Abstract
Many environmental parameters modulate the amount of the RpoS sigma factor in Escherichia coli. Temperature control of RpoS depends on the untranslated RNA DsrA. DsrA activates RpoS translation by pairing with the leader of the mRNA. We find that temperature affects both the rate of transcription initiation of the dsrA gene and the stability of DsrA RNA. Both are increased at low temperature (25°C) compared to 37 or 42°C. The combination of these results is 25-fold-less DsrA at 37°C and 30-fold less at 42°C than at 25°C. Using an adapted lacZ-based reporter system, we show that temperature control of transcription initiation of dsrA requires only the minimal promoter of 36 bp. Overall, transcription responses to temperature lead to a sixfold increase in DsrA synthesis at 25°C over that at 42°C. Furthermore, two activating regions and a site for LeuO negative regulation were identified in the dsrA promoter. The activating regions also activate transcription in vitro. DsrA decays with a half-life of 23 min at 25°C and 4 min at 37 and 42°C. These results demonstrate that the dsrA promoter and the stability of DsrA RNA are the thermometers for RpoS temperature sensing. Multiple inputs to DsrA accumulation allow sensitive modulation of changes in the synthesis of the downstream targets of DsrA such as RpoS.
Escherichia coli, a mesophile bacterium, is able to grow at temperatures from 10 to close to 50°C (21). Within this range, E. coli is able to adapt to sudden temperature shifts (cold shock or heat shock) or to maintain a physiology adapted to a constant temperature (21, 36, 49). In addition to effects of temperature on enzymatic reactions occurring in the cell, expression of many bacterial genes is specifically controlled by temperature. For instance, temperature controls the transcription of genes encoding virulence factors. These are usually expressed around 37°C, the host temperature, but are turned off at 30°C or below (16, 23, 25). This regulation operates via an interplay of regulatory proteins and DNA features (supercoiling, bending, and chemical modifications) (10, 13, 18). Temperature was also shown previously to act on the secondary structure of RNAs, resulting in long-term changes in translation efficiency (2, 22, 33), or immediately following a cold shock, when most RNAs except those encoding the cold shock proteins cannot be translated (37). Some of the RNAs for these cold shock proteins (cspA, cspI, and pnp) are also stabilized after the drop in temperature (4, 15, 48, 50).
The expression of the general stress sigma factor RpoS is controlled at the levels of transcription, translation, and protein stability. The amount of RpoS is adjusted in response to environmental signals such as osmotic pressure, temperature, and chemical modifications (pH, cell density, and nutritional limitations). Each step of RpoS expression can be affected by one or several environmental stimuli (19). How these changes in the environment are sensed and how different signals are integrated are not well understood. A stimulation of RpoS synthesis is seen in steady-state growth at low temperatures (45). This suggests an important and thus far unexplored role for some of the RpoS-dependent genes in low-temperature environments. The small untranslated regulatory RNA DsrA is necessary for the low-temperature expression of RpoS (45) and was shown previously to directly stimulate RpoS translation by interaction with the RpoS leader (30). We were interested in understanding how temperature regulates the DsrA-dependent synthesis of RpoS and have begun an in-depth analysis of the transduction of environmental signals through DsrA. We find that temperature affects both the synthesis of DsrA and its stability.
MATERIALS AND METHODS
Genetic procedures, bacterial strains, plasmids, and phage.
Standard procedures were used for growth of bacteria and bacteriophage (31, 42). The parental bacterial strain used in this study is MC4100 (6). All the other strains are lysogens of MC4100 using phage λRS45 or λRS468 (35, 43) recombined with appropriate plasmids (see below). Plasmids used are derivatives of pRS415, pRS1553, pFRΔ, or pFRα, as described in the text. pTO3 (a gift from C. Ueguchi) was described in reference 47. Isolation of plasmid DNA, digestion with restriction enzymes, and ligation with T4 DNA ligase were carried out as described by the manufacturer's protocols (New England Biolabs and Promega) or reference 41. Transformations were performed as described by Chung et al. (8) using the strain DH5α (New England Biolabs). The sequences of all the constructs described in this study (plasmids as well as fusions inserted into the chromosome) were confirmed by sequencing (National Cancer Institute, National Institutes of Health DNA Core Facility, Bethesda, Md.).
Constructions of plasmids, phages, and strains carrying transcriptional fusions for the in vivo study of dsrAp.
Sequences and other information on pFRΔ and pFRα are available on the website http://www.mimg.ucla.edu/bobs/vectors/index.htm. These plasmids were derived from pRS1553 (35). Briefly, the region carrying the sequence encoding the α peptide (α-lacZ′) in pRS1553, contained between the BamHI site and the SalI restriction site, was removed and replaced with a PCR fragment generated from pRS1553 using the primers FRαXhoI-FRΔ+trp for pFRΔ or FRαXhoI–FR-lacZ for pFRα. The sequences of all primers are listed in Table 1. These primers allowed amplification of the portion of pRS1553 from the BamHI restriction site to the end of α-lacZ′ upstream of the SalI site but replaced the initial sequence of this region (the W205 fusion joint) with sequences that delete the trp terminator (see Fig. 2). After amplification, the PCR fragment was purified on a Wizard (Promega) column (as indicated by the manufacturer's protocol), digested with BamHI and XhoI, and ligated to the purified vector pRS1553 with the BamHI and SalI fragment deleted. The ligation mixture was transformed into DH5α. Ampicillin-resistant clones were screened for the presence of a fragment of approximately 960 bp after PCR using oligonucleotides FREcoRI-415 and FRαXhoI (Table 1). This pair of oligonucleotides allows amplification of fragments inserted into pRS1553. One positive clone for each fusion was confirmed by sequencing and kept, providing the new cloning vectors pFRΔ and pFRα. For the various transcriptional fusions used here, PCR fragments were generated using the genome of MC4100 as template to provide fragments of dsrAp and b1955; the genome of the strain NT3 (from N. Trun; a lac+ version of MC4100) was used for lacUV5p. λpL was generated without template by using two partially overlapping oligonucleotides. PCR fragments were cut with EcoRI and BamHI and ligated into either pFRΔ or pFRα cut with the same enzymes. For the dsrA promoter fusions, a single reverse primer, FRdsrAp[−], encoded the transcriptional start site. At the 5′ end of the insert, the following oligonucleotide primers were used: for dsrAp205, FRdsrAp205E; for dsrAp165, FRdsrAp165E; for dsrAp64, FRdsrAp64E; for dsrAp46, FRdsrAp46E; for dsrAp46AccI, FRdsrAp46AccI; and for dsrAp36, FRdsrAp36E (Table 1). For lacUV5p and λpL, PCR fragments were obtained, respectively, with the primer pairs FRUV5[−]-FRUV5[+] and FRλ[−]-FRλ[+] (Table 1). All of these transcriptional fusions have been designed to initiate transcription at the same start point. Once cloned into pFRΔ and pFRα and confirmed by sequencing, each recombinant plasmid was introduced by transformation into MC4100 and recombined with λRS468 (43). Recombinant phages that gave blue plaques on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plus IPTG (isopropyl-β-d-thiogalactopyranoside) on the indicator strain MC4100 were chosen. After purification of each recombinant phage, they were used to lysogenize MC4100. Screening for single-copy Lac+ lysogens for each fusion was carried out as described by Powell et al. (38). One isolate of each fusion was confirmed by sequencing and kept as a strain carrying the desired fusion.
TABLE 1.
