Abstract
The expeditious growth in spatial omics technologies enables the profiling of genome-wide molecular events at molecular and single-cell resolution, highlighting a need for fast and reliable methods to characterize spatial patterns. We developed SpaGene, a model-free method to discover spatial patterns rapidly in large-scale spatial omics studies. Analyzing simulation and a variety of spatially resolved transcriptomics data showed that SpaGene is more powerful and scalable than existing methods. Spatial expression patterns identified by SpaGene reconstruct unobserved tissue structures. SpaGene also successfully discovers ligand–receptor interactions through their colocalization.
Spatial omics technologies map out organizational structures of cells along with their genomics, transcriptomics, proteomics, and epigenomics profiles, providing powerful tools for deciphering mechanisms of functional and spatial arrangements in normal development and disease pathology (Larsson et al. 2021; Longo et al. 2021; Marx 2021; Deng et al. 2022; Dhainaut et al. 2022; Ratz et al. 2022; Zhao et al. 2022). The collection of available approaches provides a wide spectrum of throughput and spatial resolution. Imaging-based approaches generally target preselected RNA or proteins at molecular and single-cell resolution, whereas sequencing-based approaches allow genome-wide profiling with limited spatial resolution (Lewis et al. 2021; Zhuang 2021). Recent advances in those approaches move the field rapidly into the direction enabling genome-wide detection with single-cell or subcellular resolution, presenting a significant computational challenge for scalable and robust methods to derive biological insights in the spatial context (Atta and Fan 2021).
One essential step in spatial omics analysis is to characterize spatial expression patterns and colocalization. Several methods have been developed to identify spatially variable genes (Edsgärd et al. 2018; Svensson et al. 2018; Sun et al. 2020a; Anderson and Lundeberg 2021; Miller et al. 2021; Zhu et al. 2021). Trendsceek uses permutation test to detect significant dependency between the spatial distribution of points and their expression levels based on marked point processes (Edsgärd et al. 2018). Sepal ranks spatially variable genes by the diffusion time with the rationale that genes with spatial patterns require more time to reach a homogenous state than those with random spatial distributions (Anderson and Lundeberg 2021). SpatialDE and SPARK both use Gaussian process regression as the underlying data generative model for spatial covariance structures. SpatialDE decomposes expression variability into spatial variance and noise, and estimates statistical significance by comparing the likelihoods with and without a spatial component (Svensson et al. 2018). SPARK extends SpatialDE via generalized linear spatial error models, with the ability to directly model raw counts and adjust for covariates (Sun et al. 2020a). SPARK-X examines the similarity of expression covariance matrix and distance covariance matrix and tests whether they are more similar than expected by chance (Zhu et al. 2021). The statistical power of such methods highly depends on spatial covariance models, that is, how well they match true underlying expression patterns. Although multiple kernels, including Gaussian, linear, and periodic kernels with different smoothness parameters, are considered to ensure identification of various spatial patterns, statistical power will be compromised substantially for identifying spatial patterns poorly modeled by those predefined kernel functions. Furthermore, spatial covariance models are built on cellular distances, which would confound true expression variances with those driven by variances in cellular densities. To take nonuniform cellular densities into consideration, MERINGUE calculates spatial autocorrelation and cross-correlation based on spatial neighborhood graphs to identify spatially variable genes and gene interactions (Miller et al. 2021). Above all, even equipped with computationally efficient algorithms, it would still take days to months for most methods to analyze large-scale spatial data with genome-wide profiling in tens of thousands of locations (Zhu et al. 2021), resulting in a high demand for scalable and robust methods for characterizing spatial expression patterns.
To address those limitations, we aim to develop a scalable and model-free method for detecting spatial patterns. Without making assumptions on spatial covariance models and data distributions, the method will have more degree of freedom and also be more computationally efficient in identifying spatial patterns than existing methods.
Results
Overview of SpaGene
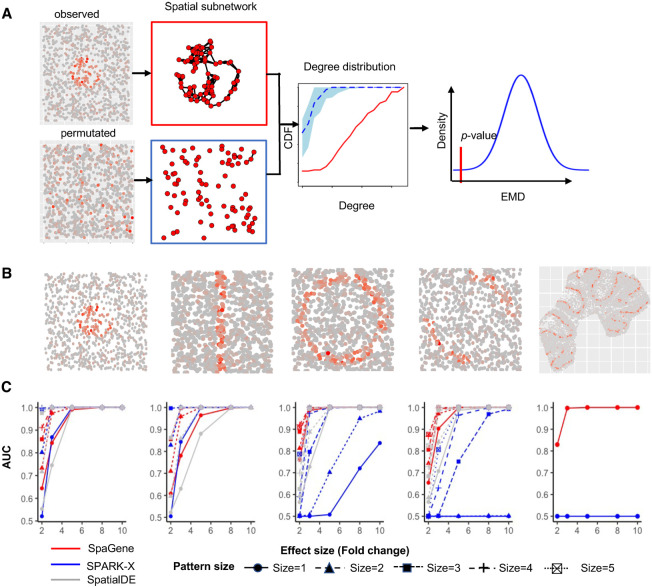
SpaGene is built on a simple intuition that spatially variable genes have uneven spatial distributions, meaning that highly expressed cells/spots tend to be more spatially connected than random. Given a set of spatial locations, SpaGene first builds the spatial network using k-nearest neighbors. For each gene, SpaGene then extracts a subnetwork comprising only cells/spots with high expression of the gene from the k-nearest neighbor graph. SpaGene quantifies the connectivity of the subnetwork by the Earth mover's distance between degree distributions of the subnetwork and a fully connected one. Finally, SpaGene compares the observed and the expected distances from random permutations. Genes with significantly shorter distances than random are identified to be spatially variable (Fig. 1A).
Figure 1.
Schematic of SpaGene and simulation results. (A) Schematic of SpaGene. (B) Visualization of five spatial patterns. (C) Area under the curve (AUC) plots of SpaGene (red), SpatialDE (gray), and SPARK-X (blue) in simulated data sets with different effect sizes (x-axis) and pattern sizes (point shapes) and 10,000 genes and 1000 cells/locations. Simulated data were generated from negative binomial distributions.
Simulation
We first applied SpaGene on two simulation data sets. One simulation was generated from negative binomial distributions following SPARK-X (Zhu et al. 2021), the other was sampled from real data following Trendsceek (Edsgärd et al. 2018). Cells/spots with higher expression (spiked cells) were located in one of those five patterns: hotspot, streak, circularity, biquarter circularity, and Purkinje layer in mouse cerebellum (Fig. 1B). The distinctness of the pattern was determined by effect sizes, which were controlled by the fold change (FC) of expression in spiked cells compared with the background. The pattern size was determined by the percentage of spiked cells. Higher effect sizes and larger pattern sizes generated more distinct and bigger patterns, which were easier to be identified. Among the simulated genes, 500 genes display spatial patterns (details in the Methods section). The area under the curve (AUC) was used to measure the ability to distinguish between spatially and nonspatially variable genes.
