table 1). Silencing a homolog of the I2 gene family in N. tabacum resulted in loss of ability to recognize P. capsici effector PcAvr3a, and subsequently compromised the resistance to the P. capsici [47]. An NLR from maize, Rxo1, conferred resistance to the bacterial streak pathogen Xanthomonas oryzae pv. oryzicola (Xoc) upon its transfer into rice plants [48,49]. Similarly, an NLR from Arabidopsis, WRR4, conferred resistance to the pathogen that causes white rust when transferred into oilseed brassica crops [50,51]. When the CcRPP1 gene from pigeon pea, which encodes an NLR, was transferred into soybean, the plant acquired resistance to the soybean rust pathogen Phakopsora pachyrhizi (Pp) [52]. In barley, the NLR resistance to Pseudomonas syringae (Rps)7 were transferred into experimentally generated susceptible barley (SusPtrit) and conferred resistance to the wheat stripe rust pathogen Puccinia striiformis [53]. An exciting prospect is apparent here. Resistance in wild rice to Xoc, or resistance to Pp in soybean has not been identified. The possibility of identifying NLRs from nonhost plants effective in host–pathogen interactions could represent a novel genetic resource for managing plant disease.
Table 1. Examples of receptors that contribute to NHR.
| Receptors | Nonhost | Pathogen | Description | References |
|---|---|---|---|---|
| NLRs | ||||
| Rxo1 | Zea mays (Maize) | Xanthomonas oryzae pv. oryzicola | Resistance in rice | [48,49] |
| WRR4 | Arabidopsis thaliana | Albugo candida | Resistance in Brassica napus and Brassica juncea | [50,51,55] |
| CcRPP1 | Cajanus cajan (pigeon pea) | Phakopsora pachyrhizi | Resistance in soybean | [52] |
| Rps7 | Hordeum vulgare (barley) | Puccinia striiformis | Resistance in susceptible barley (SusPtrit*) | [53] |
| Pm3 | Triticum aestivum | Blumeria graminis f. sp. tritici, secalis, dactylidis | Resistance in SusPtrit barley and tolerant to suppression of Svrpm3 homologs from non-adapted pathogens | [65] |
| Rpi-amr1 | Solanum americanum | Phytophthora parasitica, Phytophthora cactorum | Induce HR against Avr-amr1 homologs of non-adapted pathogens | [57] |
| Rpi-amr3 | Solanum americanum | Phytophthora parasitica, Phytophthora palmivora | Resistance in Nicotiana benthamiana | [56] |
| PBR1 | Hordeum vulgare | Pseudomonas syringae pv. phaseolicola | Recognize AvrPphB and trigger HR | [59] |
| RPS2/MR5 | Arabidopsis thaliana/Malus domestica | Pseudomonas syringae, Erwinia amylovora | Recognize both PsAvrRpt2 and EaAvrRpt2 and trigger HR | [60] |
| NtI2 | Nicotiana tabacum | Phytophthora capsici | I2-silenced plants exhibited compromised resistance to Pc and HR against PcAvr3a1 | [47] |
| PRRs | ||||
| FLS2 | Arabidopsis thaliana | Pseudomonas syrngae pv. tabaci | Suppressed by adapted pathogen, P. syringae pv. tomato, but tolerant to non-adapted P. syringae pv. tabici | [20,27,28] |
| EFR | Arabidopsis thaliana | Ralstonia solanacearum | Resistance in tomato and Nicotiana benthamiana | [29] |
| Rph22 | Hordeum bulbosum | Puccinia hordei, Puccinia tritici | Resistance in Golden SusPtrit barley | [30] |
| Rphq2 | Hordeum vulgare | Puccinia hordei-bulbosi, Puccinia tritici | Resistance in Golden SusPtrit barley | [30] |
| HvLEMK1 | Hordeum vulgare | Blumera graminis f. sp. tritici | HvLEMK1-silenced barley exhibited compromised resistance against non-adapted pathogen, and transient expression of HvLEMK1 in both wheat and barley conferred quantitative resistance to Bgt | [25] |
Physicochemical barriers of plants are generally polygenic trait in nature, and transfer of these traits between plants may prove problematic. Conversely, transferring immune receptors (PRRs or NLRs) from nonhost to host plants using biotechnological approaches, may prove more tenable. It is also an untapped genetic resource. Thus, further characterization and understanding of receptor-mediated NHR could provide promising strategies for developing durable resistant crops.
We conclude that, while only one or a few genes determine host status in some cases, NHR is generally conferred by coordination of multiple defense mechanisms. These vary from physicochemical barriers to receptor-mediated induced responses [4]. The molecular evolutionary concept could provide a plausible explanation for the context dependency of NHR. The phylogenetic distances between the host and nonhost plants may determine the quantitative nature and relative contribution of each component to NHR against pathogens [54]. Moreover, the lack of adaptation in pathogens to defense machinery of nonhost plants makes NHR effective.
