Abstract
Dilated cardiomyopathy (DCM) is a primary disorder of the cardiac muscle, characterized by dilatation of the left ventricle and contractile dysfunction. About 50% of DCM cases can be attributed to monogenic causes, whereas the etiology in the remaining patients remains unexplained. We report a family with two brothers affected by severe DCM with onset in the adolescent period. Using exome sequencing, we identified a homozygous premature termination variant in the MYZAP gene in both affected sibs. MYZAP encodes for myocardial zonula adherens protein—a conserved cardiac protein in the intercalated disc structure of cardiomyocytes. The effect of the variant was demonstrated by light and electron microscopy of the heart muscle and immunohistochemical and western blot analysis of the MYZAP protein in the heart tissue of the proband. Functional characterization using patient-derived induced pluripotent stem cell cardiomyocytes revealed significantly lower force and longer time to peak contraction and relaxation consistent with severe contractile dysfunction. We provide independent support for the role of biallelic loss-of-function MYZAP variants in dilated cardiomyopathy. This report extends the spectrum of cardiac disease associated with dysfunction of cardiac intercalated disc junction and sheds light on the mechanisms leading to DCM.
Keywords: dilated cardiomyopathy
INTRODUCTION
Dilated cardiomyopathy (DCM) is a primary disorder of the cardiac muscle characterized by dilatation of the left ventricle and left ventricular dysfunction, which is not secondary to abnormal loading conditions (hypertension, valvular diseases) or coronary artery disease (Elliott et al. 2008). DCM is the most common of the cardiomyopathies and represents the leading cause of heart failure requiring transplantation (Sweet et al. 2015). The prevalence of DCM has been estimated to be 1 per 250 in a population-based study (McKenna and Judge 2021).
Although studies indicate that up to 50% of individuals with DCM have an affected relative (Michels et al. 2010), the genetic diagnostic yield in these patients amounts to ∼30%, even using state-of-the-art approaches, such as whole-exome sequencing (Mak et al. 2018; Ramchand et al. 2020). Currently, pathogenic variants in 32 genes have been associated with various forms of DCM (PanelApp resource, Dilated cardiomyopathy—adult and teen v1.5) (Martin et al. 2019). The causative variants are identified in genes coding for structural sarcomeric proteins of the heart muscle, elements of the nuclear lamina, subunits of cardiac ion channels, and protein components of the intercellular junctions in the heart. In particular, the intercalated disc structure in cardiomyocytes has been reported as a functional unit, whose disruption is closely related to the occurrence of DCM or arrhythmogenic right ventricular cardiomyopathy (ARVC), and recently novel elements of this structure have been identified and already associated with DCM in animal models (Seeger et al. 2010).
Considering the high likelihood of genetic etiology in undiagnosed cases with DCM, we used whole-exome sequencing to identify a possible molecular cause in the MYZAP gene in a family with two brothers similarly affected by severe DCM and based on our findings, we provide independent support for its role in the recessive form of DCM.
RESULTS
Clinical Presentation and Family History
The proband is a 23-yr-old male with familial DCM. He was born to healthy and nonconsanguineous parents. His older brother died at the age of 16 because of a heart transplant rejection, one and a half years after heart transplantation because of heart failure due to DCM of unknown etiology. The proband has another 29-yr-old older brother who is healthy.
Clinical symptoms in the deceased brother started at the age of 14.5 yr when he was hospitalized because of DCM and heart failure. Before that period, he was otherwise a healthy adolescent with normal toleration of physical activity. Ten days before hospital admission, he was treated because of respiratory infection, after which he started to feel tired, and complained of nonspecific abdominal pain due to organomegaly and cough due to pulmonary edema. Echocardiography revealed dilatation of the left ventricle (LV) and low LV myocardial contractility with a reduction of ejection fraction of LV to 10%–14%. Cardiac magnetic resonance imaging (MRI) revealed extreme dilatation of the left ventricle, reduction of global systolic function to 11%, and areas of diffuse fibrosis of the left ventricle. The right ventricle (RV) showed similar, but less pronounced MRI changes with the reduction of ejection fraction to 19%. Amino-terminal-pro hormone B-type natriuretic peptide (NT-proBNP) was increased to 9324 pg/mL (normal <125 pg/mL). Myocarditis was suspected. After initial medicamentous stabilization (diuretics, digoxin, peripheral vasodilators, and sodium and water intake reduction), he was considered for heart transplantation, which was performed at the age of 14 yr and 10 mo. He was clinically stable 1 yr after transplantation. At the age of 16 yr and 3 mo, he was hospitalized because of progressive cardiac decompensation caused by acute humoral rejection requiring immediate initiation of veno-arterial extracorporal membrane oxygenation. Despite immediate therapy of acute humoral rejection (mycophenolate-mofetil, cyclosporine, plasmapheresis) clinical course was unfavorable and was further complicated by bilateral pneumonia and multiorgan failure, and the patient died after 2 mo at the age of 16 yr and 5 mo.
