Abstract
The microbiome is composed of many organisms and is impacted by an intricate exchange between genetics and environmental factors. The perinatal microbiome influences both the developing fetus and the pregnant person. The purpose of this article is to describe the tests that are currently available for laboratory analysis of the perinatal microbiome in relationship to probiotic interventions. This article focuses on the bacterial component of the microbiome. Although adverse outcomes associated with the perinatal microbiome have been studied, a comprehensive understanding of the physiologic perinatal microbiome is still emerging. Early efforts to influence the perinatal microbiome through probiotics are currently under investigation. Unique terminology is defined, and the microbial composition of perinatal microbiota is summarized. The outcomes of studies of antenatal probiotics are summarized. Microbiome testing and analysis are defined and compared. Implications for perinatal care and probiotics research are presented.
Keywords: laboratory analysis, microbiome, probiotics
The human microbiome is composed of microorganisms populating the human body, including commensals, symbionts, and pathogens.1 The microbiome is impacted by an intricate exchange between genetics and environmental factors.2 The perinatal microbiome influences nutrient absorption, the intrauterine fetal environment, immune system priming, and long-term impacts on health and illness of offspring. The diet and general health and well-being of the pregnant woman also influence the perinatal microbiome.3–6
The focus of this article is the bacterial component of the microbiome and not the viruses, bacteriophages, fungi, and archaea populating pregnant women. The term “microbiota” refers to the collection of individual microbes present in a given sample or at a specified location, as well as their genetic information or collective coding capacity. Sites studied to understand the perinatal microbiota include the maternal oral, gastrointestinal (GI) and genitourinary tracts, as well as maternal skin and breast. In the perinatal period, the microbiome has been studied in the context of pathology, such as allergies, atopy, and dysbiosis.7–11 Adverse obstetrical outcomes, such as preterm labor and birth, preterm premature rupture of membranes, intrauterine growth restriction, gestational diabetes, stillbirth, and more, are believed to be linked to the perinatal microbiome12,13; yet, a comprehensive understanding of the physiologic perinatal microbiome is still emerging. Knowledge of the genomes of microorganisms, their hosts, and relationship to each other in specific environments continues to rapidly develop.11,12,14
The purpose of this article is to describe the tests that are currently available for laboratory analysis of the perinatal microbiome in relationship to probiotic interventions. First, a broad overview of terms is presented. Second, probiotic interventions are described. Third, the microbial composition of perinatal microbiota is summarized by collection site. Studies of antenatal probiotics are presented as they relate to each of the sites. Fourth, both the microbiome testing and analysis are outlined. Finally, implications for perinatal care and probiotics research are described.
TERMINOLOGY
The taxonomical terminology associated with the microbiome and microbial analysis is essential to understanding relevant laboratory testing. Although nurses, midwives, and other perinatal clinicians are exposed to this terminology in undergraduate education, for many, it has not become a part of everyday practice. Table 1 provides terminology and definitions that serve as the foundation for an understanding of both the perinatal microbiome and probiotic interventions.
Table 1.
Terminology used in microbiome-based research
| Term | Definition |
|---|---|
| 16SrRNA | A portion of the bacterial ribosome containing variable and conserved regions. Widely used as a phylogenetic marker of a unique population, including all of its descendents.15 |
| Alpha diversity | Measures to address variation within a sample. Some measures take only richness into account, whereas others assess diversity. Common measures include Shannon,16 Simpson,17 ACE, and Chao1.18 |
| Amplicon sequence variants | Individual DNA sequences recovered from marker gene analysis following removal of spurious chimeric sequences generated during PCR amplification.19 May also be called exact sequence variants, sub-OTUs, and/or zero radius OTUs (zOTUs). |
| Archaea | Single-celled organisms structurally similar to bacteria, though evolutionarily distinct. The role of archaea in human health requires further investigations, though they likely play an important role. |
| Beta diversity | Measures how different composition is between 2 different samples, groups, or environments. Common measures include the Bray-Curtis dissimilarity,20 Jaccard distance,21 and UniFrac/Weighted UniFrac measures.22 |
| Clade | Organisms that descend and evolve from a common ancestor. |
| Commensal | Commensal organisms refer to the indigenous microorganisms present at a given anatomical site. Microorganisms that have coevolved with the host and have a symbiotic relationship with the host. Also referred to as resident microbes. |
| Coverage depth | The average number of times a genome is sequenced. Calculated by the number of bases of short reads matching a genome divided by the length of the genome. |
| Diversity | A measurement of species richness and evenness. How many species are present and how evenly distributed they are.15 |
| Evenness | Describes the relative abundance of the species present in a sample; distribution of species. |
| Facultative organisms | Those able to grow with and without oxygen. |
| Metabolomics | Study of the metabolite profiles in a sample or tissue. Can be assessed using nuclear magnetic resonance spectroscopy or mass spectrometry. Metagenomics can be used to assess the function potential.23 |
| Metagenome/metagenomics | The collection of all the genomes and genes from all the microbiota in a given environment. Metagenomics is the process of characterizing the metagenome through shotgun sequencing of DNA.23 |
| Metaproteomics | Characterization of the proteins found in a given sample or environment. Proteins are identified from microbiota, hosts, and their environment, and computational methods are used to identify biologic origin.23 |
| Metatranscriptomics | Analysis of meta-RNAs providing information on regulation and expression profiles of microbiomes.23 |
| Microbiome | The entire ecosystem of microorganisms, their genomes, and their surrounding environmental and clinical conditions. The term should not be used to describe data arising from targeted amplicon sequencing, as it does not contain reliable functional information.23 |
| Microbiota | The collection of individual microbes present in a given sample or at a specified location. The term can be used to describe composition and abundance of microbial communities.23 |
| Microflora | Refers to bacteria, fungi, and archaea. Term arose prior to the restructuring of the domains of life. This term is now considered incorrect or obsolete as microorganisms are not plants or “flora.” The term “microbiota” should be used in its place. |
| Next generation sequencing | High-throughput or massively parallel sequencing technology catch-all term used to describe modern sequencing methods. NGS has greater scalability and speed with reduced costs compared with Sanger sequencing.24 |
| Operational taxonomic units | Clusters of sequence reads based on sequence similarity typically defined as 97% similar. Each cluster is intended to represent a taxonomic unit of bacteria typically at the genus level when possible.25 |
| Prebiotic | Fermented or high-fiber foods used by probiotic bacteria to produce health benefits or to improve the balance of the microbes present.26 |
| Sampling depth | Sometimes called library size. The number of bases sequenced in a given sample. |
| Symbiont | An organism living on or within the second, larger organism. Does not require a functionally significant relationship between the organisms, though the relationship can be neutral (commensalism), mutualistic (beneficial to both), or parasitic (beneficial only to the symbiont) |
| Richness | The number of different species in a given sample.27 |
Abbreviations: ACE, abundance-based coverage estimator; NSG, next generation sequencing; OUT, operational taxonomic units.
PROBIOTICS
Probiotics are defined by the Food and Agriculture Organization of the United Nations and the World Health Organization as live microorganisms that confer a health benefit on the host when administered in adequate amounts.28 The human body naturally contains many species of probiotic bacteria. Probiotic bacteria can be used as a dietary supplement aimed at improving health. Most probiotic products contain live, freeze dried species of Lactobacillus and/or Bifidobacterium. Figure 1 shows the relationships among these 2 probiotic species to each other and to other commensals, as well as pathogenic bacteria relevant to perinatal care.
