Abstract
Argonaute 2 (AGO2) is a ubiquitously expressed protein critical for regulation of mRNA translation and vital to animal development. AGO2 protein is found in both cytoplasmic and nuclear compartments, and although its cytoplasmic role is well studied, the biological relevance of nuclear AGO2 is unclear. Here, we address this problem in vivo using spermatogenic cells as a model. We find that AGO2 transiently binds both chromatin and nucleus-specific mRNA transcripts of hundreds of genes required for sperm production during male meiosis in mice, and that germline conditional knockout (cKO) of Ago2 causes depletion of the encoded proteins. Correspondingly, Ago2 cKO males show abnormal sperm head morphology and reduced sperm count, along with reduced postnatal viability of offspring. Together, our data reveal an unexpected nuclear role for AGO2 in enhancing expression of developmentally important genes during mammalian male reproduction.
Cells control gene expression through multiple mechanisms, including regulation of chromatin state and transcription in the nucleus, mRNA export to the cytoplasm, and translation to protein. Argonaute (AGO) proteins play a major role in fine-tuning gene expression through post-transcriptional regulation (Hutvágner and Zamore 2002; Lee et al. 2004; Lim et al. 2005; Hornstein and Shomron 2006; Ebert and Sharp 2012). Among the four AGO proteins in mammals, Argonaute RISC Catalytic Component 2 (AGO2; also known as EIF2C2) is the best characterized and is essential for life, as whole-body mouse knockouts of Ago2 are embryonic lethal (Alisch et al. 2007). AGO2 canonically acts in the cytoplasm to inhibit target mRNA translation (Huang and Li 2014), but several roles for AGO2 in the nucleus have also been described. In Drosophila larvae and human fibroblasts, AGO2 mediates gene silencing by recruiting histone modifiers to target loci (Grimaud et al. 2006; Benhamed et al. 2012). AGO2 also regulates alternative splicing of pre-mRNAs in Drosophila and HeLa cells either by the coupling of RNA polymerase II (RNAPII) with histone modifications such as H3K9me3 or by an alternative unknown mechanism (Ameyar-Zazoua et al. 2012; Taliaferro et al. 2013). Lastly, AGO2 facilitates DNA double-strand break repair in Arabidopsis and cultured human cells (Wei et al. 2012). These studies point to critical nuclear roles for AGO2, but the in vivo relevance of these functions, particularly in mammals, remains poorly understood.
Mammalian spermatogenic cells represent an exceptional system to discover the in vivo nuclear contributions of AGO2. As mature sperm develop in mammals, the genome undergoes extensive chromatin reorganization and uses distinct mechanisms of transcriptional regulation. During meiosis, chromosomes condense and pair with homologs, and double-strand breaks are first induced and then repaired in a highly coordinated manner. After meiosis, the nucleus undergoes extreme compaction to form the mature sperm head, including substitution of most nucleosomes with specialized packaging proteins (Braun 2001). Spermatogenic cell nuclei therefore offer a unique opportunity to observe mechanisms of gene regulation that are latent or hard to detect in somatic cells.
The contribution of AGO2 to spermatogenesis overall remains unclear, and its nuclear activity in spermatogenic cells is completely unknown. Conditional knockout (cKO) of Ago2 in male germ cells was reported to have no phenotype: Knockout males were fertile and spermatogenesis was histologically normal, although compensation by one of the other three AGO family proteins could not be excluded (Hayashi et al. 2008). However, the AGO family member AGO4 is important for regulating entry into meiosis and silencing of sex chromosomes (Modzelewski et al. 2012), and knockout of Ago2 in mouse oocytes led to female sterility and impaired oocyte maturation (Kaneda et al. 2009), suggesting a crucial role for AGO proteins in gametes. In addition, cKO of Dicer and Drosha, which act upstream of Ago2 in the miRNA pathway, resulted in spermatogenic cell apoptosis, abnormal sperm morphology, and infertility, depending on the stage of spermatogenesis that was targeted (Hayashi et al. 2008; Maatouk et al. 2008; Romero et al. 2011; Wu et al. 2012). Together, the existing data suggest that the reported fertility of Ago2 cKO male mice may mask an undescribed role for AGO2 in spermatogenesis.