A molecular evolutionary concept of receptor-mediated NHR: recognition and durability
As previously discussed, nonhost PRRs recognize conserved PAMPs of non-adapted pathogens. It has also been demonstrated that nonhost PRRs are more tolerant to suppression by non-adapted pathogens than are host PRRs [30], which may explain the durability of PRR-mediated NHR. We assumed that NLR-mediated NHR would be similar. Therefore, we consider two questions here. First, ‘How do NLRs recognize the effectors of non-adapted pathogens, which are more diversified than PAMPs?’ Our second question is ‘Do NLRs from nonhosts confer durable resistance against immune suppression by host-adapted pathogens?’ These two questions can be partly answered from the results of recent studies on recognition and durability.
How do plants maintain receptors recognizing non-adapted pathogens?
Accumulating evidence suggest that effectors, similar to PAMPs, are conserved in closely related species of pathogens and multiple NLRs recognize homologous effectors of both non-adapted and adapted pathogens. For example, the Arabidopsis WRR4A and WRR4B NLRs recognize CCG effectors derived from both adapted and non-adapted races of Albugo candida [55]. Another example is the NLR resistance to Phytophyhora infestans (Rpi)-amr1 derived from Solanum americanum could recognize P. infestans Avramr1 as well as its homologs from Phytophthora parasitica, and Phytophthora cactorum [56,57]. Furthermore, Rpi-amr3 of S. americanum recognizes not only P. infestans Avramr3 but also Avramr3 orthologs from various Phytophthora spp. and confers resistance against P. parasitica and P. palmivora in the N. benthamiana system [58]. It may be NLRs that evolved to recognize adapted pathogen effectors also could recognize non-adapted pathogen effectors because those effectors are conserved in closely related adapted and non-adapted pathogens.
On the other hand, several NLRs reported to recognize the effectors of non-adapted pathogen, which infect evolutionarily distant host, through the homologous host targets. Arabidopsis RPS5 indirectly recognizes the Pseudomonas syringae effector AvrPphB, which cleaves the Arabidopsis host target AtPBS1. Likewise, an NLR from barley, PBR1, also recognizes AvrPphB via PBS1 orthologs of barley [59]. Moreover, Arabidopsis RPS2 indirectly recognizes both AvrRpt2 homologs of Erwinia amylovora and Pseudomonas syringae, which are non-adapted and adapted pathogens of Arabidopsis, respectively, via AtRIN4. Similarly, MR5, an NLR derived from wild apple, recognized both AvrPphB homologs through MdRIN4 [60]. Notably, MR5/RPS2 and PBR1/RPS5 exhibit no sequence homology between the functionally homologous (those recognize same or similar effectors) NLRs, but the effector targets exhibit sequence homology between AtRIN4/MdRIN4 and AtPBS1/HvPBS1. These results indicate that the ability to recognize these two effectors evolved separately in each plant.
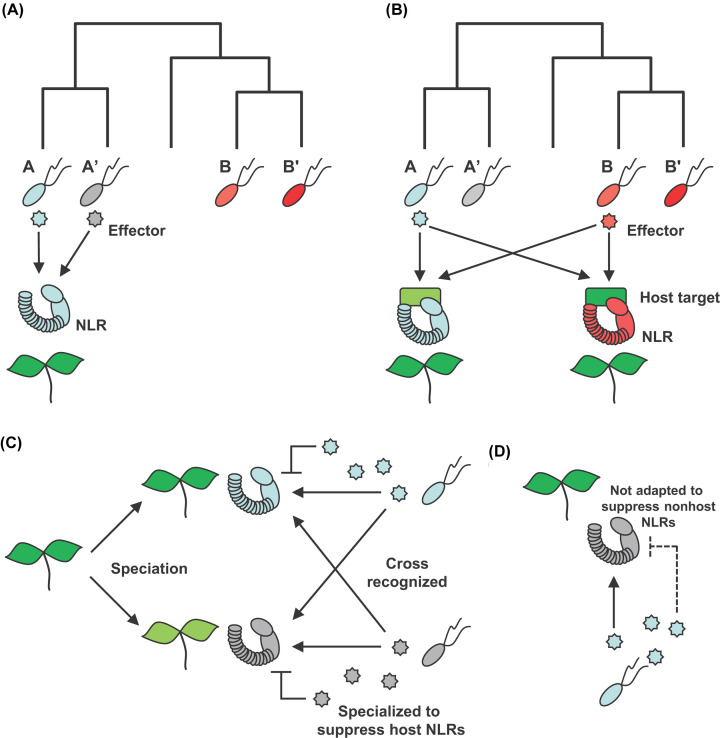
In summary, it appears that plants can recognize effectors from both adapted and non-adapted pathogens through similar NLRs because the effectors are conserved in closely related pathogens. Furthermore, even considering distantly related pathogen species, some effectors could share similar virulence strategies or host targets. Thus, plants could recognize both effectors using NLRs that surveil homologous host targets (Figure 1A,B). Therefore, if the phylogenetic distance between nonhost and host plants to a given pathogen go closer, the possibility (or a number) of cross recognition of non-adapted pathogen effectors by nonhost NLRs may be increased. This proposal is similar to the molecular evolutionary concept advanced with regard to ETI in NHR [54]. In this context, we expect NLRs of nonhost plants that are homologous to previously known host resistance genes to be promising candidates for conferring NHR.