Figure 1.
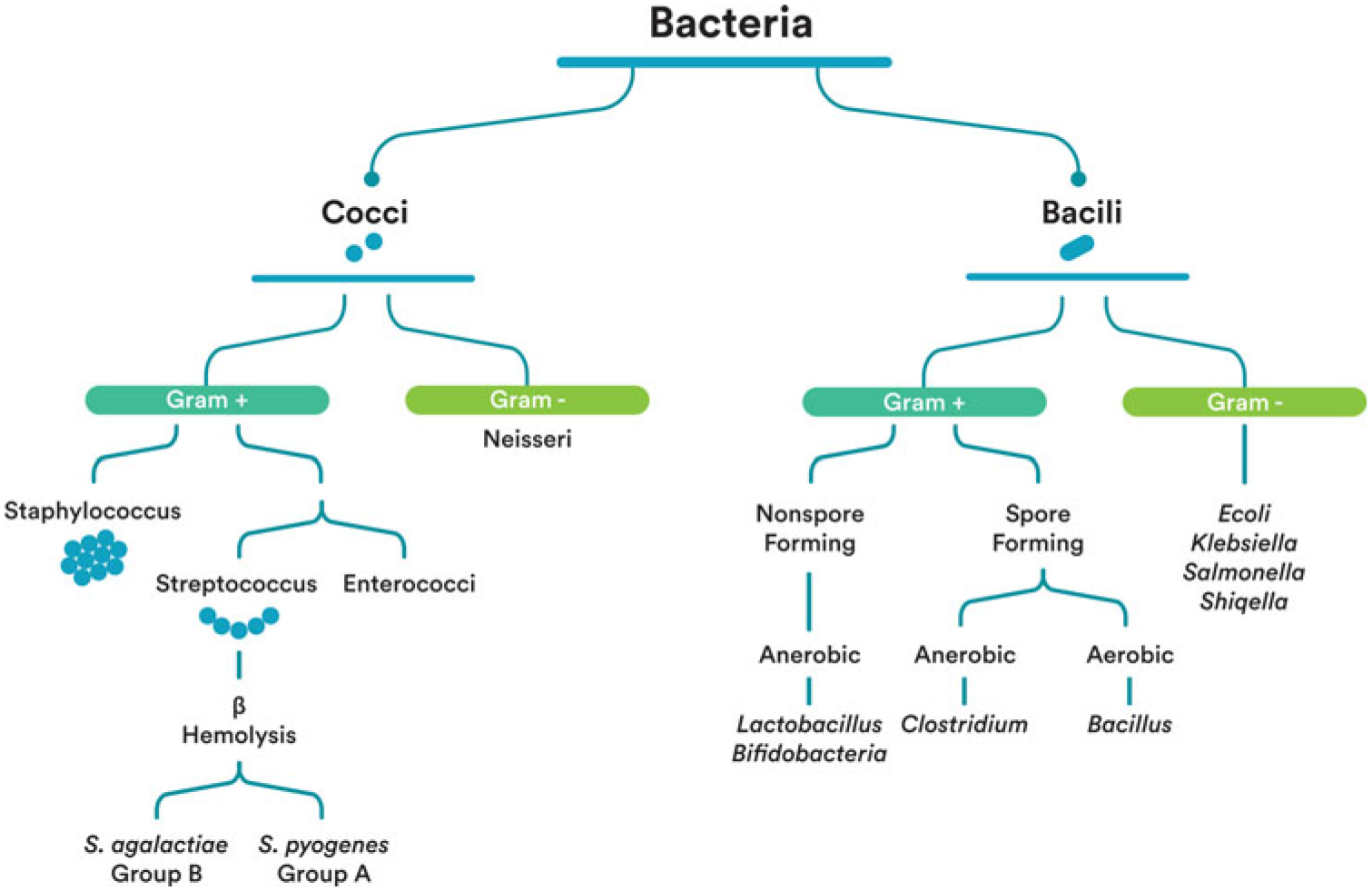
Bacteria identification. Attribution: Kara Hanson.
UNDERSTANDING PROBIOTICS
An increasingly popular microbial intervention is the use of probiotics. When discussing probiotics, the World Health Organization has guidelines for proper identification of probiotic products that are consistent with those used by the scientific community to identify microorganisms.28,29 Many have called for standardization in fast and reliable methods for probiotic identification.30,31 Probiotic strains are identified by genus, species, and subspecies with an alphanumeric designation for the specific strain; an example of this is Lactobacillus (genus) acidophilus (species) NCFM (strain).28,29 Figure 2 shows the relationship between the taxonomic levels of Lactobacillus acidophilus NCFM.
Figure 2.
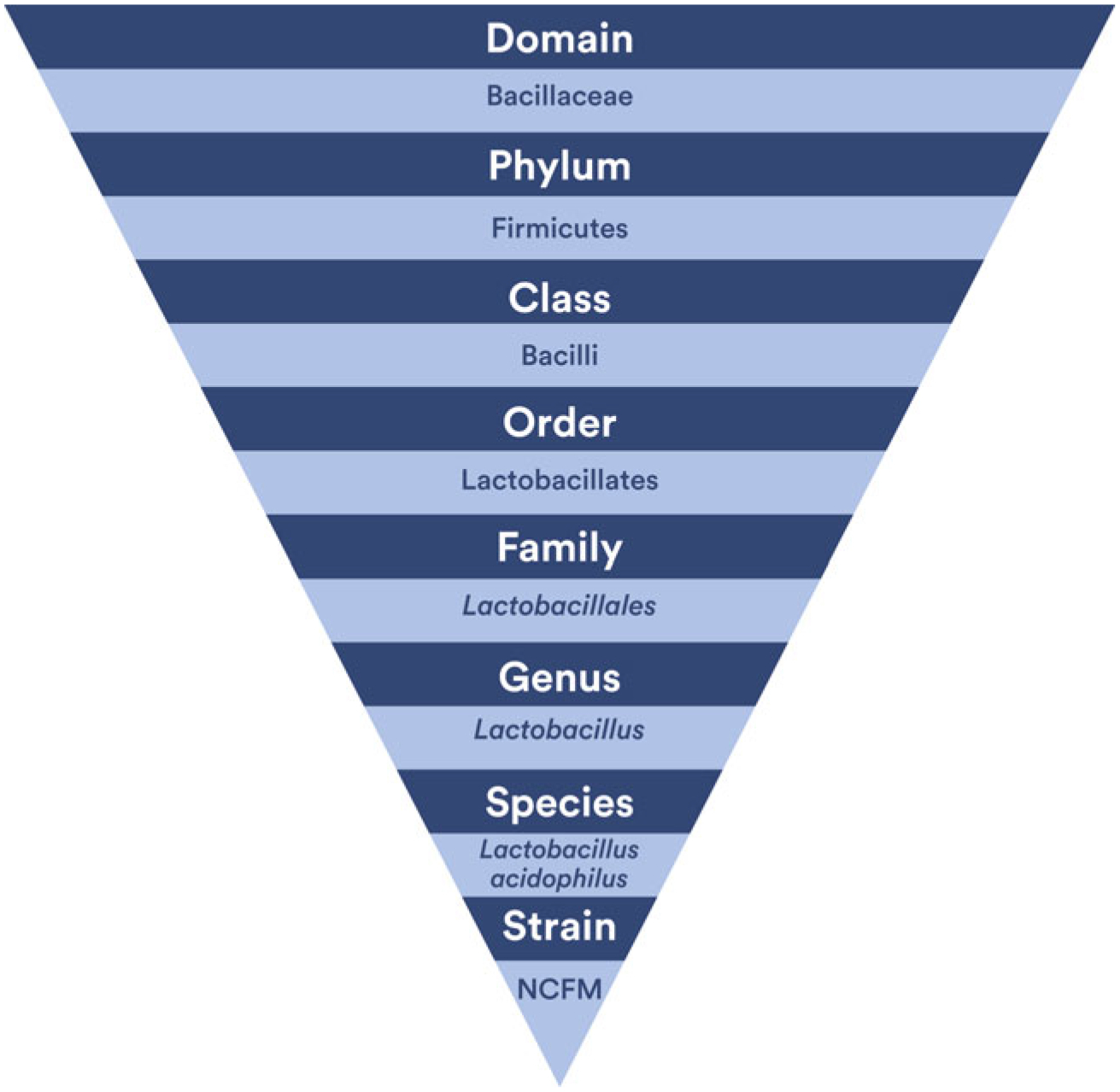
Taxonomy of Lactobacillus acidophilus NCFM. Attribution: Kara Hanson.