Oligonucleotides used
| Oligonucleotide name | Oligonucleotide sequence |
|---|---|
| Nonbiotinylated | |
| FRαXhoI | 5′-CACGCTCTCGAGAATTTCACCGCCGAAAGGCGCGGTGC-3′ |
| FRαlacZ | 5′-TGAGCGGATCCAACAATTTCACACAGGAAACAGCTATGACCATG-3′ |
| FRΔ+trp | 5′-GCATTTGGATCCGGCATTTTAACTTTCTTTATCACACAGG-3′ |
| FREcoRI-415 | 5′-GCCATAAACTGCCAGGAATTCC-3′ |
| FRdsrAp[−] | 5′-GATGTGGATCCTTCATCACCTTATCCG-3′ |
| FRdsrAp205E | 5′-GGTTGTTGAATTCAAAACATAGTCGCGCAGTACTCCTCTTACCAGG-3′ |
| FRdsrAp165E | 5′-CAGGGAATTCCTCAAAACCGCACAAATTATCAAAG-3′ |
| FRdsrAp64E | 5′-CATAAGAATTCAGCGTTAATCATTCATATGG-3′ |
| FRdsrAp46E | 5′-TCATTGAATTCGGCGAATATTTTCTTGTCAGCGAAAAAAATTGCGG-3′ |
| FRdsrAp46AccI | 5′-TCATTGAATTCGGCGAGTATACTCTTGTCAGCGAAAAAAATTGCGG-3′ |
| FrdsrAp36E | 5′-CGAATGAATTCTTGTCAGCGAAAAAAATTGCGG-3′ |
| FRUV5[−] | 5′-TCCGCGGATCCTTCCACACATTATACGAGCCGGAAGCATAAAGTG-3′ |
| FRUV5[+] | 5′-GACTGGAATTCGGGCAGTGAGCGCAACGCAATTAATGTG-3′ |
| FRλ[−] | 5′-CCTGCTGGATCCTTGTGCTCAGTATCACCGCCAGTGGTATTTATGTC-3′ |
| FRλ[+] | 5′-GATAAATGAATTCTATCTCTGGCGGTGTTGACATAAATACCAC-3′ |
| FR-dsrAHd | 5′-GATGTGAAGCTTCATCACCTTATCCG-3′ |
| FRUV5Hd[−] | 5′-TCCGCAAGCTTTTCCACACATTATACGAGCCGGAAGCATAAAGTG-3′ |
| FRλHd[−] | 5′-CCTGCAAGCTTTTGTGCTCAGTATCACCGCCAGTGGTATTTATGTC-3′ |
| Biotinylated | |
| SL-1 | 5′-TTCGTTACACCAGGAAATCTGATGTGTT-3′ |
| SL-2 | 5′-GATGAAACTTGCTTAAGCAAGAAGCACTTAAAAAATTGGTT-3′ |
| SsrA | 5′-TTGCGACTATTTTTTGCGGC-3′ |
FIG. 2.
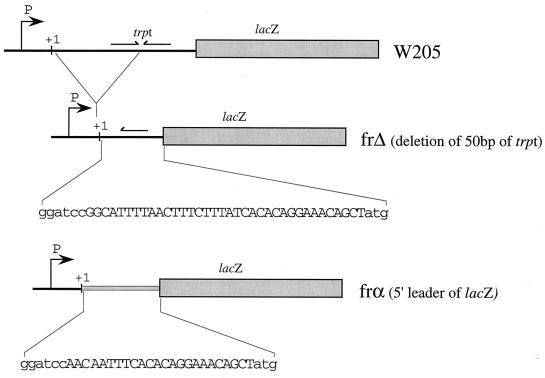
Features of frΔ and frα fusion lac-based reporter systems. The W205 fusion carries a weak cold-sensitive terminator, trpt, shown by the arrows. frΔ corresponds to a deletion of 50 bp containing half of trpt (28). The remaining sequence is shown in capital letters below the line. In frα, the sequence of W205 between the initiation start codon of lacZ and the cloning sites for the promoter (P) was replaced with the natural lacZ leader sequence (capital letters in the sequence shown). See Materials and Methods for details of the construction.
PCR conditions.
The same PCR conditions were applied to generate the fragments for cloning, for sequencing, and for the screening of recombinant clones. Briefly, a single colony was resuspended in 100 μl of water and boiled for 45 s at 95°C, and 1 μl of this treated template was used for 50 μl of PCR mixture. The following reagents were used at the final concentrations indicated: deoxynucleoside triphosphates (Boehringer Mannheim) at 0.2 mM; oligonucleotides at 0.2 μM each (synthesized by Genosys, The Woodlands, Tex.); MgCl2 at 1.6 mM; and Taq DNA polymerase (Perkin-Elmer), 1 U for 50 μl of PCR mixture. PCR conditions were adapted from the work of Repoila et al. (39), with the slight modification of 1 min for the annealing step.
Growth conditions used for β-Gal assays and RNA and plasmid extractions.
From a single colony grown at 37°C on a Lennox L broth (LB) plate, cells were grown aerobically for 18 h in 5 ml of LB medium in a 50-ml flask, at the temperature indicated in the text (preculture). When necessary, chemicals and antibiotics were added at concentrations indicated by Miller (31) or otherwise indicated in the text. A 1/1,000 dilution of the preculture was made into prewarmed and equilibrated medium for further growth and assays. With such conditions, at all temperatures used in this study, growth can be detected by measuring the optical density at 600 nm (OD600) after only three to five generations. The growth lag was estimated to be 177 min at 25°C, 54 min at 37°C, and 128 min at 42°C, with doubling times of 80, 35, and 54 min, respectively. The reason for the slower growth of this derivative of MC4100 at 42°C is not known. To monitor promoter activity with growth, OD600 was determined and β-galactosidase (β-Gal) assays were performed as described by Zhou and Gottesman (52). For each culture, successive measurements were made at about half of the doubling time. Specific activity was calculated by dividing the slope of the line (Vmax) by (OD600 × vol), where OD600 is the optical density of the culture and vol is the volume of culture used for the assay expressed in milliliters. Such activities have been estimated to be about 2.5-fold lower than Miller units. Results shown in Fig. 4 are representative results from one out of at least three different experiments performed on different clones of the same strain grown on different days. Growth conditions used provided less than 15% variation in β-Gal values for different experiments. The values reported in Table 2 represent an average of β-Gal values measured during the exponential growth phase (the range of values varies according to temperature, due to differing extents of growth before entry into stationary phase [Table 2, footnote b]).
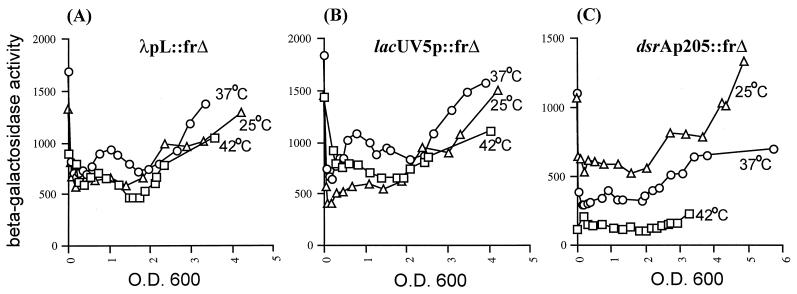
FIG. 4.
Expression of the dsrA promoter as a function of temperature. Cells carrying single-copy lysogens of either λpL::frΔ, lacUV5p::frΔ, or dsrAp205::frΔ were grown in LB at the indicated temperatures as described in Materials and Methods. β-Gal activity was measured during growth and is presented as specific activity as a function of OD600. λpL and lacUV5p were used as promoter controls for the temperature response. Graphs show results of a representative experiment.
TABLE 2.
Temperature regulation by the DsrA promoter
| Promotera | U of β-Galb at temp (°C):
|
|||
|---|---|---|---|---|
| 25 | 37 | 42 | 25/42c | |
| dsrAp36 | 37 | 15 | 11 | NAd |
| dsrAp46 | 378 | 180 | 72 | 4 |
| dsrAp64 | 380 | 210 | 84 | 4.5 |
| dsrAp165 | 600 | 419 | 193 | 3 |
| dsrAp205 | 642 | 390 | 110 | 6 |
| λpL | 715 | 850 | 579 | 1.2 |
| lacUV5p | 623 | 964 | 700 | 0.9 |
All promoter fusions are in the frΔ vector, as single-copy lysogens.
Cells were grown in LB as described in Materials and Methods. Units are specific activities in machine units (see Materials and Methods). Values are the averages of all measurements done during exponential-phase growth, starting at an OD600 of 0.35. The final sample was taken at an OD600 of 0.1 before entry into stationary phase, defined by the increase in expression of a parallel fusion of yedPp::frΔ, an RpoS-dependent fusion (data not shown). This corresponded to OD600s of 1.15 at 25°C, 1.45 at 37°C, and 0.9 at 42°C.
Ratio of activity at 25°C to activity at 42°C.
At 37 and 42°C, activity was no higher than that for the control without a promoter.
For experiments using pTO3, cells were grown in conditions identical to those described above. The induction was carried out by adding IPTG at a final concentration of 1 mM at an OD600 between 0.67 and 0.77. To measure the RNA stability, cells were grown as described above and treated with 300 μg of rifampin/ml at an OD600 between 0.67 and 0.77. Sensitivity to rifampin was confirmed by monitoring growth. The plasmids used for in vitro transcriptions were extracted from cells in late stationary phase (about 20 h after the dilution of the preculture) and previously grown as described above. Plasmids were extracted from cells using a Qiafilter-Qiagen kit (Maxi-Prep) as indicated by the manufacturer's protocol.
