Abstract
Dormant Bacillus subtilis spores germinate in response to specific nutrients called germinants, which are recognized by multisubunit receptor complexes encoded by members of the gerA family of operons, of which the gerB operon is a member. The germinant receptors are expected to be membrane associated, but there is some debate about whether they are located in the inner or outer spore membrane. In this study we have used Western blot analysis to determine the precise location of GerBA, a gerB-encoded receptor protein, in various spore fractions. GerBA was not extracted from spores by a decoating treatment that removes the coat and outer membrane but was present in lysates from decoated spores and in the insoluble fraction (termed P100) from such lysates that contained inner-membrane vesicles. GerBA was also solubilized from the P100 fraction with detergent but not with high salt. These findings suggest that GerBA is an integral membrane protein located in the spore's inner membrane. Consistent with this idea, GerBA was present in the cell membrane of the outgrowing spore, a membrane that is derived from the dormant spore's inner membrane. Based on these observations we propose that GerBA and probably the entire GerB germinant receptor are located in the inner membrane of the dormant spore. We also estimated that there are only 24 to 40 molecules of GerBA per spore, a number that is consistent with the previously reported low level of gerB operon expression and with the putative receptor function of the proteins encoded by the gerB operon.
Dormant spores of Bacillus subtilis are metabolically inert structures that are formed when growing cells are starved for particular nutrients. Despite their dormancy, these spores can detect nutrients in the environment and respond to such cues by triggering a number of reactions that result in the resumption of metabolism and growth. The initial steps in this process, called spore germination, thus represent a novel paradigm for studying the communication between a cell and its surroundings.
A variety of nutrients can induce spore germination, and in B. subtilis two such germinants have been extensively studied: l-alanine (l-Ala) and a combination of l-asparagine, d-fructose, d-glucose, and potassium ions (AFGK) (19). Previous work suggested that germinants act by binding to and activating spore receptors (9, 13, 19). This idea has been substantiated by several types of experiments which suggest that a family of homologous operons (the gerA family, which includes gerA, gerB, and gerK) encode the predicted receptors. First, mutations in gerA, gerB, and gerK block germination in a germinant-specific manner (17–19). Second, missense mutations in one family member, gerB, allow spores to recognize novel germinants (23). Finally, spores lacking all of the gerA family members show a severe defect in nutrient-induced germination (24). At the DNA sequence level, each gerA family member is a tricistronic operon that encodes three predicted proteins named A, B, and C, and there is evidence that the germinant receptor is a multisubunit complex consisting of at least the A and B proteins (23). The A and B proteins contain several putative transmembrane domains and have been suggested to be integral membrane proteins, whereas the C proteins contain a potential signal sequence for lipid modification (34). Thus, it is reasonable to postulate that the receptor complex is associated with a membrane in the dormant spore.
There are two membranes in the dormant spore—a consequence of the spore's development as a double membrane-bound endospore within a mother cell (8). The inner membrane is derived from the forespore compartment, and the outer membrane is derived from the mother cell. The two membranes are separated by two layers of peptidoglycan—the inner germ cell wall and the outer, less cross-linked cortex—and the outer membrane is overlain by a proteinaceous coat (8). The position of the outer membrane-coat in the spore make it a likely location for receptors that detect environmental cues. However, this location is not supported by the finding that detergent treatments that remove the outer membrane-coat do not abolish nutrient-induced spore germination (24, 32). Direct localization of the GerA receptor proteins has been attempted, but their location has not been determined unambiguously. One study which used immunoelectron microscopy claimed that GerAA, GerAB, and GerAC were located at the cortex-coat boundary (i.e., in the outer membrane) (27, 33), while another study which used Western blot analysis suggested that GerAA was located in the inner membrane and GerAC was located in the integument fraction (coats plus cortex) (17).
To further attempt to pinpoint the location of the germinant receptors in dormant spores, we immunolocalized a GerB receptor component, GerBA, in spore fractions. The antiserum against GerBA was produced against a fusion protein containing two-thirds of GerBA, affinity purified and shown to be specific for GerBA. The GerBA protein was not removed by detergent treatment of dormant spores, which removes the outer membrane and coat (4, 32). After disruption of dormant decoated spores, GerBA was localized in an insoluble pellet fraction which contained the inner membrane, and GerBA was located in the membrane of outgrowing spores that had shed their coat-outer membrane complex. Together, these observations indicate that GerBA is located in the inner membrane of the spore and suggest that the inner membrane is the site of germinant-receptor interaction. A very recent study also concluded that GerAA and -AC are located in the spore's inner membrane (11a).
MATERIALS AND METHODS
Construction of strains and plasmids.
The B. subtilis strains used in this study are listed in Table 1. B. subtilis transformations were performed as described previously (1), and Southern blot analysis (28) was used to confirm the proper integration of plasmids during strain construction. The Escherichia coli strain TG1 was used for plasmid manipulation (28), and strain BL21 λ(DE3) (Novagen) was used for protein expression. E. coli was transformed by the CaCl2 method (28).
TABLE 1.
B. subtilis strains useda
| Strain | Genotype | Source or reference |
|---|---|---|
| PS832 | Prototrophic derivative of strain 168 | Laboratory stock |
| FB49 | amyE::gerB | 23 |
| FB58 | PsspB::gerB | pFE135A→PS832 |
| FB60 | ΔgerB::spc | 23 |
| FB72 | ΔgerA::spc ΔgerB::cat ΔgerK::ermC | 24 |
| FB86 | ΔgerA::spc ΔgerK::ermC | 24 |
All strains were derived from PS832.
Plasmid pFE254, which was used to overexpress the N-terminal two-thirds of GerBA as a 10× His tag fusion, was derived from plasmid pET16b (Novagen). The 5′ region of the gerBA gene (from nucleotide −3 to +566 relative to the +1 translation start site of gerBA) was PCR amplified from wild-type B. subtilis chromosomal DNA with primers pETBA-5 (5′-GCATATGCAAATCGACTCTGATCTC) and pETBA-3 (5′-AGATCTCGGCCGGGGCGATATCCTGAATATAAG). The pETBA-5 primer introduced an NdeI site (underlined) at the translation start site of gerBA, while primer pETBA-3 introduced EagI and BglII sites (underlined) at the 3′ end of the amplified fragment. The PCR product was cloned into the TA vector pCR2.1 (Invitrogen) to create plasmid pFE112. A 1.4-kb EcoRV-EagI fragment containing the remainder of the gerBA gene was excised from plasmid pFE24 (23) and inserted between the same sites in plasmid pFE112. The resulting plasmid, pFE113, contained the entire gerBA open reading frame bracketed by an NdeI site at the start codon and a BglII site 500 bp downstream of the stop codon. The NdeI-BamHI (internal site in gerBA) fragment from pFE113, which encodes the N-terminal two-thirds of GerBA, was inserted between the same sites in plasmid pET16b (Novagen) to generate plasmid pFE254.
