Abstract
Background
The ongoing monkeypox virus outbreak includes at least 7553 confirmed cases in previously non-endemic countries worldwide as of July 2022. Clinical presentation has been reported as highly variable, sometimes lacking classically described systemic symptoms, and only small numbers of cutaneous lesions in most patients. The aim of this study was to compare clinical data with longitudinal qPCR results from lesion swabs, oropharyngeal swabs and blood in a well characterized patient cohort.
Methods
16 male patients (5 hospitalized, 11 outpatients) were included in the study cohort and serial testing for monkeypox virus-DNA carried out in various materials throughout the course of disease. Laboratory analysis included quantitative PCR, next-generation sequencing, immunofluorescence tests and virus isolation in cell culture.
Results
All patients were male, between age 20 and 60, and self-identified as men having sex with men. Two had a known HIV infection, coinciding with an increased number of lesions and viral DNA detectable in blood. In initial- and serial testing, lesion swabs yielded viral DNA-loads at, or above 106 cp/ml and only declined during the third week. Oropharyngeal swabs featured lower viral loads and returned repeatedly negative in some cases. Viral culture was successful only from lesion swabs but not from oropharyngeal swabs or plasma.
Discussion
The data presented underscore the reliability of lesion swabs for monkeypox virus-detection, even in later stages of the disease. Oropharyngeal swabs and blood samples alone carry the risk of false negative results, but may hold value in pre-/asymptomatic cases or viral load monitoring, respectively.
Keywords: Monkeypox virus, Clinical cohort, qPCR, Clinical specimens, Viral load kinetics
Abbreviations
- MSM
male having sex with male
- ART
anti-retroviral therapy
- IQR
inter-quartile range
1. Introduction
As of July 2022, 7553 confirmed cases of monkeypox have been reported in previously non-endemic countries worldwide [1]. In contrast to previous clusters, the ongoing outbreak appears to be driven exclusively by human-to-human transmission, with the majority of current cases reported among men who have sex with men (MSM) [2], [3], [4], [5], [6], [7]. Clinical presentation has been highly variable with patients often lacking the classically described symptoms such as fever and lymphadenopathy [8]. Lesions may be scarce, located only in the anogenital area or even limited to a single lesion [8]. Furthermore, recent reports suggest the existence of asymptomatic infections [9].
Atypical presentation entails the risk of missing cases and may also represent a challenge for diagnostics. Current WHO guidance recommends collection of two swab samples from skin lesions, while also encouraging additional oropharyngeal swabs [10]. Recent studies suggest that monkeypox virus-DNA is readily detectable in respiratory specimens and blood, though there is still insufficient data on the reliability and viral load dynamics in these specimen types throughout the course of disease [11].
In this study, we provide longitudinal quantitative PCR-data for different specimen types from a well-characterized cohort of hospitalized patients, and outpatients with confirmed monkeypox virus infection, associated with the current outbreak according to phylogenomic characterization of whole-genome sequences [12]. Further, we were able to confirm infectivity by successful viral culture in initial lesion swab samples of two patients.
2. Material and methods
2.1. Sample collection
In total, 16 patients diagnosed with monkeypox virus infection at the University Medical Center Hamburg-Eppendorf (UKE) were included in this study. Of these, 5 were hospitalized at the UKE, allowing for longitudinal viral load measurements. A further 5 were outpatients at the UKE and patient meta-data and clinical characteristics were available. 6 were external outpatients and only initial viral load data was available. For a study overview see supplementary figure 1.
Lesion swabs and oropharyngeal swabs were performed using eSwab collection kit (Copan, Italy) or VTM collection kit (Citotest, Jiangsu, China). All samples were aliquoted and inactivated by adding ≤40% guanidine hydrochloride solution in Tris–HCl prior to processing for molecular diagnostics.
The study was conducted according to the guidelines of the Declaration of Helsinki. The use of patient data and anonymized samples was approved by the ethics committee of the Medical Council of Hamburg (PV 7298 and PV5626) and additional written consent from patients was obtained for images presented in this study.