We compared SpaGene with SpatialDE and SPARK-X. SpatialDE and SPARK-X both achieved high-computational efficiency and good performance in other studies and SPARK-X is the only method applicable to data with sample size exceeding 30,000 (Zhu et al. 2021). As expected, effect sizes are the major factor affecting performance. Larger effect sizes produced more distinct patterns, which were easier to be distinguished from random spatial distributions and resulted in higher AUC values. For hotspot and streak patterns, SpaGene, SpatialDE, and SPARK-X successfully distinguished spatially from nonspatially variable genes when patterns were distinct (AUC = 1 at FC ≥ 5 for hotspot and AUC = 1 at FC ≥ 8 for streak patterns). For less distinct patterns, SpaGene performed slightly better than SpatialDE and SPARK-X for smaller patterns, which obtained AUC of 0.64, 0.52, and 0.55 for SpaGene, SPARK-X, and SpatialDE, respectively, at FC = 2 and size = 1 in hotspot patterns, although SPARK-X outperformed SpatialDE and SpaGene for bigger patterns (size > 1) (Fig. 1C). For circularity and biquarter circularity patterns, SpaGene achieved much better performance than SpatialDE and SPARK-X. For the circularity pattern, SpaGene achieved AUC of 0.99 even for the smallest pattern at FC = 3 and AUC of 1 at FC ≥ 5. In comparison, SpatialDE only obtained AUC of 0.73 at FC = 3, and SPARK-X failed to distinguish spatially from nonspatially variable genes even at FC = 5 (AUC = 0.5) for the smallest pattern (size = 1). SpaGene and SpatialDE achieved AUC of 1 whereas SPARK-X only obtained AUC of 0.72 at FC = 8 and size = 1. Although the performance of SpatialDE and SPARK-X improved with increasing pattern sizes, SpaGene was more powerful than SpatialDE and SPARK-X (Fig. 1C). For the biquarter circularity pattern, SPARK-X failed even at the largest effect size for the two small patterns (AUC = 0.5 at FC = 10, size = 1 or 2), although SpaGene achieved AUC ≥ 0.9 and SpatialDE obtained AUC of 0.7–0.83 at FC ≥ 3 for any pattern sizes (Fig. 1C). For the Purkinje layer pattern, SPARK-X failed at any effect sizes (AUC = 0.5), although SpaGene achieved AUC of 0.81 at FC = 2, 0.99 at FC = 3, and 1 at FC ≥ 5 (Fig. 1C). SpatialDE was not applied in this setting because of long computational time. To summarize, SpaGene achieved good performance for all spatial patterns, which obtained AUC ≥ 0.98 at FC ≥ 3 for relatively big patterns (size > 1) and AUC close to 1 at FC ≥ 5 for any pattern sizes. In comparison, SPARK-X seemed to be very sensitive to pattern shapes, which worked well for hotspot and streak patterns, but not for circularity, biquarter circularity, and Purkinje layer patterns even when patterns were strongly distinct from the background. Furthermore, SpaGene was more robust against pattern sizes than SpatialDE and especially SPARK-X, which sometimes showed more power to identify indistinct and large patterns than small distinct patterns. For example, SPARK-X obtained AUC of 0.8 at FC = 3 and size = 3, but AUC of 0.7 even at FC = 8 and size = 1 for circularity patterns. SpatialDE obtained AUC of 0.7 at FC = 3 and size = 1, but 0.82 at FC = 2 and size = 5 for biquarter circularity patterns. We also simulated scenarios with varying number of genes and cells/locations (Supplemental Figs. S1–S5). We found that the performance of SpaGene was less dependent on the number of cells/locations compared with SpatialDE and SPARK-X. The evaluation on the simulation data sets sampled from real data obtained similar results (Supplemental Figs. S6–S9).
In terms of time complexity, SpaGene and SPARK-X are much more computationally efficient than SpatialDE. SpatialDE requires several orders of computational time than SpaGene and SPARK-X, and its runtime increases linearly or cubically with the number of genes and the number of cells/locations (Supplemental Fig. S10A). For example, it takes SpatialDE 4045 sec to analyze a data with 10,000 genes and 5000 cells/location, although it only takes SpaGene and SPARK-X 11 sec and 22 sec, respectively. Additionally, SpaGene and SPARK-X require less memory than SpatialDE. SPARK-X and SpaGene require 0.5 G and 0.6 G memory, respectively, whereas SpatialDE demands 1.6 G memory to analyze a data with 10,000 genes and 5000 locations (Supplemental Fig. S10B).
Application to MOB by spatial transcriptomics
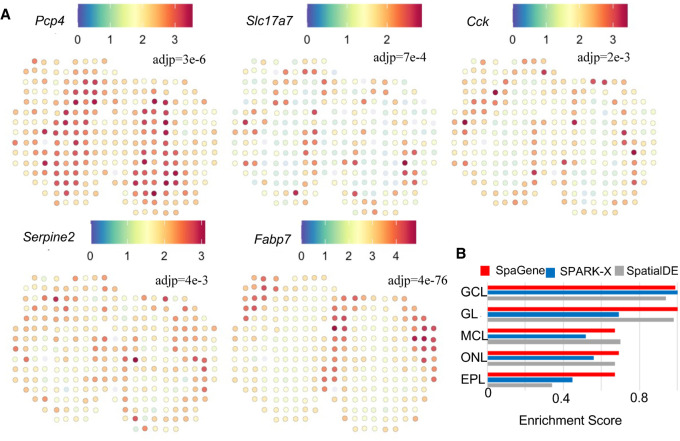
We applied SpaGene to spatial transcriptomics data from main olfactory bulb (MOB) (Ståhl et al. 2016), involving 16,218 genes measured on 262 spots. The MOB has a roughly concentric arrangement of seven-cell layers (Nagayama et al. 2014). SpaGene identified 634 spatially variable genes (adjusted P-value, adj P < 0.05), including genes known to be located in specific layers. Several examples were shown in Figure 2A, such as Pcp4 in granule cell layer (GCL) (adj P = 3×10−6) (Sangameswaran et al. 1989), Slc17a7 in mitral cell layer (MCL) (adj P = 7×10−4) (Zhang et al. 2021), Cck in glomerular layer (GL) (adj P = 2×10−3) (Sun et al. 2020b), Serpine2 in external plexiform layer (EPL) (adj P = 4×10−3) (Mansuy et al. 1993), and Fabp7 in olfactory nerve layer (ONL) (adj P = 4×10−76) (Young et al. 2013). Based on those identified spatially variable genes, SpaGene successfully reconstructed the underlying seven-layered MOB structure (Supplemental Fig. S11). To be noted, SpaGene identified a pattern corresponding to subependymal zone (SEZ) (pattern 4 in Supplemental Fig. S11). SEZ was unidentifiable by transcriptional profiles–based clustering, which only discovered five distinct clusters (Supplemental Fig. S12A). SEZ harbors neural stem cells. Sp9 is the top gene specifically located in SEZ, which is a transcription factor that regulates MOB interneuron development (Li et al. 2018).