The previously healthy proband has been under regular cardiologic observation since the age of 12 because of the positive family history mentioned above. All his findings, including echocardiography, were initially unremarkable, until the age of 16. At this point a reduction of both left and right ventricular ejection fraction (LVEF/RVEF) to 35% was recorded, whereas cardiac MRI reported fibrosis of the septum from the base to the apex along with LVEF reduction to 30%–35%. One year later, a cardiomyopathy gene panel was negative. At the age of 18, a new cardiac MRI reported LVEF of 33% and RVEF of 44%, diffuse myocardial hypocontractility, and extensive areas of myocardial fibrosis, with dominantly subendocardial but also mesocardial distribution. At age of 19, an implantable cardioverter-defibrillator device (ICD) was implanted. Because of recurrent episodes of sustained ventricular tachycardia (VT) and consequent activations of ICD, the right ventricular outflow tract (RVOT) ablation was conducted with transitory cessation of arrhythmic instabilities. At age of 21, LVEF was 30% and NT-proBNP 629 pg/ml (normal range < 125 pg/mL). Recurring activations of ICD and episodes of sustained VT prompted an attempt of a new ablation, which was unsuccessful, and further attempts were deemed infeasible. During all this time several different therapy combinations of β-blockers, sotalol, and amiodarone were attempted, but without any effect. His LVEF decreased to 22% (Fig. 1A,B) with increase of NT-proBNP to 978 pg/mL. Because all other therapeutic modalities were exhausted, at age of 21 he underwent successful heart transplantation. The immediate posttransplant period was uneventful, and he was discharged home one month after transplantation. His therapy included tacrolimus, mycophenolate mofetil, and prednisone. Five months later he was successfully treated for cytomegalovirus (CMV) colitis and CMV encephalitis, despite valganciclovir prophylaxis. Aside from this, there were no episodes of allograft rejection or other complications, and as of June 2021, at the age of 23, the patient remains well and stable.
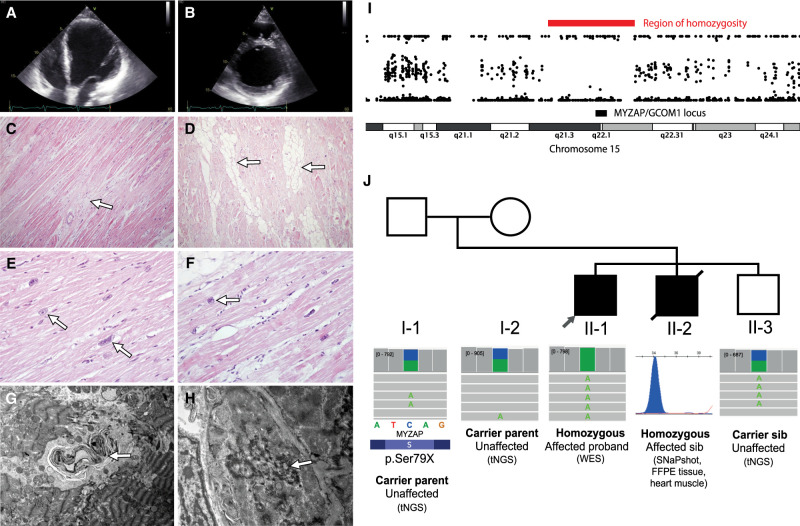
Figure 1.