2.2. Laboratory methods
Molecular diagnostics, next generation sequencing, immunofluorescence tests and viral culture were performed as described previously [13], [14], [15], [16], [17]. Methods are described in more detail in supplementary material 1.
3. Results
3.1. Patient characteristics
The first ten patients presenting with monkeypox virus infection at our center (until June 30th, 2022) were male and identified as MSM. While all patients presented with skin lesions, oral lesions were observed in only two. Lymphadenopathy occurred in five of ten patients and fever only in three, two of which also had developed bacterial superinfection of skin lesions. Patient characteristics are compiled in detail in Table 1 . Moderately elevated C-reactive protein (CrP)-levels were observed in eight of ten cases. A detailed overview of laboratory parameters is available in supplementary Table 1.
Table 1.
Clinical characteristics of hospitalized patients and outpatients. All patients were male and between 20 and 40 years old. Two patients were HIV-positive and currently under antiretroviral therapy (ART), four HIV-negative patients are taking pre-exposition prophylaxis (PrEP) and four HIV-negative patients are not taking any prophylaxis. In all cases lesions occurred anal/perianal and/or genital/perigenital. In four cases single lesions occurred in other regions of the body. Fever occurred in three patients, of which two had bacterial superinfection. In four cases inguinal lymphadenitis was described, in one case jugular lymphadenitis occurred, whereas six patients did not present with lymphadenitis. All patients received symptomatic therapy. Two patients received antibiotics due to suspected bacterial superinfection. [1] on Bictegravir, Emtricitabin and Tenofovir alafenamide, viremia 22 HIV copies/ml, CD4+ 360/µl. [2] on Dolutegravir and Lamivudin, viremia not detectable, CD4+ 279/µl.
 |
Of note, two of ten patients were HIV-positive (patients 1 and 4, both CDC Stadium A2, under ART) and presented with considerably more lesions (>30) than HIV-negative patients (patients 3,4,5–10), while also exhibiting the highest viral loads in blood (Table 1).
Of note, seroconversion was successfully demonstrated for patient 4 through immunofluorescence test (IFT) by day 34 after symptom onset (supplementary figure 2).
3.2. Initial testing results
Initial testing was performed between day three and day 17 after onset of symptoms, but median times were markedly lower in outpatients (7, [IQR: 5–9 days]) than in hospitalized patients (9, [IQR: 7–10 days]) (see supplementary figure 3A). All initial lesion swabs were positive for monkeypox virus-DNA, with the vast majority at, or above 106 cp/ml. In contrast, oropharyngeal swabs rarely exceeded 106 cp/ml, frequently fell below 103 cp/ml and some returned negative in both outpatients and hospitalized patients. (See supplementary figure 3B).
3.3. Viral DNA-load dynamics over time in lesion swabs, oropharyngeal swabs and blood
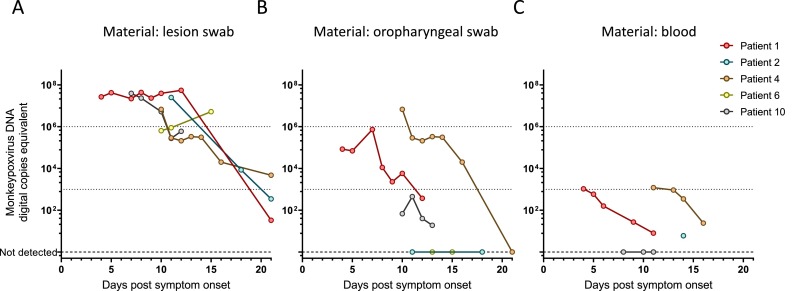
Monkeypox virus-DNA levels were observed in hospitalized patients throughout their stay and in follow-up visits (65 samples in total from five different patients). Viral DNA-loads in lesion swabs were consistently at or above 106 cp/ml during the first two weeks after symptom onset (1st week: median 3.31E+07 cp/ml; 2nd week: median 3.04E+06 cp/ml) and only declined below 103 cp/ml during the third week (median: 8.55E+03 cp/ml); however, all lesion swabs were positive for monkeypox virus-DNA over the entire time course (Fig. 1 a).
