Abstract
Background
E6 and E7 oncoproteins are considered ideal antigens of T cell therapy for human papillomavirus (HPV)-related cancers. However, little is known about the epitopes of E6 and E7 presented by HLA-A*11:01, one of the most prevalent HLA types globally, especially in Asia.
Methods
We combined in silico and experimental approaches to identify endogenously processed HLA-A*11:01-restricted epitopes of HPV16 E6 and E7. The identified epitopes were then used to screen available T cell receptors (TCRs) from healthy donors through in vitro stimulation of peripheral blood mononuclear cells (PBMCs).
Results
E693-101 (TTLEQQYNK, TTL) and E789-97 (IVCPICSQK, IVC), two novel HLA-A*11:01-restricted T cell epitopes of HPV16, were identified to be endogenously presented on tumor cells. TTL- and IVC-specific TCRs were isolated from 11 healthy donors through in vitro stimulation of PBMC. The key TTL and IVC residues involved in TCR-pMHC interactions were mapped, and the consensus sequence was “xxLEQxYNK” and “xVxPIxxxK.” The TTL- and IVC-specific TCRs with high functional avidity were used to generate TCR-engineered T cells, specifically recognizing and killing corresponding tumor cell lines in vitro and in vivo. In addition, TTL and IVC-specific TCR-T cells also recognized and killed HPV16+ patient-derived organoids.
Conclusions
The HLA-A*11:01-restricted HPV16 E6/E7 epitopes and TCRs identified in this study may provide a new strategy for HPV-related cancer immunotherapy in HLA-A*11:01+ patients.
Keywords: Immunotherapy, Adoptive; Antigens
WHAT IS ALREADY KNOWN ON THIS TOPIC
E6 and E7 oncoproteins are considered ideal antigens of T-cell therapy for human papillomavirus (HPV)-related cancers.
WHAT THIS STUDY ADDS
E693-101 (TTLEQQYNK, TTL) and E789-97 (IVCPICSQK, IVC), two novel HLA-A*11:01-restricted T-cell epitopes of HPV16, were identified. TTL-specific and IVC-specific T-cell receptor (TCR)-engineered T cells specifically recognized and killed corresponding HLA-A*11:01 and E6/E7 double-positive tumor cells in vitro and in vivo.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
The epitopes and TCRs identified in this study may provide a new strategy for HPV-related cancer immunotherapy in HLA-A*11:01+ patients.
Background
About 80% of women and men will have been exposed to human papillomavirus (HPV) by the age of 45.1 Persistent infection with oncogenic HPV increases the cancer risk of the cervix, vulva, vagina, penis, anus, mouth, tonsils, and throat.2 More than 220 different HPV types have been characterized,3 with HPV types 16 and 18 the most virulent high-risk genotypes.4 HPV16 and HPV18 are responsible for approximately 60% and 15% of cervical cancers, respectively.4 In addition, HPV16 is responsible for almost 90% of HPV+ oropharyngeal cancers.5 Although HPV-associated cancers can be reduced with prophylactic HPV vaccination distributed since late 2016, existing infections still lead to new cases each year. In 2018, approximately 690 000 HPV-attributable cancer cases were newly diagnosed worldwide.6 Especially in low-income countries, HPV-induced cancer causes a high mortality rate with a poor prognosis due to the late diagnosis. Therefore, novel treatment strategies are urgently needed, especially for patients with advanced cancer.
T-cell receptor (TCR) gene engineered T-cell therapy (TCR-T) is emerging as an effective cancer treatment, especially for solid tumor.7–10 The E6 and E7 genes of HPV are constitutively expressed and widely recognized as the primary driving factor for the carcinogenesis of epithelial cells,11 representing ideal targets for TCR-T therapy.12 13 Oncoprotein E6 binds with E6-associated binding protein (E6AP), allowing it to bind with p53. The trimeric complex E6/E6AP/p53 leads to the degradation of p53 and the uncontrolled cell proliferation.14 E7, on the other hand, binds to the retinoblastoma tumor suppressor gene product (pRb), causing its inactivation and degradation.15 Consequently, the E6 and E7 oncoproteins are, in principle, ideal targets for T cell therapy of HPV-associated cancers since they are constitutively expressed by tumor cells and absent from healthy tissues. Different HPV epitopes have been targeted with TCR gene-engineered T cells.16–18 It was reported that the TCR gene engineered T cells that recognized human leukocyte antigen (HLA)-A*02:01-restricted epitopes of HPV16 E6 (E629–38)19 or E7 (E711–19)20 can successfully target HPV+ tumor cells. Furthermore, both TCRs are currently under investigation in clinical trials.21 22 In the clinical trial of E711–19 TCR-T cells for patients with metastatic HPV-associated cancers (NCT02858310), robust tumor regression was observed with objective clinical responses in 6 of 12 patients, including 4 of 8 patients with anti-PD-1 refractory disease.22
Specific T-cell recognition of tumor cells requires T-cell epitopes presented in the context of major histocompatibility complex (MHC, also known as HLA in humans) class I or MHC class II molecules.23 24 Therefore, identifying T-cell epitopes is crucial for developing effective immunotherapies, including T cell therapy and therapeutic vaccines. HLA-A*02:01-restricted epitopes of HPV16 E6 (E629–38) and E7 (E711–19) have been intensively investigated before developing specific TCR-T under clinical trials.25–27 While HLA-A*02:01 is dominant in the Caucasian population, HLA-A*11:01 is also one of the most prevalent HLA types worldwide, especially in Asia.28 However, HLA-A*11:01-restricted epitopes of HPV16 E6 and E7 and their specific TCRs are poorly investigated.
Thus, we set out to identify HLA-A*11:01-restricted HPV16 E6 and E7 T-cell epitopes. This study identified two novel HLA-A*11:01-restricted HPV16 epitopes, E693-101 (TTLEQQYNK, TTL) and E789-97 (IVCPICSQK, IVC). Using in silico and experimental methods, we demonstrated that both epitopes were endogenously processed and presented on HPV+ tumor cells, suggesting that they are promising targets for immunotherapies. We also showed that TTL-specific and IVC-specific TCR-Ts have therapeutic potential for clinical development against HPV-related cancers.
Methods
Cell lines and plasmids
Cell lines were cultured in Roswell Park Memorial Institute (RPMI)-1640 (T2 and J.RT3-T3.5), or DMEM (293T, CaSki, and SCC90) or MEM (SK-MEL-28), supplemented with 10% Fetal Bovine Serum (Lonsera) and 1% penicillin/streptomycin (Life Technologies). T2, 293T, SK-MEL-28, and J.RT3-T3.5 cell lines were purchased from American Type Culture Collection (ATCC)CaSki and SCC90 cell lines were purchased from CTCC. CaSki is an HPV16+ cervical cancer cell line. SCC90 is an HPV16+ head and neck squamous cell carcinoma cell line. All cell lines used in the experiments were mycoplasma-free.
HPV16 E6, HPV16 E7, and HLA-A*11:01 heavy chain genes were synthesized and constructed into pLKO.1puro (addgene 8453) lentiviral vectors under EF-1α promoter. P2A-linked GFP was used as the reporter gene.
The J.RT3-T3.5 cell line was used to generate Jurkat-CD8-NFAT (JK8NF) reporter cell line. First, J.RT3-T3.5 cells were transduced with lentiviral particles carrying the pLenti-human CD8 (hCD8) transfer plasmid and sorted by FACS based on high CD8 expression (Jurkat-CD8 cells). Next, the Jurkat-CD8 cells were transduced with lentiviruses encoding the nuclear factor of activated T cells (NFAT)-inducible ZsGreen reporter genes.29 Finally, JK8NF clones were selected based on a strong ZsGreen signal following Phorbol-12-myristate-13-acetate (PMA)/ionomycin stimulation.
Flow cytometry
Staining of cell surface proteins was done by incubating the diluted antibodies for 15 min on ice. The following fluorochrome-labeled antibodies: Fluorescein Isothiocyanate (FITC) anti-human CD3 (clone UCHT1), FITC/Pacific Blue anti-human CD8a (clone HIT8a), APC anti-mouse TCRβ chain (clone H57-597), and PE/Cyanine7 anti-human CD45 (clone 2D1) were purchased from BioLegend. Phycoerthrin (PE)-streptavidin was purchased from BD Biosciences, and purified anti-MHC class I antibody (clone W6/32) was purchased from BioLegend. PE-labeled tetramers were produced in-house. All antibodies and tetramers were used at 1:100 dilution. Data were quantified using BD FACSAria III flow cytometer (BD Biosciences) and CytoFLEX S (Beckman Coulter) and analyzed with FlowJo V.10 and CytExpert software.
In silico prediction of peptide ligands
Peptide binding affinity for HLA-A*11:01 was predicted using netMHC4.0 or netMHCpan-4.1. Nonamer/decamer peptides were ranked by predicted affinity (cut-off <500 nM), and the selected peptides were synthesized by GenScript (Jiangsu, China). Homologous peptides in human proteome were predicted using Expitope V.2.030 with default parameters (the cleavage threshold=0.7, the transporter associated with antigen processing (TAP) weight=0.2, the combined score threshold=1e-4). All peptides with three or fewer mismatches were searched. Moreover, peptides with four mismatches that also contained the motifs ‘xxLxxxYNK’ or ‘xVxPIxxxK’ were searched. Peptides with HLA-A*11:01 binding affinity of <5000 nM were synthesized by GenScript.
Generation of dendritic cells (DCs)
Peripheral blood mononuclear cells (PBMCs) of HLA-A*11:01-positive, healthy donors were isolated by density-gradient centrifugation from buffy coats (Guangzhou Blood Center) and cryopreserved. CD14+ cells were obtained by magnetic separation (BioLegend) from cryopreserved PBMCs to generate DCs.