Data from the imaging, microscopy, and genetic studies in the family. Apical four-chamber (A) and parasternal short-axis (B) transthoracic echocardiography video clips of the proband at the age of 21, shortly before transplantation. Note the dilated left ventricle with global hypokinesia and severe reduction of ejection fraction. (C) Light microscopy (original magnification 100×, hematoxylin and eosin [H&E]). The arrow shows the foci of myocardial fibrosis. (D) Light microscopy (original magnification 100×, H&E). Foci of lipocytes in the myocardium (arrows). (E) Light microscopy (original magnification 400×, H&E). Degenerative changes of myocardial nuclei (arrows). (F) Light microscopy (original magnification 400×, H&E). Degenerative changes of myocardial nuclei with perinuclear vacuoles (arrow). (G) Electron microscopy (original magnification, 4400×). Myelin figure in the myocardium (arrow). (H) Electron microscopy (original magnification, 8900×). Focal disorganization of an intercalated disc (arrow). (I) The region of Chromosome 15, indicating the presence of a region of homozygosity detected in exome sequencing data. A region of homozygosity was detected on Chromosome 15, which contains the MYZAP gene locus. (J) The pedigree of the family and segregation results of the premature termination variant detected in the MYZAP gene (NM_001018100.5:c.236C > A, p.Ser79Ter) in the proband (II-1) and the affected sibling (II-2). The unaffected sibling and parents were all heterozygous carriers of the premature termination variant. The variant was initially identified in the proband using exome sequencing and confirmed in the paraffin sample of the deceased affected sibling using SNaPshot methodology and segregation performed using targeted next-generation sequencing (tNGS) in the unaffected sibling and unaffected parents.
Genomic Analyses
Exome sequencing was performed in proband II-1 affected by DCM (Fig. 1J) and the search for causative variants was performed in a set of 32 genes associated with DCM according to the PanelApp resource and the literature. No causative variants were found in this set of genes, either by the analysis of single-nucleotide variation or by using copy-number variation (CNV) analysis or mitochondrial genome analysis.
Analysis of the blocks of homozygosity revealed the presence of three regions of homozygosity larger than five megabases on Chromosomes 8, 11, and 15. We, therefore, inspected these regions for possible candidate variants in genes that had a potential association with cardiac structure and function. The region on Chromosome 15 encompassed the region of the MYZAP/GCOM1 locus (Fig. 1I). In this region, we identified the presence of a homozygous loss-of-function variant (NM_001018100.5:c.236C > A, p.Ser79Ter; Table 1) in exon 3 of 13 in MYZAP, which encodes the myocardial zonula adherens protein, a constituent of the cardiac intercalated discs (Seeger et al. 2010). No other rare candidate homozygous variants were present in the identified regions of homozygosity.
Table 1.
Variant table
| Gene | Chromo some | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | dbSNP/dbVar ID | Geno type | ClinVar ID | Parent of origin | Observed effect (if shown to be different from predicted effect) |
|---|---|---|---|---|---|---|---|---|---|---|
| MYZAP | Chr 15 | NM_001018100.5: c.236C > A | NP_001018110.1: p.Ser79Ter | Premature truncation | Substitution | rs992189342 | Homo-zygous | 523392 | Biparental | Loss of expression |
The identified variant is absent from all control populations in the gnomAD resource and was not detected in the Slovenian population database. The variant arises relatively early in the reading frame (across all transcript isoforms of MYZAP) and is thus anticipated to result in nonsense-mediated decay and complete loss of the functional protein in a homozygous state.
The SNaPshot analysis of this variant in the deceased sibling with DCM was performed in cardiac tissue, isolated from the paraffin blocks, and revealed the presence of this variant in the homozygous state. The unaffected parents and an unaffected sibling of the proband were all heterozygous carriers of this variant.