Plasmid pFE135A, which was used to overexpress the gerB operon during sporulation, was prepared by fusing the gerB operon to the promoter (PsspB) of the sspB gene, which is expressed at high levels during sporulation in a forespore-specific manner (5). The promoter (from nucleotide −191 to +10 relative to the +1 translation start site of sspB) was amplified from wild-type chromosomal DNA with primers PsspB (5′-AAGCTTTTTTTATTTCTC) and PsspBNdeI (5′-AAGCTTGGTTAGCCATATGTAAAATCTCC). Primer PsspBNdeI contains an NdeI site (underlined) at the sspB initiating codon, which can be used to fuse open reading frames directly downstream of the sspB promoter and ribosome-binding sequence. The PCR product was cloned into the TA vector pCR2.1 to create plasmid pFE136A, whose insert was confirmed by sequencing. The 220-bp HindIII fragment containing PsspB from pFE136A was inserted into the HindIII site of plasmid pUC19M (Clontech Laboratories), and one resulting plasmid in which the fragment had inserted so that its NdeI site was closer to the unique BamHI site in pUC19M was selected as plasmid pFE133. The NdeI-BglII fragment from plasmid pFE113, which contains the entire gerBA coding region, was inserted between the NdeI and BamHI sites of plasmid pFE133 to generate plasmid pFE134. The HindIII fragment from plasmid pFE134, which contains PsspB fused to 1,316 bp (about 90%) of the gerBA gene, was inserted into the HindIII site of plasmid pFE52, a pBlueScript plasmid (Stratagene) containing the spectinomycin-resistance gene (spc) derived from pJL74 (15), to create plasmid pFE135A. Plasmid pFE135A was integrated into the chromosome at the gerB locus by transformation of B. subtilis to spectinomycin resistance, resulting in the insertion of PsspB directly upstream of an intact gerB operon.
Spore preparation, cleaning, and decoating.
Spores were prepared by the nutrient exhaustion method (22) or the resuspension method (30) and cleaned by washing as previously described (22). All of the spore preparations contained >95% phase-bright spores and were essentially free of sporulating cells and cell debris. Spores were decoated by treatment for 30 min at 70°C with 0.1 M NaCl–0.1 M NaOH–1% sodium dodecyl sulfate (SDS)–0.1 M dithiothreitol, and the treated spores were washed as described previously (2). This decoating procedure also removes outer spore membrane proteins (4 and data not shown).
Preparation of spore extracts.
Spore extracts were prepared by a modification of a previously described method (2). Untreated or decoated spores (∼20 mg [dry weight]) were lyophilized and pulverized with 100 mg of glass beads in a dental amalgamator (Wig-L-Bug) for 20 pulses of 30 s each with a 30-s cooling period between pulses. Proteins were extracted from the disrupted spores with 0.5 ml of TEP buffer (50 mM Tris-HCl [pH 7.4], 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride) containing 1% SDS and 0.15 M β-mercaptoethanol by incubation at 70°C for 30 min, and the insoluble material was removed by centrifugation in a microcentrifuge (13,000 × g for 5 min at 4°C).
Preparation of spore lysates and their fractionation.
Lysates and membranes from dormant spores were prepared as described previously (2). Briefly, decoated dormant spores were resuspended at an optical density at 600 mm (OD600) of 50 to 70 in 0.5 ml of TEP buffer containing 1 mg of lysozyme, 1 μg each of RNase A and DNase I, and 20 μg of MgCl2, incubated at 37°C for 5 min, and then kept on ice for 20 min. The spore suspension was then sonicated with 100 mg of glass beads and examined microscopically for lysis. After >80% of the spores had lysed (approximately five 15-s bursts of sonication), the fluid was recovered, the glass beads were washed with 0.5 ml of TEP buffer, and the wash was pooled with the recovered fluid and centrifuged for 5 min in a microcentrifuge to remove unbroken spores and integument debris. This first supernatant fluid (termed the lysate) was centrifuged at 100,000 × g for 1 h, giving a soluble fraction (S100) and a pellet fraction (P100); the latter was resuspended in 40 to 80 μl of TEP buffer containing 1% Triton X-100. The lysate and S100 fractions were concentrated 10- to 20-fold in a centrifugal filter device (Microcon YM-3, with a 3,000-molecular-weight cutoff) as recommended by the manufacturer (Millipore, Bedford, Mass.).
Membranes from outgrowing spores were prepared as follows. Heat-activated (70°C, 30 min) spores (OD600 of 50) were germinated in 5 ml of 10 mM Tris-HCl (pH 8.4) and 10 mM l-Ala for 1 h at 37°C, diluted into 45 ml of 2× YT medium (2), and incubated with shaking at 28°C for 2 h, by which time ≥70% of the outgrowing spores had a rod-like morphology. The outgrowing spores were harvested by centrifugation at 3,000 × g, resuspended in 1 ml of TEP buffer containing 1 μg of DNase I, 1 μg of RNase A, 20 μg of MgCl2, and 1 mg of lysozyme, incubated for 3 min at 37°C followed by 30 min on ice, and then sonicated (three 15-s bursts). Cell debris and unbroken cells were removed by centrifugation in a microcentrifuge for 5 min at 13,000 × g. The resultant supernatant fluid was centrifuged at 100,000 × g for 1 h, and the pellet fraction (P100) was resuspended in 40 to 80 μl of TEP buffer.
Production and purification of antibodies.