Figure 1. Mechanistic model of the NLR-mediated NHR.
(A) NLRs evolved to recognize adapted pathogen effectors also recognize conserved homologous effectors of non-adapted pathogen. A, A′, B, and B′ indicate different strains (or species) of pathogen. (B) Effectors of distantly related non-adapted pathogens targeting similar host targets of adapted pathogens could be recognized by surveillance system of nonhost plants. (C) While non-adapted pathogens recognized by nonhost plants NLRs, non-adapted pathogens cannot efficiently suppress nonhost defense machineries, because of lack of adaptation. (D) Transfer of nonhost NLRs into susceptible host plants could be promising strategy for conferring durable resistance.
In addition, though without the sequence similarity between effectors, recent studies also suggest that conserved 3D structural features of effectors is crucial for recognition by NLRs, such as AVR1-CO39 and AVR-Pia [61,62]. Considering sequences of effectors are often not conserved even between the closely related species, recognizing conserved structurally similar effectors could be another mechanism of recognizing non-adapted pathogens.
Could immune receptors from nonhost plants be more durable?
About the PTI-mediated NHR, the following scenario is proposed. Orthologous PRRs derived from nonhost recognize the same or similar PAMPs as host PRRs, while PRRs from nonhost plants are more tolerant to suppression by non-adapted pathogens and contribute to the durability of NHR [30]. This scenario is reasonable, considering that pathogens are specialized to suppress host immune system rather than that of nonhost. In this context, pathogen machinery that is most likely to act as a suppressor of immune response would be an effector. Indeed, effector-mediated suppression of PTI/ETI is a well-known phenomenon [2,63].
We next consider the question: If homologous NLRs derived from both host and nonhost plants that recognize the same or similar effectors, would the homologous NLRs derived from nonhost plants be more tolerant to suppression by non-adapted pathogens? A suitable example for this hypothesis is the NLRs in the Poaceae family, specifically Pm3, which is a NLR family of wheat. Its corresponding effector, Avrpm3, is conserved in Blumeria graminis f. sp. tritici, secalis, and dactylidis that cause rust in wheat, rye, and wild grass (Dactylis glomerata), respectively. The Pm3 NLRs recognize all Avrpm3 effector homologs and confer resistance to the non-adapted rye mildew pathogen on wheat. Furthermore, Pm3-mediated resistance is suppressed by another effector, Svrpm3 of B. graminis f. sp. tritici [64], and Svrpm3 homologs are also conserved in B. graminis f. sp. secalis and dactylidis. Notably, while Pm3 recognizes all homologs of Avrpm3 derived from B. graminis f. sp. tritici, secalis, and dactylidis, Pm3-mediated HR cell death is not suppressed by Svrpm3 of B. graminis f. sp. secalis which is non-adapted pathogen of wheat [65]. These results imply that NLRs recognizing non-adapted pathogen effectors could be effective because non-adapted pathogen is not specialized to suppress nonhost NLR-mediated resistance while adapted pathogens have evolved suppressing mechanisms against it.
To date, a number of effectors have been identified that suppress NLRs or ETI signaling cascades, such as the Phytophthora infestans RxLR effector IPI-O4, which suppresses the wild potato-derived NLR Rpi-blb1 [66,67], and the effector PITG_15278 which suppresses Rpi-blb2 [68]. The Ralstonia solanacearum effector RipAC interacted with SGT1 and prevented mitogen-activated protein kinase (MAPK)-mediated phosphorylation [69,70]; the Xanthomonas axonopodis pv. manihotis effectors XopE4 and XopAo1 suppress the ETI-mediated HR [71]. Although a few cases such as Pm3 have been reported, multiple NLRs that contribute to NHR and ETI suppressors also have been identified. Thus, further identification of functionally homologous NLRs from host and nonhost plants that recognize the same or similar effectors of both adapted and non-adapted pathogens will be necessary for the reciprocal approaches to test whether the NLRs derived from nonhost plants could be more robust against suppression by non-adapted pathogens.
We propose that similar to PTI in NHR, nonhost plants could recognize effectors of non-adapted pathogen using NLRs. Moreover, the NLRs or ETI signaling components of nonhosts could be more tolerant to effector-mediated suppression by non-adapted pathogens due to lack of adaptation (Figure 1C,D).
Perspectives
Helper NLRs that function in the downstream of sensor NLRs contribute to multiple cases of NLR-mediated resistance [72–74]. While NLR required for cell death (NRC)-type helper NLRs are conserved in the Solanaceae plants, the helper NLRs N-requirement gene 1 (NRG1) and activated disease resistance 1 (ADR1) are broadly conserved in almost all higher plants [75]. Helper NLRs have a critical role as signaling hubs. Thus, effector-mediated suppression of helper NLRs could lead to the collapse of multiple NLR-mediated resistance. Indeed, Avrcab1b of P. infestans is reported to suppress NRC2/3 [68]. In this context, functionally homologous helper NLRs derived from nonhost plants could be tested to determine whether they are more tolerant to the suppression by non-adapted pathogens. If so, nonhost helper NLRs might reinforce plant defenses in a pleiotropic manner.