The CD14+ cells were then cultured in complete RPMI-1640 medium containing 800 IU/mLgranulocyte macrophage colony stimulating factor (PeproTech) and 500 IU/mL interleukin (IL)-4 (PeproTech). The cultures were fed with fresh medium and cytokines every 2–3 days. Mature DCs were generated by adding 10 ng/mL IL-1β, 10 ng/mL IL-6, 10 ng/mL tumor necrosis factor alpha, and 1 µg/mL PGE2 for 48 hours from day 5.
Generation of peptide-specific T cells
Mature DCs were loaded with 20 µg/mL peptide (TTL, IVC, LLI, IIL, GTT, AVC, or GIV) and 0.5 mM tris (2-carboxyethyl) phosphine (to prevent dimerization of the cysteine residues in the peptides)31 32 at 37℃ for 2 hours. CD8+ naïve T cells were enriched from autologous CD14− cells by negative selection (Stemcell). Isolated CD8+-naïve T cells were cocultured with peptide-loaded DCs in T-cell medium (TexMACS with 10% human serum, 1% penicillin/streptomycin, 60 ng/mL IL-21, 10 IU/mL IL-2, 10 IU/mL IL-7, and 10 IU/mL IL-15). After 7 days, T cells were restimulated with peptide-loaded DCs. After 14 days, T cells were stained with PE-tetramer and FITC-CD8 and analyzed by flow cytometry.
TCR sequencing and cloning
CD8+tetramer+ cells were sorted into 96-well PCR plates by single-cell sorting. TCR sequences from sorted cells were obtained by single-cell PCR as previously described.33 TCR sequences were analyzed using the IMGT/V-Quest tool (http://www.imgt.org/). TCRα/β chains were synthesized by GenScript and cloned into the lentivirus vector pLKO. The human TCR constant regions were replaced with murine TCR constant regions modified with interchain disulfide bond and hydrophobic substitution as previously described.34 35
Lentiviral production and transduction
Lentiviral particles used in this experiment were produced by transient transfection of 293 T cells with target vector plasmid and packaging plasmids (Addgene) encoding pMD2G, REV, and RRE (3:1:1:1), and concentrated with a 100 kD ultrafiltration tube before storage at −80°C.
JK8NF cells were transduced with lentiviral particles at a multiplicity of infection (MOI) of 10 to construct JK8NF-TCR cell lines. Thawed PBMCs were activated with Dynabeads human T-activator CD3/CD28 (Life Technologies). The next day, T cells were transduced twice with lentiviral particles by spinoculation at an MOI of 1 in the presence of protamine (10 µg/mL). TCR-transduced T cells were then expanded in RPMI-1640 medium supplemented with 10% Fetal Bovine Serum (Gibco) and 200 IU/mL of IL-2. The transduction efficiency was calculated by staining for mouse TCRβ and tetramers.
In vitro evaluation of TCR-transduced cells
For pulsed peptides, 2000 CD8+tetramer+ TCR-transduced T cells were cocultured with 20 000 peptide-pulsed T2-A11 cells. We harvested the supernatants after 18–24 hours and measured interferon gamma (IFN-γ) concentration by ELISA according to the manufacturer’s instructions (Invitrogen 887316). For tumor cell lines, 5000 CD8+tetramer+ T cells were cocultured with an equal number of target cells for 18–24 hours with or without 10 µg/mL anti-MHC I antibody (clone W6/32). IFN-γ production was measured via the enzyme-linked immunospot (ELISpot) assay according to the manufacturer’s instructions (BD 551849). When the ELISpot plates were dry, spots were counted automatically using a Bioreader 6000 ELISpot-reader (Bio-Sys GmbH).
SK-MEL-28-E6 (SK-E6) and SK-MEL-28-E7 (SK-E7) cell lysis was evaluated with the xCELLigence real-time cell analyzer (ACEA Biosciences), which assessed electrical impedance due to the adherent cells in each well every 15 min until the end of the experiment. Five thousand CD8+tetramer+ TCR-transduced T cells were cocultured with 5000 target cells in E-plates and monitored for 90 hours. Results are reported as cell index values. CaSki-A11 and SCC90-A11 cell lysis were assessed by the fluorescence microscopy. TCR-T cells were cocultured with target cells for 96 hours at an effector-to-target ratio of 5:1. The fluorescence intensity was analyzed and quantified by Image J.
In vivo mouse xenograft models
Female immunodeficient NOD/SCID/IL2gR−/− (NCG) mice aged 6–8 weeks were purchased from Gempharmatech. NCG mice were injected subcutaneously with 1×107 SK-E6 or SK-E7 tumor cells with Matrigel (Corning) in a single flank. When tumor volume reached 80–100 mm3, 1×107 CD8+tetramer+ T cells were administered by retro-orbital injection. The same dose of total number of cells calculated based on transfection efficiency was administered to the non-transduced T cell (TCRneg) group. At the same time, 50 000 IU IL-2 was administered to both the experimental and TCRneg groups intraperitoneally every 24 hours for 5 days. Tumor size was measured two times per week with calipers in two perpendicular dimensions, and tumor volumes were calculated using the formula: volume (mm3)=length×width2/2. All mice were sacrificed at the end of the experiment, and the tumors were excised and weighed.
For analysis of T-cell infiltration, peripheral blood, spleen, and tumor samples were collected 7 days after T-cell injection. Spleen single-cell suspensions were prepared in PBS by mechanical digestion. The tumor tissues were dissociated into single-cell suspension by enzyme digestion (Miltenyi). Red blood cells (RBC) were lysed using RBC lysis buffer (Biolegend) prior to staining. The single-cell suspension was stained with PE/Cyanine7 anti-human CD45, APC-TCRβ chain, Pacific Blue-CD8, FITC-CD3, and PE-tetramer and analyzed by flow cytometry.
Tumor samples were fixed in 10% neutral-buffered formalin and embedded in paraffin for sectioning, followed by antibody staining for human CD3 according to standard procedures. The sections were imaged using a Leica Aperio CS2 Slide Scanner. The images were processed by Aperio ImageScope software.
Patient-derived organoids (PDOs) models
Tumor organoids derived from eight patients with cervical cancer were established by K2 Oncology Co. and cultured in OrganoPro Ultra-Low Adherent Plate for Suspension Culture-24 Well plate (K2 Oncology, K2OS-24) under the conditions of OrganoPro Cervical Cancer Organoid Medium Kit (K2 Oncology, K2O-M-CC). HLA typing of organoids was done by Bfrbiotech Co. using PCR sequencing-based typing. The expression of HPV16 E6 and E7 was analyzed by real-time PCR.
HPV16+ PDO KOCC-002S4 was transduced with the HLA-A*11:01-P2A-GFP lentiviral vectors and selected with 10 µg/mL puromycin for 2 weeks. PDO cells (8×103) were cocultured with TCR-transduced T cells or TCRnegs at an effector-to-target ratio of 1:1 in 96-well plates. After 72 hours, microscopy images were captured with Leica DMi1, and the supernatant was collected and measured by IFN-γ ELISA kit (BioLegend).
Statistical analysis
D’Agostino-Pearson omnibus or Shapiro-Wilk test was used to test normality for all datasets with Prism V.8 (GraphPad Software). Differences between the two groups were compared using a two-tailed unpaired Student’s t-test with Prism V.8. Two-way analysis of variance test and Bonferroni multiple comparisons tests were used to compare the tumor growth curves between TCR-T and control T cell-treated groups with Prism V.8. EC50 values were determined using a non-linear regression curve, fit variable slope (four parameters) with Prism V.8. All data were expressed as mean±SD. P values of <0.05 were considered statistically significant.
Results
Identification of HLA-A*11:01-restricted T-cell epitopes of HPV16 E6 and E7
The main purpose of this study was to systematically identify HPV16 E6 and E7 epitopes that could (1) bind to HLA-A*11:01, (2) induce expansion of specific T cells, and (3) be presented on HPV16-positive tumor cells.
We began with in silico prediction of potential HLA-A*11:01-binding peptides derived from HPV16 E6 and E7 proteins using the netMHC 4.0 server.36 Based on whole-genome sequencing, HPV16 variants are classified into four major lineages (A–D).37 The A4/Asian type is commonly classified by the polymorphism T178G (D25E) in the E6 gene.38 All peptides except peptide DII are conserved among four major lineages. Thus, the peptide EII in A4/Asian type (D>E mutation) was also included in the in silico prediction. Predicted 9-mer and 10-mer peptides with HLA-A*11:01 binding affinity of <500 nM are listed in online supplemental table 1. These 10 peptides were synthesized and used to produce HLA-A*11:01 peptide–major histocompatibility complex (pMHC) tetramers. However, we failed to get correctly folded pMHC monomers with peptides KIS, DII, and EII. The potential HLA-A*11:01-binding peptides derived from HPV16 E6 and E7 proteins were later predicted again with netMHCpan-4.1. Interestingly, the binding affinity of peptides KIS, DII, and EII predicted by netMHCpan-4.1 was 821.51, 1832.09, and 1022.53 nM. These results indicate that the binding affinity with HLA-A*11:01 of these three peptides was probably not high enough.