Probiotic supplementation is the deliberate ingestion of potentially beneficial microorganisms. Probiotics work by lining the mucus membranes of the gut to provide a defensive barrier, promoting the host immune response, stabilizing the gut microbiota, and balancing the pro- and anti-inflammatory responses.32,33
The most common probiotics in the human body are genera Lactobacillus and Bifidobacterium.30 Lactobacillus belongs to the phylum Firmicutes, which are gram-positive, nonmotile, non–spore-forming, facultative anaerobic and fermentative bacteria. Lactobacillus is acid tolerant, so it can survive the digestive processes and is associated with mucous membranes, such as those found in the GI tract, vagina, and oral cavity.30 Bifidobacterium is gram-positive, nonmotile, non–spore-forming, anaerobic, and heterofermentative.30 The presence, abundance, and diversity of Bifidobacterium are considered a marker of GI wellness; in neonates, Bifidobacterium also is associated with gut maturation.6
The genus Lactobacillus has more than 100 species, and of these, 12 are in the Lactobacillus clade and 2 belong to the Pediococcus clade. Lactobacillus has been identified through 16S ribosomal RNA (rRNA) sequencing (a laboratory technique described later in this article).34 Lactobacillus has historically been used in food preservation in fermented foods, such as in pickled vegetables and yogurts.30 Probiotics often contain 106 to 109 (1 million to 1 billion) or more colony-forming units per milliliter (mL), and these bacteria must remain viable in order to offer maximum health benefits to the host.30 Lactobacillus cannot be identified by only colony or cell morphology, but rather by phenotyping, genotyping, or fermentation properties.30 Examples of phenotyping methods are cell motility testing and gram staining.30
Probiotics must be able to survive passage from the oral cavity through the GI system to have maximum potency and benefit.28 One study evaluated 6 different probiotic preparations including encapsulated pills and yogurt drinks and concluded that Lactobacillus is able to survive acid environments similar to the human gut.35 Although the mechanism of action is unknown, there is some evidence that even dead probiotics may offer some immunostimulation; however, it is believed that there is much greater benefit with a live probiotic.30
PERINATAL-ASSOCIATED MICROBIOTA
The physiologic changes in the perinatal microflora of the oral cavity, gut, placenta, and vagina during the perinatal period are summarized in Figure 3. The evidence from studies of probiotic interventions used to modify the microbiota in each site is also discussed.
Figure 3.
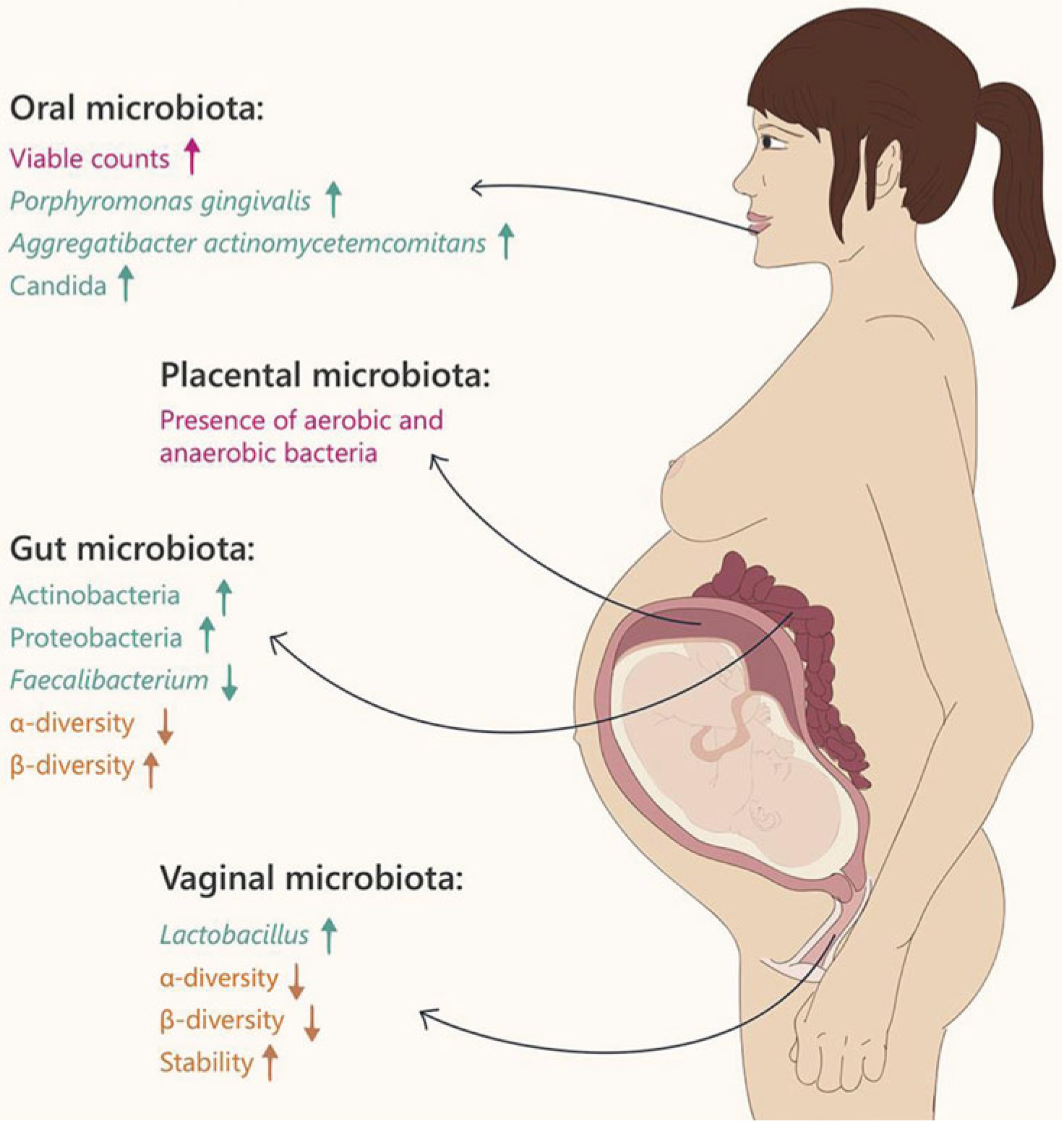
Changes in maternal microbiota during pregnancy. From Nuriel-Ohayon et al.36 Reprinted with permission.