Figure 2.
Application of SpaGene to spatial transcriptomics of main olfactory bulb (MOB) data. (A) Visualization of five known spatially variable genes located in specific MOB layers (high expression in red, and low in blue), with adjusted P-values from SpaGene. (B) Enrichment scores of markers in location-restricted cell types by SpaGene, SpatialDE, and SPARK-X.
We compared SpaGene with SPARK-X and SpatialDE. Overall, SpaGene and SpatialDE had more overlapping than SPARK-X (Supplemental Fig. S12B). The original study highlighted 15 genes differentially expressed in different domains (Ståhl et al. 2016). SpaGene detected 12 of 15 genes, whereas SPARK-X only found five and SpatialDE identified nine. Because cell clustering based on transcriptional profiles alone uncovered cell types located in MOB layers, genes highly expressed in each layer-specific cell clusters should be identified to be spatially variable. Using the top 20 markers from each of those cell clusters as the ground truth, SpaGene achieved a higher true-positive rate than SPARK-X and SpatialDE (Supplemental Fig. S12C). We also calculated scores to measure the enrichment of those top markers in SpaGene, SPARK-X, and SpatialDE. SpaGene obtained high-enrichment scores in all layers, suggesting it successfully identified all layer-specific marker genes as being very significant. In contrast, SPARK-X obtained high scores in GCL layers but low scores in other layers. SpatialDE achieved high scores in mitral cell layer, but relatively low scores in GCL and EPL layers (Fig. 2B). Moreover, we compiled the top 50 genes with enhanced expression in each layer of the MOB using the “differential search” in Allen Mouse Brain Atlas, which obtained 222 genes in total. Using the 222 genes as the ground truth, SpaGene also obtained a higher true-positive rate than SPARK-X and SpatialDE (Supplemental Fig. S12D). Finally, we ranked spatially variable genes by each method and carefully examined those genes identified to be very significant by one method but insignificant by another method. First, we ranked genes by SpaGene and listed the top six genes with inconsistent results (Supplemental Fig. S13). Kif5b, Atf5, Sorbs1, Plekhb1, and Mfap3l were detected to be very significant by SpaGene (adj P < 1×10−21), which were all specifically expressed in ONL (Supplemental Fig. S11). However, none of them were found by SPARK-X, although Atf5, Plekhb1, and Mfap3l were undiscovered by SpatialDE (Supplemental Fig. S13). Another gene, Grb2, was identified by SPARK-X but missed by SpatialDE, showing a very clear GCL pattern (Supplemental Fig. S13). Then we ranked genes by SPARK-X and checked the top six inconsistent ones (Supplemental Fig. S14). Camk2a, Psd3, Meis2, Calm2, Arf3, and Stxbp1 ranked high by SPARK-X, which displayed strong GCL patterns. All were identified by SpaGene but none by SpatialDE, indicating SpatialDE had limited power in identifying GCL-specific genes (Supplemental Fig. S14). Finally, we ranked genes by SpatialDE and examined the top six inconsistent ones (Supplemental Fig. S15). Spem1, Siglec1, and Il12a only expressed in one or two spots, which were likely to be false signals. Cck, Kif5b, and Apoe showed GL or ONL patterns, which were identified by SpaGene but missed by SPARK-X (Supplemental Fig. S15). These comparisons showed that SpaGene successfully identified genes with visually distinct patterns, whereas SPARK-X and SpatialDE missed some genes in certain layers even though they showed distinct patterns.
Application to mouse preoptic hypothalamus by MERFISH
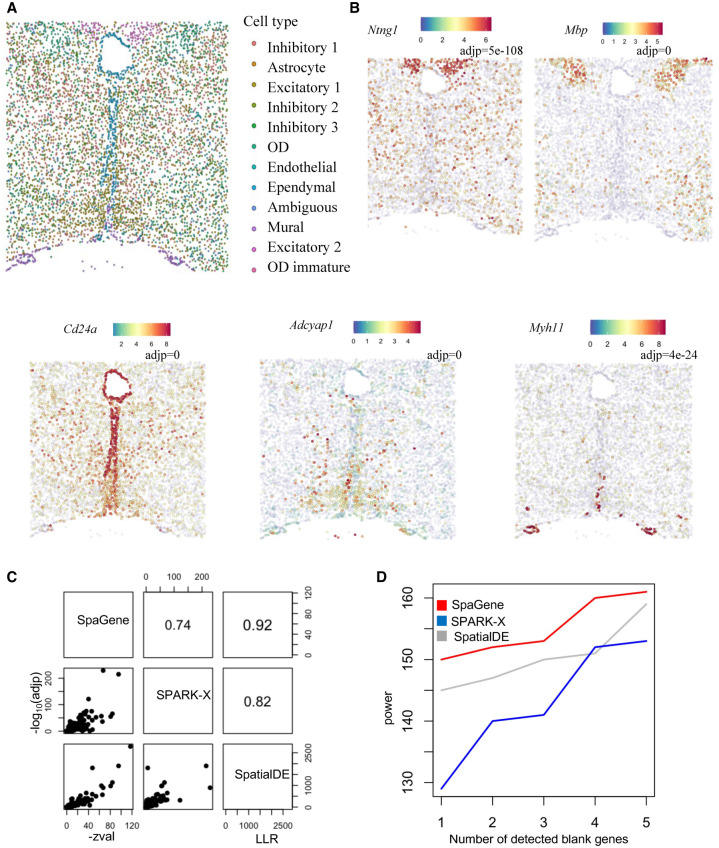
We applied SpaGene to mouse preoptic hypothalamus data by MERFISH (Moffitt et al. 2018), consisting of 161 genes measured on 5665 cells. The 161 genes include 156 preselected markers of distinct cell populations and five blank control genes. Cell clustering based on transcriptional profiles alone identified multiple cell types, most of which were spatially localized in specific regions, such as mature oligodendrocyte (OD), ependymal, mural, and some inhibitory and excitatory neuron cell types (Fig. 3A). SpaGene identified those markers from region-specific cell types as top variable genes. Some representative genes were shown in Figure 3B, such as Ntng1 in inhibitory neurons (adj P = 5×10−108), Mbp in mature OD (adj P = 0), Cd24a in ependymal (adj P = 0), Adcyap1 in excitatory neurons (adj P = 0), and Myh11 in mural cells (adj P = 4×10−24).
Figure 3.
Application of SpaGene to MERFISH of mouse preoptic hypothalamus data. (A) Cell clustering based on transcriptional profiles alone. (B) Visualization of five spatial variable genes (high expression in red and low in blue) with adjusted P-values from SpaGene. (C) Pairwise correlation of results from SpaGene, SpatialDE, and SPARK-X. (D) Power plot shows the number of genes with spatial expression pattern (y-axis) identified by SpaGene, SpatialDE, and SPARK-X versus the number of blank control genes identified at the same threshold.