jitc-2022-004790supp002.xlsx (20.4KB, xlsx)
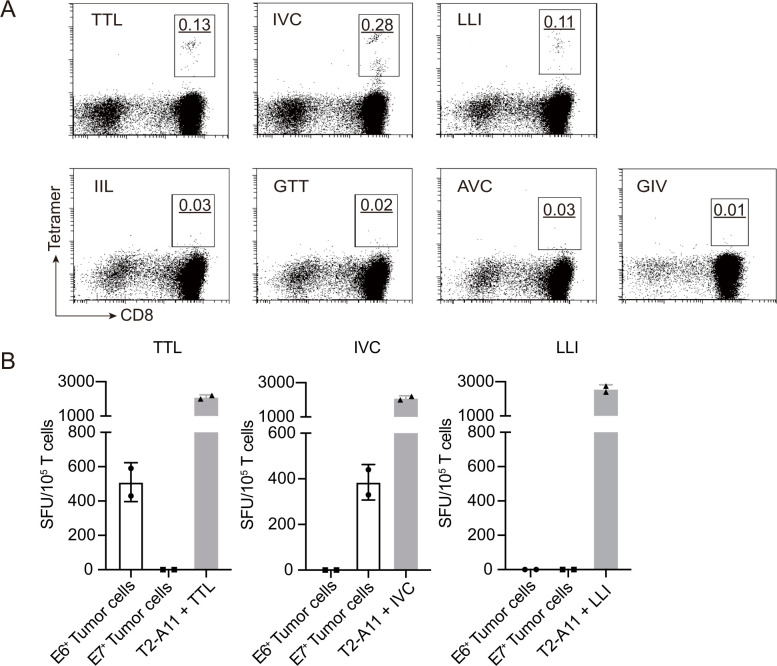
We next used HPV16 E6/E7 peptides shown to bind with HLA-A*11:01 to stimulate T cells in PBMCs. We used purified naïve CD8+ T cells from HLA-A*11:01-positive, healthy donors as responder cells and autologous peptide-pulsed monocyte-derived DCs as antigen-presenting cells. After approximately 14 days in culture, stimulated T cells were stained with the corresponding tetramers. For each peptide, at least three healthy donors were tested. The original percentages of tetramer-positive cells in naïve CD8+ T cells from healthy donors were too low to be detected for all seven peptides (data not shown). After 14 days of peptide stimulation, we could easily detect TTL, IVC, and LLI-tetramer positive CD8+ T cells in 11 out of 13, 11 out of 14 and 3 out of 6 donors, respectively. However, IIL, GTT, AVC, and GIV tetramer-positive cells were not detected. The representative tetramer staining results for each peptide are shown in figure 1A. These results indicate that TTL, IVC, and LLI derived from HPV16 E6/E7 successfully stimulated the expansion of specific T cells in peripheral blood in vitro, indicating that TTL, IVC, and LLI-specific T-cell clones could be enriched in healthy donors.
Figure 1.
Identification of HLA-A*11:01-restricted T-cell epitopes of HPV16 E6 and E7. (A) Naïve CD8+ T cells were isolated from HLA-A*11:01-positive, healthy donors and stimulated with indicated E6 or E7 peptides. After approximately 14 days in culture, stimulated T cells were stained with the corresponding tetramer. The representative tetramer staining flow plots of peptide stimulated CD8+ T cells from healthy donors are shown. The underlined number is the percentage of gated tetramer+CD8+ cells. (B) Recognition of SK-MEL-28-E6 (E6+) or SK-MEL-28-E7 (E7+) tumor cells by TTL, IVC, and LLI-specific polyclonal T cells was determined by IFN-γ ELISpot. Peptide-pulsed T2-A11 served as positive controls. IFN-γ, interferon gamma; ELISpot, enzyme-linked immunospot; SFU, spot-forming unit.
The most crucial criterion for a T-cell epitope is that the peptide is endogenously processed and presented on the cell surface.39 To investigate whether TTL, IVC, and LLI from HPV16 E6/E7 were processed in cells, E6 and E7 overexpressing tumor cell lines were used as target cells. First, E6 and E7 were separately overexpressed in HLA-A*11:01-positive SK-MEL-28 tumor cells. Then, TTL-specific, IVC-specific, and LLI-specific polyclonal T cells were cocultured with E6 and E7 overexpressing SK-MEL-28 tumor cells, and secreted IFN-γ was detected by ELISpot. The TAP-deficient T2 cell line was stably transduced with the HLA-A*11:01 heavy chain (T2-A11). TTL, IVC, and LLI-specific polyclonal T cells all recognized corresponding peptide-pulsed T2-A11 cells (figure 1B). Meanwhile, E6-derived TTL-specific T cells recognized E6-overexpressing SK-MEL-28 tumor cells, and E7-derived IVC-specific T cells recognized E7-overexpressing SK-MEL-28 tumor cells (figure 1B). However, E6-derived LLI-specific T cells failed to recognize E6-overexpressing SK-MEL-28 tumor cells. These results indicate that HPV16 E6-derived TTL peptide and E7-derived IVC peptide are processed and presented on HLA-A*11:01-positive tumor cells, making them candidate targets for T cell-based therapy.
TRAV and TRBV usage of TTL-specific and IVC-specific TCRs
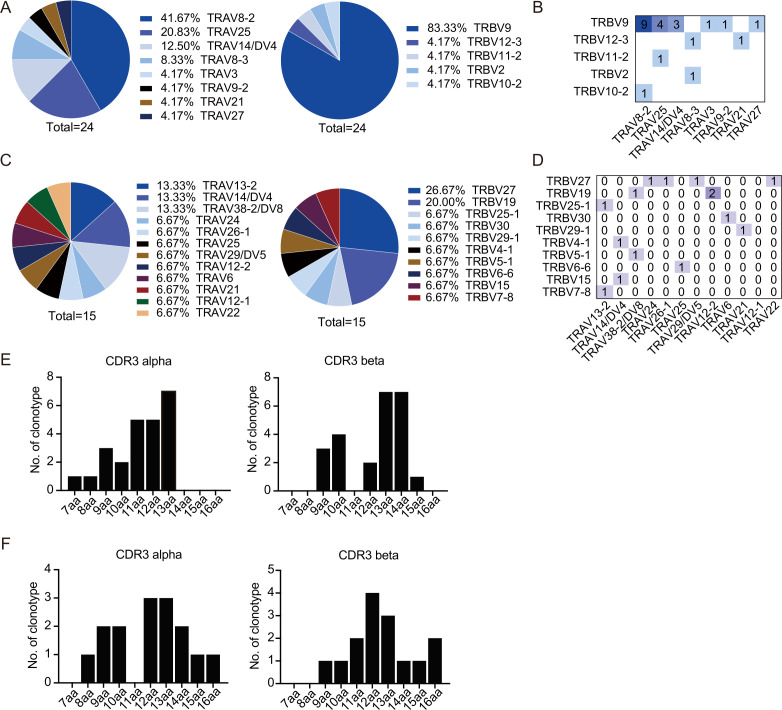
E6-derived TTL and E7-derived IVC peptides were then used to screen available TCRs for TCR-T cell immunotherapy against HPV-associated malignancies. CD8+ T cells isolated from HLA-A*11:01-positive, healthy donors were stimulated with peptide-pulsed autologous DCs for approximately 14 days. After stimulation, single CD8+ Tetramer+ T cells were sorted to obtain TCRs. In total, 24 TTL-specific TCRs and 15 IVC-specific TCRs were obtained from 11 healthy donors.
Biased TCR usage has been reported for many T cell epitopes.40 Here, T-cell receptor alpha-chain variable region (TRAV) and T-cell receptor beta-chain variable region (TRBV) usage of TTL- and IVC-specific TCRs were analyzed. Interestingly, among TTL-specific TCRs, we found a clear TRAV bias toward TRAV8-2 and TRAV25 and a strong TRBV bias toward TRBV9 (figure 2A). The clonotype with the highest frequency (9/24) was TRAV8-2/TRBV9, as shown in figure 2B. A slight TRBV bias toward TRBV27 and TRBV19 was observed among IVC-specific TCRs. However, no preferential TRAV usage was observed for IVC-specific TCRs (figure 2C, D). Lengths of CDR3α and CDR3β sequences of TTL- and IVC-specific clonotypes were also analyzed. For the TTL-specific TCR repertoire, the most common length of CDR3α and CDR3β was 13–14 amino acids (figure 2E). For the IVC-specific TCR repertoire, the most common length of CDR3α and CDR3β was 12–13 amino acids (figure 2F).
Figure 2.
TRAV and TRBV usage of TTL-specific and IVC-specific TCRs. (A, B) TRAV and TRBV usage of TTL-specific TCRs. (A) Cumulative frequencies of each TRAV (left panel) and TRBV (right panel) chains in TTL-specific T-cell repertoires. (B) Heatmaps of TRAV–TRBV combination usage for TTL-specific T-cell repertoires. The numbers of clonotypes using a given combination are indicated in the corresponding square. (C, D) TRAV and TRBV usage of IVC-specific TCRs. (C) Cumulative frequencies of each TRAV (left panel) and TRBV (right panel) chains in IVC-specific T-cell repertoires. (D) Heatmaps of TRAV–TRBV combination usage for IVC-specific T-cell repertoires. (E, F) Amino acid length distribution of CDR3α and CDR3β clonotypes in TTL-specific (E) and IVC-specific (F) T-cell repertoires. TCR, T-cell receptor; TRAV, T-cell receptor alpha-chain variable region; TRBV, T-cell receptor beta-chain variable region.
Screening of TTL-specific and IVC-specific TCRs based on functional avidity
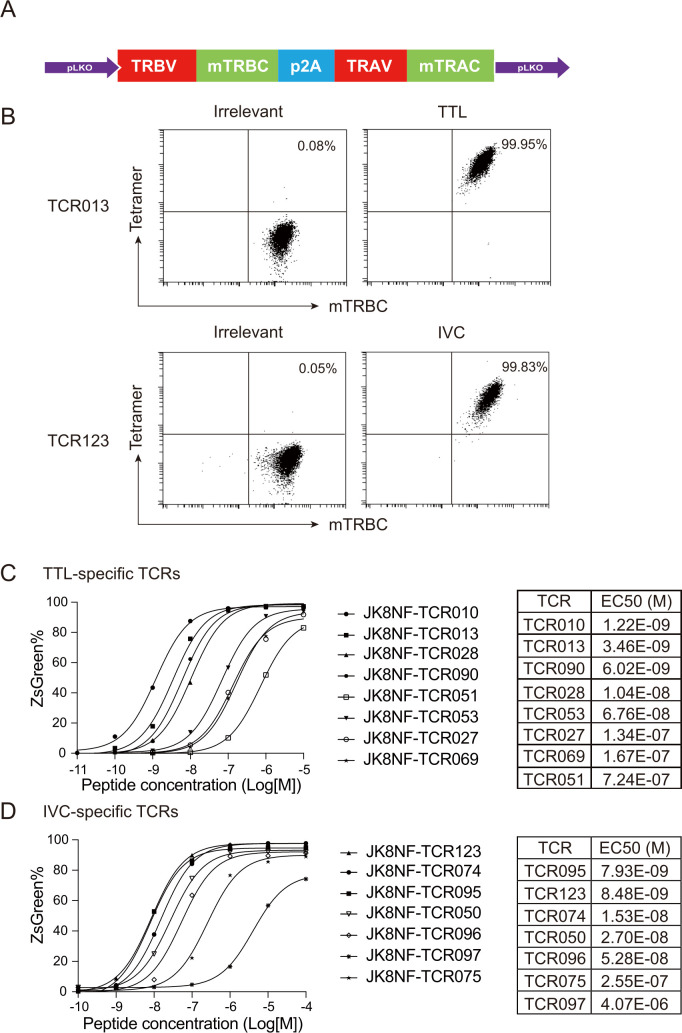
To select suitable TCRs for further TCR-T against HPV-associated malignancies, we evaluated the functional avidity of all identified TTL- and IVC-specific TCRs. Functional avidity represents T cell sensitivity and activity at different concentrations of peptide epitope and is an indicator of the properties of TCR-transduced T cells.41 Therefore, we transduced J.RT3-T3.5, a derivative of the E6-1 clone of Jurkat that lacks the beta chain of the TCR, with hCD8 and the NFAT-ZsGreen reporter gene. This JK8NF reporter cell line enables rapid flow cytometry-based readout for TCR functional avidity.