Maternal oral cavity
The oral microbiota includes facultative organisms and strict anaerobes, including Streptococcus, Bacteroides, Lactobacillus, and more.33 There is a known connection between maternal periodontal disease and birth outcomes, with higher instances of preterm labor and birth found in women with periodontal disease. There are no published studies of probiotic interventions aimed at antenatal oral health, although there is evidence of a relationship between the maternal oral cavity and the perinatal microbiome.5
Maternal gut
In microbiome research, the term “gut” is used to refer to the entirety of the GI tract. The gut microbiome has the greatest number of bacteria, estimated at more than 100 trillion.14 The human gut has a major role in both immunity and metabolism, and dysbiosis (imbalance in commensal and pathogenic bacteria) has been linked to adverse health events, such as obesity, diabetes, diarrhea, and allergic disease.37
Probiotic interventions aimed at altering the gut microflora have been studied during pregnancy. Probiotics have been shown to colonize the maternal gut during pregnancy and may also induce a beneficial proinflammatory state.38,39 A proinflammatory state naturally develops in pregnancy as labor approaches, which is protective for the maternal-neonatal dyad.39 A randomized controlled trial of prebiotics versus placebo found significantly higher levels of Bifidobacterium in the maternal gut among those in the probiotics group but no change in the neonatal gut.40 Results are conflicting regarding the impact of maternal gut colonization with probiotics on neonatal gut colonization with probiotic bacteria.37,41
The maternal gut is known to be the reservoir for bacteria that colonize the vagina, including group B Streptococcus (GBS), which has implications for neonates.42,43 More than 20 probiotic bacteria, mostly Lactobacillus species, have been studied in vitro for antagonist activity against GBS.44–48 All showed efficacy in reducing GBS in coculture, indicating that these lactobacilli inhibited the growth of GBS in laboratory testing using human epithelial cells. The mechanisms of action of the probiotic bacteria against GBS include acidification/lowering the pH and competitive exclusion. Six clinical trials of probiotics to reduce antenatal GBS have been published.49–54 The findings of 5 of these studies were promising and show some efficacy in reducing antenatal GBS colonization or colony counts.50–53 The authors of studies in which the efficacy of the probiotic intervention against GBS was not demonstrated offered possible explanations including the following: the particular probiotic was ineffective; the dosage was too low; the sample size was too small; and encountering difficulty recruiting pregnant participants.49,54 More well-controlled studies are needed to determine effectiveness of probiotic intervention to reduce antenatal GBS.
Vagina
Irrespective of hygiene or sexual practices, organisms that constitute the vaginal microbiota arise from the maternal gut.55 The GI and genitourinary tracts share some microorganisms, as in the case of GBS, which colonizes the vagina but primarily is found in the gut.12 Some populations of healthy pregnant women have less vaginal bacterial diversity than nonpregnant women, and the vagina is dominated by Lactobacillus species.12 It is possible that this decrease in diversity is protective against other pathogens.12 Vaginal birth exposes the neonate to both the maternal vaginal microbiota and the rectal microbiota, likely via a fecal-oral route, colonizing the neonatal gut.56 Neonates born vaginally have more Lactobacillus species in their meconium than those born by cesarean delivery.57 One randomized controlled trial of an antenatal probiotic intervention versus placebo showed no significant difference in vaginal microbiota between the 2 groups.58 More recently, Yang and colleagues59 compared vaginal microbiota in pregnant women who took a probiotic from 12 weeks’ gestation versus controls. They also found no difference between the groups. More research is needed to identify the possibility of modifying the vaginal microflora during pregnancy.
Placenta
The placental microbiome is controversial. Historically, the placenta and the uterine environment were considered to be sterile.60 Aagaard and colleagues61 conducted a recent study of a population-based cohort of 320 placentas, collected under sterile conditions, and conducted metagenomic studies using 16S rDNA and whole genome shotgun sequencing to compare the placentas with other body sites. They found that the placenta had a unique microbiome that is most similar to the human oral cavity.61 Despite this finding, the microbial composition of the placenta remains controversial, with some scientists arguing that microbes obtained actually represent contamination during specimen collection.60,62–64 A strong argument has been made for a low biomass (containing limited numbers of bacteria) fetal and neonatal microbiome.61,65–67 Probiotics may be able to change this biomass; Kaplas and colleagues68 identified that an antenatal probiotic intervention modified placental phospholipid fatty acids. Several authors have described the type and importance of collection methods and emphasized the need for careful collection to avoid contamination.1,5,69 Specimen collection technique is critical for the clinical staff, who are often obtaining the samples. More research is needed to understand the relationship between collection, contamination, and the placental microbiome.
Skin and breast
In physiologic births, or births allowed to proceed with no or few obstetric interventions, infants are placed skin to skin on the maternal chest and the neonate begins breastfeeding. The human axilla is known to have high microbial biomass, and the breastfed neonate is typically cradled in the mother’s arms in close proximity to the axilla while receiving breast milk.70,71 Breast milk is a synbiotic food, meaning that it contains both prebiotics and probiotics. There is evidence that probiotics are passed through breast milk, especially Lactobacillus and Bifidobacterium.37,72 Breastfed neonates have increased Bifidobacterium in their guts compared with formula-fed neonates. Formula-fed neonates have increased gut bacterial diversity, including Bacteroides, clostridia, and streptococci, and (fewer) bifidobacteria.33 Probiotic interventions aimed at altering breast milk composition have been conducted and antenatal probiotic administration resulted in an increase in antibodies and cytokines in the breast milk of probiotic group participants compared with controls.73 It has become increasingly common for infant formula to contain probiotic bacteria in an effort to confer health benefits similar to breast milk.73
Maternal-fetal dyad
The maternal-neonatal dyad is a dynamic unit; maternal microbiome changes impact the neonate, and vice versa. The impact of probiotics on the maternal-fetal dyad is a developing science. Microbiome testing has been conducted on the maternal-fetal dyad to assess similarities and differences.2 Hegde and Munshi75 sought to examine the relationship between the vaginal microbiota and the neonatal oral cavity after birth. They found that Staphylococcus epidermidis (typically a commensal skin bacterium) persisted in the neonatal oralcavity; however, by the termination of the study, the neonatal oral cavity was dominated by Staphylococcus salivarius (a commensal oral cavity bacterium).75 This study helped demonstrate that part of the pioneer microbiota of the neonate occurs following vaginal birth. As previously stated, investigators in one study found similarity between the placental microbiome and the maternal oral cavity, which may demonstrate an additional route of colonization for the early perinatal microbiome.61
One longitudinal study of 25 mother-neonate pairs conducted strain-level metagenomic profiling and followed the pairs from birth to 4 months to document the microbial transition from the mother to the neonate.2 The study sampled the maternal stool, oral cavity, vagina, skin, and breast milk and the neonatal stool and oral cavity.2 All of the neonatal pairs shared a large number of gut species with their mothers, while maternal skin and vaginal strains only temporarily colonized the neonates.2 The authors conclude that the maternal gut is the source of the largest number of neonatal microbial strains.2
PURPOSE OF MICROBIOME TESTING/ANALYSIS
Formerly, researchers relied on culture-based methods to study microbes and their interrelationships.14 As a result, many probiotic interventions available over the counter may be miscategorized. Huys and colleagues31 found that 28% of the probiotic cultures from 10 different manufacturers were mislabeled. More than half of the cultures (57%) were from the genus Lactobacillus, whereas 22% were from the genus Bifidobacterium.31
As mentioned earlier, a number of probiotic interventions have been tested in vitro for efficacy against specific organisms, such as GBS.44–48 In these studies, the probiotic bacteria are grown in a liquid broth coculture with bacteria of interest to determine whether the probiotic bacteria can reduce the viability of the targeted bacteria. This type of testing can be useful before initiating a clinical trial. It can also yield information about the mechanism of action.