In general, NHR cannot be transferred through crossing and largely depends on heterologous expression. Thus, compatibility between helper or sensor NLRs or NLR signaling components should be carefully considered for transferring NLRs. Indeed, transfer of BS2, a well-known NRC-dependent sensor NLR, from Solanaceae chili pepper to Euphorbiaceae Cassava failed to confer resistance against Xanthomonas axonopodis pv. manihotis [76]. In addition, complementation assays using EDS1-SAG101-NRG1 modules also suggest that components of Arabidopsis and N. benthamiana are not fully compatible with each other compared with their own combinations [77]. Therefore, co-transfer of nonhost helper NLRs or signaling components with identified nonhost receptors could be crucial for conferring the full potential of resistance in specific cases.
Recently, a number of studies have suggested that PTI and ETI function synergistically [78–80]. Considering that single gene-mediated NHR could be overcome by adaptation of pathogen, combinational approaches would be required to confer durable resistance for susceptible crops. Thus, more identification of genetic resources contributing to NHR would be essential for achieving this goal.
Summary
NHR is conferred by the coordination of multiple layers of resistance. These layers range from physicochemical barriers to receptor-mediated responses. The contribution of each component in any interaction depends on context. Generally, the relative contribution of each component is related to the phylogenetic distances between the host and nonhost plant species.
Receptor-mediated NHR could be transferred from nonhost to susceptible host plants.
We suggest that receptor-mediated NHR is established by recognition and durability (possible to recognize non-adapted pathogens but more tolerant to immune suppression by non-adapted pathogens). Further characterization of receptors from nonhost plants and investigation on mechanisms of durability would be required for further application of NHR.
Abbreviations
- AVR
avirulence
- EDS1
enhanced disease susceptibility 1
- ETI
effector-triggered immunity
- HR
hypersensitive response
- NHO1
nonhost 1
- NHR
nonhost resistance
- NLR
nucleotide-binding leucine-rich repeat receptor
- NRC
NLR required for cell death
- NRG1
N-requirement gene 1
- PAMP
pathogen-associated molecular pattern
- PRR
pattern-recognition receptor
- PTI
PAMP-triggered immunity
- Rpi
resistance to Phytophyhora infestans
- RPS
resistance to Pseudomonas syringae
- SAG101
senescence-associated gene 101
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the National Research Foundation of Korea [grant number 2021R1A2B5B03001613]; and the Plant Immunity Research Center, SRC [grant number 2018R1A5A1023599].
Author Contribution
S.O. and D.C. conceptualized and wrote the manuscript, S.O. created the figures.
References
- 1.Heath M.C. (2000) Nonhost resistance and nonspecific plant defenses. Curr. Opin. Plant Biol. 3, 315–319 10.1016/S1369-5266(00)00087-X [DOI] [PubMed] [Google Scholar]
- 2.Hein I., Gilroy E.M., Armstrong M.R. and Birch P.R.J. (2009) The zig-zag-zig in oomycete-plant interactions. Mol. Plant Pathol. 10, 547–562 10.1111/j.1364-3703.2009.00547.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonovics J., Boots M., Ebert D., Koskella B., Poss M. and Sadd B.M. (2013) The origin of specificity by means of natural selection: evolved and nonhost resistance in host-pathogen interactions. Evol. 67, 1–9 10.1111/j.1558-5646.2012.01793.x [DOI] [PubMed] [Google Scholar]
- 4.Lee H.A., Lee H.Y., Seo E., Lee J., Kim S.B., Oh S.et al. (2017) Current understandings of plant nonhost resistance. Mol. Plant Microbe Interact. 30, 5–15 10.1094/MPMI-10-16-0213-CR [DOI] [PubMed] [Google Scholar]
- 5.Panstruga R. and Moscou M.J. (2020) What is the molecular basis of nonhost resistance? Mol. Plant Microbe Interact. 33, 1253–1264 10.1094/MPMI-06-20-0161-CR [DOI] [PubMed] [Google Scholar]
- 6.Oh S.K., Lee S., Chung E., Park J.M., Yu S.H., Ryu C.M.et al. (2006) Insight into Types I and II nonhost resistance using expression patterns of defense-related genes in tobacco. Planta 223, 1101–1107 10.1007/s00425-006-0232-1 [DOI] [PubMed] [Google Scholar]
- 7.Uppalapati S.R., Ishiga Y., Doraiswamy V., Bedair M., Mittal S., Chen J.et al. (2012) Loss of abaxial leaf epicuticular wax in Medicago truncatula irg1/palm Mutants results in reduced spore differentiation of anthracnose and nonhost rust pathogens. Plant Cell 24, 353–370 10.1105/tpc.111.093104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delventhal R., Falter C., Strugala R., Zellerhoff N. and Schaffrath U. (2014) Ectoparasitic growth of Magnaporthe on barley triggers expression of the putative barley wax biosynthesis gene CYP96B22 which is involved in penetration resistance. BMC Plant Biol. 14, 1–14 10.1186/1471-2229-14-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Z., Shen K., Newcombe G., Fan J. and Chen Q. (2019) Leaf cuticle can contribute to non-host resistance to poplar leaf rust. Forests 10, 870 10.3390/f10100870 [DOI] [Google Scholar]