The His10-GerBA fusion protein was produced in BL21 λ(DE3) E. coli cells carrying plasmid pFE254. Fusion protein expression was induced by addition of isopropyl-β-d-thiogalactoside (IPTG) to 2 mM to an actively growing (OD600 of ∼0.6) culture at 25°C in 2× YT medium; cells were harvested 4 h after IPTG addition. The growth temperature of 25°C was chosen to increase the amount of soluble GerBA formed, because the inclusion bodies formed at 37°C were not solubilized with 8 M urea or 6 M guanidinium hydrochloride and interfered with subsequent purification. The soluble His10-GerBA protein was purified by Ni2+ affinity chromatography as specified by the supplier of the Ni2+ resin (Novagen). The purified protein had a tendency to precipitate in concentrated solution and therefore was supplied for antibody production (Pocono Rabbit Farm and Laboratory, Canadensis, Pa.) as a sonicated suspension at 0.8 mg per ml in 20 mM Tris-HCl (pH 8.0)–50 mM NaCl–0.1 mM phenylmethylsulfonyl fluoride. Anti-His10-GerBA antibodies were detected in a bleed 2 months after initial antigen injection, at which time the animals were exsanguinated.
The antiserum was purified by affinity chromatography as follows. Purified His10-GerBA (1 mg/ml) was dialyzed exhaustively against 0.1 M sodium phosphate (pH 7.4) at 4°C, solubilized with 0.5% SDS, and covalently attached to CNBr-activated Sepharose 4 Fast Flow beads (Amersham Pharmacia Biotech AB) by incubation at 4°C according to the manufacturer's specifications, and the beads were packed into a 2-ml affinity column. The antiserum (80 ml) was initially partially purified by ammonium sulfate precipitation and then affinity purified as described previously (11). The purified antiserum was concentrated to 3 mg of protein per ml in a Centriprep concentrator (Amicon) and stored at −20°C in phosphate-buffered saline. The antiserum recognized a 45- to 50-kDa protein in extracts made from E. coli cells expressing the untagged full-length GerBA protein, suggesting that at least some of the purified antibodies recognized the GerBA moiety.
Western blot analysis.
GerBA in extracts, lysates, and lysate fractions was detected by Western blot analysis. The protein preparations were suspended in 1× sample buffer (11), heated to 80°C for 15 min, and separated by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were transferred to a polyvinylidenedifluoride membrane (Immobilon) (11). The GerBA protein on the membrane was detected using a 1:1,000 dilution of the affinity-purified anti-His10-GerBA serum and a 1:10,000 dilution of goat anti-rabbit immunoglobulin G-alkaline phosphatase conjugate (Southern Biotechnology Associates, Birmingham, Ala.) in 1× Tris-buffered saline as described previously (11).
RESULTS
Detection of GerBA.
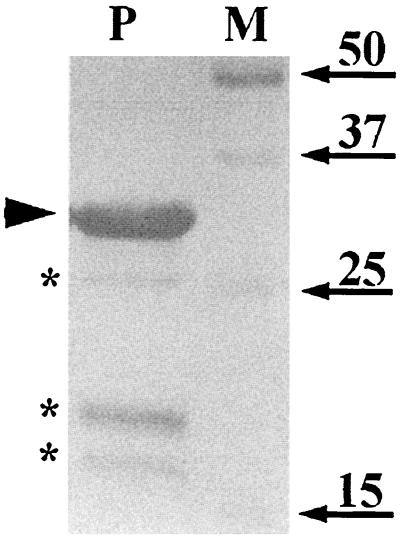
We chose to study the localization of the GerBA protein because of the strong evidence that it functions in a complex with at least the GerBB protein as a germinant receptor (23). The gerB operon is expressed at only a very low level in wild-type spores (6), and efforts to visualize GerBA by Coomassie blue staining of total spore proteins or spore membrane proteins resolved by one- or two-dimensional gel electrophoresis systems were unsuccessful (data not shown). Overexpressing the gerB operon in B. subtilis, either on a multicopy (pUB110-based) plasmid or by fusing it to the vegetative xylA or forespore-specific sspB promoters, also did not result in a detectable GerBA signal on Coomassie blue-stained gels (data not shown). Epitope tagging of GerBA was also not possible because both N- and C-terminal tags resulted in nonfunctional GerBA protein (data not shown). Therefore, we decided to detect GerBA by immunoblot analysis. While some previous attempts at germinant receptor protein localization have used antipeptide antibodies (27, 33), we decided to prepare polyclonal antiserum against GerBA itself. Although full-length GerBA would have made the ideal antigen for antiserum production, we could not overexpress this protein at detectable levels in any of several E. coli expression systems (data not shown). Consequently, we used a fusion protein, His10-GerBA, which contains the N-terminal two-thirds of GerBA fused downstream from a 10× His tag. The fusion protein was expressed at high levels (about 20% of total protein) in E. coli, and the soluble fraction was purified by Ni2+ affinity chromatography. SDS-PAGE analysis of the purified protein preparation showed that it contained a major species of 29 kDa, which is the expected size of the His10-GerBA fusion protein, and minor species of 18, 20, and 25 kDa (Fig. 1). N-terminal sequence analysis showed that the proteins in all four of the latter bands had the same His-tagged amino terminus, indicating that the smaller species are degradation products of the His10-GerBA fusion protein. The purified His10-GerBA was used to immunize naive rabbits, and the resulting antiserum was affinity purified as described in Materials and Methods.
FIG. 1.
SDS-PAGE analysis of purified His10-GerBA protein. Synthesis of the His10-GerBA protein was induced in E. coli cells of strain BL21 λ(DE3) carrying plasmid pFE254, the soluble proteins were isolated, and GerBA was purified as described in Materials and Methods. An aliquot (8 μg) of the purified protein preparation (lane P) was run on an SDS-10% PAGE gel and visualized by Coomassie blue staining. The arrowhead points to the major species that was purified, and asterisks mark the smaller species, which are degradation products of the major protein (see text). Arrows indicate the migration positions of the molecular mass markers in kilodaltons (lane M).