Each TCR alpha and beta chain pair in the β-α order was cloned into the lentiviral vector pLKO (figure 3A). The human TCR constant regions were replaced with murine TCR constant regions to ensure a preferred pairing of transgenic TCR chains. The murine constant regions were modified with a second disulfide bond34 and the LVL hydrophobic mutations in the TCRα transmembrane region.35 JK8NF cell lines were transduced with each TCR-expressing lentiviral vector at an MOI of 10 to maintain similar expression levels between different TCRs. Figure 3B shows that TTL-specific (TCR013) and IVC-specific (TCR123) TCR transduced JK8NF cells displayed expression of the murine TCR β-chain constant region (mTRBC) and binding to corresponding tetramers. To measure functional avidity, TCR transduced JK8NF cells were cocultured with T2-A11 cells pulsed with titrated concentrations of cognate peptide, and the frequency of ZsGreen-expressing cells was measured by flow cytometry. The nonlinear fit curve and mean EC50 of the representative TTL- and IVC-specific TCRs are shown in figure 3C, D. Both the TTL- and IVC-specific TCRs we identified showed a wide range of functional avidity. TTL-specific TCR010 showed the highest avidity, with a mean EC50 of about 1 nM. TCR095 showed the highest avidity among IVC-specific TCRs, with a mean EC50 of about 8 nM.
Figure 3.
Screening of TTL-specific and IVC-specific TCRs based on functional avidity. (A) Schematic of the TCR cassette that was cloned into the pLKO lentiviral vector. (B) Flow cytometry analysis of TTL-specific (TCR013) and IVC-specific (TCR123) TCR transduced JK8NF cells. HLA-A*11:01/EBV LMP2 (SSCSSCPLSK) tetramer was used as irrelevant tetramer control. (C, D) The functional avidity assay of TTL-specific and IVC-specific TCRs. TTL-specific TCR (C) and IVC-specific TCR (D) transduced JK8NF cells were cocultured with T2-A11 cells pulsed with the cognate peptides at the indicated concentration overnight. The frequency of ZsGreen expressing cells was measured by flow cytometry. EC50 values were determined using a non-linear regression curve and are shown in the right table. JK8NF, Jurkat-CD8-NFAT; mTRAC, murine T-cell receptor alpha-chain constant region; mTRBC, murine TCR beta-chain constant region; TCR, T-cell receptor; TRAV, T-cell receptor alpha-chain variable region; TRBV, T-cell receptor beta-chain variable region.
Mapping the critical residues of TTL involved in TCR–pMHC interactions
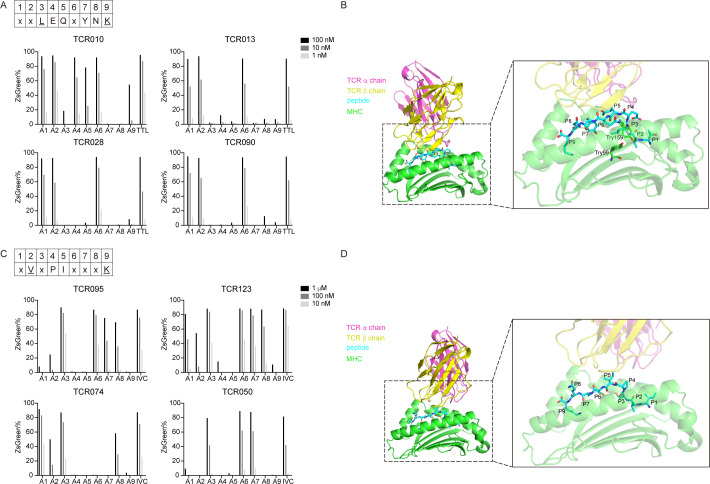
To determine residues critical for HLA binding or TCR binding, we synthesized peptides with sequential alanine substitutions of TTL or IVC and tested them for reactivity with different specific T cell clones. As shown in figure 4A, alanine substitutions of the peptide TTL at concentrations from 1 nM to 100 nM were tested for their ability to activate TCR010, TCR013, TCR028, and TCR090. Although the TCR–pMHC interaction patterns differed between these four TCRs, generic features could be summarized. In the binding of HLA-A*11:01, the peptide anchor residues are typically located at position 2 (P2) and the C-terminus.42 Substitution of the anchor residue at P9 with alanine significantly weakened activation of TTL-specific TCRs, probably because this substitution interfered with the binding of TTL to HLA-A*11:01. However, substituting the anchor residue at P2 with alanine did not affect the activation of TTL-specific TCRs. In addition, alanine substitution at P3, P7, and P8 abolished the peptide’s ability to activate TCR010, TCR013, TCR028, and TCR090. Alanine substitution at P4 and P5 only abolished the peptide’s ability to activate TCR013, TCR028, and TCR090, with nearly no effect on the activation of TCR010. Alanine substitutions at P1 and P6 were well-tolerated.
Figure 4.
Mapping the critical residues of TTL and IVC involved in TCR–pMHC interactions. (A) The alanine scanning approach for TTL-specific TCR010, TCR013, TCR028, and TCR090. JK8NF-TCR cells were cocultured with T2-A11 cells pulsed with the TTL or alanine-substituted peptides at the indicated concentration. The frequency of ZsGreen expressing cells was determined by flow cytometry. The consensus sequence of TTL is shown in the upper panel. Uppercase letters are conserved residues, underlined letters are anchor residues, and ‘X’ is the non-conserved residue. (B) The predicted 3D structure model of the TCR–pMHC complex for TTL-specific TCR013. The left panel shows the whole model of the TCR013 α chain (red), TCR013 β chain (yellow), TTL peptide (blue), and HLA-A*11:01 (green) molecules. The right panel shows the enlarged model with the transparent cartoon for TCR013 and HLA-A*11:01 molecules. Sticks illustrate amino acids of TTL peptide and Try99 and Try159 of HLA-A*11:01. (C) The alanine scanning approach for TTL-specific TCR095, TCR123, TCR074, and TCR050. The consensus sequence of IVC is shown in the upper panel. Uppercase letters are conserved residues; underlined letters are anchor residues, and ‘X’ is the non-conserved residue. (D) The predicted three-dimensional structure model of the TCR–pMHC complex for IVC-specific TCR123. The left panel shows the whole model of the TCR123 α chain (red), TCR123 β chain (yellow), IVC peptide (blue), and HLA-A*11:01 (green) molecules. The right panel shows the enlarged model with the transparent cartoon for TCR123 and HLA-A*11:01 molecules. Sticks illustrate amino acids of IVC peptides. pMHC, peptide–major histocompatibility complex; TCR, T-cell receptor.
To further investigate the orientation of TTL and IVC peptide residues, a 3D model of the TCR–pMHC complex was made using TCR model43 (http://tcrmodel.ibbr.umd.edu/). The sequence information of TCRs used in modeling is shown in online supplemental table 2. The model of TTL-specific TCR013 and HLA-A*11:01/TTL complex is shown in figure 4B. Consistent with the classic anchor residues, the residue P9-Lys was found buried deep in the peptide-binding groove of HLA-A*11:01. The residue P3-Leu was also found pointing towards the HLA-A*11:01 and forming hydrophobic interaction with the HLA-A*11:01’s Try99 and Try159, indicating that P3 is the anchor residue. The residue P7-Try also pointed to the peptide-binding groove of HLA-A*11:01. Therefore, alanine substitution at P3, P7, and P9 may significantly impede peptide loading to HLA-A*11:01. In contrast, the residues P4-Glu, P5-Gln, and P8-Asn were found pointing toward the CDR loops of TCR013, suggesting that these residues were essential for the TCR–pMHC interaction. The models of TTL-specific TCR028 and TCR090 also showed similar features, except the model of TCR010-pMHC was slightly different with the residue P7-Try pointing towards the CDR loops of TCR010 (online supplemental figure 1). According to these data, the consensus sequence of TTL, which is critical in TCR–pMHC interaction, was generated as xxLEQxYNK (figure 4A).
jitc-2022-004790supp001.pdf (678.7KB, pdf)
Mapping the critical residues of IVC involved in TCR–pMHC interactions
Alanine substitutions of the peptide IVC at concentrations from 10 nM to 1 µM were tested for their ability to activate TCR095, TCR123, TCR074, and TCR050 (figure 4C). Substitution of the anchor residue at P2 or P9 with alanine significantly impeded activation of IVC-specific TCRs, probably because these substitutions interfered with IVC binding to HLA-A*11:01. Alanine substitution at P4 or P5 also prevented the peptide from activating TCR095, TCR123, TCR074, and TCR050.