Comparing SpaGene with SPARK-X and SpatialDE, we found their results were highly correlated in terms of significance (R = 0.92 between SpaGene and SpatialDE, R = 0.74 between SpaGene and SPARK-X, and R = 0.82 between SPARK-X and SpatialDE) (Fig. 3C). We also compared the number of positive genes given the number of negative control genes identified (Fig. 3D). The results supported a higher power of SpaGene. For example, SpaGene detected 149 true positives, whereas SpatialDE discovered 144 and SPARK-X revealed 128, when one negative control was detected (one false positive). Based on those identified spatially variable genes, SpaGene successfully reconstructed the underlying spatial organization (Supplemental Fig. S16).
Application to mouse cerebellum by Slide-seqV2
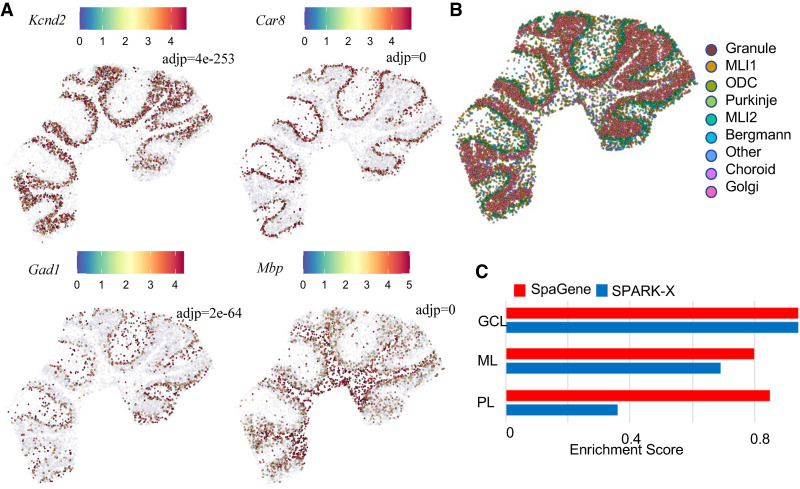
We applied SpaGene to mouse cerebellum data by Slide-seqV2 (Stickels et al. 2021), containing 20,141 genes measured on 11,626 spots. SpaGene identified 619 genes with spatial patterns (adj P < 0.05). The cerebellum is made of three layers, molecular, Purkinje, and granular layers from outer to inner, and white matter underneath. SpaGene detected genes, known to be specifically located in three layers and white matter, to be very significant, such as Kcnd2 in granular layer (adj P = 4e×10−253) (Varga et al. 2000), Car8 in Purkinje layer (adj P = 0) (Miterko et al. 2019), Gad1 in molecular layer (adj P = 2×10−64) (Kirsch et al. 2012), and Mbp in white matter (adj P = 0) (Fig. 4A; Verity and Campagnoni 1988). Based on those identified spatially variable genes, SpaGene successfully reconstructed the tightly folded layer structure of cerebellum. Patterns 1 and 3 corresponded to granular layer, patterns 2, 6, and 8 represented molecular layer, patterns 4 and 5 stood for Bergmann glia and Purkinje neurons in Purkinje layer, and pattern 7 imaged white matter (Supplemental Fig. S17).
Figure 4.
Application of SpaGene to Slide-seqV2 of mouse cerebellum data. (A) Visualization of four known spatially variable genes located in specific cerebellum layers (high expression in red, and low in blue), with adjusted P-values from SpaGene. (B) Cell clustering based on transcriptional profiles alone. (C) Enrichment scores of markers in location-restricted cell types by SpaGene and SPARK-X.
We compared SpaGene with SPARK-X but not SpatialDE because it would take hours to analyze such large-scale data. SPARK-X discovered 530 genes, whereas 230 overlapped with SpaGene (Supplemental Fig. S18). We examined carefully at those genes detected to be very significant by one method but insignificant by the other one (Supplemental Fig. S18). Those genes specifically located in Purkinje layer, such as Car8, Itpr1, Pcp2, and Pcp4, were detected as being the most significant by SpaGene (adj P = 0) but undetected by SPARK-X, suggesting SPARK-X had limited power to identify the Purkinje pattern (Supplemental Fig. S19). In comparison, Catsperd, Ifit3, and Ptprt ranked top by SPARK-X, but undetected by SpaGene, which did not seem to have obvious patterns (Supplemental Fig. S20). SpaGene estimated Mog at a significance level just below the cutoff (adj P = 0.05), whose expression seemed to be dispersed in the white matter (Supplemental Fig. S20).
Cell clustering based on transcriptional profiles alone found localized cell types, such as molecular layer neurons, Purkinje neurons in the Purkinje layer, and granule cells in the granule layer (Fig. 4B). We expected that markers in those spatially restricted cell types were identified and ranked top by the methods. The enrichment analysis found that SpaGene obtained high-enrichment scores in all three layers, although SPARK-X got a high score in granular layer, but low scores in other two layers, especially in the Purkinje layer. This result further showed that SpaGene is more robust to spatial patterns (Fig. 4C).
Although there were only 163 common genes between the 619 spatially variable and the top 2000 transcriptionally variable genes, cell clustering derived from these two gene sets were similar (Supplemental Fig. S21A). Clustering based on the spatially variable genes successfully found those cell types specifically located in the white matter, molecular, Purkinje, and granule layers (Supplemental Fig. S21B). We selected the top 2000 genes by integrating the spatially and transcriptionally variable genes. Clustering based on the integrative features improved clustering slightly, which showed a higher percentage of locations expressing cell type–specific marker genes (Supplemental Fig. S21C). The results suggested that spatially variable genes can serve as a complement to transcriptionally variable genes.
Application to MOB by HDST
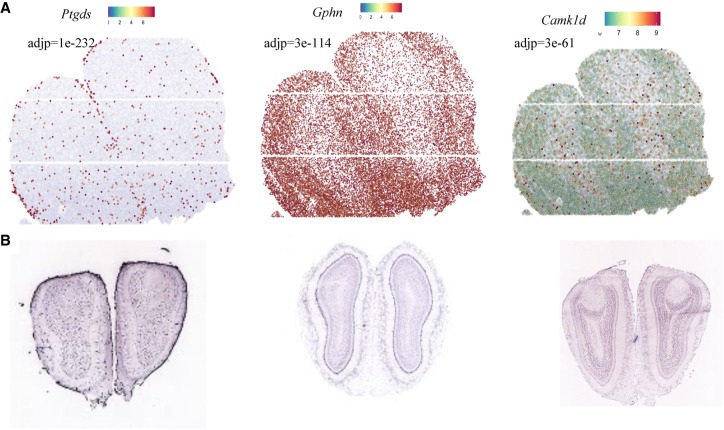
We applied SpaGene to olfactory bulb from high-definition spatial transcriptomics (HDST) (Vickovic et al. 2019), involving 19,950 genes measured on 181,367 spots. HDST is extremely sparse, in which only 21 spots have more than 50 genes detected. In this case, SpaGene used an adaptive strategy to expand the neighborhood search for genes with high sparsity. SpaGene identified 249 genes as being spatially variable. The most significant genes included Ptgds (adj P = 1×10−232), Gphn (adj P = 3×10−114), and Camk1d (adj P = 3×10−61). Although spatial patterns of those genes were not visually distinct owing to high sparsity of the HDST data (Supplemental Fig. S22), there were vague patterns showing Ptgds localized in ONL, Gphn in MCL and EPL, and Camk1d in GCL (Fig. 5A). Those specific localizations have been reported before (Rees et al. 2003; Perera et al. 2020) and validated by in situ hybridization in the Allen Brain Atlas (Fig. 5B).