- 10.Giannakopoulou A., Schornack S., Bozkurt T.O., Haart D., Ro D.K., Faraldos J.A.et al. (2014) Variation in capsidiol sensitivity between phytophthora infestans and phytophthora capsici is consistent withtheir host range. PLoS ONE 9, e107462 10.1371/journal.pone.0107462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H.A., Kim S., Kim S. and Choi D. (2017) Expansion of sesquiterpene biosynthetic gene clusters in pepper confers nonhost resistance to the Irish potato famine pathogen. New Phytol. 215, 1132–1143 10.1111/nph.14637 [DOI] [PubMed] [Google Scholar]
- 12.Osbourn A.E., Clarke B.R., Dow J.M. and Daniels M.J. (1991) Partial characterization of avenacinase from Gaeumannomyces graminis var. avenae. Physiol. Mol. Plant Pathol. 38, 301–312 10.1016/S0885-5765(05)80121-3 [DOI] [Google Scholar]
- 13.Lipka V., Dittgen J., Bednarek P., Bhat R., Wiermer M., Stein M.et al. (2005) Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310, 1180–1183 10.1126/science.1119409 [DOI] [PubMed] [Google Scholar]
- 14.Prince D.C., Rallapalli G., Xu D., Schoonbeek H., Çevik V., Asai S.et al. (2017) Albugo-imposed changes to tryptophan-derived antimicrobial metabolite biosynthesis may contribute to suppression of non-host resistance to Phytophthora infestans in Arabidopsis thaliana. BMC Biol. 15, 1–22 10.1186/s12915-017-0360-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belhaj K., Cano L.M., Prince D.C., Kemen A., Yoshida K., Dagdas Y.F.et al. (2017) Arabidopsis late blight: infection of a nonhost plant by Albugo laibachii enables full colonization by Phytophthora infestans. Cell. Microbiol. 19, e12628 10.1111/cmi.12628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones J.D. and Dangl J.L. (2006) The plant immune system. Nature 444, 323–329 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- 17.Dangl J.L., Horvath D.M. and Staskawicz B.J. (2013) Pivoting the plant immune system from dissection to deployment. Science 341, 746–751 10.1126/science.1236011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuda K. and Katagiri F. (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465 10.1016/j.pbi.2010.04.006 [DOI] [PubMed] [Google Scholar]
- 19.Navarro L., Zipfel C., Rowland O., Keller I., Robatzek S., Boller T.et al. (2004) The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135, 1113–1128 10.1104/pp.103.036749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forsyth A., Mansfield J.W., Grabov N., Torres M., Sinapidou E. and Grant M.R. (2010) Genetic dissection of basal resistance to Pseudomonas syringae pv. phaseolicola in accessions of Arabidopsis. Mol. Plant Microbe Interact. 23, 1545–1552 10.1094/MPMI-02-10-0047 [DOI] [PubMed] [Google Scholar]
- 21.Lu M., Tang X. and Zhou J.M. (2001) Arabidopsis NHO1 is required for general resistance against Pseudomonas bacteria. Plant Cell 13, 437–447 10.1105/tpc.13.2.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipka U., Fuchs R. and Lipka V. (2008) Arabidopsis non-host resistance to powdery mildews. Curr. Opin. Plant Biol. 11, 404–411 10.1016/j.pbi.2008.04.004 [DOI] [PubMed] [Google Scholar]
- 23.Maeda K., Houjyou Y., Komatsu T., Hori H., Kodaira T. and Ishikawa A. (2010) Nonhost resistance to Magnaporthe oryzae in Arabidopsis thaliana. Plant Signal. Behav. 5, 755–756 10.4161/psb.5.6.11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kemmerling B., Schwedt A., Rodriguez P., Mazzotta S., Frank M., Qamar S.A.et al. (2007) The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr. Biol. 17, 1116–1122 10.1016/j.cub.2007.05.046 [DOI] [PubMed] [Google Scholar]
- 25.Rajaraman J., Douchkov D., Hensel G., Stefanato F.L., Gordon A., Ereful N.et al. (2016) An LRR/Malectin receptor-like kinase mediates resistance to non-adapted and adapted powdery mildew fungi in barley and wheat. Front. Plant Sci. 7, 1–13 10.3389/fpls.2016.01836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senthil-Kumar M. and Mysore K.S. (2013) Nonhost resistance against bacterial pathogens: retrospectives and prospects. Annu. Rev. Phytopathol. 51, 407–427 10.1146/annurev-phyto-082712-102319 [DOI] [PubMed] [Google Scholar]