Fig. 1.
Viral DNA-load time courses were plotted for hospitalized patients with available serial measurements. A) Swabs from cutaneous lesions were taken according to established procedures; however, the exact location where swabs were taken has not been recorded. Also, swabbing procedures may entail opening a fresh lesion, which will then crust over. The indicated viral loads represent generic lesion swabs from the respective patient, not necessarily from the same lesion. (1st week: median 3.31E+07 cp/ml, range 2.19E+07 – 3.95E+07 cp/ml; 2nd week: median 3.04E+06 cp/ml, range 2.11E+05 – 5.48E+05 cp/ml). Graphs B) and C) represent oropharyngeal swabs (1st week: median 8.44E+04 cp/ml, range 6.93E+04 – 7.31E+05 cp/ml; 2nd week: median 4.04E+03 cp/ml, range 0 – 6.75E+06 cp/ml; 3rd week: median 0 cp/ml, range 0 – 2.00E+04 cp/ml) and EDTA plasma-samples (1st week: median 5.85E+02 cp/ml, range 1.58E+02 – 1.05E+03 cp/ml; 2nd week: median 7.80E+00 cp/ml, range 0 – 1.20E+03 cp/ml; 3rd week: single sample, 2.37E+01 cp/ml) respectively.
Oropharyngeal swabs were negative in two patients and exhibited consistently lower viral-DNA loads (largely below 106) and a continuous downwards trend during the entire observation period in the others. (1st week: median 8.44E+04 cp/ml; 2nd week: median 4.04E+03 cp/ml; 3rd week: median 0 cp/ml) (see Fig. 1b).
Similarly, blood samples were positive for monkeypox virus-DNA in only four of five patients, with viral DNA-loads at, or below 103 cp/ml and continuously declined throughout the observation period (1st week: median 5.85E+02 cp/ml; 2nd week: median 7.80E+00 cp/ml; 3rd week: single sample, 2.37E+01 cp/ml) (Fig. 1c).
3.4. DNA loads in lesion swab samples remain high despite evolving morphology of pustulae
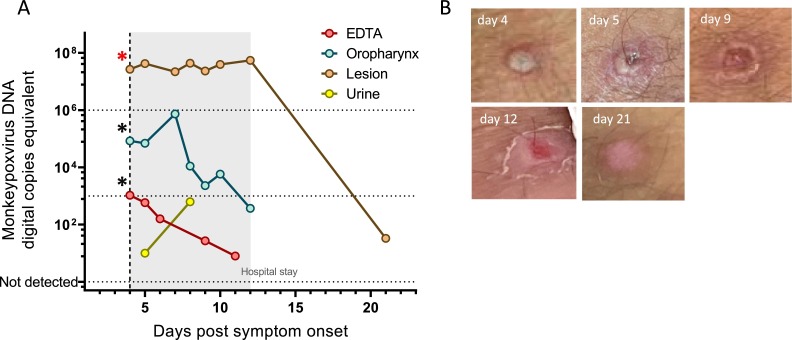
Viral DNA-loads from different specimen types were compiled for each patient (Fig. 2 a and supplementary figure 4). Photo documentation of cutaneous lesions was performed for patient 1 throughout management and images of an exemplary lesion are depicted in Fig. 2b. Despite dramatic morphological changes throughout the first two weeks, lesion swab samples received during this time were plateauing at very high levels (over 10^7 cp/ml), while DNA-loads in all other materials were gradually declining.
Fig. 2.
A) Viral DNA-loads of different specimen types are plotted for patient 1. gray area represents their stay in the hospital. Red asterisk (*) represents a sample with successful isolation of infections virus, whereas black asterisks (*) represent unsuccessful attempts at viral culture. B) The evolution of an exemplary pustula is displayed over the same timeframe.