Table 2 compares common approaches to laboratory testing of the microbiome. Significant advances have been made using laboratory methods that are independent of culture, allowing scientists a greater understanding of the human microbiome.77 Sanger sequencing was previously the most common culture independent test used to identify bacterial communities. Sanger sequencing provided laboratory reads of greater than 500 DNA pairs, whereas newer testing approaches that are described later give shorter reads of 200 to 400 pairs.14 Sanger sequencing differs from newer approaches by volume, since the Sanger technique can process only one genome sample at a time whereas next generation sequencing can simultaneously sequence millions of genes from multiple samples.
Table 2.
Common techniques used to characterize microbiota and the microbiomea
| Method | Description | Advantages | Limitations |
|---|---|---|---|
| Culture and culturomics | Isolation of microbes on selective media. May be combined with NGS approaches. | Inexpensive, semiquantitative, potential detection of low abundance organism | Less than 20% of bacteria may be identifiable by culture, though this number is increasing; labor intensive |
| qPCR | Used to amplify and quantify specific genes. | qPCR is quantitative, both are fast | Unable to identify unknown species, subject to PCR bias |
| Microarrays | Fluorochrome dyes are used to label probes that bind complementary nucleotides. The fluorescence emission pattern is detected by a laser and used for DNA identification | Semiquantitative, fast | PCR bias and cross-hybridization, poor detection of low abundance organisms |
| Targeted amplicon sequencing | Massively parallel, ultra-deep sequencing PCR products (amplicons). Typically amplification is of a marker gene such a portion of the 16S rRNA gene. | Quantitative, faster than shotgun metagenomics, can identify unknown organisms | Poor detection of low abundance organisms, PCR bias, somewhat expensive, genus-level phylogenetic information only, poor accuracy for functional predictions |
| Whole genome shotgun sequencing | Massively parallel sequencing of all of the genomic DNA in a sample. | Quantitative, high taxonomic and functional information, no PCR bias, strain-level identification possible | Most expensive, computationally intensive data analysis |
Abbreviation: NSG, next generation sequencing; PCR, polymerase chain reaction; qPCR, quantitative polymerase chain reaction.
Adapted from Fraher et al.76
Microbiome analysis is still a new and emerging field and not widely used in all areas of clinical practice.77 An exception is Fecal Microbiome Transfer that is being applied in clinical care of patients with unrelenting Clostridium difficile diarrheal infections.78
Microbiome testing is often used in probiotic intervention studies to compare treatment with control groups. In these studies, probiotics are administered to determine their efficacy at creating health outcome changes in pregnant women and their neonates.73 Microbial studies can also be used to determine the adherence to the probiotic intervention. For example, if a probiotic bacterial strain is found in the stool of a study participant, it is a potential adherence indicator if it was not present before, providing evidence that the participant took the probiotic. Locating the strain in stool also provides evidence the probiotic bacteria withstood the acidic environment of the GI tract, an essential characteristic of any oral probiotic intervention.
COMPARISON OF CONTEMPORARY APPROACHES WITH MICROBIOME TESTING
The 16S rRNA gene testing is frequently mentioned in the literature and was first introduced in 1985. This test allows for comparison between bacterial types and genes.1 This approach uses polymerase chain reaction to target and amplify (regions VI-V9) of the bacterial RNA. The pieces of RNA are called amplicons. Amplicons from separate samples are coded and pooled together and sequenced, the process where the nucleic acid order of the nucleotides is determined.77 The 16S rRNA area in bacteria allows for identification and classification of an area that is exclusive to bacteria. This is because the ribosomal subunit is a gene that is located on the 30S area of a prokaryotic ribosome; it contains 9 subregions, and each can be targeted for amplification to allow for categorization.77 16S rRNA contains a specific region that is unique to each bacterial species, meaning the bacteria can be classified and also be added to a broad categorization of the species.14 This allows for detection of new microbes and for comparison between bacterial types and locations, sometimes referred to as niches.1 The 9 specific subregions are unique, and different regions better categorize different human microbiome regions. 16S rRNA has some limitations, as the lowest level of taxonomic classification is usually the genus level, which can limit clinical utility.14 There also are several benefits to 16S rRNA use, including a low rate of false-positive results and currently it is the least costly approach for microbiome analysis.
Because 16S rRNA is the most common target region for phylogenetic identification, it is often used to identify members of the genus Lactobacillus.30 16S rRNA and 16S-23S rDNA are tools that allow for faster identification of probiotics such as Lactobacillus than was possible in the past.30 However, 16S rRNA does not provide identification of probiotic bacteria to the strain level. This is extremely important because probiotic benefits are strain dependent.30
Contemporary laboratory methods for microbiome analysis include shotgun sequencing. Unlike 16S rRNA (that uses only specific genes), shotgun sequencing uses all the DNA genomic material from a sample. This approach allows for analyses to the strain level and also allows for analysis of metabolic function and antibiotic resistance analyses. In comparison, deep sequencing provides a higher number of reads and is a better choice when samples may contain large amounts of host DNA. Deep sequencing may give more accuracy, but it is more costly.
The use of shotgun analysis is recommended for analyses of human saliva and feces, because it fragments a large genome into small random DNA fragments that are individually sequenced, compared by powerful computers for overlaps in the sequences, and then replaces them in their correct order to reconstitute the genome. There are 2 subtypes of shotgun sequencing: deep and shallow. Deep shotgun sequencing can be used for general human microbiome analyses. It provides limited coverage of bacteria, has a high risk of false positive, and is the costliest.79 Shallow shotgun sequencing has limited bacterial coverage, a high risk of false positives, and is less costly (but still more costly than 16S rRNA analysis).
DISCUSSION
A comprehensive understanding of perinatal microbiome is an evolving science. The past century has seen a rapid change in location, type, and interventions associated with birth: from the home to the hospital environment, from vaginal birth to approximately one-third of US births by cesarean section, and the use of interventions aimed at improving maternal and neonatal health such as intravenous intrapartum antibiotic prophylaxis to reduce GBS disease of the newborn. These changes may—or may not—have implications for the perinatal microbiome and for long-term human health and disease. Each aspect of the perinatal microbiome, the oral cavity, the GI tract, and the genitourinary tract, may be susceptible to the influence of microbial interventions. The use of probiotic inventions to improve health has been the focus of research during the past 20 years. More recently, perinatal researchers have been studying probiotic interventions to reduce GBS colonization, with the hopes of reducing the ongoing burden of both GBS disease of the newborn and intrapartum antibiotic use.
