Abstract
Extracellular adherence protein Eap secreted from Staphylococcus aureus was previously found to enhance the adherence of S. aureus to eukaryotic cells. This enhancement effect is due to the ability of Eap to rebind to S. aureus and to bind to eukaryotic cells and several plasma and matrix proteins. In this study we defined one potential binding target for Eap on the surface of S. aureus, a surface-located neutral phosphatase. This phosphatase lacks an LPXTG region, but around 80% is retained on the cell surface. The soluble phosphatase can form a complex with Eap at a nonrandom molar ratio, and phosphatase activity is retained. The phosphatase can also bind to fibronectin. The cell surface-located portion presumably contributes to adherence of S. aureus to fibronectin.
Staphylococcus aureus produces at least three different fibrinogen binding proteins which are secreted from the cells: coagulase (19), extracellular fibrinogen binding protein Efb (1, 17), and extracellular adherence protein Eap (2, 15). Two cell surface-associated fibrinogen binding proteins, clumping factors Clf A (10, 11) and Clf B (13), have been shown to be the major factors responsible for adherence to immobilized fibrinogen and are important in promoting endocarditis (12).
Efb (15.8 kDa) has been found to be a virulence factor in experimental wound infections because it delays wound healing (16). Antibodies against Efb have been shown to have a protective effect (9). Efb is distinct from Clf A and Clf B because it does not promote adherence of cells to fibrinogen. It binds to a different region of fibrinogen than Clf A and forms a precipitate together with fibrinogen due to its divalent binding to fibrinogen. Its interaction with fibrinogen is stimulated by Ca2+ (17).
Eap (60 kDa), the focus of this study, binds to several but not all plasma proteins, including fibrinogen, fibronectin, vitronectin, and prothrombin. Eap is secreted, but it is only partially released from cells; ca. 30% of Eap was found to be associated with cells and the rest was in the supernatant both during logarithmic growth and after overnight cultivation. We have demonstrated that externally added Eap can bind to S. aureus. To a lesser extent, Eap can bind to some other bacterial species. Eap has a strong tendency to form oligomers, and due to its affinity for S. aureus cells it causes aggregation of S. aureus, which is often seen in overnight broth cultures. Furthermore, adherence of S. aureus to fibroblasts was significantly enhanced by addition of Eap as a result of the affinity of Eap for S. aureus, for itself, and for a component(s) on the eukaryotic cell surface. We have hypothesized that Eap serves as an enhancer of bacterial adherence along with surface-located proteins, such as clumping factor, fibronectin binding protein, vitronectin binding protein, etc. (15). In several respects, Eap resembles internalin (InlB) from Listeria monocytogenes, which is responsible for internalization (3). InlB is secreted and can rebind to cells.
Eap is a member of a family of similar proteins. Eap from strain Newman exhibited homology with an S. aureus protein designated major histocompatibility complex class II analogous protein Map. The map gene of strain FDA 574 has been sequenced (8). Eap exhibits extensive homology with a p70 protein from strain Wood 46 (4, 7; National Center for Biotechnology Information accession no. Y10419). Map and p70 are highly homologous, although the C termini differ, and Eap resembles p70 more. The nucleotide sequence encoding another member of the Eap family of proteins, a protein from strain Newman, also shows that these proteins are similar (National Center for Biotechnology Information accession no. AJ132841); the deduced amino acid sequences of the N and C termini were identical to those of the N and C termini of Eap purified by us from strain Newman (14).
The observed binding of Eap to cells of S. aureus prompted us to investigate which component(s) on the staphylococcal surface is involved in Eap binding. We found that a 32-kDa neutral phosphatase (NPase) located on the surface of S. aureus is the major component to which Eap binds, but other potential docking structures were not excluded.
MATERIALS AND METHODS
Bacterial strains.
S. aureus Newman, RN3401 (20), and Phillips (18) were used in this study.
Purification of Eap and NPase.