3.5. Whole-genome sequencing of the first monkeypox cases in the series
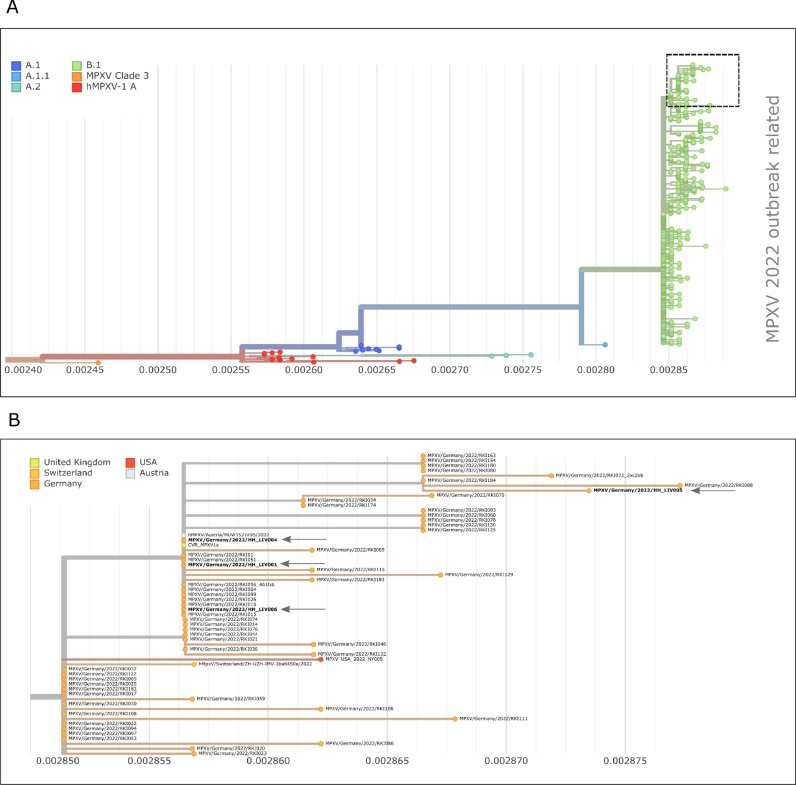
The monkeypox virus genome sequences derived from lesions of patients 1, 2, 4, 5 and 6 was confirmed by shotgun metagenome sequencing. Moreover, phylogenetic analysis of the deduced consensus sequence, derived from patient 1, with previously reported monkeypox sequences confirmed the affiliation of all sequenced cases to the ongoing multi-country monkeypox outbreak. ( Fig. 3 a, 3b and supplementary material 1)
Fig. 3.
A) phylogenetic analysis of MPXV virus sequences related to the 2022 global outbreak. Sequences used here were obtained from NCBI (as of July 21, 2022). Color coding represents the individual clades with clade B.1 containing the outbreak related sequences. B) Section of the phylogenetic tree shown in A. The sequences reconstructed from the lesion of patients 1, 4–6 of this study are marked with an arrow, MPXV/Germany/2022/HH-LIV00, MPXV/Germany/2022/HH-LIV004, MPXV/Germany/2022/HH-LIV005, MPXV/Germany/2022/HH-LIV006.). The color code represents the country from which the sequences were provided in NCBI.
3.6. Infectivity in cell-culture experiments
Viral culture was attempted in first available samples of two patients (patient 1: lesion swab, oropharyngeal swab and blood; patient 2: lesion swab and oropharyngeal swab; undiluted inoculum, see supplementary material 1). In both cases, infectious virus was successfully isolated from lesion swabs (viral DNA inoculum/well: 5.33mio copies and 4.99mio copies), but not from oropharyngeal swabs or blood (viral DNA inoculum/well: 16,872 copies and 211 copies).