- 27.Kang L., Li J., Zhao T., Xiao F., Tang X., Thilmony R.et al. (2003) Interplay of the Arabidopsis nonhost resistance gene NHO1 with bacterial virulence. Proc. Natl. Acad. Sci. U.S.A. 100, 3519–3524 10.1073/pnas.0637377100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Lin H., Zhang W., Zou Y., Zhang J., Tang X.et al. (2005) Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl. Acad. Sci. U.S.A. 102, 12990–12995 10.1073/pnas.0502425102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lacombe S., Rougon-Cardoso A., Sherwood E., Peeters N., Dahlbeck D., Van Esse H.P.et al. (2010) Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 28, 365–369 10.1038/nbt.1613 [DOI] [PubMed] [Google Scholar]
- 30.Wang Y., Subedi S., de Vries H., Doornenbal P., Vels A., Hensel G.et al. (2019) Orthologous receptor kinases quantitatively affect the host status of barley to leaf rust fungi. Nat. Plants 5, 1129–1135 10.1038/s41477-019-0545-2 [DOI] [PubMed] [Google Scholar]
- 31.Cheng J.H., Bredow M., Monaghan J. and DiCenzo G.C. (2021) Proteobacteria contain diverse flg22 epitopes that elicit varying immune responses in Arabidopsis thaliana. Mol. Plant Microbe Interact. 34, 504–510 10.1094/MPMI-11-20-0314-SC [DOI] [PubMed] [Google Scholar]
- 32.Fürst U., Zeng Y., Albert M., Witte A.K., Fliegmann J. and Felix G. (2020) Perception of Agrobacterium tumefaciens flagellin by FLS2XL confers resistance to crown gall disease. Nat. Plants 6, 22–27 10.1038/s41477-019-0578-6 [DOI] [PubMed] [Google Scholar]
- 33.Depotter J.R.L. and Doehlemann G. (2020) Target the core: durable plant resistance against filamentous plant pathogens through effector recognition. Pest Manag. Sci. 76, 426–431 10.1002/ps.5677 [DOI] [PubMed] [Google Scholar]
- 34.Vleeshouwers V.G.A.A., van Dooijeweert W., Govers F., Kamoun S. and Colon L.T. (2000) The hypersensitive response is associated with host and nonhost resistance to Phytophthora infestans. Planta 210, 853–864 10.1007/s004250050690 [DOI] [PubMed] [Google Scholar]
- 35.Huitema E., Vleeshouwers V.G.A.A., Francis D.M. and Kamoun S. (2003) Active defence responses associated with non-host resistance of Arabidopsis thaliana to the oomycete pathogen Phytophthora infestans. Mol. Plant Pathol. 4, 487–500 10.1046/j.1364-3703.2003.00195.x [DOI] [PubMed] [Google Scholar]
- 36.Lindgren P.B., Peet R.C. and Panopoulos N.J. (1986) Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity of bean plants and hypersensitivity on nonhost plants. J. Bacteriol. 168, 512–522 10.1128/jb.168.2.512-522.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vega-arreguín J.C., Jalloh A., Bos J.I., Moffett P., Thompson B., Nacional U.et al. (2014) Recognition of an Avr3a homologue plays a major role in mediating nonhost resistance to Phytophthora capsici in Nicotiana species. Mol. Plant Microbe Interact. 27, 770–780 10.1094/MPMI-01-14-0014-R [DOI] [PubMed] [Google Scholar]
- 38.Moreau M., Degrave A., Vedel R., Bitton F., Patrit O., Renou J.P.et al. (2012) EDS1 contributes to nonhost resistance of Arabidopsis thaliana against Erwinia amylovora. Mol. Plant Microbe Interact. 25, 421–430 10.1094/MPMI-05-11-0111 [DOI] [PubMed] [Google Scholar]
- 39.Peart J.R., Lu R., Sadanandom A., Malcuit I., Moffett P., Brice D.C.et al. (2002) Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. U.S.A. 99, 10865–10869 10.1073/pnas.152330599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee H., Kim S., Oh S., Yeom S., Kim S., Kim M.et al. (2014) Multiple recognition of RXLR effectors is associated with nonhost resistance of pepper against Phytophthora infestans. New Phytol. 203, 926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adlung N. and Bonas U. (2016) Non-host resistance induced by the Xanthomonas effector XopQ is widespread within the genus Nicotiana and functionally depends on EDS1. Front. Plant Sci. 7, 1796 10.3389/fpls.2016.01796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabro G., Steinbrenner J., Coates M., Ishaque N., Baxter L., David J.et al. (2011) Multiple candidate effectors from the oomycete pathogen Hyaloperonospora arabidopsidis suppress host plant immunity. PLoS Pathog. 7, e1002348 10.1371/journal.ppat.1002348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei C.F., Kvitko B.H., Shimizu R., Crabill E., Alfano J.R., Lin N.C.et al. (2007) A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 51, 32–46 10.1111/j.1365-313X.2007.03126.x [DOI] [PubMed] [Google Scholar]
- 44.Sohn K.H., Saucet S.B., Clarke C.R., Vinatzer B.A., O’Brien H.E., Guttman D.S.et al. (2012) HopAS1 recognition significantly contributes to Arabidopsis nonhost resistance to Pseudomonas syringae pathogens. New Phytol. 193, 58–66 10.1111/j.1469-8137.2011.03950.x [DOI] [PubMed] [Google Scholar]