Eap was purified from the culture supernatant of strain Newman by affinity chromatography on fibrinogen-Sepharose, followed by ion-exchange chromatography using a MonoS column (Pharmacia, Uppsala, Sweden) as described previously (15). To purify NPase, a 1-liter culture (routinely of strain RN3401) was centrifuged, washed in phosphate-buffered saline (PBS), and resuspended in 200 ml of 1 M LiCl. The proteins were extracted for 2 h at 37°C with gentle shaking. After centrifugation (5,000 × g for 15 min) the extracted proteins were dialyzed against PBS to remove LiCl and applied to an Eap-Sepharose column. Eap (5 mg) was coupled to 2 g of CNBr-activated Sepharose 4B (Pharmacia) by using the recommended procedure. The column was washed with several volumes of PBS to remove unbound proteins, and bound proteins were eluted with PBS supplemented with 1 M NaCl. The 32-kDa NPase that eluted was dialyzed against PBS and chromatographed again on Eap-Sepharose. After dialysis against 40 mM phosphate buffer (pH 6.4) (A buffer), the protein was further purified by fast protein liquid chromatography (FPLC) with a MonoS ion-exchange column. Elution was done with a gradient from A buffer to A buffer with 1 M NaCl. NPase eluted at 0.6 M NaCl.
The specificity of the Eap-Sepharose affinity chromatography procedure was assessed. LiCl extract derived from a 200-ml culture was applied several times. The effluent containing nonbound proteins was collected. The proteins bound to the Eap-Sepharose were salt extracted as described above, and the column was washed. The effluent was reapplied to the column, and this procedure was repeated eight times. The eighth salt extract contained no proteins.
Determination of amino acid sequence.
About 50 μg of NPase in PBS that was purified by two rounds of Eap-Sepharose affinity chromatography followed by FPLC was used. The sequence was determined by the Protein Analysis Center, Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden.
Capture enzyme-linked immunosorbent assay (ELISA).
Microtiter wells (Costar) were coated overnight with 100 μl of Eap, fibrinogen (Sigma Chemical Co. St. Louis, Mo.), or fibronectin (Sigma) in PBS at concentrations ranging from 1 to 180 μg/ml. Blocking was done by adding 100 μl of 5% bovine serum albumin (BSA) in PBS for 1 h at 37°C. After washing, 100 μl of NPase (20 μg/ml) was added, and the preparation was incubated for 1 h at 37°C. After washing with 50 mM Tris buffer (pH 7.0), binding of NPase was determined by adding 100 μl of para-nitrophenylphosphate (pNPP). pNPP tablets (Sigma) were dissolved in 50 mM Tris buffer (pH 7.0). Absorbance at 405 nm (A405) was measured after 20 min.
NPase (100 μl) was also immobilized in microtiter wells at concentrations ranging from 0.04 to 80 μg/ml in PBS plus 0.05% Tween 20 (PBST). Blocking was done by adding 100 μl of 5% BSA in PBS. Either 100 μl of fibronectin (10 μg/ml) or 100 μl of fibrinogen (5 μg/ml) in PBST was added, and the preparation was incubated for 1 h at 37°C. After washing with PBST, binding of fibronectin or fibrinogen was determined by adding 100 μl of horseradish peroxidase (HRP)-conjugated rabbit anti-fibronectin or anti-fibrinogen antibodies (Dako, Glostrup, Denmark) diluted 1:1,000 in PBST. The preparation was incubated for 1 h at 37°C, and after washing with PBST the color reaction was developed with OPD tablets (Dako) as recommended by the manufacturer. The results were read at 495 nm. Background values due to binding of the HRP-conjugated antibodies directly to the immobilized NPase were obtained by omitting fibronectin or fibrinogen. These values were subtracted.
Binding of human immunoglobulin G (IgG) to NPase was determined by immobilizing NPase as described above and blocking it by adding 100 μl of 5% gelatin in PBS and incubating the preparation for 1 h at 37°C. The following components (100 μl) in PBST were added consecutively with washing between additions: human IgG (10 μg/ml; Sigma), rabbit antibodies against human IgG gamma chain (diluted 1:500; Dako), and swine anti-rabbit IgG HRP-conjugated antibodies (Dako). Each preparation was incubated for 1 h at 37°C. The color reaction was developed with TMB (Dako) used according to the instructions of the manufacturer. The multiple layers of IgG enhanced the signal (range, 1.1 to 2.1), although the background level (when NPase coating was omitted) was high (1.1).