4. Discussion and conclusion
This study represents one of the first clinical case series from the ongoing monkeypox virus outbreak including serial viral DNA-load measurements in different specimen types throughout the course of disease. Different from previous monkeypox clusters outside endemic regions in Africa, most patients presented with rather mild absent systemic symptoms, which is consistent with recent reports from the 2022 outbreak ([7, 8, 18]).
Longitudinal observation of viral DNA-load kinetics demonstrated the reliability of cutaneous lesion swab samples for monkeypox virus detection, which are considered the gold standard for diagnostics ([10, 19]). In this study, lesion swabs never returned negative in infected patients, even at later stages of disease; however, very high concentrations of viral DNA and the ability to infect cell culture, especially during the first two weeks after symptom onset, may have implications for risk of contamination and personnel safety. It should be noted that viral DNA-copies are not indicative of the amount of infectious viral particles.
Other clinical material such as blood and oropharyngeal swabs were recently reported to contain detectable monkeypox virus-DNA [11]; however, throat swab samples are known to be unreliable and difficult to standardize, e.g. in SARS-CoV-2 diagnostics [20]. Blood and oropharyngeal swabs were consistently PCR-negative in 1/5 and 2/5 patients of our cohort respectively, thus making them unreliable standalone specimen types for primary diagnosis. However, their potential value for pre-/asymptomatic cases remains to be established [9]. Interestingly, the highest levels of viral-DNA in blood were detected in two HIV-positive patients (under ART) and coincided with substantially increased numbers of pustulae. Therefore, viremia as a parameter for monitoring and risk assessment, as well as potentially increased risk of severe disease in HIV patients despite adequate therapy, warrants further investigation.
CRediT authorship contribution statement
Dominik Nörz: Methodology, Investigation, Writing – original draft, Writing – review & editing. Thomas Theo Brehm: Investigation, Resources, Writing – original draft, Writing – review & editing. Hui Ting Tang: Investigation, Resources, Writing – review & editing. Ilka Grewe: Investigation, Resources, Writing – original draft, Writing – review & editing. Lennart Hermanussen: Investigation, Resources, Writing – review & editing. Hanna Matthews: Investigation, Resources, Writing – review & editing. Julia Pestel: Investigation, Resources, Writing – review & editing. Olaf Degen: Resources, Writing – review & editing. Thomas Günther: Methodology, Investigation, Resources, Writing – review & editing. Adam Grundhoff: Methodology, Resources, Writing – review & editing. Nicole Fischer: Methodology, Resources, Writing – review & editing. Marylyn M. Addo: Resources, Writing – review & editing. Sabine Jordan: Resources, Writing – review & editing. Sandra Hertling: Resources, Writing – review & editing. Stephan Unger: Resources, Writing – review & editing. Guido Schäfer: Resources, Writing – review & editing. Knud Schewe: Resources, Writing – review & editing. Christian Hoffmann: Resources, Writing – review & editing. Martin Aepfelbacher: Supervision, Writing – review & editing. Susanne Pfefferle: Methodology, Investigation, Resources, Writing – original draft, Writing – review & editing. Julian Schulze zur Wiesch: Conceptualization, Resources, Writing – review & editing, Supervision. Stefan Schmiedel: Conceptualization, Resources, Writing – review & editing, Supervision. Marc Lütgehetmann: Conceptualization, Resources, Writing – original draft, Writing – review & editing, Supervision, Project administration.
Declaration of Competing Interest
ML and DN received speaker honoraria and related travel expenses from Roche Diagnostics.All other authors declare no conflict of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2022.105254.