Figure 5.
Application of SpaGene to high-definition spatial transcriptomics (HDST) data from main olfactory bulb (MOB). Visualization of three spatially variable genes. (A) Gene-expression levels from HDST (high in red, low in blue), with adjusted P-values from SpaGene. (B) In situ hybridization results for the three genes obtained from the Allen Brain Atlas.
We compared SpaGene with SPARK-X but not SpatialDE because it would take months to analyze such large-scale data. SPARK-X detected 133 genes, which overlapped significantly with SpaGene (90 in common). Among the 40 genes most associated with each MOB layer (top five genes in eight patterns in Supplemental Fig. S11), SpaGene found 12 genes (Ptgds, Fabp7, Gad1, Vtn, Kctd12, Kif5b, Apod, Pcp4, Gpsm1, Slc1a2, Nrgn, and Map1b), whereas SPARK-X only detected six (Ptgds, Fabp7, Kctd12, Kif5b, Apod, and Pcp4).
Identification of spatially colocalized ligand–receptor pairs
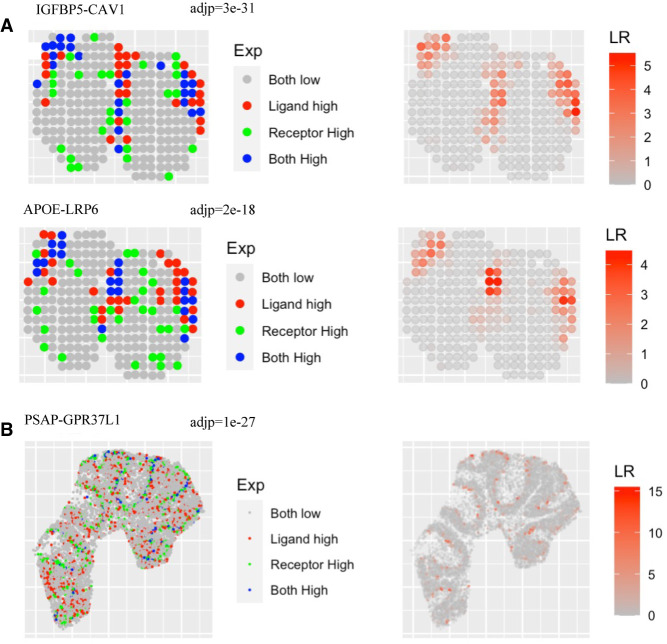
We extended SpaGene to identify cell–cell communications mediated by colocalized ligand and receptor pairs. SpaGene found 35 ligand–receptor interactions from the MOB data by spatial transcriptomics. The two most significant ligand–receptor pairs were IGFBP5-CAV1 (adj P = 3×10−31) and APOE-LRP6 (adj P = 2×10−18), both happening between ONL and GL. Apoe is known to be enriched in ONL and GL and also identified to be very significant by SpaGene (adj P = 1×10−50). Most spots with high Apoe expression were surrounded with spots with high Lrp6 expression (Fig. 6A), suggesting potential interactions between them. However, a number of spots with high Lrp6 expression were not adjacent to those with high Apoe expression, indicating other ligands might colocalize with Lrp6 as well. APOE-LRP6 mediates Wnt signaling, which is important for the regulation of synaptic integrity and cognition (Zhao et al. 2018). The identification of APOE-LRP6 between ONL and GL layers might be suggestive of the potential regulation of Wnt signaling in the establishment of periphery–CNS olfactory connections.
Figure 6.
Extension of SpaGene to identify ligand–receptor interactions. (A) Visualization of IGFBP5-CAV1 and APOE-LRP6 interactions for ST MOB data, with adjusted P-values from SpaGene. (B) Visualization of the PSAP-GPR37L1 interaction for Slide-seqV2 mouse cerebellum data, with the adjusted P-value from SpaGene. Left is the relative expression of the ligand and the receptor; right is the interaction strength.
SpaGene found 13 ligand–receptor interactions from the mouse cerebellum data by Slide-seqV2. The most significant pair was PSAP-GPR37L1 (adj P = 1×10−27) (Fig. 6B). Gpr37l1 was known to be strongly expressed in Purkinje layer and also identified by SpaGene (adj P = 8×10−130). Psap, in contrast, was not as specifically localized as Gpr37l1 (adj P = 6×10−8). PSAP-GPR37L1 protects neural cells from cellular damage (Li et al. 2017). The identification of PSAP-GPR37L1 between Purkinje layer and surrounding layers further supports its important role in brain function. Additionally, PTN-PTPRZ1, identified as the only interaction by MERINGUE (Miller et al. 2021), ranked the top four by SpaGene (adj P = 2×10−7).
Discussion
Recent advances in spatial omics technologies increase the demand for scalable and robust methods to characterize spatially variable patterns. Here, we developed SpaGene, a fast and model-free method to identify spatially variable genes. SpaGene has been extensively evaluated on seven data sets generated from a variety of spatial technologies, ranging from low to high throughput and spatial resolution. Additional analyses on breast cancer from spatial transcriptomics, mouse brain from 10x Visium, and olfactory bulb from Slide-seqV2 were shown in Supplemental Figures S23–S33. The results consistently showed that SpaGene successfully identified known spatially variable genes and also markers in spatially restricted cell clusters. Simple factor analysis on those identified genes reconstructed underlying tissue structures, further demonstrating the ability of SpaGene to characterize spatial patterns.
SpaGene builds on a simple intuition that spatially variable genes show uneven spatial distributions. As a model-free and distribution-free method, SpaGene is more robust to pattern shapes, data distribution and sparsity, nonuniform cellular densities, and the number of spatial locations than existing approaches. The power of SpatialDE, SPARK, and SPARK-X highly depend on spatial covariance models, that is, how well those predefined kernel functions match the true underlying spatial patterns. Moreover, SpatialDE and SPARK use parametric modeling based on the assumption of spatial data following Gaussian or Poisson distributions. Therefore, their performance would be compromised significantly for those genes whose expression misalign the model defined by those kernel functions and whose distribution violate Gaussian or Poisson distributions. SpaGene, in contrast, is a model-free and distribution-free method. Without any assumption, SpaGene is able to identify any spatial patterns and applied on any spatial omics data, such as identification of spatially localized clones and histone markers in spatial genomics and epigenomics data. The significance from SpaGene reflects the distinctness of spatial patterns rather than the extent of match to the defined model. SpaGene uses neighborhood graphs to represent spatial connections, making it more robust to nonuniform cellular densities common in tissues. Furthermore, SpaGene is highly computationally efficient in terms of runtime and memory requirement. It only took SpaGene seconds to minutes to analyze large-scale spatial transcriptomics data (Supplemental Fig. S10C), which required hours, days, or even months for most methods (Zhu et al. 2021).