The 3D model of IVC-specific TCR123 and HLA-A*11:01/IVC complex is shown in figure 4D. The residues P2-Val and P9-Lys were found buried deep in the peptide-binding groove of HLA-A*11:01, indicating both are the anchor residues. The residues P4-Pro and P5-Ile point toward CDR loops of TCR095, which is consistent with the alanine scanning, indicating that these residues are likely involved in the TCR–pMHC interaction. The models of IVC-specific TCR095, 074, and 050 also showed similar features (online supplemental figure 2). Although the side chain of P8-Gln points outward from the peptide-binding groove, as shown in the 3D model, the responses of alanine substitution at P8 differed among the four TCRs, with activation abolished only in TCR050. The CDRβ loop of TCR050 was more biased toward the peptide’s C terminus than the CDRβ loop of the other three TCRs. This difference may explain why alanine substitution at P8 only abolished activity in TCR050. Thus, the consensus sequence of IVC was generated as xVxPIxxxK (figure 4C).
TTL-specific TCR-T cells specifically recognized and killed HLA-A*11:01+E6+ tumor cells in vitro and in vivo
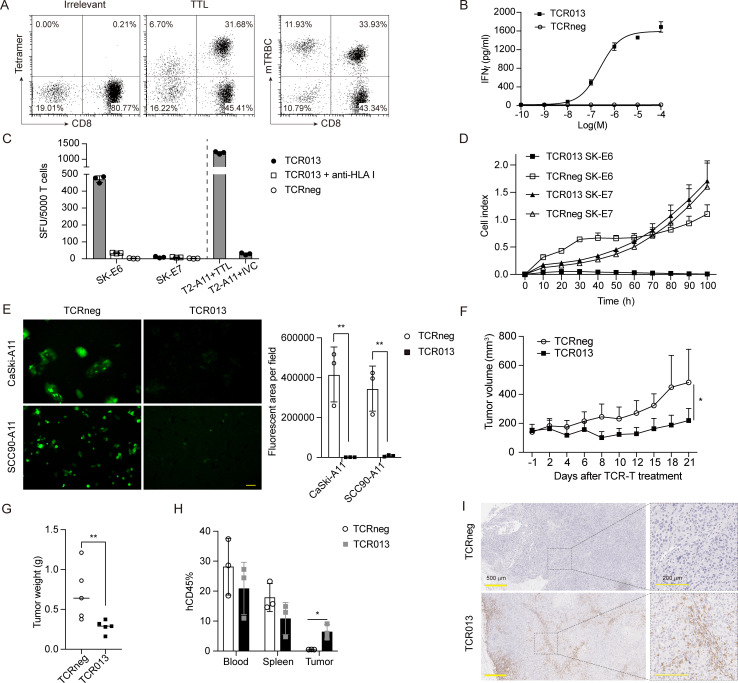
We further investigated the functionality of TTL-specific TCRs in genetically engineered primary T cells. PBMCs from healthy donors were transduced with TCR013 lentiviral vectors, then stained for TTL-tetramer and mouse β-chain constant region. As shown in figure 5A, TCR013 was successfully expressed on the cell membrane of primary T cells and stained positive for TTL-tetramer but not irrelevant tetramer. TCR013-transduced T cells were cocultured with the TTL peptide-pulsed T2-A11 overnight, and the released IFN-γ was detected by ELISA. Figure 5B shows that TCR013 but not TCRnegs recognized exogenously pulsed peptides.
Figure 5.
TTL-specific TCR-T cells specifically recognized and killed HLA-A*11:01+E6+ tumor cells in vitro and in vivo. (A) Flow cytometry analysis of TTL-specific TCR013-transduced T cells. HLA-A*11:01/EBV LMP2 (SSCSSCPLSK) tetramer was used as irrelevant tetramer control. (B) Dose–response curves for TCR013-transduced T cells. IFN-γ secretion was determined by ELISA 24 hours after coculture with T2-A11 cells pulsed with different concentrations of the TTL peptide. The relative individual values and the nonlinear regression curve are shown. (C) The TCR013-transduced T cells or TCRneg were cocultured with indicated tumor cell lines overnight, and the released IFN-γ was determined by ELISpot. Anti-HLA class I (clone W6/32) antibodywas added to block the TCR–pMHC interaction. T2-A11 cells pulsed with TTL/IVC peptide were included as the positive/negative control. The y-axis shows SFU produced by 5000 CD8+tetramer+ cells. (D) TCR013-transduced T cells or TCRneg were cocultured with SK-E6 or SK-E7 tumor cell lines at an effector-to-target ratio of 1:1. Tumor cell growth was monitored using a real-time impedance-based cytolysis assay (xCELLigence RTCA). (E) TCR013-transduced T cells or TCRneg were cocultured with CaSki-A11 or SCC90-A11 tumor cell lines at an effector-to-target ratio of 5:1 for 96 hours. The fluorescence microscopy images (left panels) and quantitative results analyzed by Image J (right panel) are shown. Scale bar, 50 µm. **P<0.01. (F–I) Antitumor activity of TTL-specific TCR T cells in vivo. SK-E6 tumor cells were subcutaneously injected into NCG mice and then treated with a single retro-orbital injection of 1×107 TCR013-transduced T cells or TCRneg as control. The tumor growth curve (F) and the tumor weight measured at the end of the experiment (G) are shown. *P<0.05, **P<0.01. (H) The peripheral blood, spleen, and tumor samples were collected 7 days after T-cell injection. The percentage of hCD45 was analyzed by flow cytometry. *P<0.05. (I) Tumor sections on day 7 after T-cell injection were stained for CD3. The right panels are the enlarged images. Scale bar, 500 µm (left panels) or 200 µm (right panels). ELISpot, enzyme-linked immunospot; IFN-γ, interferon gamma; RTCA, real-time cell analyzer; SFU, spot-forming unit; TCR, T-cell receptor; TCRneg, non-transduced T cell.
TCR013-transduced T cells were next cocultured with different tumor cell lines to assess whether TTL-specific TCRs could recognize the endogenously processed antigen. The HLA-A*11:01-positive SK-MEL-28 cell line was transduced with the E6 or E7 genes (SK-E6/E7). As measured by IFN-γ ELISpot, TCR013-transduced T cells recognized HLA-A*11:01+E6+ SK-E6 cells but not SK-E7 cells. In addition, recognition of the HLA-A*11:01+E6+ tumor cells by TCR013 was inhibited by antibody blockade of HLA class I molecules, indicating that target recognition is HLA class I restricted (figure 5C). SK-E6 tumor cell lines cocultured with the TCR013-transduced T cells were killed gradually, while those cocultured with the TCRneg cells were unaffected. The growth of SK-E7 cells was also not affected by TCR013-transduced T cells, indicating the specificity of TCR013 (figure 5D). HPV-positive cervical cancer cell line CaSki and HPV-positive oral and pharyngeal cancer cell line SCC90 were transduced with the HLA-A*11:01-P2A-GFP gene (CaSki-A11 and SCC90-A11). As shown in figure 5E, TCR013-transduced T cells but not TCRneg cells killed CaSki-A11 and SCC90-A11 tumor cells. These data indicate that the TCR013 can specifically recognize and kill HPV E6-expressing tumor cells in an HLA-A*11:01-restricted manner.
Having observed a specific T-cell response against E6+ tumor cells in vitro, we tested TTL-specific TCRs in vivo to further assess their therapeutic potential. For this purpose, we generated a xenograft model by subcutaneously injecting SK-E6 tumor cells into immunodeficient mice. All TCRneg cell-treated mice showed progressive tumor growth, while groups treated with TCR013 showed significantly delayed tumor growth, with tumor shrinking starting at 4 days after T-cell treatment (figure 5F and online supplemental figure 3A). All mice were sacrificed at the end of the experiment, and tumors were excised. Consistent with the tumor volume, the tumor mass of mice in the TTL-specific TCR-T group was significantly less than in the control group (figure 5G).
TTL-specific TCR-T cells successfully infiltrated into the tumor microenvironment
One of the main challenges in cancer immunotherapy is the T-cell infiltration into solid tumor tissues. We collected peripheral blood, spleen, and tumor samples 7 days after T-cell injection to examine TTL-specific TCR-T cells’ infiltration. The single-cell suspension was prepared and analyzed by flow cytometry. The percentage of hCD45+ cells in peripheral blood and spleen was similar between TCR013-transduced and control T-cell groups (figure 5H). However, hCD45+ cells were detected only in TCR013-treated tumor tissues but not in the control T-cell group. Meanwhile, tumor tissue sections were analyzed by immunohistochemistry. As shown in figure 5I, CD3+ T cells were detected in the tumor microenvironment of TCR013-treated mice. However, nearly no positive signal was seen in the control group. Importantly, TCR013-transduced T cells infiltrated both the marginal and central regions of the tumor tissue, indicating that TTL-specific TCR-T cells can successfully infiltrate the tumor microenvironment. Together, these data demonstrate that TTL-specific TCRs could be used to treat E6-expressing tumors.
IVC-specific TCR-T cells specifically recognized and killed HLA-A*11:01+E7+ tumor cells in vitro and in vivo
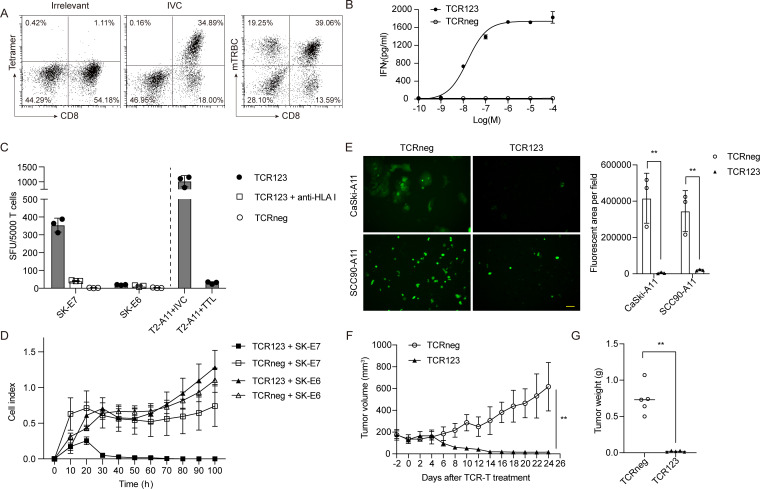
Similarly, the functionality of IVC-specific TCRs was investigated in genetically engineered primary T cells. As shown in figure 6A, TCR123 was successfully expressed on the cell membrane of primary T cells. Figure 6B shows that TCR123-transduced T cells but not TCRneg cells recognized exogenously pulsed peptides.