Northern blots.
Total RNAs were extracted using the Qiagen kit or Trizol (Life Technologies, Gaithersburg, Md.) reagent as recommended by the manufacturer's protocols. Both methods provided identical results. The Northern procedure was performed as previously described by Majdalani et al. (29). Probes used were 5′ biotinylated oligonucleotides, as listed in Table 1 (for DsrA, SL-1 and SL-2). Values provided for the decay represent an average of the values obtained for at least three different RNA preparations. Signals were quantified by serial dilutions of samples to ensure linearity of the detection method. Detection was performed with the Eagle Eye II system (Stratagene).
Construction of plasmids for in vitro transcription experiments.
All of the plasmids used in this study are derivatives of pRLG770 carrying the rrnBp1 promoter (a gift from D. Jin, described in reference 40). rrnBp1 is flanked by the restriction sites EcoRI and HindIII. Those sites were used to replace rrnBp1 with PCR fragments carrying either the various dsrAp fragments tested, lacUV5p, or λpL. For such PCRs, the genomes of strains carrying the corresponding frΔ fusions were used as template. As the forward primer, FR.EcoRI(415) was used. The corresponding reverse primers were, for the various fragments of the dsrA promoter, FR-dsrAHd, for lacUV5p, FRUV5Hd[−], and for λpL, FRλHd[−] (Table 1). The oligonucleotides used for lacUV5p and λpL introduce two extra bases into the transcript (172 nucleotides [nt]) compared to the transcript generated with dsrAp (170 nt).
In vitro transcription assays.
Assays were adapted from the method described by Zhou and Jin (53) for single-round transcription. Briefly, all compounds of the reaction were warmed up for 5 min at 37°C. At the start of the reaction, 2.5 nM supercoiled plasmid DNA was mixed with 40 mM Tris-glutamate (pH 8.0), 5 mM magnesium glutamate, 1 mM dithiothreitol, 100 μg of acetylated bovine serum albumin/ml, 200 mM potassium glutamate, and 10 nM RNA polymerase (0.1 U; Boehringer Mannheim). After 2 min, a solution of heparin and nucleoside triphosphates was added at final concentrations of 0.1 μg/μl of heparin; 200 μM for ATP, GTP, and CTP; and 20 μM for UTP (including [α-32P]UTP, 800 Ci/mmol; Pharmacia). This elongation step mixture was incubated for 4 min and stopped by adding 0.2 volume of stop solution (50% glycerol, 250 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue). Transcripts were run on 8% sequencing gels. Similar results were obtained with different preparations of the same plasmids. Initial transcription experiments were carried out with purified core RNA polymerase and added ς70, obtained from D. Jin. Results were similar to those described here with commercial RNA polymerase.
RESULTS
The amount of DsrA varies with the growth temperature.
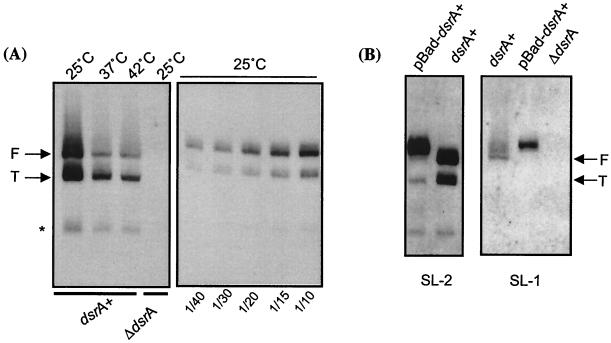
To determine if DsrA amount varies with temperature, RNAs were extracted from cells grown at 25, 37, and 42°C. Amounts of DsrA were quantitated in Northern blots, using a probe complementary to the second stem-loop of the RNA (probe SL-2) (30). Two strong signals were obtained, with a much weaker third signal (Fig. 1 A). All three signals were absent in a host carrying a deletion of dsrA (Fig. 1A, fourth lane). The two strong signals were detected consistently even when different RNA extraction methods were used, indicating that they were unlikely to be an artifact due to the RNA isolation (data not shown). The larger band (called “F” for “full length”) was estimated to be at 85 to 87 nt, which agrees with the predicted size of full-length DsrA (44). The smaller band (“T” for “truncated”) was estimated to be 60 or 61 nt long. In parallel, each sample was also probed for SsrA, a small RNA used as an internal control and known to be stable, abundant, and expressed equally at 30 and at 43°C (7). Equivalent amounts of SsrA were detected in the samples from 25, 37, and 42°C, confirming that synthesis of this RNA is temperature independent and is a useful control (data not shown). This confirms that the variations observed for DsrA amounts are specific. To quantitate the differences in DsrA amounts with changing temperature, we used a serial dilution of the 25°C RNA sample to perform Northern blotting (Fig. 1A, right panel) to confirm that quantitation of the RNA was in the linear range for these gels. Our results indicate that the full-length DsrA decreases by 25-fold at 37°C and 30-fold at 42°C, compared to the amount at 25°C. The T form of DsrA shows less variation with temperature than does the full-length DsrA (Fig. 1A). At 37 and 42°C, this form was present at similar levels, about four- and eightfold less than that seen at 25°C. Therefore, one major effect of temperature is to control the amounts of full-length DsrA in the cell and, to a lesser extent, all forms of DsrA.
FIG. 1.
Steady-state levels of DsrA as a function of temperature. (A) DsrA amounts at 25, 37, and 42°C. Northern blotting was performed using the SL-2 probe on RNAs extracted from MC4100 (dsrA+) or the ΔdsrA isogenic strain (FR299) grown in LB to an OD600 of ∼0.67 to 0.72 at 25, 37, and 42°C (left panel). Fifteen micrograms of total RNA was loaded in each lane. F and T forms of DsrA are indicated. A minor band detected by the probe is indicated by a star. In the right panel, dilutions of the sample extracted from cells grown at 25°C and shown in the left panel were loaded on a parallel gel. Dilutions are indicated at the bottom of the gel. (B) Comparison of the signals from probes SL-2 and SL-1. At the top of the lanes, dsrA+ and ΔdsrA correspond to the RNAs extracted from MC4100 and FR299, respectively. pBAD-dsrA corresponds to RNA extracted from FR299 carrying pNM13, the pBAD-dsrA+ plasmid, grown in LB-ampicillin and 30 μM arabinose. The exposures in the left and right panels are different; in general, the SL-2 probe detects DsrA much more efficiently than does the SL-1 probe.
In order to define the nature of the truncated form of DsrA, we used a second probe (SL-1) complementary to the first stem-loop of DsrA. This probe detected only the full-length DsrA (Fig. 1B, right panel), suggesting that the first stem-loop of DsrA is absent in the T form. Therefore, T could be generated either from processing of the F form of DsrA or from an internal transcription start site. In vitro transcription assays using purified RNA polymerase and a supercoiled template carrying DsrA under the control of its own promoter provided a single transcript at 85 to 87 nt, as expected for the full size of DsrA. No signal was detected at 60 nt (data not shown), consistent with an in vivo processing event to produce the truncated form of DsrA seen in vivo. In addition, a plasmid (pNM13) (30) expressing DsrA under the control of an inducible promoter (pBAD) was introduced into a strain (FR312) carrying a deletion of dsrA (30). RNAs extracted from FR312 were used for Northern blots probed with either the SL-1 or SL-2 probe. The full-length transcript made from pNM13 is 3 or 4 nt longer than the chromosomal transcript, suggesting that the transcription from the arabinose promoter initiates upstream from the normal start site of DsrA. However, as with DsrA expressed from the chromosome, two transcripts (and a small amount of a third transcript) were detected with SL-2, while only the longer transcript was seen with SL-1 (Fig. 1B). The T form from pBAD-dsrA+ is identical in size to that from the chromosome (Fig. 1B, left panel). This is inconsistent with a second promoter, since there is not enough DNA (25 bp) between a possible start site within the dsrA coding sequence and the pBAD promoter to carry such a full second promoter on pNM13. We did note that less of the truncated form was seen from cells carrying pNM13. It is possible that overexpression of DsrA titrates an RNase necessary for the cleavage.
Our direct measurements of DsrA levels in Northern blots could reflect a combination of changes in synthesis, processing, and stability with temperature. We have separately assessed the effects of temperature on the synthesis and on the stability of DsrA.