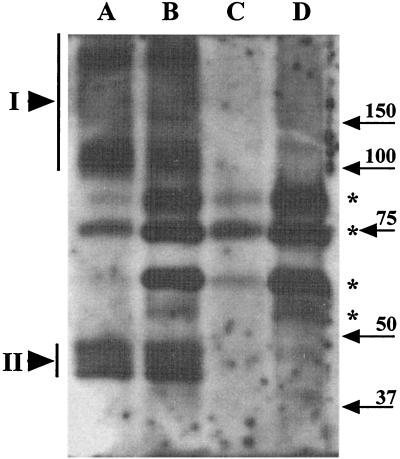
To test the specificity of the antiserum towards GerBA, total SDS-soluble proteins from disrupted spores (henceforth referred to as “extracts”) of strains lacking the gerB operon (FB60) or overexpressing it (FB58, about 500-fold overexpression; see below) under the control of the strong, forespore-specific, sspB promoter (PsspB::gerB) were run on an SDS-PAGE gel and subjected to Western blot analysis (Fig. 2, lanes A and C). Although a number of bands were detected in both the ΔgerB and the PsspB::gerB spore extracts (Fig. 2, marked with asterisks), these same bands were also detected with preimmune serum (data not shown). However, a set of bands was detected which was specific to the PsspB::gerB spore extract (Fig. 2, lane A). This latter set included a series of four bands in the 40- to 50-kDa range (Fig. 2, lane A; region labeled II); since the predicted molecular mass of GerBA is ∼54 kDa, this set of bands very likely represents GerBA. Additional bands (Fig. 2, lane A; region labeled I) were observed at higher positions (≥100 kDa) in the gel; a putative GerBA degradation product was also seen much lower (30 kDa) in the gel (data not shown). In contrast to the specific GerBA signal in extracts from the PsspB::gerB spores, we failed to detect any GerBA signal in extracts from wild-type spores (data not shown). This is probably due to the extremely low level of gerB expression in wild-type spores (6) because, as described below, we could detect GerBA in specific fractions from wild-type spores.
FIG. 2.
GerBA in extracts from untreated and decoated spores. Extracts from identical amounts (∼3 OD600 units) of disrupted spores without (lanes A and C) or with (lanes B and D) prior decoating treatment as described in Materials and Methods were run on an SDS-PAGE gel and transferred to an Immobilon membrane, and GerBA was detected as described in Materials and Methods. The spores were from strains FB58 (PsspB::gerB; lanes A and B) and FB60 (ΔgerB; lanes C and D). Bands marked with an asterisk are nonspecific background bands, whereas those in regions marked I and II represent GerBA-specific signals; bands in region II lie close to the predicted GerBA molecular mass (53 kDa), whereas those in region I probably contain highly modified or complexed forms of GerBA. Migration positions of molecular mass markers in kilodaltons are indicated with arrows.
Effect of decoating on levels of GerBA in spores.
A proteinaceous coat and an underlying outer membrane form the outermost layers of the spore integument—a possible site for germinant receptor location. To determine if GerBA is located in these outer layers, we subjected spores to a decoating treatment (see Materials and Methods) which removes the coat and outer membrane (4, 14, 32) and examined treated spores for GerBA. Extracts (see above and Materials and Methods) from untreated or decoated spores of a ΔgerB strain (FB60) and a PsspB::gerB strain (FB58) were run on an SDS-PAGE gel and analyzed for GerBA by Western blotting (Fig. 2). As noted above, some nonspecific bands (Fig. 2, marked with asterisks) were seen in extracts from PsspB::gerB and ΔgerB spores and a subset of these bands appeared to be stronger in the extracts from decoated spores. In contrast, the GerBA signal was similar in the extracts from untreated or decoated PsspB::gerB spores (Fig. 2, lanes A and B) and, as expected, absent in extracts from ΔgerB spores (Fig. 2, lanes C and D). Thus, GerBA was not removed by treatments that remove the spore coat and outer membrane. Consistent with this observation, we did not detect any GerBA in the coat extracts themselves (data not shown). Thus, GerBA is not extracted by treatments that solubilize the outer layers of the spore integument.
One might argue that the validity of the above conclusion is contingent upon the overexpressed GerBA being both functional and correctly located. To address these issues, we first determined if the PsspB::gerB operon was functional by examining the ability of the overexpressed GerB proteins to induce spore germination in response to AFGK. Spores of strain FB58, in which PsspB::gerB represents the only complete copy of the gerB operon, germinated similarly to wild-type (PS832) spores in AFGK (data not shown) while spores lacking the gerB operon do not (19, 24). Since the PsspB::gerB construct complemented the disrupted gerB operon in strain FB58, this suggests that at least a portion of the overexpressed GerB proteins is functional and therefore likely to be correctly located. In addition, the absence of any overexpressed GerBA in coat extracts did not suggest an outer membrane location for this protein and other experiments (see below) showed that the location of the overexpressed GerBA protein mirrored the location of the endogenous protein. Thus, although we cannot completely rule out that some of the overexpressed GerBA protein was localizing incorrectly, at least the great majority of the overexpressed GerBA protein did localize correctly.
Location of GerBA protein in spore lysates.
Because GerBA was not removed from spores by a decoating treatment, it seemed likely that GerBA is located within the spore, presumably in the inner membrane. To more precisely locate GerBA within spores, we made lysates from decoated spores by treatment with lysozyme. These detergent-free lysates, which contain spore core proteins as well as inner membrane vesicles, were concentrated 20-fold by filtration and subjected to Western blot analysis (Fig. 3). A GerBA signal was detected in the lysates from spores of wild-type (PS832) and ΔgerA ΔgerK (FB86) strains but not in those of the ΔgerB (FB60) or ΔgerA ΔgerB ΔgerK (FB72) strain. The size and distribution of the 40- to 50-kDa GerBA signal in the lysates from wild-type spores (Fig. 3, region labeled II) were essentially identical to those observed with extracts from disrupted spores of the PsspB::gerB strain (Fig. 2, bands in region labeled II). This observation supports the earlier conclusion that the 40- to 50-kDa signal represents the GerBA protein. The lysates also contained several background bands that were distinct from those seen in the extracts (compare Fig. 2 and 3); consequently, a ΔgerB lysate was included as a control in subsequent experiments to facilitate unambiguous identification of GerBA. Note that the background bands could not be attributed to the homologous proteins encoded by the gerA or gerK operons (Fig. 3, lanes C and D).
FIG. 3.