- 45.Inoue Y., Vy T.T.P., Yoshida K., Asano H., Mitsuoka C., Asuke S.et al. (2017) Evolution of the wheat blast fungus through functional losses in a host specificity determinant. Science 357, 80–83 10.1126/science.aam9654 [DOI] [PubMed] [Google Scholar]
- 46.Niks R.E., Qi X. and Marcel T.C. (2015) Quantitative resistance to biotrophic filamentous plant pathogens: concepts, misconceptions, and mechanisms. Annu. Rev. Phytopathol. 53, 445–470 10.1146/annurev-phyto-080614-115928 [DOI] [PubMed] [Google Scholar]
- 47.Vega-Arreguín J.C., Shimada-Beltrán H., Sevillano-Serrano J. and Moffett P. (2017) Non-host plant resistance against Phytophthora capsici is mediated in part by members of the I2 R gene family in Nicotiana spp. Front. Plant Sci. 8, 1–11 10.3389/fpls.2017.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao B.Y., Ardales E., Brasset E., Claflin L.E., Leach J.E. and Hulbert S.H. (2004) The Rxo1/Rba1 locus of maize controls resistance reactions to pathogenic and non-host bacteria. Theor. Appl. Genet. 109, 71–79 10.1007/s00122-004-1623-y [DOI] [PubMed] [Google Scholar]
- 49.Zhao B., Lin X., Poland J., Trick H., Leach J. and Hulbert S. (2005) A maize resistance gene functions against bacterial streak disease in rice. Proc. Natl. Acad. Sci. U.S.A. 102, 15383–15388 10.1073/pnas.0503023102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borhan M.H., Gunn N., Cooper A., Gulden S., Tör M., Rimmer S.R.et al. (2008) WRR4 encodes a TIR-NB-LRR protein that confers broad-spectrum white rust resistance in Arabidopsis thaliana to four physiological races of Albugo candida., Mol. Plant Microbe Interact. 21, 757–768 10.1094/MPMI-21-6-0757 [DOI] [PubMed] [Google Scholar]
- 51.Borhan M.H., Holub E.B., Kindrachuk C., Omidi M., Bozorgmanesh-frad G. and Rimmer S.R. (2010) WRR4, a broad-spectrum TIR-NB-LRR gene from Arabidopsis thaliana that confers white rust resistance in transgenic oilseed brassica crops. Mol. Plant Pathol. 11, 283–291 10.1111/j.1364-3703.2009.00599.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawashima C.G., Guimarães G.A., Nogueira S.R., MacLean D., Cook D.R., Steuernagel B.et al. (2016) A pigeonpea gene confers resistance to Asian soybean rust in soybean. Nat. Biotechnol. 34, 661–665 10.1038/nbt.3554 [DOI] [PubMed] [Google Scholar]
- 53.Bettgenhaeuser J., Herna I., Green P., Taylor J., Smoker M., Ferguson J.N.et al. (2021) The barley immune receptor Mla recognizes multiple pathogens and contributes to host range. Nat. Commun. 12, 1–14 10.1038/s41467-021-27288-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schulze-lefert P. and Panstruga R.A. (2011) Molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 16, 117–125 10.1016/j.tplants.2011.01.001 [DOI] [PubMed] [Google Scholar]
- 55.Redkar A., Cevik V., Bailey K., Furzer O.J., Fairhead S., Borhan M.H.et al. (2021) The Arabidopsis WRR4A and WRR4B paralogous NLR proteins both confer recognition of multiple Albugo candida effectors. bioRxi 10.1101/2021.03.29.436918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Witek K., Jupe F., Witek A.I., Baker D., Clark M.D. and Jones J.D.G. (2016) Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nat. Biotechnol. 34, 656–660 10.1038/nbt.3540 [DOI] [PubMed] [Google Scholar]
- 57.Witek K., Lin X., Karki H.S., Jupe F., Witek A.I., Steuernagel B.et al. (2021) A complex resistance locus in Solanum americanum recognizes a conserved Phytophthora effector. Nat. Plants 7, 198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin X., Olave-Achury A., Heal R., Witek K., Karki H.S., Song T.et al. (2021) Rpi-amr3 confers resistance to multiple Phytophthora species by recognizing a conserved RXLR effector. bioRxi 10.1101/2021.06.10.447899 [DOI] [PubMed] [Google Scholar]
- 59.Carter M.E., Helm M., Chapman A.V.E., Wan E., Sierra A.M.R., Innes R.W.et al. (2019) Convergent evolution of effector protease recognition by arabidopsis and barley. Mol. Plant Microbe Interact. 32, 550–565 10.1094/MPMI-07-18-0202-FI [DOI] [PubMed] [Google Scholar]
- 60.Prokchorchik M., Choi S., Chung E.H., Won K., Dangl J.L. and Sohn K.H. (2020) A host target of a bacterial cysteine protease virulence effector plays a key role in convergent evolution of plant innate immune system receptors. New Phytol. 225, 1327–1342 10.1111/nph.16218 [DOI] [PubMed] [Google Scholar]