Here, we asked how AGO2 functions in the nucleus to control gene expression and what role this activity plays in male germ cell development. We analyzed protein, RNA, and chromatin interactions for AGO2 in the nucleus compared with the cytoplasmic compartment during spermatogenic development. We then investigated how conditional loss of AGO2 in male germ cells impacts expression of genes targeted by its nuclear activity. Finally, we examined the phenotypic effects of conditional AGO2 loss and the extent to which they depend on nuclear AGO2 function. We reveal that AGO2 activity contributes to normal male germ cell development and uncover an important and surprising role for nuclear AGO2 in positive regulation of gene expression during spermatogenesis.
Results
AGO2 interacts with distinct protein partners in male germ cell nuclei
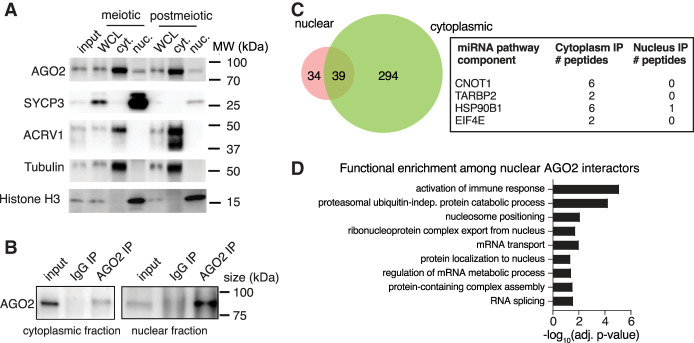
To determine if male germ cells offer a promising system for understanding AGO2 nuclear function, we first examined the subcellular localization of AGO2 protein in spermatogenic cells. We collected purified populations of cells at two stages of spermatogenesis: meiotic prophase (pachytene spermatocytes, hereafter “meiotic”) and following completion of meiotic divisions (round spermatids, hereafter “postmeiotic”) from adult mouse testes by gravity sedimentation (STA-PUT) (Bellvé et al. 1977; Bellvé 1993). Purity was estimated at 80%–85% for meiotic and 90% for postmeiotic populations based on visual examination of brightfield and Hoechst-stained cells (Supplemental Fig. S1A) as well as western blot for meiotic and postmeiotic markers (Supplemental Fig. S1B). We isolated nuclear and cytoplasmic fractions from each population and confirmed AGO2 protein localization in both the nucleus and cytoplasm of meiotic and postmeiotic male germ cells, in agreement with previous observations in other mammalian tissues (Fig. 1A; Kim et al. 2011; Gagnon et al. 2014; Ravid et al. 2020; Chu et al. 2021), suggesting that AGO2 can function in both the nucleus and cytoplasm at multiple steps of spermatogenic development.
Figure 1.
Nucleus-specific protein function for AGO2 in male germ cells. (A) Western blot for AGO2 protein in whole-cell lysate (WCL), cytoplasmic fractions, or nuclear fractions of meiotic (pachytene spermatocyte, SYCP3+) and postmeiotic (round spermatid, ACRV1+) male germ cells. Tubulin and histone H3 are found in cytoplasmic and nuclear fractions, respectively. Round spermatids have not yet begun histone-to-protamine exchange and therefore have similar levels of histone H3 compared with meiotic cells. (B) Immunoprecipitation of AGO2 for IP-MS in cytoplasmic and nuclear germ cell fractions validated by western blotting with anti-AGO2 antibody. One percent of total lysate was used as input. IP with mouse IgG is a negative control. (C) Common and compartment-specific AGO2 protein interactors in the nucleus and cytoplasm of wild-type postmeiotic cells. Box shows detected miRNA/siRNA pathway components along with the number of unique peptides identified in each compartment. (D) Statistically enriched Gene Ontology (GO) terms for AGO2 protein interactors in the nuclear compartment.
