To the Editor
Orviz et al. recently described the outbreak of monkeypox in Spain, which is part of a larger European outbreak, in this journal.1 The outbreak has been epidemiologically linked to potential superspreader events in Europe earlier in the year 2022 and over a short period has spread across 75 countries causing over 20,000 infections as on 30 July 2022, with the World Health Organization subsequently declaring the outbreak as a public health emergency of international concern in July 2022.2 Following the outbreak, the wide availability of monkeypox genome sequences in the public domain provided a unique opportunity to understand the genetic epidemiology as well as the evolution of the pathogen.
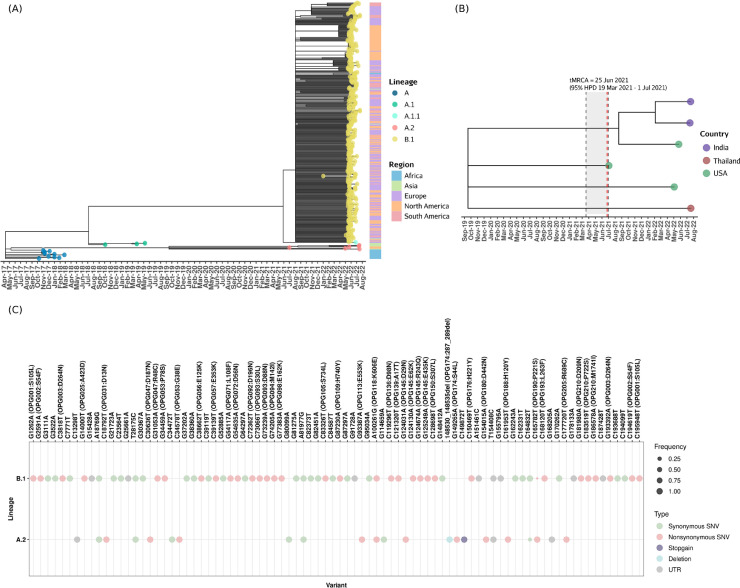
With heightened surveillance and molecular diagnosis across the globe, several genomes of the monkeypox virus are being deposited in public databases like GISAID.3 As early epidemiologic studies linked the outbreak of monkeypox in 2022 to superspreader event(s) in Europe, it was therefore not surprising that a majority of the genomes available on GISAID cluster together (clade IIb). Further reports suggest that the early transmission in this outbreak was largely amongst gays, bisexuals and other men who have sex with men (MSM) with some exceptions.4 , 5 However, the recently deposited genome sequences from the United States of America, Thailand and much more recently from India suggest a distinctly different phylogenetic cluster of genomes within clade IIb, classified by Nextclade as lineage A.2, in contrast to the cluster encompassing the majority of genomes (N = 547) that are classified as lineage B.1 (Fig. 1 A).6 , 10 , 11 The A.2 lineage comprises 9 genomes from 6 unique clinical isolates, with the earliest genome belonging to this lineage (collected in July 2021) being deposited from the state of Texas, USA. The two genome isolates from India cluster closely with a genome isolate from Florida (hMpxV/USA/FL-DHCPPCDC-001/2022) on the phylogenetic tree, while the isolates from Texas, Virginia and Thailand mapped to separate sub-clusters (Fig. 1B).
Fig. 1.
(A) Phylogenetic tree of genome isolates from GISAID belonging to the hMPXV-1 clade of monkeypox virus constructed using Nextstrain.6 (B) Phylogenetic tree of the genomes belonging to A.2 monkeypox lineage. (C) Comparison of frequencies of variations present in >90% genomes of lineages A.2 (N = 6) and B.1 (N = 547).
The genomes belonging to the A.2 lineage have 16 distinct genetic variations which are not found in other lineages, of which 9 are nonsynonymous, 3 are synonymous and 1 is a stopgain variation, along with a deletion of 3-amino acid deletion in the gene OPG174 (Fig. 1C). Variants found at a minimum frequency of 90% in the respective lineages A.2 and B.1 were compared and are summarised in Fig. 1C.