Figure 6.
IVC-specific TCR-T cells specifically recognized and killed HLA-A*11:01+E7+ tumor cells in vitro and in vivo. (A) Flow cytometry analysis of IVC-specific TCR123-transduced T cells. HLA-A*11:01/EBV LMP2 (SSCSSCPLSK) tetramer was used as irrelevant tetramer control. (B) Dose–response curves for TCR123-transduced T cells. IFN-γ secretion was determined by ELISA 24 hours after coculture with T2-A11 cells pulsed with different concentrations of the IVC peptide. The relative individual values and the non-linear regression curve are shown. (C) The TCR123-transduced T cells or TCRneg were cocultured with indicated tumor cell lines overnight, and the released IFN-γ was determined by ELISpot. Anti-HLA class I (clone W6/32) antibody was added to block the TCR–pMHC interaction. T2-A11 cells pulsed with IVC/TTL peptide were included as the positive/negative control. The y-axis shows SFU produced by 5000 CD8+Tetramer+ cells. (D) TCR123-transduced T cells or TCRneg were cocultured with SK-E6 or SK-E7 tumor cell lines at an effector-to-target ratio of 1:1. Tumor cell growth was monitored using a real-time impedance-based cytolysis assay (xCELLigence RTCA). (E) TCR123-transduced T cells or TCRneg were cocultured with CaSki-A11 or SCC90-A11 tumor cell lines at an effector-to-target ratio of 5:1 for 96 hours. The representative fluorescent microscopy images (left panels) and quantitative results analyzed by Image J (right panel) are shown. Scale bar, 50 µm. **P<0.01. (F, G) Antitumor activity of IVC-specific TCR T cells in vivo. SK-E7 tumor cells were subcutaneously injected into NCG mice and then treated with a single retro-orbital injection of 1×107 TCR123-transduced T cells or TCRneg as a control. The tumor growth curve (F) and the tumor weight measured at the end of the experiment (G) are shown. **P<0.01. ELISpot, enzyme-linked immunospot; IFN-γ, interferon gamma; RTCA, real-time cell analyzer; SFU, spot-forming unit; TCR, T-cell receptor; TCRneg, non-transduced T cell.
TCR123-transduced T cells were then cocultured with different tumor cell lines to assess whether IVC-specific TCRs could recognize the endogenously processed antigen. As measured by IFN-γ ELISpot, TCR123-transduced T cells successfully recognized HLA-A*11:01+E7+ SK-E7 but not SK-E6 cells. Besides, recognition of the HLA-A*11:01+E7+ tumor cells by TCR123 was also inhibited by antibody blockade of HLA class I molecules, indicating that target recognition was restricted to HLA class I (figure 6C). Furthermore, the growth of SK-E7 tumor cell lines was significantly inhibited by the TCR123-transduced T cells, while SK-E6 tumor cells were not affected (figure 6D). Fluorescence microscopy images also showed that TCR123-transduced T cells but not TCRneg cells killed CaSki-A11 and SCC90-A11 tumor cells, which express the natural HPV16 E7 gene (figure 6E). These data indicate that TCR123-transduced T cells can specifically recognize and kill HPV E7-expressing tumor cells in an HLA-A*11:01-restricted manner.
The therapeutic potential of IVC-specific TCRs was also assessed in vivo. SK-E7 tumor cells were subcutaneously injected into immunodeficient NCG mice. All TCRneg T cell-treated mice showed progressive tumor growth. In contrast, IVC-specific TCR-T cell-treated group showed complete tumor remission, with tumor-shrinking beginning between days 4 and 6 after T-cell application (figure 6F and online supplemental figure 3B). On day 24, all mice were sacrificed, and the tumors were excised. Consistent with tumor volume, the tumor mass of mice in the IVC-specific TCR-T group was significantly reduced compared with the TCRneg group (figure 6G).
TTL-specific and IVC-specific TCR-T cells specifically recognized and killed HPV16+ PDOs
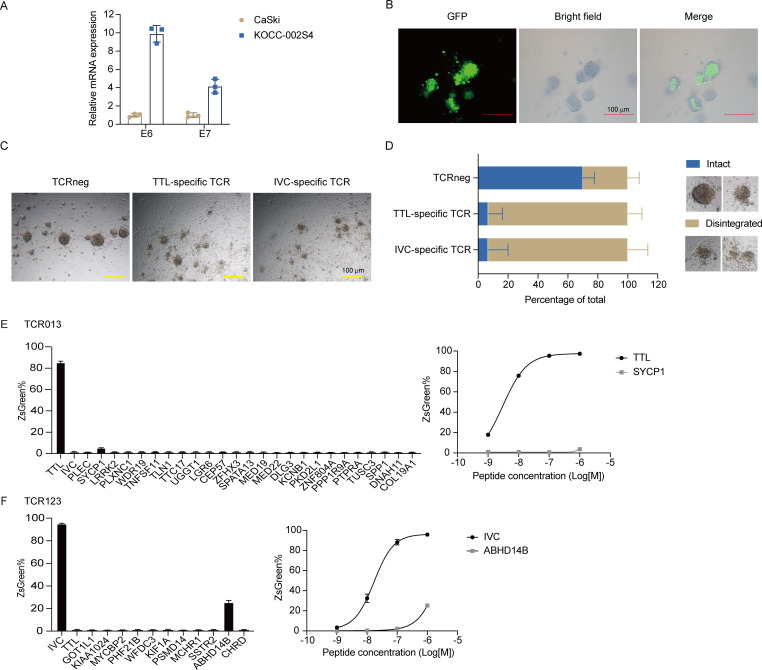
To evaluate the TTL and IVC-specific TCR-T cell activities towards primary tumor cells, we screened for HPV16+ cervical PDOs. PDOs have been reported as a promising model to mimic the biological characteristics of the primary tumors.44 The PDO KOCC-002S4 was found to be HPV16 E6 and E7 positive (figure 7A). The original HLA-A type of KOCC-002S4 is A*02:01/A*02:01. Thus, KOCC-002S4 organoids were transduced with the HLA-A*11:01-P2A-GFP gene and used as targets to analyze the TTL and IVC-specific TCR-T cell activities (figure 7B). We noticed that both TTL and IVC-specific TCR-T treatments changed the shape of cystic PDO structures (figure 7C). There were significantly reduced intact PDO structures in TTL and IVC-specific TCR-T groups than in the TCRneg group (figure 7D and online supplemental figure 4). Meanwhile, TTL and IVC-specific TCR-Ts also secreted more IFN-γ than TCRnegs after coculture with KOCC-002S4 PDOs (online supplemental figure 5). These data demonstrate that TTL and IVC-specific TCRs recognized patient-derived tumor cells expressing natural HPV16 E6 and E7.
Figure 7.
(A–D) TTL and IVC-specific TCR-T cells specifically recognized and killed HPV16+ patient-derived organoids. (A) Expression of HPV16 E6 and E7 in CASKICaSki cell lines and PDO KOCC-002S4 analyzed by real-time quantitative PCR. (B) KOCC-002S4 organoids were transduced with the HLA-A*11:01-P2A-GFP gene. The representative fluorescent and corresponding bright field images are shown. Scale bar, 100 µm. (C) The TTL-specific (TCR013) T cell, IVC-specific (TCR123) T cell, or TCRneg were cocultured with A11-transduced KOCC-002S4 organoids at an effector-to-target ratio of 1:1 for 72 hours. The representative microscopy images are shown. Scale bar, 100 µm. (D) The percentage of intact or disintegrated organoids in (C). The representative images of intact or disintegrated structures are shown in the right panels. (E,F) Cross-reactivity evaluation of TCR013 (E) and TCR123 (F) to human homologous peptides. JK8NF-TCR cells were cocultured with T2-A11 cells pulsed with the TTL/IVC or human homologous peptides at 1 µM (left panels) or the indicated concentration (right panels). The frequency of ZsGreen expressing cells was determined by flow cytometry. JK8NF, Jurkat-CD8-NFAT; TCR, T-cell receptor; TCRneg, non-transduced T cell.
Cross-reactivity evaluation of TTL and IVC-specific TCRs to human homologous peptides
Finally, the cross-reactivity of TTL and IVC-specific TCRs to human homologous peptides was evaluated. We used Expitope V.2.030 to predict homologous peptides in the human proteome. All peptides with three or fewer mismatches and with HLA-A*11:01 binding affinity of <5000 nM were synthesized as candidates. Moreover, the motifs ‘xxLxxxYNK’ and ‘xVxPIxxxK’ were used to search for peptides with four mismatches and with HLA-A*11:01 binding affinity of <5000 nM. The homologous peptide information is shown in online supplemental tables 3,4.
An initial screen for cross-reactivity against these peptides was performed using a high-peptide concentration (1 µM) loaded onto T2-A11 cells. TTL-specific TCR013 showed minimal cross-reactivity against selected candidate peptides except for the SYCP1 peptide (figure 7E). We then performed the assay with titrated concentrations of the SYCP1 peptide. TCR013 did not recognize SYCP1 peptides at a concentration of 100 nM or less. IVC-specific TCR123 showed minimal cross-reactivity against selected candidate peptides except for the ABHD14B peptide (figure 7F). Further experiments showed that TCR123 did not cross-react with the ABHD14B peptide at a concentration of 100 nM or less. These results suggested a low probability of these TCRs to cross-react against human proteins.