Functional Analyses
Immunohistochemistry and Western Blot
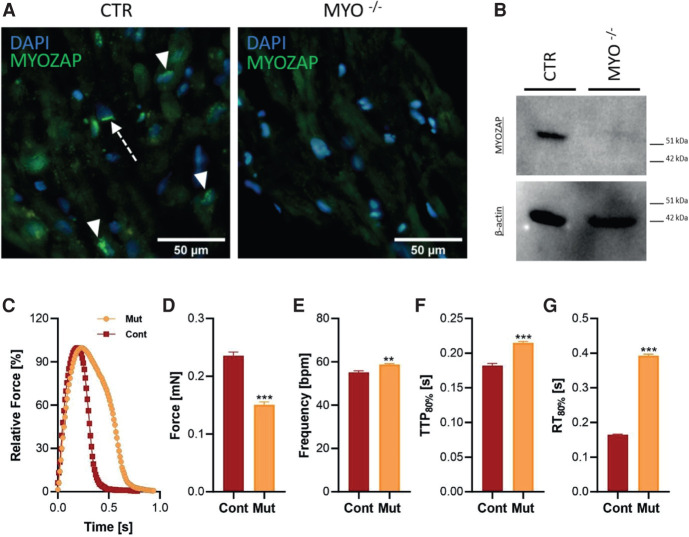
Immunohistochemical (IHC) analysis demonstrated the complete absence of the fluorescent signal corresponding to the carboxy-terminal fragment epitope of the MYZAP protein in the proband tissue sample (Fig. 2A). In contrast, a diffused green perinuclear signal (Fig. 2A, arrowhead) and dense green fluorescent bands perpendicular to myocyte orientation (Fig. 2A arrow) were observed in the control sample by the same procedure. A complete absence of MYZAP in the same tissue was confirmed with western blot (WB) using β-actin (Fig. 2B) and total protein fluorescence (Supplemental Results, Fig. S1) as a loading control.
Figure 2.
Immunohistochemical and functional studies. The absence of the MYZAP protein in proband tissue was demonstrated by catalyzed fluorescent reporter deposition immunohistochemistry and western blot (WB). (A) In the control sample (CTR), epitopes corresponding to the carboxy-terminal end of the MYZAP protein were distributed in the perinuclear cytoplasm (green signal; arrowhead) and in dense bands perpendicular to myocyte orientation (green signal; arrow). The signal was completely absent in the proband tissue (MYZAP−/−). (B) The absence of the MYZAP signal in the proband tissue was demonstrated by western blot. A β-actin signal on the same membrane was used for internal control (total protein loading control is provided in Supplemental Fig. S1). The bottom panels contain the results of the functional characterization of the engineered heart tissues (EHTs) of patient-derived human induced pluripotent stem cell cardiomyocytes (hiPSC-CMs). (C) The average contraction peaks of EHTs in 1.8 mM Ca2+ Tyrode's solution under 1-Hz pacing at 37°C. (D–G) Functional parameters of absolute force (D), frequency (beats per minute, bpm) (E), time to peak 80% (TTP80%, F), and relaxation time (RT80%, G) measured in 1.8 mM Ca2+ Tyrode's solution under 1-Hz pacing at 37 °C. n = 8 patient EHTs and 11 control EHTs. Statistical calculations were carried out by a two-tailed Student's t-test. (**) P < 0.01, (***) P < 0.001.
Functional Analysis of Patient hiPSC-Derived Cardiomyocytes Using the Engineered Heart Tissue Model
To explore the functional consequences of the MYZAP variant, we first reprogrammed patient and healthy (independent control cell line) skin fibroblasts into human induced pluripotent stem cell (hiPSCs) and differentiated cardiomyocytes (CMs). These hiPSC-CMs were then used to generate engineered heart tissues (EHTs) to study contractile behavior (Fig. 2C). Interestingly, contraction traces of mutant EHTs paced at 1 Hz showed significantly lower (by ∼35%) peak force (Fig. 2D), whereas beating frequency (up by ∼7%), time to peak (TTP80%, up by ∼17%), and relaxation time (RT80%, up by ∼2.2-fold) were significantly higher in mutant EHTs compared to the control EHTs used in this study (Fig. 2E–G). These data suggest severe contractile dysfunction and a relaxation deficit as an underlying disease phenotype observed in patient-derived EHTs.
DISCUSSION
Genetics of dilated cardiomyopathy remains a significant diagnostic challenge with a high rate of patients with unexplained etiology, even among familial cases. Despite technological advancements in sequencing and joint international efforts, only a few novel genes associated with DCM have been reported in the last decade. We provide independent support for the role of MYZAP in a recessive form of DCM, caused by biallelic variants in MYZAP.
In a family with two siblings similarly affected by DCM, we performed exome sequencing and identified a rare homozygous loss-of-function variant in MYZAP that segregated with the DCM phenotype. MYZAP encodes for the myocardial zonula adherens protein, which is transcribed from the upstream part of the GRINL1A complex transcription unit or GCOM1.