Lac-based reporter systems to study temperature control in vivo: features of the cloning vectors pFRΔ and pFRα.
We undertook the study of the dsrA promoter (dsrAp) using the widely used lacZ reporter system described by Simons et al. (43). This system allows one to clone a promoter in a high-copy-number plasmid, pRS415, in front of the W205 trp-lac fusion (32). Using the λ phage RS45, the transcriptional fusion is then inserted in a single copy at the attachment site of the phage in the chromosome. However, during our initial studies, two problems were encountered. (i) dsrAp is a strong promoter, and strong promoters (lacUV5 or λpL, for instance) cloned into pRS415 have toxic effects on the cell, most likely as a result of the high expression level of β-Gal. In our case, we found that some versions of the dsrA promoter, when cloned into pRS415, yielded only mutant plasmids that had 1 bp deleted from the spacer region. To avoid this strong selection for promoter mutants, we made use of the pRS415 derivative, pRS1553 (provided by R. Simons). In pRS1553, the cloned promoter drives the expression of the lacZ-α peptide (35). Alpha complementation allows screening for the correct insert without the toxicity observed in pRS415 (R. Simons, personal communication). (ii) Both pRS415 and pRS1553 carry the trp-lac fusion W205 located downstream of the promoter cloning site, including the lac translation initiation sites. Using the promoter of the E. coli spc operon, Liang et al. (28) demonstrated that the transcription terminator trpt, present in the W205 fusion in front of lacZ, is cold sensitive, resulting in efficient termination at high temperatures and very little termination at low temperatures. We observed a similar temperature effect, in vivo and in vitro, on the expression of our W205 fusions with the dsrA promoters, as well as a variety of others, including lacUV5, rcsA, rrnBp1, λpL, and λpR′. As previously reported (28), we also have noticed other effects that seem to be dependent on the combination of a specific promoter and the W205 fusion (i.e., influence on weak DNA activating sites and transcription termination efficiency). To avoid these complications in interpreting the effects of temperature on the dsrA promoter, we constructed two pRS1553 derivatives, vectors pFRΔ and pFRα (see Materials and Methods), that remove the trpt functional sequence (Fig. 2). The frΔ fusion carries the deletion of the first 50 bp of trpt described by Liang et al. (28). According to these authors, the β-Gal activity from this fusion reflects the transcriptional activity of the promoter cloned upstream and no longer shows different expression levels with temperature. In the fusion frα, the 75 bp of trpt was replaced with a sequence almost identical to the natural 5′ lacZ leader mRNA (Fig. 2). pFRΔ and pFRα can both be recombined onto the λ phage λRS468, used for recombination with pRS1553 (35), and single lysogens carrying the transcriptional fusion can be isolated. Using frΔ and frα fusions, the activities of two control promoters, lacUV5 and λpL, and five derivatives of dsrAp were tested at 25, 37, and 42°C. Results were identical with the parallel frΔ and frα fusions. Below, we report results obtained with the frΔ fusions.
Analysis of the dsrA promoter in vivo.
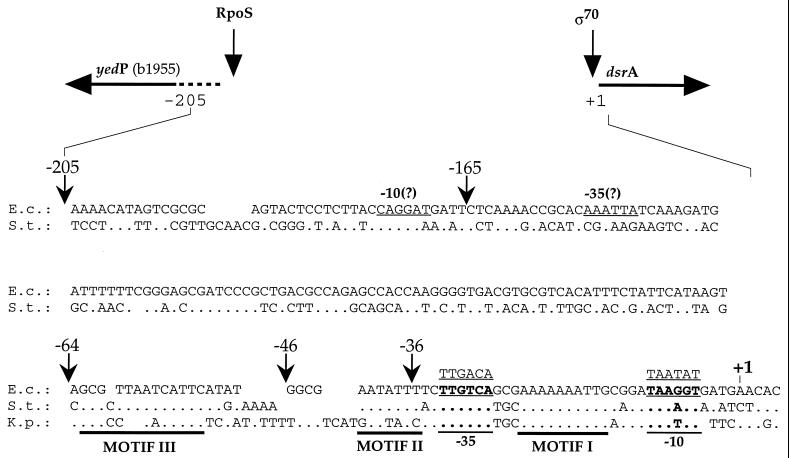
dsrA is a single expression unit located at 43.6 min on the E. coli map. The transcription start site was previously determined, and the putative −35 and −10 boxes of the promoter were assigned (44). We undertook a deletion analysis of the dsrA promoter (dsrAp) by cloning the promoter and various lengths of the upstream region in front of lacZ. All fusions extended to +2 of the DsrA transcript (Fig. 3). The upstream endpoints were chosen by aligning the dsrAp region for E. coli with that for Klebsiella pneumoniae and Salmonella enterica serovar Typhi. This alignment suggested that, in addition to the −35 and −10 boxes, three regions of the promoter were particularly well conserved (Fig. 3): (i) a portion of the spacer region (marked as motif I), (ii) the region between the −35 box and position −46 (motif II), and (iii) the region between −46 and −64 (motif III). Because it seemed possible that this interspecies conservation reflected the importance of these motifs for promoter regulation, we constructed fusions with breakpoints after each of these regions: dsrAp36::frΔ contains the minimal promoter, with motif I only; dsrAp46::frΔ contains sequences through −46, including motifs I and II; and dsrAp64::frΔ contains sequences through −64 carrying motifs I, II, and III. Two additional fusions were constructed: dsrAp205::frΔ, carrying the whole intergenic region between dsrA and the upstream divergent open reading frame, yedP (see below), and dsrAp165::frΔ, having the most distant 40 nt deleted and therefore not carrying the predicted promoter for yedP. Once introduced as a single copy on the bacterial chromosome, the expression of each transcriptional fusion was monitored by measuring the β-Gal activity in cells growing at various temperatures.
FIG. 3.
Sequence of the dsrA promoter region. The top of the figure shows the organization of the dsrA locus and the divergent yedP (b1955). The expression of yedP is strictly dependent on RpoS; dsrA expression is not affected by rpoS mutants and is apparently ς70 dependent. The lower part of the figure shows the alignment of the promoter regions from E. coli (E.c.), Salmonella enterica serovar Typhi (S.t.), and K. pneumoniae (K.p.). The conserved bases are indicated by dots. Very little conservation was found upstream of −64 for Klebsiella, and yedP is not detected in the sequences available for this organism. Sequences indicated as motifs I, II, and III were found to be conserved among these three organisms. Promoter elements (−10 and −35) of dsrA are indicated in boldface and underlined, with the ς70 consensus sequence above. For the divergent gene to dsrA, yedP, the predicted translation start site is indicated and the putative −10 and −35 boxes are underlined. Transcriptional dsrAp::lacZ fusions (frΔ) used are indicated by arrows with the numbers referring to distances from the transcription start site of the dsrA gene (+1); the fusions extend to +2 of the dsrA promoter.
(i) The dsrA promoter is more active at low temperature.
We first compared the activity of the longest dsrA promoter construct (dsrAp205) to the activities of the lacUV5p and λpL promoters, used as controls, in cells growing in LB medium at 25, 37, and 42°C (Fig. 4). lacUV5 and λpL had similar activities at 25 and 42°C, and were less than twofold higher at 37°C, at all stages of growth (Fig. 4A and B; Table 2). Thus, in the range tested, lacUV5p and λpL activities were not significantly affected by temperature. In contrast, the activity for dsrAp205 was highest at 25°C, decreased by close to half at 37°C, and decreased another fourfold from 37 to 42°C (Fig. 4C; Table 2). Therefore, compared to lacUV5p and λpL, the dsrA promoter is temperature sensitive. In order to identify the sequences within the dsrA205 promoter necessary for temperature control, we tested the temperature response for all the other dsrAp::frΔ fusions. All of them show temperature regulation similar to that seen for dsrAp205 (Table 2).
The minimal promoter (dsrAp36) shows about a twofold decrease in activity from 25 to 37°C, indicating that it still carries sufficient information for the temperature response. However, its activities at 37 and 42°C were similar to the level of the frΔ fusion inserted in the chromosome without a promoter (≤15 U at 25, 37, or 42°C). Thus, at 37°C or above, dsrAp36 does not have enough activity to be measured (Table 2).