Appendix. Supplementary materials
References
- 1.ECDC Monkeypox multi-country outbreak, first update –8 July 2022. ECDC Website. 2022 [Google Scholar]
- 2.Kozlov M. Monkeypox goes global: why scientists are on alert. Nature. 2022;606:15–16. doi: 10.1038/d41586-022-01421-8. [DOI] [PubMed] [Google Scholar]
- 3.Adalja A., Inglesby T. A novel international monkeypox outbreak. Ann. Intern. Med. 2022 doi: 10.7326/M22-1581. doi. [DOI] [PubMed] [Google Scholar]
- 4.Antinori A., Mazzotta V., Vita S., Carletti F., Tacconi D., Lapini L.E., D'Abramo A., Cicalini S., Lapa D., Pittalis S.,., Puro V., Rivano Capparuccia M., Giombini E., Gruber C.E.M., Garbuglia A.R., Marani A., Vairo F., Girardi E., Vaia F., Nicastri E., Group tIM Epidemiological, clinical and virological characteristics of four cases of monkeypox support transmission through sexual contact, Italy, May 2022. Eurosurveillance. 2022;27 doi: 10.2807/1560-7917.ES.2022.27.22.2200421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vivancos R., Anderson C., Blomquist P., Balasegaram S., Bell A., Bishop L., Brown C.S., Chow Y., Edeghere O., Florence I., Logan S., Manley P., Crowe W., McAuley A., Shankar A.G., Mora-Peris B., Paranthaman K., Prochazka M., Ryan C., Simons D., Vipond R., Byers C., Watkins N.A., team UMIM. Welfare W., Whittaker E., Dewsnap C., Wilson A., Young Y., Chand M., Riley S., Hopkins S. Community transmission of monkeypox in the United Kingdom, April to May 2022. Eurosurveillance. 2022;27 doi: 10.2807/1560-7917.ES.2022.27.22.2200422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez Duque M., Ribeiro S., Martins J.V., Casaca P., Leite P.P., Tavares M., Mansinho K., Duque L.M., Fernandes C., Cordeiro R., Borrego M.J., Pelerito A., de Carvalho I.L., Núncio S., Manageiro V., Minetti C., Machado J., Haussig J.M., Croci R., Spiteri G., Casal A.S., Mendes D., Souto T., Pocinho S., Fernandes T., Firme A., Vasconcelos P., Freitas G. Ongoing monkeypox virus outbreak, Portugal, 29 April to 23 May 2022. Eurosurveillance. 2022;27 doi: 10.2807/1560-7917.ES.2022.27.22.2200424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reed K.D., Melski J.W., Graham M.B., Regnery R.L., Sotir M.J., Wegner M.V., Kazmierczak J.J., Stratman E.J., Li Y., Fairley J.A., Swain G.R., Olson V.A., Sargent E.K., Kehl S.C., Frace M.A., Kline R., Foldy S.L., Davis J.P., Damon I.K. The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 2004;350:342–350. doi: 10.1056/NEJMoa032299. [DOI] [PubMed] [Google Scholar]
- 8.WHO Multi-country monkeypox outbreak: situation update, 27 June 2022. WHO Website. 2022 [Google Scholar]
- 9.De Baetselier I., Van Dijck C., Kenyon C., Coppens J., Van den Bossche D., Smet H., Liesenborghs L., Vanroye F., de Block T., Rezende A., Florence E., Vercauteren K., Van Esbroeck M. Asymptomatic monkeypox virus infections among male sexual health clinic attendees in Belgium. medRxiv. 2022 doi: 10.1101/2022.07.04.22277226:2022.07.04.22277226. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health O Laboratory testing for the monkeypox virus: interim guidance, 23 May 2022. World Health Organization, Geneva. 2022 [Google Scholar]