We next sought to determine if AGO2 has a specialized role in the germ cell nucleus compared with the cytoplasm. First, we performed AGO2 immunoprecipitation mass spectrometry (IP-MS) in cytoplasmic and nuclear fractions of postmeiotic cells to identify unique and common interactors in each compartment (Fig. 1B; Supplemental Table S1). As expected, four components of the miRNA/siRNA pathway (CNOT1, TARBP2, HSP90B1, and EIF4E) were detected as cytoplasmic AGO2 interactors (Fig. 1C), along with 329 other proteins enriched for numerous biological processes, including RNA localization (Supplemental Table S1). Of the 73 proteins that interacted with AGO2 in the nucleus, more than half were also cytoplasmic interactors (Fig. 1C). However, no known components of the miRNA/siRNA pathway precipitated with nuclear AGO2. Instead, the set of proteins that exclusively bound nuclear, but not cytoplasmic, AGO2 was enriched for splicing, ribonucleoprotein nuclear export, and nucleosome positioning (Fig. 1D; Supplemental Fig. S1C). We confirmed interaction between AGO2 and PTBP2 (a splicing factor), and between AGO2 and SMARCA5 (a chromatin remodeler) by coimmunoprecipitation in nuclear fractions from whole testis (Supplemental Fig. S1D). These data suggest that AGO2 operates via distinct pathways in the nuclear and cytoplasmic compartments.
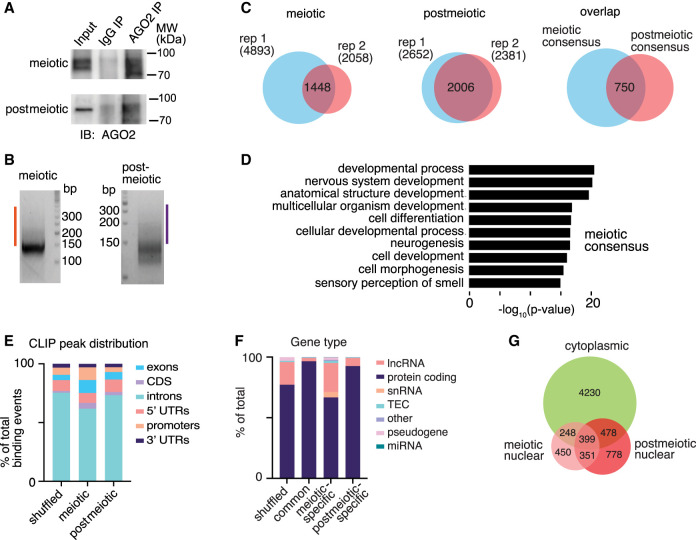
Nuclear AGO2 binds introns of unspliced mRNAs in nuclei of male germ cells
Given the known molecular role of AGO2 as an RNA-binding protein, we next asked if it interacts with specific RNA species in the nucleus. To identify AGO2 RNA-binding targets, we performed enhanced UV crosslinking and immunoprecipitation (eCLIP) (Van Nostrand et al. 2016) in nuclear fractions of meiotic and postmeiotic cells (Fig. 2A,B). AGO2 robustly bound 1448 and 2006 RNA targets in meiotic and postmeiotic nuclei, respectively (Fig. 2C; Supplemental Table S2). AGO2-bound nuclear transcripts were enriched for a variety of developmental and signaling functions in both cell types (Fig. 2D; Supplemental Fig. S2A). Seven hundred fifty RNAs were bound by AGO2 in both meiotic and postmeiotic nuclear samples (Fig. 2C; Supplemental Table S2), and these stable targets were enriched for a similar set of broad developmental and molecular functions (Supplemental Fig. S2B). Unlike cytoplasmic AGO2, which primarily binds to 3′ UTRs of target transcripts (Chi et al. 2009; Hafner et al. 2010), most nuclear AGO2 binding sites were located in intronic regions of unspliced transcripts (62% of CLIP peaks in meiotic cells and 74% of CLIP peaks in postmeiotic cells), consistent with a recent study in a human cancer cell line (Fig. 2E; Chu et al. 2021). Most nuclear RNA targets corresponded to protein-coding genes in both meiotic and postmeiotic cells (Fig. 2F).
Figure 2.