At 25°C, dsrAp46::frΔ and dsrAp64::frΔ have an activity 10-fold higher than that of the minimal promoter (dsrAp36) (Table 2). Therefore, extending the minimal promoter to contain the 10 bp upstream of the −35 box introduces an activator element. To determine if the conserved motif II present in this region was the activator element, we replaced AATATTT with the sequence AGTATAC. This new promoter (dsrAp46AccI) shows an activity about twofold lower than that of dsrAp46 (164 and 66 β-Gal U, respectively, at 25 and 37°C), which is consistent with a partial inactivation of the activator function for motif II. No differences in activity were seen in a comparison of the 46-bp and 64-bp promoter fusions. Therefore, the activating element associated with motif II appears to be entirely contained within 46 bp (Table 2).
Fusions dsrAp205::frΔ and dsrAp165::frΔ had activities at 25 and 37°C that were consistently about twofold higher than that of dsrAp64::frΔ (Table 2). This suggests the presence of an additional weak activator element between −165 and −64 bp. At 42°C, dsrAp165::frΔ still is expressed at a level twofold higher than that of dsrAp64::frΔ (193 versus 84 U), suggesting that activation of the promoter by this element occurs at all temperatures. However, dsrAp205 decreases in activity from 37 to 42° (390 to 110 U), losing the twofold elevation seen at lower temperatures. At 42°C, its activity is close to the level of dsrAp64::frΔ at 42°C (84 U). Thus, at high temperature, the region between −205 and −165 bp of dsrAp appears to have a negative effect about equal to and opposite from the modest activator effect of the region between −64 and −165.
The demonstration of temperature-sensitive expression of the dsrAp fusions suggests that temperature controls the synthesis of DsrA at the level of transcription initiation. However, it is possible that, in the natural context, temperature might affect elongation through DsrA as well, contributing to differences in the amount of full-length RNA. To examine this possibility, we constructed frΔ fusions containing, in addition to the dsrA205 promoter, sequences encoding SL-1, SL-1 and SL-2, or the entire DsrA molecule. The construct containing dsrAp205 and SL-1::frΔ gave an activity around twofold higher than that of dsrAp205::frΔ at any temperature, indicating that SL-1 alone does not lead to temperature regulation of elongation (data not shown). The two other constructs did not express β-Gal activity at any temperature tested (data not shown). This suggests that SL-2 and/or SL-3 may lead to termination, instability of the message, and/or interference with lacZ translation. Efficient transcription termination after SL-3 is not surprising and has been noted before (45). However, if SL-2 acts as a terminator in its normal context, we might have expected to detect such a truncated RNA in vivo or in vitro; none has been seen. Therefore, unless this truncated RNA is extremely unstable, SL-2 may interfere with lacZ translation. In summary, there is no evidence for an effect of temperature on the elongation of DsrA.
(ii) Additional regulatory inputs for DsrA: effect of LeuO.
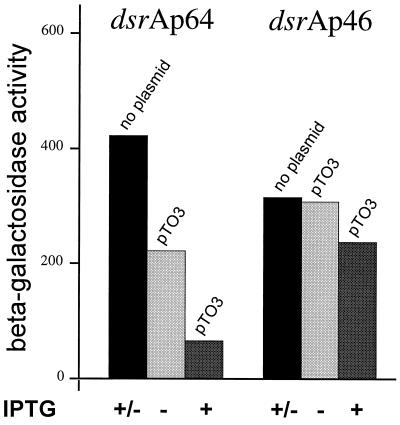
LeuO is a LysR-like protein regulator. When overexpressed, LeuO was shown to strongly reduce the expression of a translational rpoS::lacZ fusion as well as a dsrA72::lacZ fusion, suggesting that the effect on RpoS was mediated through repression of DsrA expression (24). In order to define sites necessary for LeuO action, we introduced the plasmid pTO3 (ptac-leuO) (47), into strains carrying the various dsrAp::frΔ fusions. λpL::frΔ was used as a promoter control. Even without induction of ptac, pTO3 had an inhibitory effect on the expression of the dsrAp205, dsrAp165, and dsrAp64::frΔ fusions but not on dsrAp46, dsrAp36, or λpL::frΔ (data for dsrAp64 and dsrAp46 are shown in Fig. 5). Therefore, the region between −64 and −46 bp of dsrAp that carries motif III (Fig. 3) was necessary for the LeuO effect (Fig. 5). Because ptac is a leaky promoter (5), we assume that LeuO is produced at modest levels by the uninduced plasmid. When the expression of LeuO was induced (by adding IPTG), a strong repression of dsrAp64::frΔ fusion was observed while no effect was observed for dsrAp46::frΔ (Fig. 5). Therefore, directly or indirectly, LeuO repression of the dsrA promoter requires sequences in the region between −64 and −46, the location of conserved motif III (Fig. 3). We have not detected an effect of the chromosomal mutation (leuO::cam) (47) on dsrAp205::frΔ in LB medium at any temperature (data not shown). The mapping of the regions responsible for the temperature control and the mapping of the LeuO regulatory site in the dsrA promoter separately from the site necessary for LeuO action confirm the earlier conclusion of Klauck et al. (24) that LeuO is not involved in temperature control of dsrA.
FIG. 5.
Effect of LeuO overexpression on the dsrA promoter. The fusion construct present as a single-copy lysogen in MC4100 is indicated at the top of each of the bar graphs. Each bar is labeled to indicate the presence or absence of the plasmid pTO3 (ptac-leuO). IPTG was added to growing cells (OD600 of ∼0.67 to 0.77) at a final concentration of 1 mM. “±” indicates that results were the same with or without IPTG. Values provided represent averages of at least three independent experiments performed at 37°C, when the OD600 of the culture is about 2.7 to 3, although the effect of LeuO on dsrAp64 is detectable as soon as 20 min after the addition of IPTG. The presence of pTO3 with or without IPTG did not affect the growth of the strain (data not shown).
Analysis of the dsrA promoter in vitro.
The features of the dsrA promoter (−10 and −35 regions and a spacer sequence of 17 bp) are close to the consensus sequences for RNA polymerase-ς70 holoenzyme (Eς70) (Fig. 3) (17). In vivo, mutations in the RpoS sigma factor, which recognizes promoter sequences close to those used by Eς70 (19), do not affect the expression of dsrAp (data not shown). Thus, we would expect transcription of dsrA to be initiated efficiently by Eς70. We examined the nature of the activator elements defined in vivo in in vitro transcription assays and whether the temperature response of dsrAp could be duplicated with only purifed Eς70 and dsrAp.
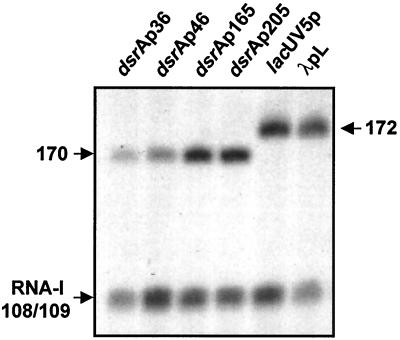
In vitro transcription experiments were performed using purified Eς70 and supercoiled plasmids carrying the portions of dsrAp used in vivo (dsrAp36, dsrAp46, dsrAp165, and dsrAp205). Single-round transcription conditions were used. Figure 6 shows the in vitro transcripts generated from these plasmids. The minimal promoter, dsrAp36, provides a signal of the expected size (170 bases). dsrAp46, carrying an additional upstream 10 bp, provides a signal that is about twofold stronger than that of the minimal promoter. This is consistent with the increased expression from this promoter in vivo, although the extent of stimulation is not as great as the 10-fold increase seen in vivo. This also suggests that the activating effect of the extra 10 bp in dsrAp46 is effective with Eς70 alone. A further increase was observed for dsrAp165 compared to dsrAp46, indicating that a second activator region affecting the transcription efficiency by Eς70 is present in this longer promoter fragment (Fig. 6). This is consistent with the positive effect observed in vivo for the sequence contained between −64 and −165 bp of dsrAp (Table 2). Transcription levels detected with dsrAp205 were comparable to those seen with dsrAp165. Therefore, two of the upstream elements defined in vivo are active in an in vitro system containing only Eς70. Note that neither of these upstream elements was necessary for temperature regulation in vivo.
FIG. 6.
In vitro analysis of the dsrA promoter. Supercoiled plasmids extracted from cells growing at 37°C were used as template. At the top of each lane, the fragment of the dsrA promoter carried by the plasmid used is indicated. On the left of the gel, “170” corresponds to the size of the transcript generated from dsrAp constructs, and “RNA-I 108/109” indicates the transcript generated from the plasmid replication origin. The two lanes on the right show the signals provided under the same reaction conditions by λpL and lacUV5 promoters, which generate 172-nt transcripts.