Binding of NPase to S. aureus cells.
Cells of S. aureus Newman (5 × 105 to 8 × 106 CFU) that were washed overnight in 50 mM Tris buffer (pH 7.0) were mixed with 10 μg of NPase in 1 ml (total volume) of 50 mM Tris buffer (pH 7.0). The mixtures were incubated at 37°C for 1 h with shaking. The bacteria were then washed four times with Tris buffer and resuspended in 120 μl of Tris buffer. The bacteria were transferred to microtiter wells, and pNPP substrate was added. The A405 was determined after 20 min. In control experiments, in which NPase was omitted, the endogenous phosphatase activity of the bacteria was determined and was found to be negligible compared to the amounts added.
Binding of radioactive Eap to S. aureus cells.
Iodinated Eap (1 μg) was added to 108 CFU of S. aureus Newman in 750 μl of PBS. Various amounts of nonradioactive Eap or NPase were added simultaneously. The mixtures were incubated at room temperature for 1 h. The cells were washed five times, and the radioactivity associated with the cells was determined with a gamma counter.
Determination of the Kd for NPase and Eap.
Microtiter wells were coated with 100 μl of Eap (80 μg/ml in PBS) overnight and blocked by adding 100 μl of 2% BSA in PBS and incubating the preparation for 1 h. After washing, increasing concentrations of NPase, ranging from 0.2 to 1.9 μM in 100 μl, were added, and the preparations were incubated for 2 h. The unbound fractions of NPase were determined by transferring the preparations to wells in another microtiter plate, in which the samples were serially diluted. Phosphatase activity was determined by adding pNPP substrate as described above. The dilution required to obtain a certain absorbance value was determined, and by using standardized serial dilutions of NPase the specific phosphatase activity per mole was determined. After the plates were washed, the fractions of NPase which had bound to Eap were determined with pNPP by comparison with the standardized dilutions. The amount of nonspecific binding of NPase to wells coated only with 2% BSA was below the limit of detection. A Scatchard plot was obtained by plotting the number of bound molecules per unbound concentration as a function of the number of bound molecules. The dissociation constant (Kd) was calculated by dividing the number of bound molecules by the number of bound molecules per unbound concentration where the best-fit line intercepted the x axis and y axis. In separate experiments we found that Eap does not inhibit NPase.
RESULTS
Identification of NPase as a binding target for Eap.
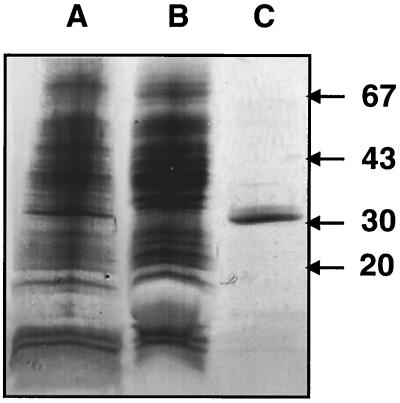
In order to find a cell surface target for Eap, an LiCl extract of S. aureus RN3401 was applied to an Eap-Sepharose column, and the affinity-purified material was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE). A 32-kDa protein was the major protein found by PAGE (Fig. 1, lane C). This could have been due to a high affinity, a high level of the protein, or both factors. The effluent from the Eap-Sepharose column was collected and reelectrophoresed eight times with salt elution and reequilibration of the column between cycles. No detectable proteins were bound to the Eap-Sepharose column after the last electrophoresis. The eighth effluent was devoid of the 32-kDa protein, as shown in Fig. 1, lane B. The purified 32-kDa protein could be further purified by reapplying it, after dialysis, to the Eap-Sepharose column, which verified its affinity to Eap. The 32-kDa protein was also found in strains Newman and Phillips but in smaller amounts (data not shown).
FIG. 1.