GerBA in spore lysates. Lysates from equal amounts (15 OD600 units) of spores of strains PS832 (wild type; lane A), FB60 (ΔgerB; lane B), FB86 (ΔgerA ΔgerK; lane C), and FB72 (ΔgerA ΔgerB ΔgerK; lane D) were run on an SDS-PAGE gel and transferred to an Immobilon membrane, and GerBA was detected as described in Materials and Methods. The arrowhead points to GerBA bands in region II as described in the legend to Fig. 2. Background bands and molecular mass markers are labeled as in Fig. 2. Note that the samples run in Fig. 2 were from spores that overexpressed GerBA while the samples run in this experiment were from spores that did not overexpress GerBA. That difference and the low exposure level of the Western blot shown here account for the relative lack of higher molecular mass GerBA observed in this figure compared to Fig. 2 and 6.
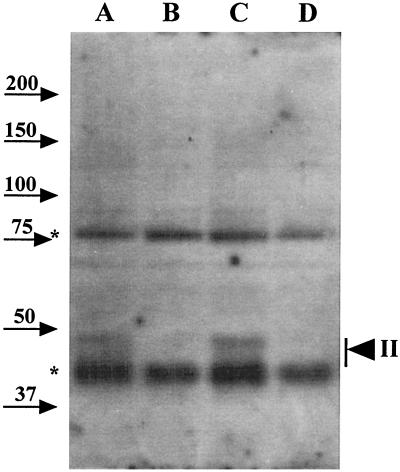
To further define the location of GerBA, the lysates were fractionated by centrifugation at 100,000 × g into supernatant (S100) and pellet (P100) fractions. The P100 fraction is expected to contain large nucleoprotein complexes as well as inner membrane vesicles (25, 26, 31), and YhcN, a putative inner spore membrane protein, has been found in this fraction (2). The total lysate and S100 and P100 fractions from equal amounts of spores were subjected to Western blot analysis, and most of the GerBA protein from the total lysate was recovered in the P100 fraction (Fig. 4, lanes A, B, E, and F) with no detectable GerBA signal in the S100 fraction (Fig. 4, lanes C and D). Since the lysates had been treated with nucleases prior to centrifugation to minimize the level of nucleoprotein complexes in the P100 fraction (25, 26), these observations suggest that GerBA is associated with the spore inner membrane.
FIG. 4.
GerBA in fractionated spore lysates. Lysates from equal amounts (15 OD600 units) of decoated spores of strains PS832 (wild type; lanes A, C, and E) and FB60 (ΔgerB; lanes B, D, and F) were subjected to centrifugation at 100,000 × g for 1 h. The initial lysate (lanes A and B), S100 supernatant fractions (lanes C and D), and P100 pellet fractions (lanes E and F) were run on an SDS-PAGE gel, proteins were transferred to an Immobilon membrane, and GerBA was detected as described in Materials and Methods. The arrowheads denoting GerBA bands, the background bands, and molecular mass markers are labeled as in Fig. 2.
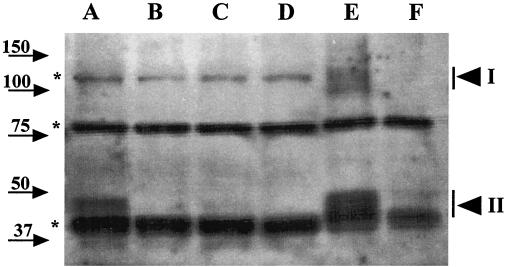
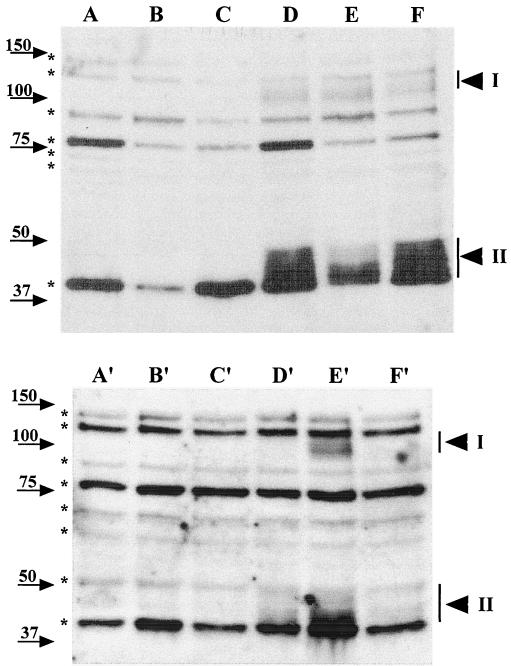
To further characterize the location of GerBA, we determined if detergent and salt treatment affected GerBA partitioning into the P100 fraction. Lysates from ΔgerB or wild-type spores were divided into aliquots that were subjected to no treatment or incubated with 1% Triton X-100 or 0.5 M NaCl for 30 min on ice. Each aliquot was centrifuged at 100,000 × g for 1 h, and GerBA in the P100 and S100 fractions was analyzed by Western blot analysis (Fig. 5). Triton X-100 treatment markedly reduced (by about 70%) the amount of GerBA protein recovered in the P100 fraction (Fig. 5, lanes D and E) with a comparable increase in GerBA in the S100 fraction (Fig. 5, lanes D′ and E′). In contrast, treatment with high salt did not affect GerBA location (Fig. 5, lanes D, F, D′, and F′). These data suggest that GerBA is an integral membrane protein, which agrees with the hydropathy profile of the predicted GerBA protein (17, 34).
FIG. 5.
Effects of detergent and salt treatments on GerBA fractionation. Lysates from decoated spores (15 OD600 units) of strain FB60 (ΔgerB; lanes A to C and A′ to C′) or strain PS832 (wild type; lanes D to F and D′ to F′) were incubated with no additions (lanes A, D, A′, and D′), with 1% Triton X-100 (lanes B, E, B′, and E′), or with 0.5 M NaCl (lanes C, F, C′, and F′) for 30 min on ice, followed by centrifugation at 100,000 × g to obtain the P100 pellet (lanes A to F) and S100 supernatant (lanes A′ to F′) fractions. The P100 fractions from 10 OD600 units of spores and S100 fractions from 7 to 8 OD600 units of spores were run on an SDS-PAGE gel and transferred to an Immobilon membrane, and GerBA was detected as described in Materials and Methods. The arrowheads denoting GerBA bands, the background bands, and molecular mass markers are labeled as in Fig. 2. Note that detergent and salt treatments also affect fractionation of some of the background bands.