- 11.Adler H., Gould S., Hine P., Snell L.B., Wong W., Houlihan C.F., Osborne J.C., Rampling T., Beadsworth M.B.J., Duncan C.J.A., Dunning J., Fletcher T.E., Hunter E.R., Jacobs M., Khoo S.H., Newsholme W., Porter D., Porter R.J., Ratcliffe L., Schmid M.L., Semple M.G., Tunbridge A.J., Wingfield T., Price N.M., Abouyannis M., Al-Balushi A., Aston S., Ball R., Beeching N.J., Blanchard T.J., Carlin F., Davies G., Gillespie A., Hicks S.R., Hoyle M.-.C., Ilozue C., Mair L., Marshall S., Neary A., Nsutebu E., Parker S., Ryan H., Turtle L., Smith C., van Aartsen J., Walker N.F., Woolley S., Chawla A., Hart I., Smielewska A., et al. Clinical features and management of human monkeypox: a retrospective observational study in the UK. The Lancet Infect. Dis. 2022;22(8):1153–1162. doi: 10.1016/S1473-3099(22)00228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isidro J., Borges V., Pinto M., Sobral D., Santos J.D., Nunes A., Mixão V., Ferreira R., Santos D., Duarte S., Vieira L., Borrego M.J., Núncio S., de Carvalho I.L., Pelerito A., Cordeiro R., Gomes J.P. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat. Med. 2022 doi: 10.1038/s41591-022-01907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nörz D., Tang H.T., Emmerich P., Giersch K., Fischer N., Addo M.M., Aepfelbacher M., Pfefferle S., Lütgehetmann M. Rapid adaptation of established high-throughput molecular testing infrastructure for detection of monkeypoxvirus. Emerg Infect Dis. 2022 doi: 10.3201/eid2809.220917. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norz D., Pfefferle S., Brehm T.T., Franke G., Grewe I., Knobling B., Aepfelbacher M., Huber S., Klupp E.M., Jordan S., Addo M.M., Schulze Zur Wiesch J., Schmiedel S., Lutgehetmann M., Knobloch J.K. Evidence of surface contamination in hospital rooms occupied by patients infected with monkeypox, Germany, June 2022. Euro Surveill 27. 2022 doi: 10.2807/1560-7917.ES.2022.27.26.2200477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunther T., Haas L., Alawi M., Wohlsein P., Marks J., Grundhoff A., Becher P., Fischer N. Recovery of the first full-length genome sequence of a parapoxvirus directly from a clinical sample. Sci. Rep. 2017;7:3734. doi: 10.1038/s41598-017-03997-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Olson V.A., Laue T., Laker M.T., Damon I.K. Detection of monkeypox virus with real-time PCR assays. J. Clin. Virol. 2006;36:194–203. doi: 10.1016/j.jcv.2006.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shchelkunov S.N., Shcherbakov D.N., Maksyutov R.A., Gavrilova E.V. Species-specific identification of variola, monkeypox, cowpox, and vaccinia viruses by multiplex real-time PCR assay. J. Virol. Methods. 2011;175:163–169. doi: 10.1016/j.jviromet.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minhaj F.S., Ogale Y.P., Whitehill F., Schultz J., Foote M., Davidson W., Hughes C.M., Wilkins K., Bachmann L., Chatelain R., Donnelly M.A.P., Mendoza R., Downes B.L., Roskosky M., Barnes M., Gallagher G.R., Basgoz N., Ruiz V., Kyaw N.T.T., Feldpausch A., Valderrama A., Alvarado-Ramy F., Dowell C.H., Chow C.C., Li Y., Quilter L., Brooks J., Daskalakis D.C., McClung R.P., Petersen B.W., Damon I., Hutson C., McQuiston J., Rao A.K., Belay E., McCollum A.M. Monkeypox Outbreak - Nine States. May 2022. MMWR Morb Mortal Wkly Rep. 2022;71:764–769. doi: 10.15585/mmwr.mm7123e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li D., Wilkins K., McCollum A.M., Osadebe L., Kabamba J., Nguete B., Likafi T., Balilo M.P., Lushima R.S., Malekani J., Damon I.K., Vickery M.C.L., Pukuta E., Nkawa F., Karhemere S., Tamfum J.-.J.M., Okitolonda E.W., Li Y., Reynolds M.G. Evaluation of the GeneXpert for human monkeypox diagnosis. The Am. Soc. Tropical Med. Hygiene. 2017;96:405–410. doi: 10.4269/ajtmh.16-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsang N.N.Y., So H.C., Ng K.Y., Cowling B.J., Leung G.M., Ip D.K.M. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: a systematic review and meta-analysis. The Lancet Infect. Dis. 2021;21:1233–1245. doi: 10.1016/S1473-3099(21)00146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.