Sodium dodecyl sulfate-PAGE. Lane A, proteins in an LiCl extract before electrophoresis through an Eap-Sepharose column; lane B, material in the flowthrough after eight rounds of affinity chromatography on Eap-Sepharose, with elution between cycles; lane C, NPase recovered from the Eap-Sepharose column after the first round of affinity chromatography (the material was concentrated more than the material in lanes A and B to show purity). The positions of molecular weight markers (in thousands) are indicated on the right.
Purification of Eap from strain RN3401 by the fibrinogen-Sepharose method followed by FPLC, as described in Materials and Methods, showed that strain RN3401 also produces Eap but produces less than strain Newman (data not shown).
The 32-kDa protein was subjected to NH2-terminal amino acid analysis; 12 amino acids were identified (NH2-KSSAEVQQTQQA), and these amino acids exhibited identity with the amino acids of a previously identified NPase (22). A search of a database (The Institute for Genomic Research; http://www.tigr.org/tdb/tdb.html) showed that the NPase is synthesized with a typical 31-amino-acid signal sequence for secretion but no LPXTG for attachment to the peptidoglycan (21). The sequence shows that there is an open reading frame encoding a 30,165-Da protein.
The protein obtained with the Eap-Sepharose column was chromatographed through a cation-exchange column, and the elution profile for 0.6 M NaCl was compatible with previous findings (22). As shown below, this protein had phosphatase activity. Thus, we concluded that our 32-kDa protein was an NPase that was described previously (22).
The proportions of cell surface-associated NPase from strain RN3401 and released NPase were determined by comparing the amounts obtained from LiCl extracts and from culture supernatants. Approximately 80% of all NPase was found to be cell surface associated (i.e., in the LiCl extract) (data not shown). Although less NPase was produced by the other strains, the proportion of cell surface-associated NPase was the same for all strains.
Binding of NPase to Eap, fibrinogen, and fibronectin.
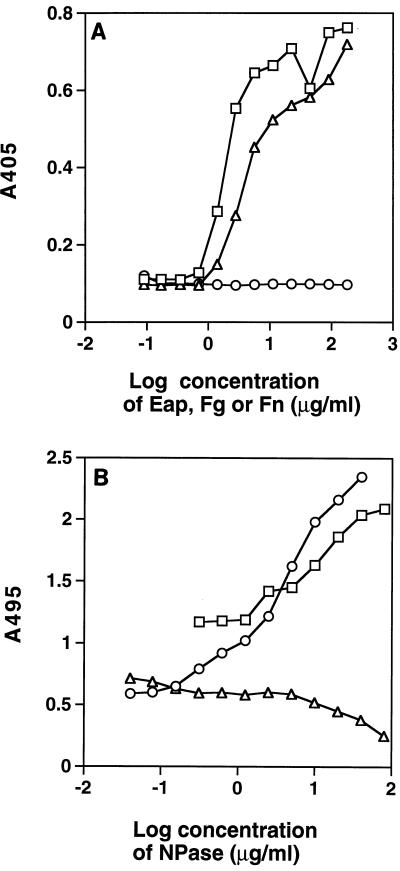
The binding of NPase to Eap was confirmed by a capture ELISA in which various concentrations of Eap, fibronectin, or fibrinogen were immobilized in microtiter wells and then NPase was added. Figure 2A shows the binding of NPase, measured as bound phosphatase activity, to Eap and fibronectin. In a similar assay in which the components were added in the reverse order, binding of fibronectin to various amounts of immobilized NPase was determined (Fig. 2B). The antibodies used to detect fibronectin binding resulted in minimal background binding to NPase in the control in which fibronectin was omitted, although binding of IgG to NPase has been reported previously (22). Such binding was demonstrated by adding higher amounts of IgG to immobilized NPase, as shown in Fig. 2B. Varying the amount of fibronectin or IgG revealed a dose-response relationship (data not shown). No interaction between NPase and fibrinogen was evident in any of the ELISA tests. Instead, binding of the sticky fibrinogen molecules in the microtiter wells was blocked by the NPase (Fig. 2B).
FIG. 2.