As an understanding of the laboratory techniques aimed at evaluating probiotic interventions develop, the interventions may be targeted to more precisely impact specific areas of the perinatal microbiome. For example, if specific strains of probiotics are found to be more capable of reducing GBS colonization in clinical trials, the intervention could be applied in clinical practice. Currently, probiotics may be found at grocery stores, health food stores, and pharmacies and seem to be increasingly popular. However, probiotics are not available to women who lack the financial resources to purchase over-the-counter supplements. Probiotics may be an innovative strategy to improve perinatal health; yet, rigorous studies and methodologies to evaluate their potency and effects are necessary before clinicians can recommend them.
Laboratory analysis has developed quickly and continues to be increasingly precise and to offer a greater scope of identification. Although not currently part of routine clinical practice, clinicians must understand laboratory analysis techniques, both to comprehend the literature and to discuss the perinatal microbiome with pregnant women and their families.
IMPLICATIONS FOR PERINATAL CLINICIANS AND RESEARCHERS
It is important that perinatal clinicians and researchers become familiar with the laboratory analysis associated with the microbiome. Clinicians such as nurses and midwives will likely be the scientists collecting the specimens and will benefit from an understanding of the analysis techniques. Furthermore, clinicians will be the point of contact for pregnant people and therefore will need to understand what is known and what is unknown about the perinatal microbiome and which factors influence it.
At this time, antenatal probiotic interventions are still under investigation and not widely applied to clinical practice. More well-controlled studies are needed to determine the effectiveness. Ames and colleagues14 state that microbiome research is currently a descriptive science and few interventions based on this research are available. However, interventions are emerging; in the perinatal world, one example of new microbiome research is of probiotics in pregnancy to reduce GBS. As this science continues to develop, perinatal nurses and midwives may be able to encourage interventions such as probiotics to improve short- and long-term health.
CONCLUSION
A scientific understanding of the perinatal microbiome is emerging and has the potential to impact human health and disease. Laboratory analysis of the perinatal microbiome has made great advances in recent years and is continuing to evolve. As this science develops, interventions such as the use of probiotics to improve perinatal health will be better understood and therefore may be recommended by perinatal clinicians.
Acknowledgments
Emily Malloy (PhD student), Lisa Hanson (PI), Leona VandeVusse (Co-I), Lauren Watson, and Nasia Safdar (Co-I) are currently conducting an NIH-funded study “The Efficacy of Probiotics to Reduce Antepartum Group B Colonization” (R21HD095320).
Ashley Kates is supported by an NLM training grant to the Computation and Informatics in Biology and Medicine Training Program (NLM5T15LM007359).
Footnotes
Disclosure: The authors have disclosed that they have no significant relationships with, or financial interest in, any commercial companies pertaining to this article.
Each author has indicated that he or she has met the journal’s requirements for Authorship.
References
- 1.Jordan S, Baker B, Dunn A, et al. Maternal-child microbiome: specimen collection, storage and implications for research and practice. Nurs Res. 2017;66(2):175–183. doi: 10.1097/nnr.0000000000000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferretti P, Pasolli E, Tett A, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018;24(1):133–145.e5. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Combellick JL, Shin H, Shin D, et al. Differences in the fecal microbiota of neonates born at home or in the hospital. Sci Rep. 2018;8(1):15660. doi: 10.1038/s41598-018-33995-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valentine G, Chu DM, Stewart CJ, Aagaard KM. Relationships between perinatal interventions, maternal-infant micro-biomes, and neonatal outcomes. Clin Perinatol. 2018;45(2): 339–355. doi: 10.1016/j.clp.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Stearns JC, Simioni J, Gunn E, et al. Intrapartum antibiotics for GBS prophylaxis alter colonization patterns in the early infant gut microbiome of low risk infants. Sci Rep. 2017;7(1):16527. doi: 10.1038/s41598-017-16606-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huurre A, Laitinen K, Rautava S, Korkeamaki M, Isolauri E. Impact of maternal atopy and probiotic supplementation during pregnancy on infant sensitization: a double-blind placebo-controlled study. Clin Experiment Allergy. 2008;38:1342–1348. doi: 10.1111/j.1365-2222.2008.03008.x. [DOI] [PubMed] [Google Scholar]
- 8.Cox MJ, Huang YJ, Fujimura KE, et al. Lactobacillus casei abundance is associated with profound shifts in the infant gut microbiome. PLoS One. 2010;5(1):e8745. doi: 10.1371/journal.pone.0008745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dotterud CK, Storro O, Johnsen R, Oien T. Probiotics in pregnant women to prevent allergic disease: a randomized, double-blind trial. Br J Dermatol. 2010;163(3):616–623. doi: 10.1111/j.1365-2133.2010.09889.x. [DOI] [PubMed] [Google Scholar]
- 10.Vangay P, Ward T, Gerber JS, Knights D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe. 2015;17(5):553–564. doi: 10.1016/j.chom.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez NB, Dorsen C, Squires A. Dysbiosis of the gut microbiome: a concept analysis. J Holist Nurs. 2019. doi: 10.1177/0898010119879527. [DOI] [PubMed] [Google Scholar]
- 12.Solt I The human microbiome and the great obstetrical syndromes: a new frontier in maternal-fetal medicine. Best Pract Res Clin Obstet Gynaecol. 2015;29(2):165–175. doi: 10.1016/j.bpobgyn.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 13.Dunn AB, Hanson L, VandeVusse L, Leslie S. Through the microbial looking glass: premature labor, preeclampsia, and gestational diabetes: a scoping review. J Perinat Neonatal Nurs. 2019;33(1):35–51. doi: 10.1097/jpn.0000000000000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ames NJ, Ranucci A, Moriyama B, Wallen GR. The human microbiome and understanding the 16S rRNA gene in translational nursing science. Nurs Res. 2017;66(2):184–197. doi: 10.1097/nnr.0000000000000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarangi AN, Goel A, Aggarwal R. Methods for studying gut microbiota: a primer for physicians. J Clin Exp Hepatol. 2019;9(1):62–73. doi: 10.1016/j.jceh.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27(3):379–423. [Google Scholar]
- 17.Simpson EH. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- 18.Colwell RK, Coddington JA. Estimating terrestrial biodiversity through extrapolation. Philos Trans R Soc Lond B Biol Sci. 1994;345(1311):101–118. doi: 10.1098/rstb.1994.0091. [DOI] [PubMed] [Google Scholar]
- 19.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bray JR, Curtis JT. An ordination of upland forest communities of southern Wisconsin. Ecol Monogr. 1957;27:325–349. [Google Scholar]
- 21.Jaccard P The distribution of the flora in the alpine zone. New Phytol. 1912;11(2):37–50. doi: 10.1111/j.1469-8137.1912.tb05611.x. [DOI] [Google Scholar]
- 22.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. 2015;3(1):31. doi: 10.1186/s40168-015-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behjati S, Tarpey PS. What is next generation sequencing? Arch Dis Child Educ Pract Ed. 2013;98(6):236–238. doi: 10.1136/archdischild-2013-304340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blaxter M, Mann J, Chapman T, et al. Defining operational taxonomic units using DNA barcode data. Philos Trans R Soc Lond B Biol Sci. 2005;360(1462):1935–1943. doi: 10.1098/rstb.2005.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberfroid M Prebiotics: the concept revisited. J Nutr. 2007;137(3)(suppl 2):830S–837S. [DOI] [PubMed] [Google Scholar]
- 27.Gotelli NJ, Colwell RK. Estimating species richness. In: Magurran AE, McGill BJ, eds. Biological Diversity: Frontiers of Measurement & Assessment. New York, NY: Oxford University Press; 2011:39–54. [Google Scholar]
- 28.Food and Agriculture Organization of the United Nations (FAO), World Health Organization (WHO). Guidelines for the evaluation of probiotics in food. Report of a joint FAO/WHO working group on drafting guidelines for the evaluation of probiotics in food. http://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf. Published May 1, 2002. Accessed March 21, 2020.