Abundance of GerBA in spores.
Our ability to detect GerBA in extracts from disrupted spores of a strain that overexpressed the gerB operon but not in those from wild-type spores suggested that GerBA might be a low-abundance protein. Indeed, studies with a gerB-lacZ fusion have indicated that the gerB operon is expressed at a rather low level (6). To estimate the number of GerBA molecules per spore, the GerBA signal in the P100 fraction from a known quantity of spores was compared to that obtained from a known amount of the purified His10-GerBA fusion protein. With the P100 fraction, the intensity of each of the four GerBA bands in the 40- to 50-kDa region appeared similar in intensity to the signal generated by 3 × 10−14 mol of the His10-GerBA protein (data not shown). Since the P100 fraction was obtained from 15 OD600 units of spores (∼3 × 109 spores), these findings suggest that there are about 24 molecules of GerBA per spore (4 × 3 × 10−14 × 6 × 1023/3 × 109 = 24). In other experiments (data not shown), this number ranged between 24 and 40 molecules per spore. While this value may well be a slight underestimate due to losses during P100 isolation, it clearly shows that GerBA is a very-low-abundance protein.
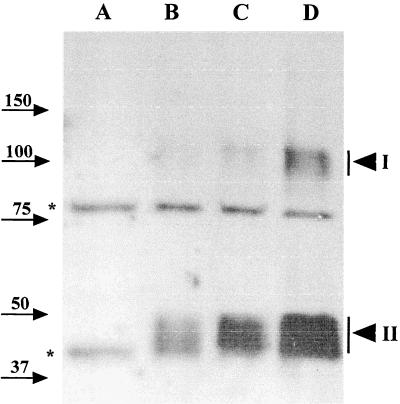
We also compared the relative abundance of GerBA in spores from strains that carried different doses of the gerB operon. Western blot analysis was used to compare the amounts of GerBA protein in P100 fractions from equivalent amounts of spores of strains FB60, PS832, and FB49, which contain zero, one, or two copies of gerB, respectively, and of strain FB58, which contains the PsspB::gerB construct (Fig. 6). As expected, the strength of the GerBA signal increased with increasing dosage or expression of the gerB operon. Comparison of the strength of the GerBA signal in dilutions of the P100 fractions from PsspB::gerB spores and wild-type spores showed that the former contained at least 500-fold more GerBA than wild-type spores (data not shown). These findings further indicate that bands in regions I and II are due to the GerBA protein and also suggest that the overexpressed GerBA mirrors the normal level of this protein in its inner membrane localization.
FIG. 6.
GerBA signal in spores with different levels of expression of gerB. The P100 pellet fractions of spores (15 OD600 units) from strains FB60 (ΔgerB; lane A), PS832 (wild type; lane B), FB49 (PS832 amyE::gerB; lane C), or FB58 (PsspB::gerB; lane D) were run on an SDS-PAGE gel, transferred to an Immobilon membrane, and probed for GerBA as described in Materials and Methods. The arrowheads labeled I and II denote the GerBA-specific signals in the 100-kDa and 40- to 50-kDa ranges, respectively. Background bands and molecular mass markers are labeled as in Fig. 2.
GerBA location in outgrowing spores.
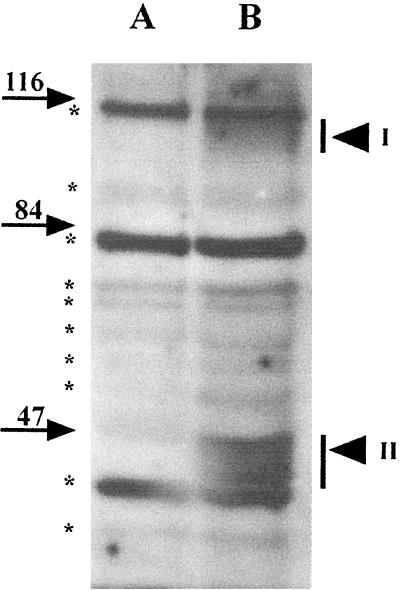
The inner membrane of the dormant spore becomes the cell membrane in the outgrowing spore, whereas the outer membrane and coat layers of the dormant spore are shed during spore germination. Thus, inner membrane proteins persist in the outgrowing spore, unlike those of the outer membrane and coat layers. If, as suggested above, GerBA is located in the inner membrane, then assuming that there is no significant degradation of GerBA during spore germination and outgrowth, GerBA should be present in outgrowing spores (29). To test this prediction, we germinated spores of ΔgerB (FB60) and wild-type (PS832) strains in l-Ala and allowed them to outgrow in rich medium. When the spores were predominantly rod-shaped, they were harvested and used to prepare lysates and P100 fractions which were examined for GerBA by Western blot analysis (Fig. 7). GerBA was detected in the P100 fraction of outgrowing wild-type spores 2 h after initiation of spore germination (Fig. 7), and the amount recovered was comparable to that from a similar quantity of decoated wild-type spores (data not shown). This finding is again consistent with an inner membrane location for GerBA. Note that the GerBA signal was unlikely to arise from spores that had failed to shed their coats because the coats would be removed along with cell wall debris in the low-speed centrifugation of lysates prior to the ultracentrifugation used to pellet the membranes.
FIG. 7.
GerBA in outgrowing spores. P100 fractions from outgrowing (10 OD600 units) spores of strains FB60 (ΔgerB; lane A) and PS832 (wild type; lane B) prepared as described in Materials and Methods were run on an SDS-PAGE gel, proteins were transferred to an Immobilon membrane, and GerBA was detected as described in Materials and Methods. Arrowheads labeled I and II denote GerBA bands in regions I and II, respectively, and background bands and molecular mass markers are labeled as in Fig. 2.
DISCUSSION
Dormant B. subtilis spores detect specific nutrients in the environment and respond to that stimulus by germinating. Previous work showed that this response is mediated by germinant receptor proteins that are encoded by the gerA family of operons (17, 19). In this study we used Western blot analysis to follow one gerB-encoded protein, GerBA, in spore fractions and our observations showed that GerBA is located in the inner membrane of the dormant spore. Previous studies (23) have suggested that the GerB receptor is a membrane-associated, multisubunit complex consisting of GerBA, GerBB, and probably other proteins. Thus, in light of our observations with GerBA, we propose that the entire GerB receptor is located in the inner membrane of the dormant spore. Future studies addressing the location of GerBB and other putative receptor components, for example GerBC, will be needed to fully test this hypothesis.