(A) Capture of NPase with immobilized fibrinogen, fibronectin, and Eap. Different concentrations of fibrinogen (○), fibronectin (▵), or Eap (□) were used to coat microtiter wells to which NPase (20 μg/ml) was added. Bound NPase activity was measured based on the phosphatase activity. The results of a typical experiment are shown. (B) Capture of fibronectin, fibrinogen, and IgG with immobilized NPase. Different concentrations of NPase were used to coat microtiter wells to which fibronectin (10 μg/ml), fibrinogen (5 μg/ml), or human IgG (10 μg/ml) was added. Binding of these proteins was measured as described in Materials and Methods. Background values for fibronectin binding (obtained by omitting fibronectin) were subtracted, and the background value for IgG binding (obtained by omitting NPase coating) was 1.1. The results of a typical experiment are shown. Symbols: ○, fibronectin; ▵, fibrinogen; □, IgG.
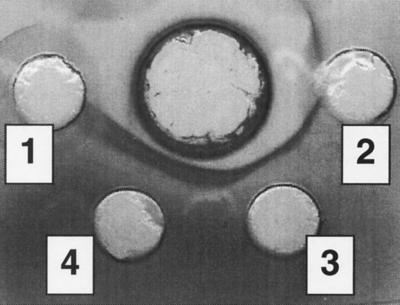
Binding between NPase and Eap was also demonstrated in a double-diffusion test performed with agarose. A precipitation band was observed between wells filled with Eap and NPase, and the distance between the precipitation band and the central well containing NPase was smaller with a higher concentration of Eap (Fig. 3). This means that the interaction between Eap and NPase results in a lattice and can take place only in an ordered form with a nonrandom molar ratio, as is the case with an antibody-antigen reaction. Precipitation bands were also observed between Eap and fibronectin but not between NPase and fibrinogen or fibronectin (data not shown).
FIG. 3.
Double diffusion in agarose. The central well contained NPase (120 μg/ml), and the peripheral wells contained Eap (well 1, no Eap; well 2, 350 μg/ml; well 3, 700 μg/ml; well 4, 1,400 μg/ml).
Adding increasing concentrations of Eap, fibrinogen, or fibronectin to a constant amount of immobilized or soluble NPase did not reduce the phosphatase activity (data not shown).
Binding of NPase to cells of S. aureus.
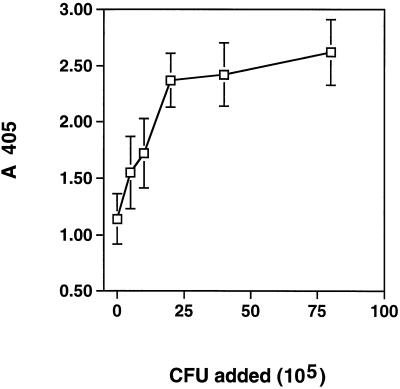
As mentioned above, the major portion of NPase is cell surface associated. The interaction between cells of S. aureus and NPase was shown by adding NPase to S. aureus Newman. By measuring the phosphatase activity bound to the cells, we found that externally added NPase, in addition to the endogenous NPase, could bind to the cells (Fig. 4). Endogenous phosphatase activity was not detectable in this assay when there was excess exogenous NPase (data not shown).
FIG. 4.
Binding of NPase to S. aureus. Different amounts of washed S. aureus cells were incubated with NPase (10 μg/ml). The bacteria were washed, and the cell-associated phosphatase activity was determined. The endogenous phosphatase activity of the cells was insignificant. Mean values from four experiments and standard errors are shown.
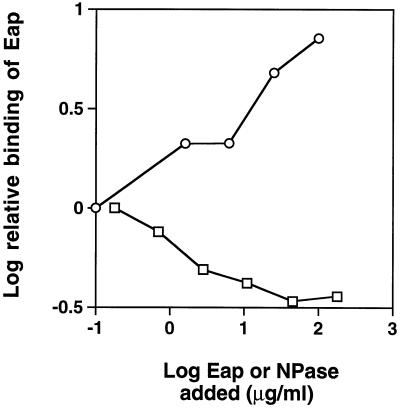
Binding of radiolabeled Eap to S. aureus could be competitively inhibited by unlabeled Eap (15). However, addition of NPase enhanced the binding of Eap, as shown in Fig. 5.