The effect of the variant was supported by light and electron microscopy analysis of the heart muscle and IHC and WB analysis of the MYZAP protein in the heart tissue of the proband. In addition, by demonstrating dysfunctional contractility of engineered heart tissue generated from patient-derived induced pluripotent stem cell cardiomyocytes, we provided insight into the pathogenesis of this novel form of DCM.
The role of MYZAP protein in cardiac structure and function was initially established by the bioinformatic screening of transcripts with predominant expression in the heart (Seeger et al. 2010). In mice, its expression was found to be primarily confined to cardiac muscle tissue by birth. Further IHC analyses performed by Seeger et al. have shown this protein to be colocalized with N-cadherin and positioned at the intercalated disc structure at the intercellular junctions between cardiomyocytes (Seeger et al. 2010). In addition, the gene was also found to be highly conserved in mammals, indicating its critical role in normal physiology.
Disruption of the structure and function of intercalated discs in cardiomyocytes has been reported in several genetic forms of DCM as well as ARVC (Perriard et al. 2003; Rampazzo et al. 2014). Genes encoding several components of the intercalated discs have already been linked to monogenic forms of DCM and ARVC, including DES, DSC2, DSG2, DSP, JUP, PKP2, and VCL genes. The MYZAP protein was shown to localize in the intercalated disc structure, interact with desmoplakin and zonula adherens proteins, and regulate expression in cardiomyocytes, which further supports its close functional association with the established genetic causes of cardiomyopathy.
The MYZAP gene is a part of a broader and complex transcriptional unit—the GRINL1A complex transcription unit—that contains exonic sequences of MYZAP and POLR2M genes (Roginski et al. 2004). The transcription of the GRINL1A locus is regulated by an upstream and a downstream promotor. Transcription from the upstream promotor is associated with the production of two transcript forms—one containing only the MYZAP exons and the second transcript (GCOM1) that contains exonic regions from both the MYZAP and the POLR2M. Conversely, the transcription from the downstream promotor produces a transcript containing exonic regions only belonging to the POLR2M gene (Roginski et al. 2004; Heliö et al. 2021). RNA sequencing data in the GTEx resource indicate that the exonic regions of MYZAP are predominantly and highly expressed in the adult cardiac tissue, whereas exonic regions specific to the GCOM1 and POLR2M isoforms are not, further supporting MYZAP as the gene related to cardiac function within this locus (Lonsdale et al. 2013).
A recent publication reports biallelic pathogenic loss-of-function variants in the GRINL1A locus unit that similarly introduce a premature termination in the MYZAP reading frame in two unrelated Finnish families with multiple individuals with recessive DCM. Consistent with the findings in our study, the patients with homozygous premature termination variants presented with DCM and heart failure. Similarly, the heterozygous carriers of MYZAP variants were not reported to have a cardiac phenotype (Heliö et al. 2021). Furthermore, studies in animal models are also consistent with the role of MYZAP in cardiomyopathy. In a zebrafish model, the knockdown of MYZAP homolog led to severe cardiomyopathy and pericardial edema (Seeger et al. 2010). Moreover, Myzap deficiency in genetically engineered knockout mice led to a maladaptive response to pressure overload because of transverse aortic constriction associated with cardiac remodeling, heart failure, and premature death (Rangrez et al. 2016). Interestingly, a meta-analysis of genome-wide association studies (GWASs) of atrial fibrillation among 29,502 cases and 767,760 controls from Iceland and the UK Biobank has shown a suggestive association between the p.Gln254Pro missense variant in MYZAP and the risk of atrial fibrillation (Thorolfsdottir et al. 2018).
The EHT model is a valuable tool to assess the functional consequences of cardiac genes (Mannhardt et al. 2016). We used patient and healthy skin fibroblasts to reprogram into hiPSCs, which were then differentiated into cardiomyocytes. These hiPSC-CMs were used to generate EHTs for functional evaluation that revealed severe contractile abnormalities in patient-derived EHTs. A striking observation was the dramatic increase in relaxation time pointing toward relaxation dysfunction, which likely contributes to the DCM phenotype of this patient.
Limitations of our study include that we cannot rule out additional disease-causing genetic variants as well as the lack of an isogenic control for the patient-derived hiPSCs, as an attempt to correct the identified variant via CRISPR–Cas9 yielded no positive clones.
In summary, we present novel evidence to support the role of biallelic loss-of-function MYZAP variants in severe DCM. The presented functional characterization, immunohistochemistry, WB studies, properties of the variant and evidence from animal models, all strongly support MYZAP as a cause of DCM in humans.