Discussion
In this study, we identified two novel HLA-A*11:01-restricted T-cell epitopes of HPV16 E6 and E7, E693-101 and E789-97, that were able to (1) bind to HLA-A*11:01, (2) induce expansion of specific T cells and (3) be presented on HPV16+ tumor cells. TTL-specific and IVC-specific TCRs were isolated from 11 healthy donors through in vitro stimulation of PBMC. Like many other T-cell epitopes, biased TCR chain usage was observed among TTL-specific and IVC-specific TCRs. In addition, TTL-specific and IVC-specific TCRs with high functional avidity were identified. These TCR-engineered T cells specifically recognized and killed corresponding HLA-A*11:01 and E6/E7 double-positive tumor cells in vitro and in vivo. The HLA-A*11:01-restricted HPV16 E6/E7 epitopes and TCRs identified in this study may provide a new strategy for HPV-related cancer immunotherapy.
Identifying T-cell epitopes is an essential but challenging task. The perfect T-cell epitopes must (1) bind to the specific HLA, (2) elicit a functional T-cell response, and (3) be present on tumor cells. Several studies have investigated HLA-A*11:01-restricted HPV E6/E7 epitopes.45–48 However, none of these studies clarified whether these epitopes were present on HPV-associated tumor cells. Here, among 10 peptides with predicted HLA-A*11:01 binding affinity of <500 nM, TTL from E6 and IVC from E7 were endogenously processed and presented on tumor cells (figure 1). TTL-specific and IVC-specific TCR engineered T cells specifically inhibited the growth of corresponding HLA-A*11:01 and E6/E7 double-positive tumor cell lines in vitro and in vivo (figures 5 and 6). Furthermore, they also recognized and killed A11-transduced HPV16+ PDOs (figure 7). It should be noted that natural A1101+HPV16+ PDOs as targets would be ideal. We will continue to screen such double-positive PDOs in further investigation. To our knowledge, this study provides the first evidence that E6-derived TTL and E7-derived IVC epitopes could be potential therapeutic targets of TCR-T treatment for patients with HLA-A*11:01-positive HPV-related cancer.
We retrospectively analyzed the found epitopes with the most current prediction tools. Researchers are using machine learning to improve the prediction models for T-cell epitopes. NetMHCpan-4.1 improved predictions of HLA ligand by concurrent motif deconvolution and integration of mass spectrometry-eluted ligand data.49 PRIME (a predictor of immunogenic epitopes) improved predictions of peptide immunogenicity by combining molecular properties of both antigen presentation and TCR recognition.50 The potential HLA-A*11:01-binding peptides derived from HPV16 E6 and E7 proteins were predicted again with netMHCpan-4.1 and PRIME (online supplemental table 5). Interestingly, the binding affinity of peptides KIS, DII, and EII predicted by netMHCpan-4.1 was 821.51, 1832.09, and 1022.53 nM. As shown in online supplemental table 1, the predicted binding affinity of these peptides was <500 nM using the netMHC V.4.0 server. According to our experiments, we cannot get the properly folded pMHC complexes in vitro. These results suggest that the netMHCpan-4.1 may be better than the netMHC V.4.0 server. Furthermore, the predicted immunogenicity using PRIME for both TTL and IVC epitopes was among the top 0.5% strong HLA-A*11:01 binders (0.039% and 0.235%, respectively), indicating that E6-derived TTL and E7-derived IVC epitopes could be prospective therapeutic targets.
IVC-specific TCRs appeared to have greater efficacy than TTL-specific TCRs in vivo. However, the functional avidity of TTL-specific TCRs (eg, TCR013EC50=3.46 nM) was higher than that of IVC-specific TCRs (eg, TCR123EC50=8.48 nM). IVC-specific TCR095 (EC50=7.73 nM) engineered T cells could also control the growth of SK-E7 tumor cells at the dose of 1×107 tetramer+ cells per mouse (online supplemental figure 6) and even at a low dose of 5×105 tetramer+ cells per mouse (online supplemental figure 7). We also found that TTL-specific TCR028 with lower avidity (EC50=10.4 nM) could significantly inhibit tumor growth of SK-E6 in vivo (online supplemental figure 8). Other groups have found a similar phenomenon in different models.51–54 In attempts to improve the efficacy of tumor vaccines, investigators have found that optimal in vivo T-cell responses occur for ligands with intermediate TCR/pMHC half-lives, both in CD8+ T cells51 and in CD4+ T cells.52 Wu et al studied two α-fetoprotein (AFP)-specific CD8+ T cells (AFP499 and AFP212) with different antitumor effects and found that Tet499 cells with weaker TCR signaling expanded more than Tet212 cells when re-encountering the antigen and generated more potent antitumor effects in vivo.54 Recently, Shakiba et al developed a cancer mouse model in which tumor-specific CD8 T cells encounter tumor antigens with varying TCR signal strength.55 They found that neither high-signal strength nor low-signal strength interactions led to tumor control in vivo because high signal strength drove dysfunction, while low signal strength resulted in functional inertness. We hypothesize that IVC-specific TCRs with intermediate affinity prevent T cells from exhaustion and apoptosis after encountering the tumor antigen in vivo. Other explanations are possible; for example, IVC peptide may be presented better than TTL peptide by HLA-A*11:01 on the cell membrane. The amount of each HLA-peptide complex on HPV+ tumor cell surface should be compared. Besides, one limitation of this study is that only subcutaneous tumor models were used to investigate the efficacy of TCR-Ts. It would also be interesting to analyze the phenotype of the transferred engineered T cells in the tumor microenvironment to determine if the cells with higher avidity are more prone to exhaustion. Thus, which TCRs are better for IVC and TTL requires more exploration, especially in clinical trials.
In this study, all TCR sequences were identified from healthy donors. Most TCRs under clinical trials are isolated from patients’ tumor-infiltrating lymphocytes (TILs). For example, the clinical trial NCT02280811 uses the TCR targeting E629-38 isolated from TILs of one patient with metastatic anal cancer.21 The clinical trial NCT02858310 uses the TCR targeting E711-19 isolated from cervix-infiltrating lymphocytes from one patient with HPV16+ cervical intraepithelial neoplasia II/III.22 One possible reason is that the percentage of tumor-reactive T cells is higher in TILs, making it easier to discover tumor-specific TCRs. Given the high prevalence of HPV-positive cancers and the HLA-A*11:01 allele in the population, it is worthwhile to screen TTL and IVC-specific high-avidity TCRs in patients with HLA-A*11:01+ HPV-related cancer in the future.
Acknowledgments
We thank Yingxia Wu and Ping Li for their help with flow cytometry cell sorting. We thank Yucong Sun for her assistance with T-cell receptor cloning.
Footnotes
Correction notice: This article has been corrected since it was first published online. The abstract of this article has now been included in the published version.
Contributors: LC, ZW, and XW designed the study. LC, CX, LH, HK, CW, XZ, SL, BW, JL, and XW performed the experiments and collected data. HS performed three-dimensional modeling and analysis of the pMHC-TCR complex. LC, ZW, CX, and XW wrote the manuscript. ZW and LC are responsible for the overall content as guarantor.
Funding: National Natural Science Foundation of China (81903156, 81972690).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
All animal handling and tumor experiments were approved by Guangzhou Medical University Animal Care Committee (Ref: S2020-062).
References
- 1. Chesson HW, Dunne EF, Hariri S, et al. The estimated lifetime probability of acquiring human papillomavirus in the United States. Sex Transm Dis 2014;41:660–4. 10.1097/OLQ.0000000000000193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brianti P, De Flammineis E, Mercuri SR. Review of HPV-related diseases and cancers. New Microbiol 2017;40:80–5. [PubMed] [Google Scholar]
- 3. HPV reference clones. Available: https://www.hpvcenter.se/human_reference_clones/ [Accessed May 2022].