FIG. 5.
Effect of Eap or NPase on Eap binding to S. aureus. Radioactive Eap was added to S. aureus in the presence of different concentrations of nonradioactive Eap or NPase. The relative amounts of labeled Eap bound to the cells were then determined. Symbols: ○, NPase; □, Eap.
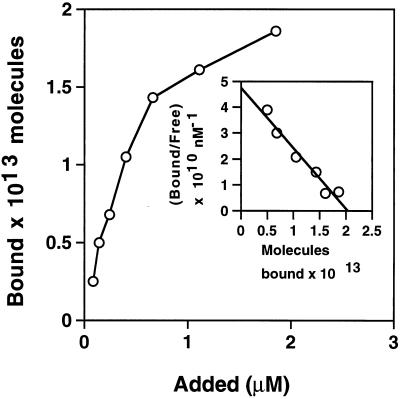
Determination of the Kd.
Increasing amounts of NPase were added to immobilized Eap. The number of bound molecules was plotted against the amount added (Fig. 6). A Scatchard analysis of the data obtained gave a Kd value of approximately 0.5 μM (Fig. 6, inset). This value is close to the Kd reported for clumping factor and fibrinogen (11).
FIG. 6.
Determination of Kd. Different concentrations of NPase were added to immobilized Eap, and the number of bound molecules was calculated. The inset shows a Scatchard plot of the data.
DISCUSSION
Adherence of S. aureus directly to eukaryotic cells is considered an important feature of this organism. We have previously shown that adherence of S. aureus to eukaryotic cells is enhanced by a secreted protein, Eap, which can bind both to S. aureus and to cells acting as adherence mediators. We therefore set out to identify a possible target structure for Eap on the bacterial cell surface that explains its rebinding. The number of externally added Eap molecules which could bind per bacterial cell was found to be more than 50,000, and because of a marked tendency of Eap to form aggregates, it is conceivable that clusters of Eap molecules bind to cells at an undefined site(s). Such a site(s) could be either a highly specialized receptor or a structure(s) to which the sticky Eap molecule could bind by nonspecific electrostatic forces. Our approach for identifying a target structure was to use affinity chromatography of surface-associated proteins using Sepharose-coupled Eap. If such an affinity column was not saturated, different amounts of proteins with different affinities would bind. If instead overloading was allowed, the protein with the strongest binding and/or the most abundant protein would win in a competition for a limited number of Eap sites on the column. In the latter competitive situation, Eap was the best Eap binder from strain Newman, but when strain RN3401 was examined, a 32-kDa protein was the best Eap binder. However, both of these strains contain both proteins, although they contain different amounts. Reelectrophoresis of the flowthrough several times resulted in depletion from the LiCl extract mainly of the NPase, as shown in Fig. 1 (compare lanes A and B). Thus, we concluded that a major target structure for Eap on the S. aureus surface is the 32-kDa protein, although there may also be other targets not found in this study. The identity of the 32-kDa protein and a previously described NPase was shown by the identical NH2 termini, the phosphatase activity, the binding to human IgG, the molecular size, and the elution profile obtained during ion-exchange chromatography.
The interaction between Eap and the NPase was demonstrated in a capture ELISA (Fig. 2A) and by precipitation bands formed in a double-diffusion test (Fig. 3). A precipitation band was formed only when the two proteins were mixed at an optimal concentration, showing that the binding between Eap and NPase is not random aggregation or a monovalent interaction. Both Eap and NPase have high isoelectric points (14, 22), which means that the binding is not due to a charge interaction but rather is more specific. The Kd (0.5 μM) is close to the value that has been reported for the interaction between Clf and fibrinogen (11). Addition of external NPase, which bound to the cells (Fig. 4), stimulated association of Eap with the cells (Fig. 5), further suggesting that NPase is a mediator of Eap attachment to the cell surface.