Thus far, we have not been able to recreate in vitro the differential response to temperature seen in vivo for any of the dsrAp DNAs. A number of different in vitro conditions and ratios of polymerase to template were tested; no effects specific for dsrA promoters, compared to the control lacUV5 and λpL promoters, were detected (data not shown). Thus, either a factor not present in the basic transcription system or a condition not present must account for temperature sensitivity.
DsrA stability changes with temperature.
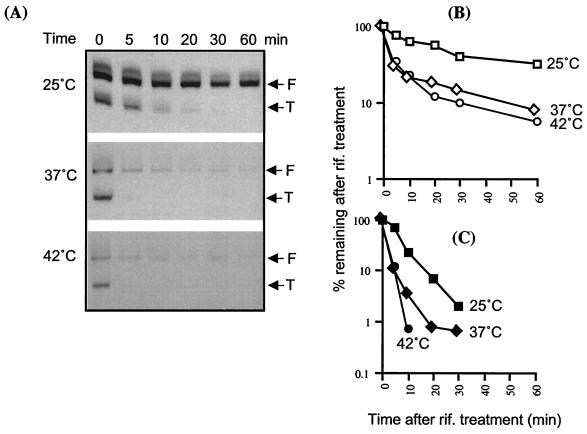
In addition to the temperature effect on DsrA synthesis, variations observed in the amount of DsrA at different temperatures (Fig. 1A) could also be due to changes in the stability of the RNA. We measured the stability of DsrA in cells growing at 25, 37, and 42°C. Northern blotting was performed at various time after treatment of the cells with rifampin, an antibiotic that blocks transcription (Fig. 7). Each sample was probed with SL-2, to allow a comparison of the stability of both forms of DsrA (F and T). At 25°C, we measured half-lives of 23 min for F and 5 min for T (Fig. 7). At both 37 and 42°C, F and T forms had half-lives of 4 and 2 min, respectively. However, the decay of the F form seems to have two components, an initial rapid component (4′ half-life) followed by a slower degradation of the remaining fraction (about 20%). As an internal control, SsrA was monitored and found to be stable at 25°C after 1 h of treatment with rifampin; at 37 and 42°C, its half-life was estimated to be around 40 min (data not shown). Thus, SsrA appears much more stable than DsrA at all temperatures.
FIG. 7.
DsrA stability as a function of temperature. (A) Northern blots were probed with SL-2 to detect both forms of DsrA (F and T). Cells were grown at the indicated temperature to an OD600 of ∼0.67 to 0.77, and rifampin was added to a final concentration of 300 μg/ml. Numbers indicate the time in minutes after the addition of rifampin. Two micrograms of total RNA per lane was loaded for 25°C, and 10 μg was loaded for 37 and 42°C. Different exposures are shown for each temperature. (B and C) Graphs show the decay of the full-length (F) form (B) and truncated (T) form (C) of DsrA at three temperatures. Values are averages from at least three different experiments. rif., rifampin.
Our measurements of stability for DsrA show that both forms of the molecule are more stable at low (25°C) than at high (37 or 42°C) temperatures. However, temperature is known to generally affect the rate of mRNA degradation in E. coli and other bacteria (1, 4). The growth rate of E. coli varies as a simple function of temperature that agrees with the Arrhenius law in a range of temperature from 21 to 37°C (21). In such a range, changes in metabolism result from the difference in the growth temperature (21). We therefore normalized the half-lives of the DsrA species to the doubling times of the cells at 25 and 37°C. The decrease in the half-life of T when the temperature changes from 25 to 37°C is proportional to the difference in the doubling times at these temperatures (ratios of 0.062 and 0.057, calculated from doubling times of 80 and 35 min, and half-lives of 5 and 2 min, at 25 and 37°C, respectively). In contrast, F is threefold more stable at 25°C than would be expected from its stability at 37°C (ratios of 0.3 to 0.1, for half-lives of 23 min at 25°C and 4 min at 37°C). Thus, besides the temperature control of DsrA synthesis, changes in stability of the full-length DsrA RNA are also under temperature regulation and therefore will contribute to the temperature control of RpoS.
DsrA synthesis and stability as a function of growth phase.
RpoS amounts increase dramatically as cells enter stationary phase (19). As previously reported (45), we found that the expression of RpoS requires DsrA during both exponential and stationary growth at 25°C. At 37°C, RpoS expression was DsrA independent (data not shown; see Discussion). This differs somewhat from our previous findings (45) where different growth conditions were used. Northern blots on RNA samples extracted from cells growing at 25 and 37°C at various phases of cell growth did not show any significant variation in the synthesis and the stability of DsrA (data not shown). This result indicates that the marked changes in RpoS amounts as cells enter stationary phase do not reflect changes in the levels of DsrA.
DISCUSSION
Environmental factors control the expression of RpoS at the level of transcription, translation, and proteolysis (20). Thus far, relatively few of the molecular links between environmental stimuli and the steps leading to changes in RpoS expression have been explained. We have analyzed how temperature controls the level of RpoS in the cell via the small regulatory RNA, DsrA. DsrA is necessary for the temperature control of RpoS and stimulates its translation (30, 45). We have found two major components of temperature regulation of RpoS by DsrA: (i) temperature-sensitive synthesis of the RNA and (ii) temperature-sensitive degradation. The net effect is a 25- and 30-fold decrease in full-length DsrA at 37 and 42°C, respectively, compared to 25°C (Fig. 1; summarized in Table 3).
TABLE 3.
Summarized data for DsrAa
| Temp (°C) | Relative amt of DsrAb (%)
|
F/T ratiob | dsrAp activityc (%) | Half-life of DsrA (min)d
|
RpoS::LacZ activitye
|
|||
|---|---|---|---|---|---|---|---|---|
| F | T | F | T | dsrA+ | ΔdsrA | |||
| 25 | 100 | (100) | 4 | 100 | 23 | 5 | 16 | 2 |
| 37 | 4 | 25 | 0.5 | 60 | 4 | 2 | 6 | 5 |
| 42 | 3.3 | 12.5 | 0.9 | 17 | 4 | 2 | <2 | <2 |
All data are from parallel experiments performed on cells growing in rich medium at an OD600 between 0.65 and 0.8.
Data from quantitation of Northern blots in Fig. 1, normalized to amounts at 25°C.
Data from experiments whose results are shown in Fig. 7.
Data from assay of RpoS::LacZ fusion as described in the text.
Temperature regulation of DsrA synthesis appears to be responsible for an important portion of the differences in DsrA accumulation, particularly as the temperature increases from 37 to 42°C. While two control promoters, λpL and lacUV5, increase slightly in activity at 37 compared to 25°C, the dsrA promoter fusions were about twofold less active at 37 than at 25°C (Fig. 4 and Table 2). There is a further 2.5- to 3.5-fold decrease in the interval from 37 to 42°C; overall, synthesis from this promoter at 42°C is only 17% of what it is at 25°C (Tables 2 and 3). The minimal promoter contained within dsrAp36 is responsible for the majority of this temperature regulation; a second regulatory element located between −165 and −205 of the dsrA promoter contributes further (Table 2). We note that λpL has previously been described as a temperature-sensitive promoter (14). The work on λpL utilized fusions containing the W205 trp-lac fusion with its temperature-dependent terminator; we believe that the behavior of this terminator explains those observations.
What is responsible for the temperature regulation of the dsrA promoter? Our inability to recreate temperature regulation in vitro leaves open the possibility that a trans-acting factor might be necessary for sensing temperature. However, if so, it must interact with the minimal promoter, which exhibits significant temperature regulation (Table 2). Replacing the conserved spacer sequence of AAAAAAATTG with TCTAGAATTG did not change promoter strength or temperature sensitivity, inconsistent with an essential binding site for a protein in this region of the promoter. We favor instead changes in the structure of the promoter itself, possibly mediated by more general changes in the cellular milieu. The high AT content of the spacer, although not essential, may contribute to poising the system to facilitate melting during open complex formation. We have observed that the spacer is exquisitely sensitive to changes in size. The addition or deletion of 1 nt in the spacer decreases the activity of the promoter drastically, even at low temperatures. Other properties of the minimal promoter may help the dsrA promoter to compete more effectively for RNA polymerase at low temperatures. For instance, the high-AT spacer, combined with other sequences, might allow open complex formation at 25°C more effectively than for other promoters. At higher temperatures, these features would be less of an advantage, and the dsrA promoter would compete less well. Whether these features act independently, by directly promoting interactions with RNA polymerase, or respond to global changes in DNA topology, for instance, or chemical modifications, we do not yet know. For instance, the osmE promoter is activated by osmotic shock, apparently leading to changes in supercoiling and therefore changes in promoter activation (9). Such a requirement for a change in topology might explain our inability to see in vitro temperature regulation in a purified system and is being further investigated.