- 29.Baldassarre ME. Probiotic genera/species identification is insufficient for evidence-based medicine. Am J Gastroenterol. 2018;113(10):1561. doi: 10.1038/s41395-018-0236-z. [DOI] [PubMed] [Google Scholar]
- 30.Herbel SR, Vahjen W, Wieler LH, Guenther S. Timely approaches to identify probiotic species of the genus Lactobacillus. Gut Pathog. 2013;5(1):27. doi: 10.1186/1757-4749-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huys G, Vancanneyt M, D’Haene K, Vankerckhoven V, Goossens H, Swings J. Accuracy of species identity of commercial bacterial cultures intended for probiotic or nutritional use. Res Microbiol. 2006;157(9):803–810. doi: 10.1016/j.resmic.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Reid G, Bocking A. The potential for probiotics to prevent bacterial vaginosis and preterm labor. Am J Obstet Gynecol. 2003;189(4):1202–1208. doi: 10.1067/s0002-9378(03)00495-2. [DOI] [PubMed] [Google Scholar]
- 33.Isolauri E, Sütas Y, Kankaanpää P, Arvilommi H, Salminen S. Probiotics: effects on immunity. Am J Clin Nutr. 2001;73(2)(suppl):444S–450S. doi: 10.1093/ajcn/73.2.444s. [DOI] [PubMed] [Google Scholar]
- 34.Neville BA, O’Toole PW. Probiotic properties of Lactobacillus salivarius and closely related Lactobacillus species. Future Microbiol. 2010;5(5):759–774. doi: 10.2217/fmb.10.35. [DOI] [PubMed] [Google Scholar]
- 35.Fleming D, Kesey J, Rumbaugh K, Dissanaike S. Comparing the survivability of Lactobacillus species in various probiotic delivery vehicles. JPEN J Parenter Enteral Nutr. 2017;41(8):1411–1413. doi: 10.1177/0148607116672266. [DOI] [PubMed] [Google Scholar]
- 36.Nuriel-Ohayon M, Neuman H, Koren O. Microbial changes during pregnancy, birth, and infancy. Front Microbiol. 2016;7: 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naaktgeboren CA. Effects of maternal probiotic exposure during pregnancy and lactation on the mother and infant. Int J Probiotics Prebiotics. 2010;5(3):113–124. https://0-search-proquest-com.libus.csd.mu.edu/docview/757449059?accountid=100. Accessed March 15, 2020. [Google Scholar]
- 38.Lahtinen SJ, Boyle RJ, Kivivuori S, et al. Prenatal probiotic administration can influence Bifidobacterium microbiota development in infants at high risk of allergy. J Allergy Clin Immunol. 2009;123(2):499–501. doi: 10.1016/j.jaci.2008.11.034. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, Li Z, Tye KD, et al. Probiotic supplementation during human pregnancy affects the gut microbiota and immune status. Front Cell Infect Microbiol. 2019;9:254. doi: 10.3389/fcimb.2019.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shadid R, Haarman M, Knol J, et al. Effects of galactooligosaccharide and long-chain fructooligosaccharide supplementation during pregnancy on maternal and neonatal microbiota and immunity—a randomized, double-blind, placebo-controlled study. Am J Clin Nutr. 2007;86(5):1426–1437. doi: 10.1093/ajcn/86.5.1426. [DOI] [PubMed] [Google Scholar]
- 41.Schultz M, Göttl C, Young RJ, Iwen P, Vanderhoof JA. Administration of oral probiotic bacteria to pregnant women causes temporary infantile colonization. J Pediatr Gastroenterol Nutr. 2004;38(3):293–297. doi: 10.1097/00005176-200403000-00012. [DOI] [PubMed] [Google Scholar]
- 42.Picard FJ, Bergeron MG. Laboratory detection of group B Streptococcus for prevention of perinatal disease. Eur J Clin Microbiol Infect Dis. 2004;23:665–671. doi: 10.1007/s10096-004-1183-8. [DOI] [PubMed] [Google Scholar]
- 43.Manning SD, Neighbors K, Tallman PA, et al. Prevalence of group B Streptococcus colonization and potential for transmission by casual contact in healthy young men and women. Clin Infect Dis. 2004;39(3):380–388. doi: 10.1086/422321. [DOI] [PubMed] [Google Scholar]
- 44.Martin V, Cárdenas N, Ocaña S, et al. Rectal and vaginal eradication of Streptococcus agalactiae (GBS) in pregnant women by using Lactobacillus salivarius CECT 9145, a target-specific probiotic strain. Nutrients. 2019;11(4):E810. doi: 10.3390/nu11040810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marziali G, Foschi C, Parolin C, Vitali B, Marangoni A. In-vitro effect of vaginal lactobacilli against group B Streptococcus. Microb Pathog. 2019;136:103692. doi: 10.1016/j.micpath.2019.103692. [DOI] [PubMed] [Google Scholar]
- 46.Patras KA, Derieux J, Al-Bassam MM, et al. Group B Streptococcus biofilm regulatory protein a contributes to bacterial physiology and innate immune resistance. J Infect Dis. 2018;218(10):1641–1652. doi: 10.1093/infdis/jiy341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ephraim E, Schultz RD, Duster M, Warrack S, Spiegel CA, Safdar N. In-vitro evaluation of the antagonistic effects of the probiotics Lactobacillus rhamnosus HN001 and Florajen 3 against group B streptococci. Int J Probiotics Prebiotics. 2012;7(3/4):113–120. https://www.researchgate.net/publication/261870832_International_Journal_of_Probiotics_and_Prebiotics. Accessed March 15, 2020. [Google Scholar]
- 48.Zárate G, Nader-Macias ME. Influence of probiotic vaginal lactobacilli on in vitro adhesion of urogenital pathogens to vaginal epithelial cells. Lett Appl Microbiol. 2006;43(2):174–180. doi: 10.1111/j.1472-765X.2006.01934.x. [DOI] [PubMed] [Google Scholar]
- 49.Sharpe M, Shah V, Freire-Lizama T, et al. Effectiveness of oral intake of Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 on group B Streptococcus colonization during pregnancy: a midwifery-led double-blind randomized controlled pilot trial. J Matern Fetal Neonatal Med. 2019:1–8. doi: 10.1080/14767058.2019.1650907. [DOI] [PubMed] [Google Scholar]
- 50.Martin V, Cárdenas N, Ocaña S, et al. Rectal and vaginal eradication of Streptococcus agalactiae (GBS) in pregnant women by using Lactobacillus salivarius CECT 9145, a target-specific probiotic strain. Nutrients. 2019;11(4):E810. doi: 10.3390/nu11040810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ho M, Chang YY, Chang WC, et al. Oral Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 to reduce group B Streptococcus colonization in pregnant women: a randomized controlled trial. Taiwan J Obstet Gynecol. 2016;55(4):515–518. doi: 10.1016/j.tjog.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 52.Hanson L, Vandevusse L, Duster M, Warrack S, Safdar N. Feasibility of oral prenatal probiotics against maternal group B Streptococcus vaginal and rectal colonization. J Obstet Gynecol Neonatal Nurs. 2014;43(3):294–304. doi: 10.1111/1552-6909.12308. [DOI] [PubMed] [Google Scholar]