In contrast to our evidence for an inner membrane location of the GerB receptor, previous studies which used antisera against GerA peptides and immunoelectron microscopy led to the claim that the GerA receptor proteins were located at the cortex-coat boundary in dormant spores and were thus likely to be in the outer membrane (27, 33). The results of these studies also suggested that the GerAB protein redistributes throughout the cortex shortly after the initiation of spore germination (27). Although it is possible that the GerA and GerB receptors are located in different places, this seems unlikely given the high degree of homology between the proteins encoded by the gerA and gerB operons. Indeed, it is possible that the observations which suggested that the GerA proteins are located in the outer membrane-coat were compromised by some reactivity of the antisera used with proteins other than GerA proteins (27, 33), and the specificity of those antibodies was never unambiguously demonstrated. In addition, if we assume that the number of GerAB molecules per spore is similar to that for GerBA, then the immunoelectron micrographs of spore sections often detected more molecules of GerAB than the expected total number in the entire spore (27). Finally, a separate study using antipeptide antisera to detect GerA proteins in spore fractions by Western blot analysis also reported that GerAA was located in the inner membrane (17), and this has been confirmed for both GerAA and GerAC in a recent study (11a). Taking all of the available data into account, we would argue that as is almost certainly the case with the GerB receptor, the GerA receptor and by analogy the GerK receptor are also located in the spore's inner membrane.
The possible inner membrane localization of GerB and other germinant receptors raises several interesting mechanistic questions. First, how do germinants reach receptors in the inner membrane, which lies below the germ cell wall, cortex, outer membrane, and coat layers? The peptidoglycan cortex and germ cell wall are thought to be freely permeable to small germinants (14), and the integrity of the outer membrane, which is ill-defined in electron micrographs, has been questioned (7). However, biochemical studies which measured the spore volume that is accessible for diffusion of small molecules suggested that a permeability barrier may exist for molecules such as glucose and ribose in the coat-outer membrane region (14). If true, this would require the existence of special systems that allow germinants to traverse this barrier and reach the inner spore membrane. Although no such permeability systems have been unequivocally identified, recent work described mutations in the gerP operon that blocked germination in intact but not in decoated spores (3). Thus, a gerP-dependent system might allow germinants to traverse the coat-outer membrane barrier. A second question raised by an inner membrane location of the germinant receptor concerns how the germinant-binding signal is transduced outward into the cortex where the cortex-lytic enzymes (CLEs) such as SleB and CwlJ are most likely located (12, 20). The CLEs are necessary for hydrolysing the cortex during germination but their activity is repressed during dormancy; consequently, a signal from the site of the germinant-receptor interaction must ultimately activate these quiescent enzymes. One might imagine that the activated receptors locally change inner-membrane permeability and thereby affect the hydration level in the spore core. However, no specific second messenger or mechanism has yet been identified or proposed to explain the transduction of this signal to the CLEs in the spore cortex. An interesting possibility is suggested by the finding that some CLEs act only on the spore cortex as it exists in situ and have minimal activity on isolated spore cortex (10, 16, 20, 21). Thus, receptor-triggered changes in the inner membrane might stress the cortex by stimulating core rehydration and thereby trigger cortex hydrolysis. Although the precise mechanism(s) of signal transduction during spore germination remains to be elucidated and confirmed, the inner membrane is certainly a plausible location for the receptors for nutrient germinants.
The observations presented here also present new strategies to study germinant receptor biochemistry, which has lagged behind the genetic analysis of these spore components. We observed that GerBA appeared as a series of four or more bands in the 40- to 50-kDa range on SDS-PAGE, suggesting that the protein is subject to either modification or degradation or both. There is also a significant amount of GerBA signal at higher molecular mass on SDS-PAGE, suggesting that some GerBA can be either covalently attached to another molecule or associated in a very strong complex. Indeed, genetic studies have suggested that the receptor is a multicomponent system (23), but questions concerning the orientation, transport, and breakdown of the receptor proteins remain to be addressed. In this study, we found that GerBA can be appreciably overexpressed in spores and is also found in outgrowing spore membranes, which are easier to prepare than membranes from dormant spores. These findings may aid in the biochemical analysis of germinant receptors by overcoming some of the problems, in particular receptor abundance, that have limited such research in the past.
ACKNOWLEDGMENTS
We thank members of this laboratory for their comments and criticisms on the manuscript and are grateful to A. Moir for sharing results prior to their publication.