Within the upstream regulatory region for dsrA is the promoter for a divergent gene of unknown function (yedP or b1955). Using a fusion for this promoter containing the whole region between yedP and dsrA, we found that the yedP promoter is induced upon the entry into stationary phase and that its activity is not controlled by temperature. The expression of yedPp::frΔ strictly depends on RpoS (data not shown). The presence of this RpoS-dependent gene suggested the possibility of coupling between the transcription of dsrA and yedP. However, the activity of dsrAp205 was not affected in an rpoS mutant background (rpoS::Tn10) during growth at 25, 37, or 42°C (data not shown), conditions where the divergent yedP promoter is no longer active. It remains possible that the activity of this promoter or of factors regulating it affects the dsrA promoter under some conditions. We note the twofold negative effect of the far-upstream region (believed to contain the promoter) found at 42°C. Whether this is ever important for dsrA and therefore RpoS regulation is unknown.
Two regions of the dsrA promoter stimulate synthesis in vivo and in vitro, the region from −46 to the −36 box (Fig. 3) which contains conserved motif II and the region from −64 to −165. The sequence of motif II (AATATTT) is close to that proposed for the distal part of an UP element known to interact with the carboxy-terminal part of the α subunit (α-CTD) of the RNA polymerase (AAA[A/T][A/T]T[A/T]TTTT) (12). Mutations in motif II (AATATTT changed to AGTATAC) reduce the stimulating effect of this sequence. Although the position of the sequence for dsrA is closer to the −35 box than usually found, the stimulatory activity of this region in vitro (Fig. 6) is most consistent with its action as a UP element. We have not characterized further the less conserved activator element(s) within the region from −64 to −165. The distance between the minimal promoter and region −64/−165 suggests that it is likely that the flexible carboxy-terminal part of the α subunit of the RNA polymerase would also be involved in these interactions. Whether use of either of these activating signals changes with growth conditions is not known.
LeuO, a regulator in the LysR family, down-regulates DsrA synthesis when overexpressed (24). We find that LeuO is acting in a conserved region of the dsrA promoter (motif III, Fig. 3). Conditions affecting either the synthesis or the activity of LeuO or another LysR family regulator may significantly perturb DsrA synthesis and therefore synthesis of RpoS. This regulation is entirely independent of temperature. Thus, assuming that this conserved site in the dsrA promoter and the effect of LeuO overexpression reflect a physiologically relevant regulatory signal, at least one other regulatory signal for DsrA synthesis besides temperature must exist.
While temperature provides a sixfold difference in the activity of the DsrA promoter between 25 and 42°C, the steady-state level of DsrA at 42°C is 30-fold less than at 25°C. The differences in the stability of DsrA at low and high temperature must contribute significantly to these differences in amounts, reflected in turn in differences in DsrA-dependent stimulation of RpoS (Table 3). Furthermore, we found two different forms of DsrA, a full-length molecule (F) and a truncated molecule (T), probably missing the first stem-loop; the relative amounts of F and T also vary with temperature (Fig. 1 and Table 3). In independent work, Sledjeski et al. measured a half-life for full-length DsrA of 6 to 30 min (depending on the method for measuring the RNA) at 30°C and also noted the truncated species (46). Since sequences within SL-1 are essential for RpoS translation (30), we expect only the F form to be active for stimulation of RpoS; the decay of this form is particularly sensitive to temperature (Table 3). Therefore, the degradation of the RNA molecule and the regulation of its synthesis will both participate in enhancing the differences in the steady-state levels of DsrA at low and high temperature.
Our data are most consistent with the T form of DsrA arising through cleavage of the full-length molecule (see above). In addition, T is relatively less abundant when DsrA is overproduced from a plasmid compared to the chromosome (Fig. 1 and Table 3), suggesting that the cleavage of DsrA to the T form may be limiting. If so, it may happen very quickly, possibly even before transcription has finished, because we did not observe the full-length molecule chasing into the truncated form after rifampin treatment (Fig. 7). A sequence consistent with the RNase E cleavage site, GAAUUU, is present in DsrA at the base of SL-2, where we predict the cleavage to occur (11). This region will be unpaired as the rest of the second stem-loop of DsrA is synthesized; it is also predicted to be unpaired from in vitro studies of the DsrA structure (26).
The ratio between the two forms of DsrA (F/T) varies with temperature, with relatively more T form at higher temperatures (Table 3). Processing of DsrA to the T form (in addition to general degradation) should act to block the formation of the RpoS-stimulating RNA but would allow accumulation of a form which may well be active on other targets. DsrA has been shown elsewhere to negatively regulate HNS synthesis (27, 44). A deletion of the first stem-loop of DsrA, leading to the expression of a mutant form similar to the T form, is still able to regulate HNS while losing RpoS regulation (30). Possibly, processing allows DsrA activity on targets such as HNS to be less affected by temperature than regulation of RpoS (or other targets dependent on the first stem-loop). Thus, while degradation of the intact form of DsrA contributes to its overall temperature sensitivity, the specific cleavage of DsrA may allow channeling of the molecule to different targets.
In addition to the contributions of temperature to synthesis, degradation, and processing of DsrA, it is possible that DsrA activity is itself regulated by temperature. At 37 or 42°C, full-size DsrA was detected (Fig. 1), but the expression of rpoS::lacZ is not dependent on DsrA (Table 3). This suggest either that levels of DsrA at 37°C are below the threshold needed to stimulate RpoS translation or that temperature controls the activity of the molecule, either directly (changing the secondary structure of DsrA) or indirectly (acting on a factor controlling the pairing of DsrA with RpoS mRNA). Evidence does exist that the activity of DsrA on RpoS expression can change dramatically due to environmental changes. It has long been known that an osmotic shock increases translation of RpoS (34). This lab has recently demonstrated that this increase requires DsrA but does not need an increase in DsrA amount (29). Therefore, osmotic shock allows more efficient use of DsrA to stimulate RpoS translation and leads to DsrA-dependent translation at 37°C, even though, under other conditions, DsrA is not needed at 37°C (Table 3). Temperature could affect DsrA activity in a similar manner. We tried to address this issue by synthesizing DsrA from a foreign promoter, pBAD. While induction of RpoS by DsrA was independent of temperature for this promoter, a great deal more DsrA was synthesized from pBAD at 42 than at 25°C; this lack of correlation between DsrA amounts and DsrA activity is again consistent with some temperature regulation of DsrA activity (data not shown).
The requirement for small, trans-acting RNAs such as DsrA to stimulate RpoS translation allows the sensing of a variety of physiological conditions to result in major changes in the cell's capacity to react to stress. Two components of the temperature sensor for RpoS translation have been identified: the dsrA promoter and the degradation of DsrA. Changes in synthesis and overall degradation with environmental variations such as temperature will affect RpoS translation as well as other targets of DsrA. HNS has been shown to be a target, and a number of other targets have been proposed (i.e., argR, ilvIH, and rbsD) (27). In addition, changes in the efficiency of the processing, as we have demonstrated here, should differentially affect the balance between these targets.
We now know that DsrA is only one of at least three small RNAs affecting RpoS translation. The other two small RNAs, RprA and OxyS, are synthesized in response to other signals (3, 29, 51), significantly increasing the number of signals that can affect RpoS availability. It is becoming apparent that this is a paradigm for a new and significant level of cellular regulation. Multifunctional RNAs such as DsrA are likely to exist in all organisms. They allow integration of multiple environmental signals to coordinately regulate multiple outputs. Such a coordination can be added to independently operating transcriptional, translational, and posttranslational controls, providing even more responsiveness for the cell.
ACKNOWLEDGMENTS
We thank J. Cabrera, N. Janabi, and D. Jin for their advice and technical help with the in vitro transcription experiments. We thank M. Cashel, A. J. Carpousis, C. Gutierrez, N. Majdalani, E. Massé, O. Sand, and Y.-N. Zhou for valuable comments. We are very grateful to R. Simons for providing us pRS1553 and λRS468 and to C. Ueguchi for pTO3.
The NATO Science Program is acknowledged for a Collaborative Research Grant (CRG 972150).