Identification of AGO2 mRNA targets in nuclei of male germ cells by eCLIP. (A) Immunoblot analysis of AGO2 immunoprecipitation. (B) Size selection of cDNA (orange and purple bars) following AGO2 IP, linker (147 nt) ligation, and PCR amplification. (C) Overlap of AGO2 targets from two eCLIP replicates from meiotic (left) and postmeiotic (center) cells, and transcripts identified in both replicates of both cell types (right). (D) GO analysis of meiotic consensus targets. (E) Genomic distribution of all AGO2 eCLIP binding regions. (F) Distribution of categories of bound transcript in both cell types (common), meiotic cells only, or postmeiotic cells only. Shuffled control distributions were generated by randomly shuffling a representative CLIP peak set across all annotated transcripts. (TEC) To be experimentally confirmed. (G) Overlap between cytoplasmic and nuclear AGO2 eCLIP targets.
To determine if nuclear AGO2 mRNA targets are distinct from cytoplasmic targets, we also collected eCLIP data from cytoplasmic fractions of whole testes, where the majority of cells correspond to either our meiotic or postmeiotic populations (Soumillon et al. 2013), and compared mRNAs bound by AGO2 in the cytoplasm and nucleus (Supplemental Fig. S2C,D). As expected, cytoplasmic AGO2 binding was enriched at 3′ UTRs and exons, compared with intronic enrichment for nuclear AGO2 (Supplemental Fig. S2E). The majority of nuclear AGO2-associated transcripts (64% of meiotic, 62% of postmeiotic, and 47% of shared targets) were not bound by AGO2 in the cytoplasm, although a substantial minority were associated with AGO2 in both compartments (Fig. 2G). These data support distinct nuclear and cytoplasmic AGO2–mRNA interactions in germ cells.
AGO2 interacts with open chromatin in meiotic male germ cells
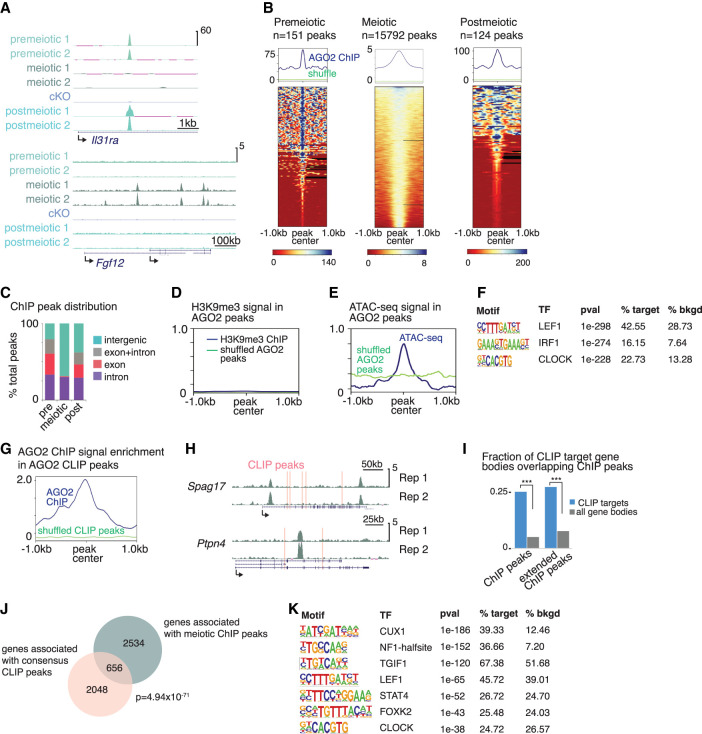
The association of AGO2 with introns in the nucleus prompted us to ask if it interacts with chromatin near sites of active transcription, consistent with reports in other cell types (Cernilogar et al. 2011; Moshkovich et al. 2011; Nazer et al. 2018). We first validated a chromatin immunoprecipitation followed by sequencing (ChIP-seq) protocol for AGO2 in mouse embryonic stem cells (mESCs) using two different antibodies to endogenous AGO2 (Supplemental Fig. S3A). We then evaluated AGO2 chromatin interactions by ChIP-seq in premeiotic (differentiating spermatogonia), meiotic, and postmeiotic cells (Fig. 3A,B; Supplemental Table S3). We further validated the specificity of our AGO2 antibody by performing ChIP-seq in Ago2 cKO male germ cells, confirming that ChIP signal was almost completely abrogated compared with the control (Fig. 3A; Supplemental Fig. S3B,C). In mESCs and in pre- and postmeiotic germ cells, we found relatively few peaks (148 peaks in mESCs, 151 peaks in premeiotic cells, and 124 peaks in postmeiotic cells) (Fig. 3B). In contrast, we identified thousands of broader, weaker peaks in meiotic cells (Fig. 3A,B; Supplemental Table S3). Notably, peaks were frequently shared across pre- and postmeiotic germ cells, but these locations rarely overlapped with binding sites identified in meiotic cells (Fig. 3A,B; Supplemental Fig. S3B). Meiotic peaks were more frequently intergenic, whereas premeiotic and postmeiotic peaks were most often found in gene bodies (Fig. 3C). Together, these data suggest that AGO2 interacts with chromatin in a developmentally dynamic manner during spermatogenesis.