Albeit the limited number of sequences available for the A.2 lineage, we attempted to compute the time to the most recent common ancestor (tMRCA) for A.2 and the nucleotide substitution rates for lineages A.2 and B.1 using BEAST v1.10.4.7 The tMRCA was calculated following a coalescent growth rate model with a strict molecular clock and the HKY+Γ substitution model. MCMC was run for 50 million steps and the initial 1% steps were discarded as burn-in. The tMRCA of the A.2 lineage was computed as 25 June 2021 (95% HPD 19 March 2021 to 1 July 2021). The A.2 lineage had a mean nucleotide substitution rate of 5.53 × 10−5 (95% HPD 3.39 × 10−5 to 7.46 × 10−5) substitutions per base/year, suggesting a modest rate of substitution compared to the substitution rate of 1.13 × 10−4 (95% HPD 9.33 × 10−5 to 1.33 × 10−4) substitutions per base/year for the larger B.1 lineage of genomes. Accelerated evolution of the B.1 lineage has been observed recently.8
Limited demographic information could be linked to the members of the A.2 cluster and has been primarily compiled from the metadata associated with the genome sequences as well as reports in the public domain. The two genomes from Kerala, India were isolated from men who had a travel history to the United Arab Emirates, while the genome from Thailand was isolated from a male traveller from Nigeria. The genome from Texas, United States of America, the earliest in the cluster, was also isolated from a male traveller from Nigeria suggesting a wider geographic area with ongoing transmission of the virus beyond regions in Central and Eastern Africa where the virus is endemic.9
Put together, the evidence suggests that this unique and distinct phylogenetic cluster of genomes, therefore, represents sustained and previously uncharacterized human-human transmission events spanning multiple countries. The tMRCA dating to mid-2021 and the earliest genome dating to July 2021 suggests that this sustained transmission event possibly preceded the outbreak in 2022 in Europe and has remained largely undetected. The distinct genomic signatures suggest that this transmission chain may not be linked to the large outbreak of monkeypox which occurred in 2022 and has been potentially uncovered due to heightened awareness, surveillance and the wider availability of diagnostics.
This report, therefore, re-affirms the unique and significant value of genomic surveillance of emerging pathogens in uncovering potential new insights and leads for epidemiological investigations. The distinctive finding in this report may have a significant impact on public health policies, surveillance as well as public-health communication.
Funding
This work was supported by the Council of Scientific and Industrial Research (CSIR), India. The funders had no role in the analysis of data, preparation of the manuscript or decision to publish.
Declaration of Competing Interest
The authors report no potential conflicts of interest.
Acknowledgement
BJ acknowledges research fellowships from the Council of Scientific and Industrial Research (CSIR), India. The authors acknowledge Division of High Consequence Pathogens and Pathology (DHCPP, CDC) - the United States of America, Indian Council of Medical Research (ICMR), National Institute of Virology (NIV) - India, National Institute of Health - Thailand, Thai Red Cross Emerging Infectious Diseases Clinical Center, Faculty of Medicine (Chulalongkorn University) - Thailand, and other investigators for the data deposited in the public domain. A detailed list of acknowledgements is available at https://github.com/banijolly/Phylovis-MPX. The authors also acknowledge Dr. Sandhya Pulukool and Vishu Gupta for offering suggestions and insights for enriching the manuscript.
References
- 1.Eva O, Anabel N, Oskar A, Ana V, Ana M-G, Sara M, et al. Monkeypox outbreak in Madrid (Spain): clinical and virological aspects. J Infect. 2022 doi: 10.1016/j.jinf.2022.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Director-General declares the ongoing monkeypox outbreak a Public Health Emergency of International Concern. Available at https://www.who.int/europe/news/item/23-07-2022-who-director-general-declares-the-ongoing-monkeypox-outbreak-a-public-health-event-of-international-concern. Accessed July 30, 2022.
- 3.Yuelong S, John MC. GISAID: global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22(13) doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. 2022 Monkeypox outbreak: global trends. Available at https://worldhealthorg.shinyapps.io/mpx_global/. Accessed July 30, 2022
- 5.Thornhill JP, Sapha B, Sharon W, Juergen R, Andrea A, Harrison Luke B, et al. Monkeypox virus infection in humans across 16 countries - April-June 2022. N Engl J Med. 2022 doi: 10.1056/NEJMoa2207323. [DOI] [PubMed] [Google Scholar]
- 6.James H, Colin M, Sidney MB, John H, Barney P, Charlton C, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suchard MA, Philippe L, Guy B, Ayres DL, Drummond AJ, Andrew R. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4(1) doi: 10.1093/ve/vey016. vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joana I, Vítor B, Miguel P, Daniel S, Dourado SJ, Alexandra N, et al. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat Med. 2022:1–4. [Google Scholar]
- 9.Rao AK, Joann S, Tai-Ho C, Hughes CM, Whitni D, Neff JM, et al. Monkeypox in a traveler returning from Nigeria — Dallas, Texas, July 2021. MMWR Surveill Summ. 2022;71(14):509. doi: 10.15585/mmwr.mm7114a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Happi Christian, Adetifa Ifedayo, Mbala Placide, Njouom Richard, Nakoune Emmanuel, Happi Anise, et al. Urgent need for a non-discriminatory and non-stigmatizing nomenclature for monkeypox virus. PLOS Biology. 2022;20(8) doi: 10.1371/journal.pbio.3001769. e3001769, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aksamentov Ivan, Roemer Cornelius, Hodcroft Emma B., Neher Richard A. Nextclade: clade assignment, mutation calling and quality control for viral genomes. The Journal of Open Source Software. 2021 doi: 10.21105/joss.03773. In press. [DOI] [Google Scholar]