REFERENCES
- 1.Aguilar P S, Lopez P, de Mendoza D. Transcriptional control of the low-temperature-inducible des gene, encoding the Δ5 desaturase of Bacillus subtilis. J Bacteriol. 1999;181:7028–7033. doi: 10.1128/jb.181.22.7028-7033.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altuvia S, Kornitzer D, Teff D, Oppenheim A B. Alternative mRNA structures of the cIII gene of bacteriophage lambda determine the rate of its translation initiation. J Mol Biol. 1989;210:265–280. doi: 10.1016/0022-2836(89)90329-x. [DOI] [PubMed] [Google Scholar]
- 3.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 4.Brandi A, Pietroni P, Gualerzi C O, Pon C L. Post-transcriptional regulation of CspA expression in Escherichia coli. Mol Microbiol. 1996;19:231–240. doi: 10.1046/j.1365-2958.1996.362897.x. [DOI] [PubMed] [Google Scholar]
- 5.Brosius J, Erfle M, Storella J. Spacing of the −10 and −35 regions in the tac promoter. Effects on its in vivo activity. J Biol Chem. 1985;260:3539–3541. [PubMed] [Google Scholar]
- 6.Casadaban M J, Cohen S N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA. 1979;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chauhan A K, Apirion D. The gene for a small stable RNA (10Sa RNA) of Escherichia coli. Mol Microbiol. 1989;3:1481–1485. doi: 10.1111/j.1365-2958.1989.tb00133.x. [DOI] [PubMed] [Google Scholar]
- 8.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conter A, Menchon C, Gutierrez C. Role of DNA supercoiling and RpoS sigma factor in the osmotic and growth phase-dependent induction of the gene osmE of Escherichia coli K12. J Mol Biol. 1997;273:75–83. doi: 10.1006/jmbi.1997.1308. [DOI] [PubMed] [Google Scholar]
- 10.Drlica K, Perl-Rosenthal N R. DNA switches for thermal control of gene expression. Trends Microbiol. 1999;7:425–426. doi: 10.1016/s0966-842x(99)01614-5. [DOI] [PubMed] [Google Scholar]
- 11.Ehretsmann C P, Carpousis A J, Krisch H M. Specificity of Escherichia coli endoribonuclease RNase E: in vivo and in vitro analysis of mutants in a bacteriophage T4 mRNA processing site. Genes Dev. 1992;6:149–159. doi: 10.1101/gad.6.1.149. [DOI] [PubMed] [Google Scholar]
- 12.Estrem S T, Gaal T, Ross W, Gourse R L. Identification of an UP element consensus sequence for bacterial promoters. Proc Natl Acad Sci USA. 1998;95:9761–9766. doi: 10.1073/pnas.95.17.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giladi H, Goldenberg D, Koby S, Oppenheim A B. Enhanced activity of the bacteriophage λ PL promoter at low temperature. Proc Natl Acad Sci USA. 1995;92:2184–2188. doi: 10.1073/pnas.92.6.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldenberg D, Azar I, Oppenheim A B. Differential mRNA stability of the cspA gene in the cold-shock response of Escherichia coli. Mol Microbiol. 1996;19:241–248. doi: 10.1046/j.1365-2958.1996.363898.x. [DOI] [PubMed] [Google Scholar]
- 16.Harel J, Martin C. Virulence gene regulation in pathogenic Escherichia coli. Vet Res. 1999;30:131–155. [PubMed] [Google Scholar]
- 17.Harley C B, Reynolds R P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heithoff D M, Sinsheimer R L, Low D A, Mahan M J. An essential role for DNA adenine methylation in bacterial virulence. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 19.Hengge-Aronis R. The general stress response in Escherichia coli. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 161–178. [Google Scholar]
- 20.Hengge-Aronis R. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr Opin Microbiol. 1999;2:148–152. doi: 10.1016/S1369-5274(99)80026-5. [DOI] [PubMed] [Google Scholar]
- 21.Herendeen S L, VanBogelen R A, Neidhardt F C. Levels of major proteins of Escherichia coli during growth at different temperatures. J Bacteriol. 1979;139:185–194. doi: 10.1128/jb.139.1.185-194.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoe N P, Goguen J D. Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J Bacteriol. 1993;175:7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurme R, Rhen M. Temperature sensing in bacterial gene regulation—what it all boils down to. Mol Microbiol. 1998;30:1–6. doi: 10.1046/j.1365-2958.1998.01049.x. [DOI] [PubMed] [Google Scholar]
- 24.Klauck E, Bohringer J, Hengge-Aronis R. The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol Microbiol. 1997;25:559–569. doi: 10.1046/j.1365-2958.1997.4911852.x. [DOI] [PubMed] [Google Scholar]
- 25.Konkel M E, Tilly K. Temperature-regulated expression of bacterial virulence genes. Microbes Infect. 2000;2:157–166. doi: 10.1016/s1286-4579(00)00272-0. [DOI] [PubMed] [Google Scholar]
- 26.Lease R A, Belfort M. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc Natl Acad Sci USA. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lease R A, Cusick M, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang S-T, Dennis P P, Bremer H. Expression of lacZ from the promoter of the Escherichia coli spc operon cloned into vectors carrying the W205 trp-lac fusion. J Bacteriol. 1998;180:6090–6100. doi: 10.1128/jb.180.23.6090-6100.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majdalani N, Chen S, Murrow J, St. John K, Gottesman S. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- 30.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 32.Mitchell D H, Reznikoff W S, Beckwith J R. Genetic fusions defining trp and lac operon regulatory elements. J Mol Biol. 1975;93:331–350. doi: 10.1016/0022-2836(75)90281-8. [DOI] [PubMed] [Google Scholar]
- 33.Morita M T, Tanaka Y, Kodama T S, Kyoguoku Y, Yanagi H, Yura T. Translational induction of heat shock transcription factor ς32: evidence for a built-in RNA thermosensor. Genes Dev. 1999;13:655–665. doi: 10.1101/gad.13.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muffler A, Traulsen D D, Lange R, Hengge-Aronis R. Posttranscriptional osmotic regulation of the ςS subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1996;178:1607–1613. doi: 10.1128/jb.178.6.1607-1613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pepe C M, Suzuki C, Laurie C, Simons R W. Regulation of the “tetCD” genes of transposon Tn10. J Mol Biol. 1997;270:14–25. doi: 10.1006/jmbi.1997.1094. [DOI] [PubMed] [Google Scholar]
- 36.Phadtare S, Alsina J, Inouye M. Cold-shock response and cold-shock proteins. Curr Opin Microbiol. 1999;2:175–180. doi: 10.1016/S1369-5274(99)80031-9. [DOI] [PubMed] [Google Scholar]
- 37.Phadtare S, Yamanaka K, Inouye M. The cold shock response. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 33–46. [Google Scholar]
- 38.Powell B S, Rivas M P, Court D L, Nakamura Y, Rivas M P, Turnbough C L J. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Repoila F, Tetart F, Bouet J Y, Krisch H M. Genomic polymorphism in the T-even bacteriophages. EMBO J. 1994;13:4181–4192. doi: 10.1002/j.1460-2075.1994.tb06736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross W, Thompson J F, Newlands J T, Gourse R L. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 43.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 44.Sledjeski D, Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sledjeski D D, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 46.Sledjeski D D, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueguchi C, Ohta T, Seto C, Suzuki T, Mizuno T. The leuO gene product has a latent ability to relieve bgl silencing in Escherichia coli. J Bacteriol. 1998;180:190–193. doi: 10.1128/jb.180.1.190-193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang N, Yamanaka K, Inouye M. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J Bacteriol. 1999;181:1603–1609. doi: 10.1128/jb.181.5.1603-1609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yura T, Kanemori M, Morita M T. The heat shock response: regulation and function. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 3–18. [Google Scholar]
- 50.Zangrossi S, Briani F, Ghisotti D, Regonesi M E, Tortora P, Deho G. Transcriptional and post-transcriptional control of polynucleotide phosphorylase during cold acclimation in Escherichia coli. Mol Microbiol. 2000;36:1470–1480. doi: 10.1046/j.1365-2958.2000.01971.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The oxyS regulatory RNA represses rpoS translation by binding Hfq (HF-1) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Y-N, Gottesman S. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J Bacteriol. 1998;180:1154–1158. doi: 10.1128/jb.180.5.1154-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Y-N, Jin D-J. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2908–2913. doi: 10.1073/pnas.95.6.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]