- 4. Schiffman M, Doorbar J, Wentzensen N, et al. Carcinogenic human papillomavirus infection. Nat Rev Dis Primers 2016;2:16086. 10.1038/nrdp.2016.86 [DOI] [PubMed] [Google Scholar]
- 5. Kreimer AR, Clifford GM, Boyle P, et al. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev 2005;14:467–75. 10.1158/1055-9965.EPI-04-0551 [DOI] [PubMed] [Google Scholar]
- 6. de Martel C, Georges D, Bray F, et al. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health 2020;8:e180–90. 10.1016/S2214-109X(19)30488-7 [DOI] [PubMed] [Google Scholar]
- 7. Li D, Li X, Zhou W-L, et al. Genetically engineered T cells for cancer immunotherapy. Signal Transduct Target Ther 2019;4:35. 10.1038/s41392-019-0070-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morgan RA, Dudley ME, Wunderlich JR, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science 2006;314:126–9. 10.1126/science.1129003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Robbins PF, Morgan RA, Feldman SA, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol 2011;29:917–24. 10.1200/JCO.2010.32.2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Robbins PF, Kassim SH, Tran TLN, et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin Cancer Res 2015;21:1019–27. 10.1158/1078-0432.CCR-14-2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2002;2:342–50. 10.1038/nrc798 [DOI] [PubMed] [Google Scholar]
- 12. Wang C, Xiong C, Hsu Y-C, et al. Human leukocyte antigen (HLA) and cancer immunotherapy: HLA-dependent and -independent adoptive immunotherapies. Ann Blood 2020;5:14–70. 10.21037/aob-20-27 [DOI] [Google Scholar]
- 13. Tashiro H, Brenner MK. Immunotherapy against cancer-related viruses. Cell Res 2017;27:59–73. 10.1038/cr.2016.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomas M, Pim D, Banks L. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene 1999;18:7690–700. 10.1038/sj.onc.1202953 [DOI] [PubMed] [Google Scholar]
- 15. Giarrè M, Caldeira S, Malanchi I, et al. Induction of pRb degradation by the human papillomavirus type 16 E7 protein is essential to efficiently overcome p16INK4a-imposed G1 cell cycle arrest. J Virol 2001;75:4705–12. 10.1128/JVI.75.10.4705-4712.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scholten KBJ, Turksma AW, Ruizendaal JJ, et al. Generating HPV specific T helper cells for the treatment of HPV induced malignancies using TCR gene transfer. J Transl Med 2011;9:147. 10.1186/1479-5876-9-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lorenz FKM, Wilde S, Voigt K, et al. Codon optimization of the human papillomavirus E7 oncogene induces a CD8+ T cell response to a cryptic epitope not harbored by wild-type E7. PLoS One 2015;10:e0121633. 10.1371/journal.pone.0121633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lorenz FKM, Ellinger C, Kieback E, et al. Unbiased identification of T-cell receptors targeting immunodominant peptide-MHC complexes for T-cell receptor immunotherapy. Hum Gene Ther 2017;28:1158–68. 10.1089/hum.2017.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Draper LM, Kwong MLM, Gros A, et al. Targeting of HPV-16+ epithelial cancer cells by TCR gene engineered T cells directed against E6. Clin Cancer Res 2015;21:4431–9. 10.1158/1078-0432.CCR-14-3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jin BY, Campbell TE, Draper LM, et al. Engineered T cells targeting E7 mediate regression of human papillomavirus cancers in a murine model. JCI Insight 2018;3. 10.1172/jci.insight.99488. [Epub ahead of print: 19 04 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Doran SL, Stevanović S, Adhikary S, et al. T-Cell receptor gene therapy for human papillomavirus-associated epithelial cancers: a first-in-human, phase I/II study. J Clin Oncol 2019;37:2759–68. 10.1200/JCO.18.02424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nagarsheth NB, Norberg SM, Sinkoe AL, et al. TCR-engineered T cells targeting E7 for patients with metastatic HPV-associated epithelial cancers. Nat Med 2021;27:419–25. 10.1038/s41591-020-01225-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kessler JH, Beekman NJ, Bres-Vloemans SA, et al. Efficient identification of novel HLA-A(*)0201-presented cytotoxic T lymphocyte epitopes in the widely expressed tumor antigen PRAME by proteasome-mediated digestion analysis. J Exp Med 2001;193:73–88. 10.1084/jem.193.1.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peters B, Nielsen M, Sette A. T cell epitope predictions. Annu Rev Immunol 2020;38:123–45. 10.1146/annurev-immunol-082119-124838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ressing ME, Sette A, Brandt RM, et al. Human CTL epitopes encoded by human papillomavirus type 16 E6 and E7 identified through in vivo and in vitro immunogenicity studies of HLA-A*0201-binding peptides. J Immunol 1995;154:5934–43. [PubMed] [Google Scholar]
- 26. Evans M, Borysiewicz LK, Evans AS, et al. Antigen processing defects in cervical carcinomas limit the presentation of a CTL epitope from human papillomavirus 16 E6. J Immunol 2001;167:5420–8. 10.4049/jimmunol.167.9.5420 [DOI] [PubMed] [Google Scholar]
- 27. Riemer AB, Keskin DB, Zhang G, et al. A conserved E7-derived cytotoxic T lymphocyte epitope expressed on human papillomavirus 16-transformed HLA-A2+ epithelial cancers. J Biol Chem 2010;285:29608–22. 10.1074/jbc.M110.126722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou F, Cao H, Zuo X, et al. Deep sequencing of the MHC region in the Chinese population contributes to studies of complex disease. Nat Genet 2016;48:740–6. 10.1038/ng.3576 [DOI] [PubMed] [Google Scholar]
- 29. Mann SE, Zhou Z, Landry LG, et al. Multiplex T cell stimulation assay utilizing a T cell activation Reporter-Based detection system. Front Immunol 2020;11:633. 10.3389/fimmu.2020.00633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jaravine V, Mösch A, Raffegerst S, et al. Expitope 2.0: a tool to assess immunotherapeutic antigens for their potential cross-reactivity against naturally expressed proteins in human tissues. BMC Cancer 2017;17:892. 10.1186/s12885-017-3854-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen W, Yewdell JW, Levine RL, et al. Modification of cysteine residues in vitro and in vivo affects the immunogenicity and antigenicity of major histocompatibility complex class I-restricted viral determinants. J Exp Med 1999;189:1757–64. 10.1084/jem.189.11.1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schnurr M, Chen Q, Shin A, et al. Tumor antigen processing and presentation depend critically on dendritic cell type and the mode of antigen delivery. Blood 2005;105:2465–72. 10.1182/blood-2004-08-3105 [DOI] [PubMed] [Google Scholar]
- 33. Pasetto A, Gros A, Robbins PF, et al. Tumor- and Neoantigen-Reactive T-cell receptors can be identified based on their frequency in fresh tumor. Cancer Immunol Res 2016;4:734–43. 10.1158/2326-6066.CIR-16-0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cohen CJ, Li YF, El-Gamil M, et al. Enhanced antitumor activity of T cells engineered to express T-cell receptors with a second disulfide bond. Cancer Res 2007;67:3898–903. 10.1158/0008-5472.CAN-06-3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haga-Friedman A, Horovitz-Fried M, Cohen CJ. Incorporation of transmembrane hydrophobic mutations in the TCR enhance its surface expression and T cell functional avidity. J Immunol 2012;188:5538–46. 10.4049/jimmunol.1103020 [DOI] [PubMed] [Google Scholar]
- 36. Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 2016;32:511–7. 10.1093/bioinformatics/btv639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burk RD, Harari A, Chen Z. Human papillomavirus genome variants. Virology 2013;445:232–43. 10.1016/j.virol.2013.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hang D, Yin Y, Han J, et al. Analysis of human papillomavirus 16 variants and risk for cervical cancer in Chinese population. Virology 2016;488:156–61. 10.1016/j.virol.2015.11.016 [DOI] [PubMed] [Google Scholar]
- 39. Kessler JH, Melief CJM. Identification of T-cell epitopes for cancer immunotherapy. Leukemia 2007;21:1859–74. 10.1038/sj.leu.2404787 [DOI] [PubMed] [Google Scholar]
- 40. Gras S, Kjer-Nielsen L, Burrows SR, et al. T-cell receptor bias and immunity. Curr Opin Immunol 2008;20:119–25. 10.1016/j.coi.2007.12.001 [DOI] [PubMed] [Google Scholar]
- 41. Morimoto S, Fujiki F, Kondo K, et al. Establishment of a novel platform cell line for efficient and precise evaluation of T cell receptor functional avidity. Oncotarget 2018;9:34132–41. 10.18632/oncotarget.26139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chujoh Y, Sobao Y, Miwa K, et al. The role of anchor residues in the binding of peptides to HLA-A*1101 molecules. Tissue Antigens 1998;52:501–9. 10.1111/j.1399-0039.1998.tb03080.x [DOI] [PubMed] [Google Scholar]
- 43. Gowthaman R, Pierce BG. TCRmodel: high resolution modeling of T cell receptors from sequence. Nucleic Acids Res 2018;46:W396–401. 10.1093/nar/gky432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wensink GE, Elias SG, Mullenders J, et al. Patient-derived organoids as a predictive biomarker for treatment response in cancer patients. NPJ Precis Oncol 2021;5:30. 10.1038/s41698-021-00168-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kast WM, Brandt RM, Sidney J, et al. Role of HLA-A motifs in identification of potential CTL epitopes in human papillomavirus type 16 E6 and E7 proteins. J Immunol 1994;152:3904–12. [PubMed] [Google Scholar]
- 46. Zhang QJ, Lindquist Y, Levitsky V, et al. Solvent exposed side chains of peptides bound to HLA A*1101 have similar effects on the reactivity of alloantibodies and specific TCR. Int Immunol 1996;8:927–38. 10.1093/intimm/8.6.927 [DOI] [PubMed] [Google Scholar]
- 47. Alexander J, Oseroff C, Sidney J, et al. Derivation of HLA-A11/Kb transgenic mice: functional CTL repertoire and recognition of human A11-restricted CTL epitopes. J Immunol 1997;159:4753–61. [PubMed] [Google Scholar]
- 48. Krishna S, Ulrich P, Wilson E, et al. Human Papilloma Virus Specific Immunogenicity and Dysfunction of CD8+ T Cells in Head and Neck Cancer. Cancer Res 2018;78:6159–70. 10.1158/0008-5472.CAN-18-0163 [DOI] [PubMed] [Google Scholar]
- 49. Reynisson B, Alvarez B, Paul S, et al. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res 2020;48:W449–54. 10.1093/nar/gkaa379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schmidt J, Smith AR, Magnin M, et al. Prediction of neo-epitope immunogenicity reveals TCR recognition determinants and provides insight into immunoediting. Cell Rep Med 2021;2:100194. 10.1016/j.xcrm.2021.100194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McMahan RH, McWilliams JA, Jordan KR, et al. Relating TCR-peptide-MHC affinity to immunogenicity for the design of tumor vaccines. J Clin Invest 2006;116:2543–51. 10.1172/JCI26936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Corse E, Gottschalk RA, Krogsgaard M, et al. Attenuated T cell responses to a high-potency ligand in vivo. PLoS Biol 2010;8. 10.1371/journal.pbio.1000481. [Epub ahead of print: 14 Sep 2010]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhong S, Malecek K, Johnson LA, et al. T-cell receptor affinity and avidity defines antitumor response and autoimmunity in T-cell immunotherapy. Proc Natl Acad Sci U S A 2013;110:6973–8. 10.1073/pnas.1221609110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu S, Zhu W, Peng Y, et al. The Antitumor Effects of Vaccine-Activated CD8+ T Cells Associate with Weak TCR Signaling and Induction of Stem-Like Memory T Cells. Cancer Immunol Res 2017;5:908–19. 10.1158/2326-6066.CIR-17-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shakiba M, Zumbo P, Espinosa-Carrasco G, et al. TCR signal strength defines distinct mechanisms of T cell dysfunction and cancer evasion. J Exp Med 2022;219. 10.1084/jem.20201966. [Epub ahead of print: 07 02 2022]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-004790supp002.xlsx (20.4KB, xlsx)
jitc-2022-004790supp001.pdf (678.7KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information.