METHODS
Genetic Analyses
In 2015, in proband II-1 at age of 17, a molecular genetic analysis utilizing the Blueprint Genetics Core Cardiomyopathy Panel did not show the presence of any pathogenic or likely pathogenic variants nor did it show the presence of any variants of uncertain significance.
Exome sequencing was performed in the affected proband (II-1, Fig. 1J) using Nextera Coding Exome capture targeting 37 Mb of exonic coding sequences (214,405 targets in RefSeq gene models). Sequencing of the captured library was performed on the HiSeq 2500 platform in the 2×100 reads paired-end sequencing mode. A median on target coverage exceeding 60× was reached, assuring a sufficient variant detection rate across most of the targeted regions (>98% of regions were covered at 20×). The coverage table is available in the Supplemental Information (Supplemental Table 1).
Sequencing results were analyzed using an in-house analysis pipeline, with the alignment to hg19 reference performed by the Burrows–Wheeler (BWA) aligner (v0.7.2) and the GATK v4.1.2 framework was used for variant calling (DePristo et al. 2011). Further details on the exome analysis are available in Supplemental Methods.
The identified MYZAP gene variant was submitted to Clinvar under accession number SCV000747431.1. Patient consent to deposit raw sequencing data was not granted.
Light and Electron Microscopy and Immunohistochemical Studies
After explantation, the heart muscle of the proband was analyzed by light and electron microscopy. Light microscopy examination showed diffused foci of myocardial fibrosis and lipocyte infiltration and nuclear degeneration with perinuclear vacuoles (Fig. 1C–F). Electron microscopy showed myelin figures in the myocardium and focal disorganization of the intercalated disc (Fig. 2G,H). Tissue submitted for electron microscopy was fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer, then processed in 1% osmium tetroxide, dehydrated in an acetone gradient, and embedded in an epoxy embedding medium. One-micrometer-thick sections were cut and stained with toluidine blue stain. Ultrathin sections were stained with UranyLess and lead citrate and examined with a FEI-MORGAGNI 268D electron microscope.
The details of the western blotting experiments and catalyzed reporter deposition immunohistochemistry are available in the Supplemental Methods.
Functional Analyses
Generation and Culture of hiPSC-Derived Cardiomyocytes
A skin biopsy from the patient was taken, washed in PBS, minced, and placed in a six-well plate in fibroblast medium [DMEM supplemented with 10% FBS, 2 mM l-glutamine, and 0.5% penicillin and streptomycin (all Life Technologies)]. Fibroblasts growing out of the explants were harvested, passaged, and expanded for cryopreservation. Fibroblasts at passage 5 were reprogrammed using the CytoTune-iPS 2.0 Sendai Reprogramming Kit (Thermo Fisher Scientific) following the manufacturer's instructions. Three of the clones of hiPSCs were karyotyped to validate the absence of chromosomal aberrations and differentiated into cardiomyocytes (hiPSC-CM) using a monolayer culture method as previously described (Mosqueira et al. 2018). For functional analysis, hiPSC-CMs were used to generate EHTs in a 24-well format, as reported before (Mosqueira et al. 2018). Briefly, EHTs were formed of one million hiPSC-CMs/EHT in a fibrin matrix consisting of 10 µL/100 µL Matrigel (BD Biosciences 256235), 5 mg/mL bovine fibrinogen (from the stock of 200 mg/mL in NaCl 0.9% [Sigma-Aldrich F4753] plus 0.5 µg/mg aprotinin [Sigma-Aldrich A1153], 2× DMEM, 10 µM Y-27632, and 3 U/mL thrombin [Biopur BP11101104]). EHTs were then maintained in culture at 37°C in 7% CO2 and 40% O2 (with media change on Monday, Wednesday, and Friday) in DMEM (10% heat-inactivated FCS [Gibco], 0.1% insulin [Sigma-Aldrich], 0.5% penicillin/streptomycin [Gibco], 0.1% aprotinin [Sigma-Aldrich]). Silicone racks (C001) and Teflon spacers (C002) needed for EHT generation and EHT analysis equipment (A001) were purchased from EHT Technologies GmbH.