SpaGene uses equal weights by default. Its power can be further improved if we adjust the weight parameter to assign unequal weights to different degrees (Supplemental Figs. S34A,B). Because clustered connections are more informative than scattered ones in defining spatial patterns, putting more weights on higher degrees strengthen the ability of SpaGene to distinguish visually distinct patterns from vague ones. For example, Nppa displayed a more distinct expression pattern than Smim36 (also known as Gm45716). Nppa is locally expressed in a specific region, whereas Smim36 is expressed everywhere. SpaGene with unequal weights successfully ranked Nppa much more statistically significant (adj P = 1×10−35) to be spatially variable than Smim36 (adj P = 1×10−8). SpaGene with equal weights, however, ranked the opposite (Supplemental Fig. S34C). Another example on Wfdc2 and Zfp235 was given in Supplemental Figure S34D. In general, the performance of SpaGene is insensitive to the parameter k to build the nearest-neighbor graph. The results were highly correlated across four different k-values (4, 8, 24, and 48) on three large-scale spatial transcriptomics data (Supplemental Fig. S35). For very sparse data, SpaGene provides an option to tune k-values automatically based on the expression sparsity of each gene. Moreover, SpaGene can incorporate the cell type information to find spatially variable genes within the same cell type. For example, SpaGene identified Aldoc as the most spatially variable gene within the Purkinje layer (adj P = 4×10−90) (the function SpaGene_CT was provided in the package), which has been demonstrated to show a regional enrichment pattern that was consistent with the known paths of parasagittal stripes across individual lobules (Kozareva et al. 2021). Furthermore, SpaGene was easily extended to find colocalized gene pairs. It successfully identified Psap-Gpr37l1 and Ptn-Ptprz1 in mouse cerebellum, and FN1-CD44 in invasive breast cancer regions (Supplemental Fig. S33). The default neighborhood search regions could be further adjusted to identify those long-distance interactions. Finally, potential extensions of SpaGene to find common and specific spatial patterns across multiple samples would further expand its application. SpaGene provides two functions FindPattern_Multi and PlotPattern_Multi to detect and visualize common and different patterns across samples. An example on two mouse brain data sets from anterior and posterior regions is provided at GitHub (see Software availability).
Although SpaGene is powerful in characterizing localized and colocalized patterns, it has some limitations. SpaGene binarizes gene expression into high and low, which increases the speed but loses the quantitative information of expression abundances. The binarization might underpower its performance on the identification of patterns with a gradient. SpaGene is able to identify long-distance interactions with a large k-value. However, it lacks the ability of modeling the diffusivity properties of ligands and receptors and their activity range.
Methods
Identification of spatially variable genes
Spatially variable genes show uneven spatial distribution of expression, in which cells/spots with high expression are more likely to be spatially connected than random. SpaGene constructs the k-nearest neighbor graph based on spatial locations. For each gene, SpaGene extracts a subnetwork comprising only cells/spots with high expression of the gene from the k-nearest neighbor graph. SpaGene quantifies the connectivity of the subnetwork using the Earth mover's distance between degree distributions of the subnetwork and a fully connected one. The degree distribution is more powerful and flexible than the total number of connections (Ren et al. 2020) to define spatial connectivity. The reason is that sparsely scattered connections are less informative and important than clustered ones in defining spatial patterns. For example, it is hard to shape a spatial pattern from a number of scattered connections. The utilization of degree distribution allows us to assign different weights to different degrees rather than treating them equally.
Earth mover's distance (EMDg) quantifies the distance from the observed degree distribution of the subnetwork of the gene g to a distribution from a fully connected network (Equation 1). Therefore, shorter EMD distances indicate higher spatial connectivity. The degree distribution is defined to be the fraction of cells/spots with degree of i in the subnetwork for the gene g, wi is the weight assigned to the degree of i, and k is the number of nearest neighbors to build the spatial network. Because clustered connections are more important than scattered ones in defining spatial patterns, at least equal or more weights should be assigned to higher degrees, that is, wi ≤ wj, if i ≤ j. EMD with equal weights (wi = 1, i = 0, 1,…2*k) is reduced to the average number of nonconnections.
| (1) |
To generate the null distribution of EMD, the same number of cells/spots is randomly sampled and the spatial connection of those cells/spots is quantified as EMD′. The mean and the standard deviation of EMD′ are estimated after random permutations (default: 500). The observed EMD is compared with the null distribution of EMD′ to evaluate its significance. The Benjamini–Hochberg procedure is used to adjust P-values for FDR control.
Identification of spatial patterns
Non-negative matrix factorization is applied on spatially variable genes detected by SpaGene to identify distinct spatial patterns. NMF is implemented by the RcppML R package (DeBruine et al. 2021). It is challenging to choose the optimal number of NMF factors. Although several approaches have been proposed (Brunet et al. 2004; Frigyesi and Höglund 2008; Hutchins et al. 2008), the computation is very lengthy and results from different approaches are inconsistent. Therefore, selecting the number of ranks based on the prior knowledge of the tissue structure is recommended. For example, the number of ranks of eight to 12 is recommended for ST MOB data with a rough arrangement of seven layers. The Spearman's correlation between expression of spatially variable genes and cells/spots factor matrix from NMF is used to find the most representative genes in each pattern.
Adaptive strategy to tune neighborhood search regions
SpaGene uses an adaptive strategy to expand neighborhood search regions in very sparse data sets, in which a single k-value to build the nearest neighbor graph will not work well for all genes. To improve sensitivity, SpaGene increases the k-value for genes with high sparsity. SpaGene groups genes into different bins (bj, j = 1, 2…J) based on the number of cells/spots with detected expression, in which different bins bj correspond to different k-values. In this way, SpaGene chooses the k-value automatically based on the sparsity level of the gene.
where J is the number of bins, determined by the maximum and the minimum number of cells/spots with detected expression that users set (nmax and nmin). bj is the bin j that one gene is assigned to by the number of cells/spots with the gene expression detected and kj is the corresponding k-value for the bin j. For example, if one gene has the number of cells/spots with detected expression greater than nmax, this gene is grouped into b1 with k1 = 8.
Identification of ligand–receptor interactions
SpaGene is extended to identify ligand–receptor interactions. For each ligand–receptor pair, SpaGene estimates the spatial connectivity of the subnetwork comprising only connections between cells/spots with both high expression of the ligand and the receptor. SpaGene uses the Earth mover's distance based on the degree distribution of the subnetwork to quantify its spatial connectivity.