- 61.de Guillen K., Ortiz-Vallejo D., Gracy J., Fournier E., Kroj T.et al. (2015) Structure analysis uncovers a highly diverse but structurally conserved effector family in phytopathogenic fungi. PLoS Pathog. 11, e1005228 10.1371/journal.ppat.1005228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo L., Cesari S., de Guillen K., Chalvon V., Mammri L.et al. (2018) Specific recognition of two MAX effectors by integrated HMA domains in plant immune receptors involves distinct binding surfaces. Proc. Natl. Acad. Sci. U.S.A. 115, 11637–11642 10.1073/pnas.1810705115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martel A., Ruiz-Bedoya T., Breit-McNally C., Laflamme B., Desveaux D. and Guttman D.S. (2021) The ETS-ETI cycle: evolutionary processes and metapopulation dynamics driving the diversification of pathogen effectors and host immune factors. Curr. Opin. Plant Biol. 62, 102011 10.1016/j.pbi.2021.102011 [DOI] [PubMed] [Google Scholar]
- 64.Bourras S., McNally K.E., Ben-David R., Parlange F., Roffler S., Praz C.R.et al. (2015) Multiple avirulence loci and allele-specific effector recognition control the Pm3 race-specific resistance of wheat to powdery mildew. Plant Cell 27, 2991–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bourras S., Kunz L., Xue M., Praz C.R., Müller M.C., Kälin C.et al. (2019) The AvrPm3-Pm3 effector-NLR interactions control both race-specific resistance and host-specificity of cereal mildews on wheat. Nat. Commun. 10, 1–16 10.1038/s41467-019-10274-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Champouret N., Bouwmeester K., Rietman H., van der Lee T., Maliepaard C., Heupink A.et al. (2009) Phytophthora infestans isolates lacking class I ipiO variants are virulent on Rpi-blbl potato. Mol. Plant Microbe Interact. 22, 1535–1545 10.1094/MPMI-22-12-1535 [DOI] [PubMed] [Google Scholar]
- 67.Chen Y., Liu Z. and Halterman D.A. (2012) Molecular determinants of resistance activation and suppression by Phytophthora infestans effector IPI-O. PLoS Pathog. 8, e1002595 10.1371/journal.ppat.1002595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Derevnina L., Contreras M.P., Adachi H., Upson J., Cruces A.V., Xie R.et al. (2021) Plant pathogens convergently evolved to counteract redundant nodes of an NLR immune receptor network. PLoS Biol. 19, e3001136 10.1371/journal.pbio.3001136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu G., Xian L., Xue H., Yu W., Rufian J.S., Sang Y.et al. (2020) A bacterial effector protein prevents mapk-mediated phosphorylation of sgt1 to suppress plant immunity. PLoS Pathog. 16, 1–30 10.1371/journal.ppat.1008933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakano M., Ichinose Y. and Mukaihara T. (2021) Ralstonia solanacearum Type III effector RipAC targets SGT1 to suppress effector-triggered immunity. Plant Cell Physiol. 61, 2067–2076 10.1093/pcp/pcaa122 [DOI] [PubMed] [Google Scholar]
- 71.Medina C.A., Reyes P.A., Trujillo C.A., Gonzalez J.L., Bejarano D.A., Montenegro N.A.et al. (2018) The role of type III effectors from Xanthomonas axonopodis pv. manihotis in virulence and suppression of plant immunity. Mol. Plant Pathol. 19, 593–606 10.1111/mpp.12545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu C.H., Abd-El-Haliem A., Bozkurt T.O., Belhaj K., Terauchi R., Vossen J.H.et al. (2017) NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. U.S.A. 114, 8113–8118 10.1073/pnas.1702041114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu C.H., Belhaj K., Bozkurt T.O., Birk M.S. and Kamoun S. (2015) Helper NLR proteins NRC2a/b and NRC3 but not NRC1 are required for Pto-mediated cell death and resistance in Nicotiana Benthamiana. New Phytol. 209, 1344–1352 [DOI] [PubMed] [Google Scholar]
- 74.Qi T., Seong K., Thomazella D.P.T., Ryan J., Pham J., Seo E.et al. (2018) NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc. Natl. Acad. Sci. U.S.A. 115, E10979–E10987 10.1073/pnas.1814856115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Collier S.M., Hamel L. and Moffett P. (2011) Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant Microbe Interact. 24, 918–931 10.1094/MPMI-03-11-0050 [DOI] [PubMed] [Google Scholar]
- 76.Díaz-tatis P.A., Ochoa J.C., García L., Chavarriaga P., Bernal A.J. and López C.E. (2019) Interfamily transfer of Bs2 from pepper to cassava (Manihot esculenta Crantz). Trop. Plant Pathol. 44, 225–237 10.1007/s40858-019-00279-y [DOI] [Google Scholar]
- 77.Lapin D., Kovacova V., Sun X., Dongus J.A., Bhandari D., Von Born P.et al. (2019) A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 31, 2430–2455 10.1105/tpc.19.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ngou B.P.M., Ahn H.K., Ding P. and Jones J.D.G. (2021) Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115 [DOI] [PubMed] [Google Scholar]
- 79.Pruitt R.N., Locci F., Wanke F., Zhang L., Saile S.C., Joe A.et al. (2021) The EDS1–PAD4–ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature 598, 495–499 10.1038/s41586-021-03829-0 [DOI] [PubMed] [Google Scholar]
- 80.Yuan M., Jiang Z., Bi G., Nomura K., Liu M., Wang Y.et al. (2021) Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]