Analysis of Contractile Force in EHTs
EHTs generated as detailed in the previous section were used for the analysis of contractile force as previously described (Mosqueira et al. 2018). In brief, at the age of 26-d-old, the 24-well plate carrying the EHTs was placed inside a transparent chamber of EHT analysis equipment where homeostatic temperature (37°C), CO2 (5%), and O2 (40%) were maintained. The beating of the EHTs was analyzed by a video camera placed above the chamber using automated video-optical recordings of polydimethylsiloxane (PDMS) postdeflection. The contractile force was determined using customized software (CTMV) based on the known mechanical properties of the PDMS. The contraction peaks were analyzed for force amplitude, time to peak (contraction time, TTP80%), and relaxation time (RT80%) at 20% above baseline.
Statistics
Functional parameters of EHTs were compared with a two-tailed Student's t-test. The threshold of significance (α) was set to 5%.
ADDITIONAL INFORMATION
Data Deposition and Access
The identified MYZAP gene variant was submitted to Clinvar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession number SCV000747431.1. Details of the variant identified in this study are also provided in Supplemental Table 1. Patient consent to deposit raw sequencing data was not granted.
Ethics Statement
The patient gave a written informed consent to participate in the study. All the procedures were in accordance with the ethical standards of the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The institutional ethics board at the University Medical Centre Ljubljana approved the publication of this case report.
Acknowledgments
A tissue sample used as a control for western blot and catalyzed reporter deposition was kindly provided by Prof. Frane Paic from University of Zagreb School of Medicine, Zagreb, Croatia. T.Ž. and I.B. are members of the European Reference Network for Rare Hereditary Metabolic Disorders (MetabERN)—Project ID No 739543.
Author Contributions
A.M. and B.P. conceptualized the project. A.M., A.Y.R., J.H., U.S., S.D.L, A.H., N.F., and T.Ž. established the methodology. A.M., A.Y.R., J.H., U.S., S.D.L, A.H., N.F., and T.Ž. performed the formal analysis. B.P., I.B., and N.F. acquired the funding. A.M., A.Y.R., M.Ć., D.Š., I.Š., M.U., M.Z., J.H., U.S., S.D.L, A.H., N.F., and T.Ž. investigated. A.M., A.Y.R., J.H., U.S., S.D.L, A.H., and T.Ž. visualized the project. B.P. supervised. A.M., A.Y.R., N.F., T.Ž., and I.B. wrote the original draft. All the authors critically reviewed and contributed to the present manuscript.
Funding
This study was supported by a Slovenian National Research Agency grant (P3-0326) and partly funded by the financial support of the University of Zagreb to I.B. for 2019, and a start-up grant received from medical faculty of the Christian-Albrechts University of Kiel to A.Y.R.
Competing Interest Statement
The authors have declared no competing interest.
Referees
Juha W. Koskenvuo
Anonymous
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- DePristo MA, Banks E, Poplin R, Garimella K V, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–501. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott P, Andersson B, Arbustini E, Bilinska Z, Cecchi F, Charron P, Dubourg O, Kühl U, Maisch B, McKenna WJ, et al. 2008. Classification of the cardiomyopathies: a position statement from the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J 29: 270–276. 10.1093/eurheartj/ehm342 [DOI] [PubMed] [Google Scholar]
- Heliö K, Mäyränpää MI, Saarinen I, Ahonen S, Junnila H, Tommiska J, Weckström S, Holmström M, Toivonen M, Nikus K, et al. 2021. GRINL1A complex transcription unit containing GCOM1, MYZAP, and POLR2M genes associates with fully penetrant recessive dilated cardiomyopathy. Front Genet 12: 2358. 10.3389/fgene.2021.786705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, et al. 2013. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–585. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak TSH, Lee Y-K, Tang CS, Hai JSH, Ran X, Sham P-C, Tse H-F. 2018. Coverage and diagnostic yield of whole exome sequencing for the evaluation of cases with dilated and hypertrophic cardiomyopathy. Sci Rep 8: 10846. 