- 53.Di Pierro F, Parolari A, Brundu B, Nigro R. Positive clinical outcomes derived from using a proprietary mixture of selected strains during pregnancy. Acta Biomed. 2016;87(3):259–265. [PMC free article] [PubMed] [Google Scholar]
- 54.Olsen P, Williamson M, Traynor V, Georgiou C. The impact of oral probiotics on vaginal group B streptococcal colonisation rates in pregnant women: a pilot randomised control study. Women Birth. 2018;31(1):31–37. doi: 10.1016/j.wombi.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 55.Bolton M, van der Straten A, Cohen CR. Probiotics: potential to prevent HIV and sexually transmitted infections in women. Sex Transm Dis. 2008;35(3):214–225. doi: 10.1097/OLQ.0b013e31815b017a. [DOI] [PubMed] [Google Scholar]
- 56.Dominguez-Bello MG, Godoy-Vitorino F, Knight R, Blaser MJ. Role of the microbiome in human development. Gut. 2019;68(6):1108–1114. doi: 10.1136/gutjnl-2018-31750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagpal R, Tsuji H, Takahashi T, et al. Sensitive quantitative analysis of the meconium bacterial microbiota in healthy term infants born vaginally or by cesarean section. Front Microbiol. 2016;7:1997. doi: 10.3389/fmicb.2016.01997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Husain S, Allotey J, Drymoussi Z, et al. Effects of oral probiotic supplements on vaginal microbiota during pregnancy: a randomised, double-blind, placebo-controlled trial with microbiome analysis. BJOG. 2020;127:275–284. doi: 10.1111/1471-0528.15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang S, Reid G, Challis JRG, et al. Effect of oral probiotic Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 on the vaginal microbiota, cytokines and chemokines in pregnant women. Nutrients. 2020;12(2):E368. doi: 10.3390/nu12020368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bushman FD. De-discovery of the placenta microbiome. Am J Obstet Gynecol. 2019;220(3):213–214. doi: 10.1016/j.ajog.2018.11.1093. [DOI] [PubMed] [Google Scholar]
- 61.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6(237):237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leiby JS, McCormick K, Sherrill-Mix S, et al. Lack of detection of a human placenta microbiome in samples from preterm and term deliveries. Microbiome. 2018;6(1):196. doi: 10.1186/s40168-018-0575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perez-Muñoz ME, Arrieta MC, Ramer-Tait AE, Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 2017;5(1):48. doi: 10.1186/s40168-017-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lauder AP, Roche AM, Sherrill-Mix S, et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome. 2016;4(1):29. doi: 10.1186/s40168-016-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martinez KA, Romano-Keeler J, Zackular JP, et al. Bacterial DNA is present in the fetal intestine and overlaps with that in the placenta in mice. PLoS One. 2018;13(5):e0197439. doi: 10.1371/journal.pone.0197439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prince AL, Chu DM, Seferovic MD, Antony KM, Ma J, Aagaard KM. The perinatal microbiome and pregnancy: moving beyond the vaginal microbiome. Cold Spring Harb Perspect Med. 2015;5(6):a023051. doi: 10.1101/cshperspect.a023051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Antony KM, Ma J, Mitchell KB, Racusin DA, Versalovic J, Aagaard K. The preterm placental microbiome varies in association with excess maternal gestational weight gain. Am J Obstet Gynecol. 2015;212(5):653.e1–e16. doi: 10.1016/j.ajog.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaplas N, Isolauri E, Lampi AM, Ojala T, Laitinen K. Dietary counseling and probiotic supplementation during pregnancy modify placental phospholipid fatty acids. Lipids. 2007;42(9):865–870. doi: 10.1007/s11745-007-3094-9. [DOI] [PubMed] [Google Scholar]
- 69.Dunn AB, Jordan S, Baker BJ, Carlson NS. The maternal infant microbiome: considerations for labor and birth. MCN Am J Matern Child Nurs. 2017;42(6):318–325. doi: 10.1097/nmc.0000000000000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Urban J, Fergus DJ, Savage AM, et al. The effect of habitual and experimental antiperspirant and deodorant product use on the armpit microbiome. PeerJ. 2016;4:e1605. doi: 10.7717/peerj.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singleton ML. Group B Strep prophylaxis: what are we creating? Midwifery Today Int Midwife. 2007;81:18–20. [PubMed] [Google Scholar]
- 72.Quin C, Estaki M, Vollman DM, Barnett JA, Gill SK, Gibson DL. Probiotic supplementation and associated infant gut microbiome and health: a cautionary retrospective clinical comparison. Sci Rep. 2018;8(1):8283. doi: 10.1038/s41598-018-26423-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.VandeVusse L, Hanson L, Safdar N. Perinatal outcomes of prenatal probiotic and prebiotic administration: an integrative review. J Obstet Gynecol Neonatal Nurs. 2013;27(4):288–301. doi: 10.1097/JPN.0b013e3182a1e15d. [DOI] [PubMed] [Google Scholar]
- 74.Bertelsen RJ, Jensen ET, Ringel-Kulka T. Use of probiotics and prebiotics in infant feeding. Best Pract Res Clin Gastroenterol. 2016;30(1):39–48. doi: 10.1016/j.bpg.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 75.Hegde S, Munshi AK. Influence of the maternal vaginal microbiota on the oral microbiota of the newborn. J Clin Pediatr Dent. 1998;22(4):317–321. [PubMed] [Google Scholar]
- 76.Fraher MH, O’Toole PW, Quigley EM. Techniques used to characterize the gut microbiota: a guide for the clinician. Nat Rev Gastroenterol Hepatol. 2012;9(6):312–322. doi: 10.1038/nrgastro.2012.44. [DOI] [PubMed] [Google Scholar]
- 77.Claesson MJ, Clooney AG, O’Toole PW. A clinician’s guide to microbiome analysis. Nat Rev Gastroenterol Hepatol. 2017;14(10):585–595. doi: 10.1038/nrgastro.2017.97. [DOI] [PubMed] [Google Scholar]
- 78.Luo Y, Lucas AL, Grinspan AM. Fecal transplants by colonoscopy and capsules are cost-effective strategies for treating recurrent Clostridioides difficile infection. Dig Dis Sci. 2020;65(4):1125–1133. doi: 10.1007/s10620-019-05821-1. [DOI] [PubMed] [Google Scholar]
- 79.Hillmann B, Al-Ghalith GA, Shields-Cutler RR, et al. Evaluating the information content of shallow shotgun metagenomics. mSystems. 2018;3(6):e00069–18. doi: 10.1128/mSystems.00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]