The interaction between Eap and NPase has at least two biological implications. First, surface-associated NPase clearly serves as a target for Eap on the bacterial cell surface, although Eap might also bind to other bacterial surface structures not identified here. Second, NPase not associated with the cell surface (in the cultures used in this study this was about 20% of all NPase) can dock with extracellular Eap; Eap is mainly found extracellularly (15). The NPase-Eap complex has multiple functions; it adheres to fibronectin (both NPase and Eap adhere), fibrinogen, prothrombin, and immunoglobulins, and it has phosphatase activity. Since Eap strongly enhances the interaction between S. auerus and the host cell (15), a phosphatase attached to the Eap molecule could influence subsequent events and affect the host cell surface, which in turn could affect cell signaling, phagocytosis, and bacterial internalization.
A double mutant of S. aureus lacking both genes for fibronectin binding (fnbA and fnbB) was constructed (5). We have observed residual binding to fibronectin of this strain (16), and we also have observed that the amount of fibronectin binding protein on the cell surface does not correlate with the capacity to bind to fibronectin (6). We suggest that this is due to the presence of different amounts of NPase and bound Eap on the cell surface.
Furthermore, NPase has been shown to bind to rat and human IgG, IgM, and IgA (22), and Yousif et al. speculated that NPase could be relevant for postinfectious sequelae. This effect could be further enhanced by the binding of NPase to fibronectin reported here.
ACKNOWLEDGMENTS
This work was supported by The Swedish Medical Research Council (grant K2000-16X-12218-04B) and by Biostapro AB.
REFERENCES
- 1.Bodén M, Flock J-I. Cloning and characterization of a gene for a 19 kDa fibrinogen binding protein from Staphylococcus aureus. Mol Microbiol. 1994;12:599–606. doi: 10.1111/j.1365-2958.1994.tb01046.x. [DOI] [PubMed] [Google Scholar]
- 2.Bodén M, Flock J-I. Evidence for three different fibrinogen-binding proteins with unique properties from Staphylococcus aureus strain Newman. Microb Pathog. 1992;12:289–298. doi: 10.1016/0882-4010(92)90047-r. [DOI] [PubMed] [Google Scholar]
- 3.Braun L, Dramsi S, Dehoux P, Bierne H, Lindahl G, Cossart P. In1B: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol Microbiol. 1997;25:285–294. doi: 10.1046/j.1365-2958.1997.4621825.x. [DOI] [PubMed] [Google Scholar]
- 4.Fujigaki Y, Yousif Y, Morioka T, Batsford S, Vogt A, Hishida A, Miyasaka M. Glomerular injury induced by cationic 70-kD staphylococcal protein; specific immune response is not involved in early phase in rats. J Pathol. 1998;184:436–445. doi: 10.1002/(SICI)1096-9896(199804)184:4<436::AID-PATH1225>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 5.Greene C, McDevitt D, Francois P, Vaudaux P, Lew D P, Foster T. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin binding proteins and studies on the expression of fnb genes. Mol Microbiol. 1995;17:1143–1152. doi: 10.1111/j.1365-2958.1995.mmi_17061143.x. [DOI] [PubMed] [Google Scholar]
- 6.Hienz S A, Palma M, Flock J-I. Insertional inactivation of the gene for collagen-binding protein has a pleiotropic effect on the phenotype of Staphylococcus aureus. J Bacteriol. 1996;178:5327–5329. doi: 10.1128/jb.178.17.5327-5329.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jahreis A, Yousif Y, Rump J A, Dräger R, Vogt A, Peter H H, Schlesier M. Two novel cationic staphylococcal proteins induce IL-2 secretion, proliferation and immunoglobulin synthesis in peripheral blood mononuclear cells (PBMC) of both healthy controls and patients with common variable immunodeficiency (CVID) Clin Exp Immunol. 1995;100:406–411. doi: 10.1111/j.1365-2249.1995.tb03714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jönsson K, McDevitt D, McGavin M H, Patti J M, Höök M. Staphylococcus aureus expresses a major histocompatibility complex class II analog. J Biol Chem. 1995;270:21457–21460. doi: 10.1074/jbc.270.37.21457. [DOI] [PubMed] [Google Scholar]