10.1038/s41598-018-29263-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannhardt I, Breckwoldt K, Letuffe-Brenière D, Schaaf S, Schulz H, Neuber C, Benzin A, Werner T, Eder A, Schulze T, et al. 2016. Human engineered heart tissue: analysis of contractile force. Stem Cell Rep 7: 29–42. 10.1016/j.stemcr.2016.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Williams E, Foulger RE, Leigh S, Daugherty LC, Niblock O, Leong IUS, Smith KR, Gerasimenko O, Haraldsdottir E, et al. 2019. PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat Genet 51: 1560–1565. 10.1038/s41588-019-0528-2 [DOI] [PubMed] [Google Scholar]
- McKenna WJ, Judge DP. 2021. Epidemiology of the inherited cardiomyopathies. Nat Rev Cardiol 18: 22–36. 10.1038/s41569-020-0428-2 [DOI] [PubMed] [Google Scholar]
- Michels VV, Moll PP, Miller FA, Tajik AJ, Chu JS, Driscoll DJ, Burnett JC, Rodeheffer RJ, Chesebro JH, Tazelaar HD. 2010. The frequency of familial dilated cardiomyopathy in a series of patients with idiopathic dilated cardiomyopathy. N Engl J Med 326: 77–82. 10.1056/NEJM199201093260201 [DOI] [PubMed] [Google Scholar]
- Mosqueira D, Mannhardt I, Bhagwan JR, Lis-Slimak K, Katili P, Scott E, Hassan M, Prondzynski M, Harmer SC, Tinker A, et al. 2018. CRISPR/Cas9 editing in human pluripotent stem cell-cardiomyocytes highlights arrhythmias, hypocontractility, and energy depletion as potential therapeutic targets for hypertrophic cardiomyopathy. Eur Heart J 39: 3879–3892. 10.1093/eurheartj/ehy249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriard J-C, Hirschy A, Ehler E. 2003. Dilated cardiomyopathy: a disease of the intercalated disc? Trends Cardiovasc Med 13: 30–38. 10.1016/S1050-1738(02)00209-8 [DOI] [PubMed] [Google Scholar]
- Ramchand J, Wallis M, Macciocca I, Lynch E, Farouque O, Martyn M, Phelan D, Chong B, Lockwood S, Weintraub R, et al. 2020. Prospective evaluation of the utility of whole exome sequencing in dilated cardiomyopathy. J Am Heart Assoc 9: e013346. 10.1161/JAHA.119.013346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampazzo A, Calore M, van Hengel J, van Roy F. 2014. Intercalated discs and arrhythmogenic cardiomyopathy. Circ Cardiovasc Genet 7: 930–940. 10.1161/CIRCGENETICS.114.000645 [DOI] [PubMed] [Google Scholar]
- Rangrez AY, Eden M, Poyanmehr R, Kuhn C, Stiebeling K, Dierck F, Bernt A, Lüllmann-Rauch R, Weiler H, Kirchof P, et al. 2016. Myozap deficiency promotes adverse cardiac remodeling via differential regulation of mitogen-activated protein kinase/serum-response factor and β-catenin/GSK-3β protein signaling. J Biol Chem 291: 4128–4143. 10.1074/jbc.M115.689620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roginski RS, Mohan Raj BK, Birditt B, Rowen L. 2004. The human GRINLIA gene defines a complex transcription unit, an unusual form of gene origanization in eukaryotes. Genomics 84: 265276. 10.1016/j.ygeno.2004.04.004 [DOI] [PubMed] [Google Scholar]
- Seeger TS, Frank D, Rohr C, Will R, Just S, Grund C, Lyon R, Luedde M, Koegl M, Sheikh F, et al. 2010. Myozap, a novel intercalated disc protein, activates serum response factor-dependent signaling and is required to maintain cardiac function in vivo. Circ Res 106: 880–890. 10.1161/CIRCRESAHA.109.213256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet M, Taylor MRG, Mestroni L. 2015. Diagnosis, prevalence, and screening of familial dilated cardiomyopathy. Expert Opin Orphan Drugs 3: 869–876. 10.1517/21678707.2015.1057498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorolfsdottir RB, Sveinbjornsson G, Sulem P, Nielsen JB, Jonsson S, Halldorsson GH, Melsted P, Ivarsdottir E V, Davidsson OB, Kristjánsson RP, et al. 2018. Coding variants in RPL3L and MYZAP increase risk of atrial fibrillation. Commun Biol 1: 68. 10.1038/s42003-018-0068-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The identified MYZAP gene variant was submitted to Clinvar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession number SCV000747431.1. Details of the variant identified in this study are also provided in Supplemental Table 1. Patient consent to deposit raw sequencing data was not granted.