The work was supported by a grant from the National Institutes of Health, GM19698.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:74–76. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagyan I, Noback M, Bron S, Paidhungat M, Setlow P. Characterization of yhcN, a new forespore-specific gene of Bacillus subtilis. Gene. 1998;212:179–188. doi: 10.1016/s0378-1119(98)00172-3. [DOI] [PubMed] [Google Scholar]
- 3.Behravan J, Chirakkal H, Masson A, Moir A. Mutations in the gerP locus of Bacillus subtilis and Bacillus cereus affect access of germinants to their targets in spores. J Bacteriol. 2000;182:1987–1994. doi: 10.1128/jb.182.7.1987-1994.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchanan C E, Neyman S L. Correlation of penicillin-binding protein composition with different functions of two membranes in Bacillus subtilis forespores. J Bacteriol. 1986;165:498–503. doi: 10.1128/jb.165.2.498-503.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connors M J, Mason J M, Setlow P. Cloning and nucleotide sequence of genes for three small acid-soluble proteins of Bacillus subtilis spores. J Bacteriol. 1986;166:417–425. doi: 10.1128/jb.166.2.417-425.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corfe B M, Sammons R L, Smith D A, Mauël C. The gerB region of the Bacillus subtilis 168 chromosome encodes a homologue of the gerA spore germination operon. Microbiology. 1994;140:471–478. doi: 10.1099/00221287-140-3-471. [DOI] [PubMed] [Google Scholar]
- 7.Crafts-Lighty A, Ellar D J. The structure and function of the spore outer membrane in dormant and germinating spores of Bacillus megaterium. J Appl Bacteriol. 1980;48:135–145. doi: 10.1111/j.1365-2672.1980.tb05215.x. [DOI] [PubMed] [Google Scholar]
- 8.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster S J, Johnstone K. Pulling the trigger: the mechanism of bacterial spore germination. Mol Microbiol. 1990;4:137–141. doi: 10.1111/j.1365-2958.1990.tb02023.x. [DOI] [PubMed] [Google Scholar]
- 10.Foster S J, Johnstone K. Purification and properties of a germination-specific cortex-lytic enzyme from spores of Bacillus megaterium KM. Biochem J. 1987;242:573–579. doi: 10.1042/bj2420573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 11a.Hudson, K. D., B. M. Corfe, E. H. Kemp, I. M. Feavers, P. J. Coote, and A. Moir. 2001. Localization of GerAA and GerAC germination proteins in the Bacillus subtilis spore. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 12.Ishikawa S, Yamane K, Sekiguchi J. Regulation and characterization of a newly deduced cell wall hydrolase gene (cwlJ) which affects germination of Bacillus subtilis spores. J Bacteriol. 1998;180:1375–1380. doi: 10.1128/jb.180.6.1375-1380.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnstone K. The trigger mechanism of spore germination: current concepts. J Appl Bacteriol Symp Suppl. 1994;76:17S–24S. doi: 10.1111/j.1365-2672.1994.tb04354.x. [DOI] [PubMed] [Google Scholar]
- 14.Koshikawa T, Beaman T C, Pankratz H S, Nakashio S, Corner T R, Gerhardt P. Resistance, germination, and permeability correlates of Bacillus megaterium spores successively divested of integument layers. J Bacteriol. 1984;159:624–632. doi: 10.1128/jb.159.2.624-632.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeDeaux J R, Grossman A D. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J Bacteriol. 1995;177:166–175. doi: 10.1128/jb.177.1.166-175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyata S, Moriyama R, Sugimoto K, Makino S. Purification and partial characterization of a spore cortex-lytic enzyme of Clostridium perfringens S40 spores. Biosci Biotech Biochem. 1995;59:514–515. doi: 10.1271/bbb.59.514. [DOI] [PubMed] [Google Scholar]
- 17.Moir A, Kemp E H, Robinson C, Corfe B M. The genetic analysis of bacterial spore germination. J Appl Bacteriol Symp Suppl. 1994;76:9S–16S. [PubMed] [Google Scholar]
- 18.Moir A, Lafferty E, Smith D A. Genetic analysis of spore germination mutants of Bacillus subtilis 168: the correlation of phenotype and map location. J Gen Microbiol. 1979;111:165–180. doi: 10.1099/00221287-111-1-165. [DOI] [PubMed] [Google Scholar]
- 19.Moir A, Smith D A. The genetics of bacterial spore germination. Annu Rev Microbiol. 1990;44:531–553. doi: 10.1146/annurev.mi.44.100190.002531. [DOI] [PubMed] [Google Scholar]
- 20.Moriyama R, Fukuoka H, Miyata S, Kudoh S, Hattori A, Kozuka S, Yasuda Y, Tochikubo K, Makino S. Expression of a germination-specific amidase, SleB, of Bacilli in the forespore compartment of sporulating cells and its localization on the exterior side of the cortex in dormant spores. J Bacteriol. 1999;181:2373–2378. doi: 10.1128/jb.181.8.2373-2378.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriyama R, Kudoh S, Miyata S, Nonobe S, Hattori A, Makino S. A germination-specific spore cortex-lytic enzyme from Bacillus cereus spores: cloning and sequencing of the gene and molecular characterization of the enzyme. J Bacteriol. 1996;178:5330–5332. doi: 10.1128/jb.178.17.5330-5332.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholson W L, Setlow P. Sporulation, germination, and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons Ltd.; 1990. pp. 391–450. [Google Scholar]
- 23.Paidhungat M, Setlow P. Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant d-alanine. J Bacteriol. 1999;181:3341–3350. doi: 10.1128/jb.181.11.3341-3350.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paidhungat M, Setlow P. Role of Ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J Bacteriol. 2000;182:2513–2519. doi: 10.1128/jb.182.9.2513-2519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Racine F M, Vary J C. Improved method for purification of membranes from spores of Bacillus megaterium. Prep Biochem. 1982;12:265–273. doi: 10.1080/00327488208065567. [DOI] [PubMed] [Google Scholar]
- 26.Racine F M, Vary J C. Isolation and properties of membranes from Bacillus megaterium spores. J Bacteriol. 1980;143:1208–1214. doi: 10.1128/jb.143.3.1208-1214.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakae Y, Yasuda Y, Tochikubo K. Immunoelectron microscopic localization of one of the spore germination proteins, GerAB, in Bacillus subtilis spores. J Bacteriol. 1995;177:6294–6296. doi: 10.1128/jb.177.21.6294-6296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Santo L Y, Doi R H. Ultrastructural analysis during germination and outgrowth of Bacillus subtilis spores. J Bacteriol. 1974;120:475–481. doi: 10.1128/jb.120.1.475-481.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sterlini J M, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swerdlow R D, Setlow P. Isolation and characterization of two distinct fractions from the inner membrane of dormant Bacillus megaterium spores. J Bacteriol. 1984;158:9–15. doi: 10.1128/jb.158.1.9-15.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vary J C. Germination of Bacillus megaterium spores after various extraction procedures. J Bacteriol. 1973;95:1327–1334. doi: 10.1128/jb.116.2.797-802.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasuda Y, Sakae Y, Tochikubo K. Immunological detection of the GerA spore germination proteins in the spore integuments of Bacillus subtilis using scanning electron microscopy. FEMS Microbiol Lett. 1996;139:235–238. doi: 10.1111/j.1574-6968.1996.tb08208.x. [DOI] [PubMed] [Google Scholar]
- 34.Zuberi A R, Moir A, Feavers I M. The nucleotide sequence and gene organization of the gerA spore germination operon of Bacillus subtilis 168. Gene. 1987;51:1–11. doi: 10.1016/0378-1119(87)90468-9. [DOI] [PubMed] [Google Scholar]