- 9.Mamo W, Bodén M, Flock J-I. Vaccination with Staphylococcus aureus fibrinogen binding proteins (FgBPs) reduces colonization of S. aureus in a mouse mastitis model. FEMS Immunol Med Microbiol. 1994;10:47–54. doi: 10.1111/j.1574-695X.1994.tb00010.x. [DOI] [PubMed] [Google Scholar]
- 10.McDevitt D, Francois P, Vaudaux P, Foster T J. Identification of the ligand-binding domain of the surface-located fibrinogen receptor (clumping factor) of Staphylococcus aureus. Mol Microbiol. 1995;16:895–907. doi: 10.1111/j.1365-2958.1995.tb02316.x. [DOI] [PubMed] [Google Scholar]
- 11.McDevitt D, Nanavaty T, House-Pompeo K, Bell E, Turner N, McIntire L, Foster T J, Höök M. Characterization of the interaction between the Staphylococcus aureus clumping factor (ClfA) and fibrinogen. Eur J Biochem. 1997;247:416–424. doi: 10.1111/j.1432-1033.1997.00416.x. [DOI] [PubMed] [Google Scholar]
- 12.Moreillon P, Entenza J M, Francioli P, McDevitt D, Foster T J, Francois P, Vaudaux P. Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect Immun. 1995;63:4738–4743. doi: 10.1128/iai.63.12.4738-4743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ni Eidhin D, Perkins S, Francois P, Vaudaux P, Höök M, Foster T. Clumping factor B (ClfB), a new surface located fibrinogen-binding adhesin of Staphylococcus aureus. Mol Microbiol. 1998;30:245–257. doi: 10.1046/j.1365-2958.1998.01050.x. [DOI] [PubMed] [Google Scholar]
- 14.Palma M. Fibrinogen binding proteins from Staphylococcus aureus. Ph.D. thesis. Stockholm, Sweden: Karolinska Institutet; 1999. [Google Scholar]
- 15.Palma M, Haggar A, Flock J-I. Adherence of Staphylococcus aureus is enhanced by an endogenous secreted protein with broad binding specificity. J Bacteriol. 1999;181:2840–2845. doi: 10.1128/jb.181.9.2840-2845.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palma M, Nozohoor S, Schennings T, Heimdahl A, Flock J-I. Lack of the extracellular 19-kilodalton fibrinogen-binding protein from Staphylococcus aureus decreases virulence in experimental wound infection. Infect Immun. 1996;64:5284–5289. doi: 10.1128/iai.64.12.5284-5289.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palma M, Wade D, Flock M, Flock J-I. Multiple binding sites in the interaction between fibrinogen and an extracellular fibrinogen binding protein from Staphylococcus aureus. J Biol Chem. 1998;273:13177–13181. doi: 10.1074/jbc.273.21.13177. [DOI] [PubMed] [Google Scholar]
- 18.Patti J M, Bremell T, Krajewska-Pietrasik D, Abdelnour A, Tarkowski A, Ryden C, Höök M. The Staphylococcus aureus collagen adhesion is a virulence determinant in experimental septic arthritis. Infect Immun. 1994;62:152–161. doi: 10.1128/iai.62.1.152-161.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phonimdaeng P, O'Reilly M, Nowlan P, Bramley A J, Foster T J. The coagulase of Staphylococcus aureus 8325-4. Sequence analysis and virulence of site-specific coagulase-deficient mutants. Mol Microbiol. 1990;4:393–404. doi: 10.1111/j.1365-2958.1990.tb00606.x. [DOI] [PubMed] [Google Scholar]
- 20.Poston S M, Glancey G R, Wyatt J E, Hogan T, Foster T J. Co-elimination of mec and spa genes in Staphylococcus aureus and the effect of agr and protein A production on bacterial adherence to cell monolayer. J Med Microbiol. 1993;39:422–428. doi: 10.1099/00222615-39-6-422. [DOI] [PubMed] [Google Scholar]
- 21.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yousif Y, Schiltz E, Okada K, Batsford S, Vogt A. Staphylococcal neutral phosphatase; a highly cationic molecule with binding properties for immunoglobulin. APMIS. 1994;102:891–900. [PubMed] [Google Scholar]