Enrichment analysis of cell type–specific marker genes
Cell clustering based on transcriptional profiles alone discovers cell types localized in specific spatial regions. Therefore, marker genes in those spatially restricted cell types should be identified as spatially variable genes. The gene set is built from the top markers based on the fold change between the expression in the cell type compared with others. Top 20 are selected for ST MOB, whereas top 50 are chosen for other data sets. The results from SpaGene, SpatialDE, and SPARK-X are ranked from the most to the least significant. Unweighted gene set enrichment analysis (Subramanian et al. 2005) is implemented to evaluate the enrichment of the gene set in the high ranking of preranked gene lists of SpaGene, SpatialDE, and SPARK-X.
Simulation designs
We followed simulation designs of SPARK-X and Trendsceek. Briefly we generated two data sets with five spatial expression patterns: local hotspot, streak, circularity, biquarter circularity, and mouse Purkinje layer. For the first four patterns, spatial locations of cells were generated by a random-point-pattern Poisson process. The spatial locations of the pattern of mouse Purkinje layer was obtained from Slide-seqV2 mouse cerebellum data. The expression values were either generated from negative binomial distributions following SPARK-X or bootstrap-sampled from spatial transcriptomics MOB data following Trendsceek. Simulation data sets varied on a number of parameters: (1) the number of genes varied from 1000, 3000, and 10,000, of which 500 genes are spatially variable; (2) the number of cells varied from 300, 1000, 2000, and 5000 except for the Purkinje layer pattern; (3) the fold change of expression in the spatial region compared with those in the background. For the negative binomial distribution, the fold change varied from 2, 3, 5, 8 to 10. For the resampled real data set, the expression of spiked cells were generated from 65%, 70%, 80% to 90% quantile of the expression distribution; (4) the number of spiked cells except for the Purkinje layer pattern. For the hotspot and the streak patterns, the percentage of spiked cells varied from 5%, 10%, 20% to 30%. For the circularity and biquarter circularity patterns, the width of circularity varied between 0.05, 0.075, 0.1, 0.125, and 0.15.
Spatial transcriptomics data sets
SpaGene was applied on seven spatial transcriptomics data sets, covering a variety of platforms with low and high throughput and spatial resolution. Two spatial transcriptomics data from mouse olfactory bulb and human breast cancer contained genome-wide expression profiles on only hundreds of spots (low spatial resolution) (Ståhl et al. 2016). MERFISH on the mouse preoptic region of the hypothalamus targeted only 160 genes at single-cell resolution (Moffitt et al. 2018). 10x Visium on the mouse brain comprised whole transcriptomics on thousands of spots with a spatial resolution of 55 µm, which can be downloaded from the 10x Genomics website (https://support.10xgenomics.com/spatial-gene-expression/datasets). Two Slide-seqV2 from mouse cerebellum and olfactory bulb contained whole transcriptomics on tens of thousands of spots with a spatial resolution of 10 μm (Stickels et al. 2021). HDST from mouse olfactory bulb measured whole transcriptomics on hundreds of thousands of spots with a spatial resolution of 2 µm (Vickovic et al. 2019).
Software availability
SpaGene, an R package (R Core Team 2021), is freely available at GitHub (https://github.com/liuqivandy/SpaGene). Source codes and seven transcriptomics data are also available as Supplemental Code. Vignettes on seven spatial transcriptomics data with raw data, codes and results, including spatial variable genes identification, pattern identification and visualization, colocalized ligand–receptor pairs identification and visualization, are also available at the SpaGene GitHub repository.
Supplementary Material
Acknowledgments
This work is supported by National Cancer Institute Grants (U2C CA233291 and U54 CA217450), National Institutes of Health Grant (P01 AI139449), and Cancer Center Support Grant (P30CA068485).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276851.122.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Anderson A, Lundeberg J. 2021. sepal: identifying transcript profiles with spatial patterns by diffusion-based modeling. Bioinformatics 37: 2644–2650. 10.1093/bioinformatics/btab164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atta L, Fan J. 2021. Computational challenges and opportunities in spatially resolved transcriptomic data analysis. Nat Commun 12: 5283. 10.1038/s41467-021-25557-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet JP, Tamayo P, Golub TR, Mesirov JP. 2004. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci 101: 4164–4169. 10.1073/pnas.0308531101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruine ZJ, Melcher K, Triche TJ. 2021. Fast and robust non-negative matrix factorization for single-cell experiments. bioRxiv 10.1101/2021.09.01.458620 [DOI]
- Deng Y, Bartosovic M, Kukanja P, Zhang D, Liu Y, Su G, Enninful A, Bai Z, Castelo-Branco G, Fan R. 2022. Spatial-CUT&Tag: spatially resolved chromatin modification profiling at the cellular level. Science 375: 681–686. 10.1126/science.abg7216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhainaut M, Rose SA, Akturk G, Wroblewska A, Nielsen SR, Park ES, Buckup M, Roudko V, Pia L, Sweeney R, et al. 2022. Spatial CRISPR genomics identifies regulators of the tumor microenvironment. Cell 185: 1223–1239.e20. 10.1016/j.cell.2022.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edsgärd D, Johnsson P, Sandberg R. 2018. Identification of spatial expression trends in single-cell gene expression data. Nat Methods 15: 339–342. 10.1038/nmeth.4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigyesi A, Höglund M. 2008. Non-negative matrix factorization for the analysis of complex gene expression data: identification of clinically relevant tumor subtypes. Cancer Inform 6: 275–292. 10.4137/CIN.S606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins LN, Murphy SM, Singh P, Graber JH. 2008. Position-dependent motif characterization using non-negative matrix factorization. Bioinformatics 24: 2684–2690. 10.1093/bioinformatics/btn526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch L, Liscovitch N, Chechik G. 2012. Localizing genes to cerebellar layers by classifying ISH images. PLoS Comput Biol 8: e1002790. 10.1371/journal.pcbi.1002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozareva V, Martin C, Osorno T, Rudolph S, Guo C, Vanderburg C, Nadaf N, Regev A, Regehr WG, Macosko E. 2021. A transcriptomic atlas of mouse cerebellar cortex comprehensively defines cell types. Nature 598: 214–219. 10.1038/s41586-021-03220-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson L, Frisén J, Lundeberg J. 2021. Spatially resolved transcriptomics adds a new dimension to genomics. Nat Methods 18: 15–18. 10.1038/s41592-020-01038-7 [DOI] [PubMed] [Google Scholar]
- Lewis SM, Asselin-Labat ML, Nguyen Q, Berthelet J, Tan X, Wimmer VC, Merino D, Rogers KL, Naik SH. 2021. Spatial omics and multiplexed imaging to explore cancer biology. Nat Methods 18: 997–1012. 10.1038/s41592-021-01203-6 [DOI] [PubMed] [Google Scholar]
- Li X, Nabeka H, Saito S, Shimokawa T, Khan MSI, Yamamiya K, Shan F, Gao H, Li C, Matsuda S. 2017. Expression of prosaposin and its receptors in the rat cerebellum after kainic acid injection. IBRO Rep 2: 31–40. 10.1016/j.ibror.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang C, Zhang Z, Wen Y, An L, Liang Q, Xu Z, Wei S, Li W, Guo T, et al. 2018. Transcription factors Sp8 and Sp9 coordinately regulate olfactory bulb interneuron development. Cereb Cortex 28: 3278–3294. 10.1093/cercor/bhx199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo SK, Guo MG, Ji AL, Khavari PA. 2021. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat Rev Genet 22: 627–644. 10.1038/s41576-021-00370-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansuy IM, van der Putten H, Schmid P, Meins M, Botteri FM, Monard D. 1993. Variable and multiple expression of Protease Nexin-1 during mouse organogenesis and nervous system development. Development 119: 1119–1134. 10.1242/dev.119.4.1119 [DOI] [PubMed] [Google Scholar]
- Marx V. 2021. Method of the Year: spatially resolved transcriptomics. Nat Methods 18: 9–14. 10.1038/s41592-020-01033-y [DOI] [PubMed] [Google Scholar]
- Miller BF, Bambah-Mukku D, Dulac C, Zhuang X, Fan J. 2021. Characterizing spatial gene expression heterogeneity in spatially resolved single-cell transcriptomic data with nonuniform cellular densities. Genome Res 31: 1843–1855. 10.1101/gr.271288.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miterko LN, White JJ, Lin T, Brown AM, O'Donovan KJ, Sillitoe RV. 2019. Persistent motor dysfunction despite homeostatic rescue of cerebellar morphogenesis in the Car8 waddles mutant mouse. Neural Dev 14: 6. 10.1186/s13064-019-0130-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C, et al. 2018. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362: eaau5324. 10.1126/science.aau5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagayama S, Homma R, Imamura F. 2014. Neuronal organization of olfactory bulb circuits. Front Neural Circuits 8: 98. 10.3389/fncir.2014.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera SN, Williams RM, Lyne R, Stubbs O, Buehler DP, Sauka-Spengler T, Noda M, Micklem G, Southard-Smith EM, Baker CVH. 2020. Insights into olfactory ensheathing cell development from a laser-microdissection and transcriptome-profiling approach. Glia 68: 2550–2584. 10.1002/glia.23870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratz M, von Berlin L, Larsson L, Martin M, Westholm JO, La Manno G, Lundeberg J, Frisén J. 2022. Clonal relations in the mouse brain revealed by single-cell and spatial transcriptomics. Nat Neurosci 25: 285–294. 10.1038/s41593-022-01011-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rees MI, Harvey K, Ward H, White JH, Evans L, Duguid IC, Hsu CC, Coleman SL, Miller J, Baer K, et al. 2003. Isoform heterogeneity of the human gephyrin gene (GPHN), binding domains to the glycine receptor, and mutation analysis in hyperekplexia. J Biol Chem 278: 24688–24696. 10.1074/jbc.M301070200 [DOI] [PubMed] [Google Scholar]
- Ren X, Zhong G, Zhang Q, Zhang L, Sun Y, Zhang Z. 2020. Reconstruction of cell spatial organization from single-cell RNA sequencing data based on ligand-receptor mediated self-assembly. Cell Res 30: 763–778. 10.1038/s41422-020-0353-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangameswaran L, Hempstead J, Morgan JI. 1989. Molecular cloning of a neuron-specific transcript and its regulation during normal and aberrant cerebellar development. Proc Natl Acad Sci 86: 5651–5655. 10.1073/pnas.86.14.5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, et al. 2016. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353: 78–82. 10.1126/science.aaf2403 [DOI] [PubMed] [Google Scholar]
- Stickels RR, Murray E, Kumar P, Li J, Marshall JL, Di Bella DJ, Arlotta P, Macosko EZ, Chen F. 2021. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat Biotechnol 39: 313–319. 10.1038/s41587-020-0739-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhu J, Zhou X. 2020a. Statistical analysis of spatial expression patterns for spatially resolved transcriptomic studies. Nat Methods 17: 193–200. 10.1038/s41592-019-0701-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Liu X, Starr ER, Liu S. 2020b. CCKergic tufted cells differentially drive two anatomically segregated inhibitory circuits in the mouse olfactory bulb. J Neurosci 40: 6189–6206. 10.1523/JNEUROSCI.0769-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson V, Teichmann SA, Stegle O. 2018. SpatialDE: identification of spatially variable genes. Nat Methods 15: 343–346. 10.1038/nmeth.4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga AW, Anderson AE, Adams JP, Vogel H, Sweatt JD. 2000. Input-specific immunolocalization of differentially phosphorylated Kv4.2 in the mouse brain. Learn Mem 7: 321–332. 10.1101/lm.35300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verity AN, Campagnoni AT. 1988. Regional expression of myelin protein genes in the developing mouse brain: in situ hybridization studies. J Neurosci Res 21: 238–248. 10.1002/jnr.490210216 [DOI] [PubMed] [Google Scholar]
- Vickovic S, Eraslan G, Salmén F, Klughammer J, Stenbeck L, Schapiro D, Äijö T, Bonneau R, Bergenstråhle L, Navarro JF, et al. 2019. High-definition spatial transcriptomics for in situ tissue profiling. Nat Methods 16: 987–990. 10.1038/s41592-019-0548-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JK, Heinbockel T, Gondré-Lewis MC. 2013. Astrocyte fatty acid binding protein-7 is a marker for neurogenic niches in the rat hippocampus. Hippocampus 23: 1476–1483. 10.1002/hipo.22200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hernandez VS, Gerfen CR, Jiang SZ, Zavala L, Barrio RA, Eiden LE. 2021. Behavioral role of PACAP signaling reflects its selective distribution in glutamatergic and GABAergic neuronal subpopulations. eLife 10: e61718. 10.7554/eLife.61718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Liu CC, Qiao W, Bu G. 2018. Apolipoprotein E, receptors, and modulation of Alzheimer's disease. Biol Psychiatry 83: 347–357. 10.1016/j.biopsych.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Chiang ZD, Morriss JW, LaFave LM, Murray EM, Del Priore I, Meli K, Lareau CA, Nadaf NM, Li J, et al. 2022. Spatial genomics enables multi-modal study of clonal heterogeneity in tissues. Nature 601: 85–91. 10.1038/s41586-021-04217-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Sun S, Zhou X. 2021. SPARK-X: non-parametric modeling enables scalable and robust detection of spatial expression patterns for large spatial transcriptomic studies. Genome Biol 22: 184. 10.1186/s13059-021-02404-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X. 2021. Spatially resolved single-cell genomics and transcriptomics by imaging. Nat Methods 18: 18–22. 10.1038/s41592-020-01037-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.