Figure 3.
AGO2 interacts with chromatin and binds the corresponding nuclear transcripts in a developmentally dynamic manner. (A) Genome browser tracks (mm10) showing representative AGO2 ChIP peaks in two replicates from each cell type and from Ago2 cKO mixed meiotic and postmeiotic cells. (B) Enrichment of AGO2 on chromatin in premeiotic, meiotic, and postmeiotic male germ cells. (Top) Metagenes showing average ChIP signal at AGO2 peaks, with reference point set to the peak center. “Shuffle” shows ChIP signal when the same number of length-matched peaks were randomized across the genome. (Bottom) Heatmaps showing ChIP signal enrichment at each peak. (C) Distribution of AGO2 peaks in introns, exons, and intergenic regions. (D) Metagene of H3K9me3 ChIP signal at AGO2 peaks in meiotic cells. (E) Metagene of ATAC-seq signal at AGO2 peaks in meiotic cells. (F) Top motifs enriched in AGO2 ChIP peaks in meiotic cells. (G) AGO2 ChIP enrichment signal at eCLIP peaks in meiotic cells relative to shuffled eCLIP peaks. (H) Genome browser tracks (mm10) showing representative genes associated with both ChIP signal and eCLIP peaks (pink bars). (I) Fraction of meiotic CLIP target genes for which the gene body overlaps a meiotic ChIP peak. “Extended ChIP peaks” represent the same analysis performed with ChIP peaks extended by 10 kb in both directions. (***) P < 10−15, hypergeometric test. (J) Overlap between genes associated with meiotic ChIP peaks and genes containing eCLIP peaks in both meiotic and postmeiotic cells; P-value, hypergeometric test. (K) Motifs enriched in AGO2 ChIP peaks associated with genes that also contain eCLIP peaks. Only motifs matching a transcription factor expressed in meiotic cells are shown.
We next sought to determine if AGO2–chromatin interactions correlate with the transcriptional state of the nearby genome. AGO2 and other AGO family proteins can associate with heterochromatin and the repressive histone modification H3K9me3 in other organisms (Zilberman et al. 2003; Verdel et al. 2004; Ameyar-Zazoua et al. 2012). However, we found that H3K9me3 was not associated with AGO2 binding sites in our data (Fig. 3D), and there was no change in global levels of H3K9me3 in Ago2 cKO meiotic or postmeiotic cells (Supplemental Fig. S3D; H3K9me3 data from Liu et al. 2019). We then tested whether AGO2 binding sites were preferentially found in regions of transcriptional activity. We evaluated ATAC-seq signal strength, an indicator of open chromatin, at AGO2 peaks and found a moderate enrichment for open chromatin (Fig. 3E; ATAC-seq data from Maezawa et al. 2018). Although most expressed genes were not associated with AGO2 peaks, a substantial fraction of AGO2-associated genes were expressed (744/3190 (23%), TPM > 5), supporting a relationship between AGO2 and active transcription. Correspondingly, AGO2 binding was also enriched at short interspersed nuclear elements (SINEs) (Supplemental Fig. S3E), which gain the activation-associated histone modification H3K4me2 during spermatogenesis (Lambrot et al. 2019) and are associated with active regulatory elements (Faulkner et al. 2009; Tashiro et al. 2011; Nakajima et al. 2017; Policarpi et al. 2017).
In meiotic prophase, open chromatin is found both at regions of transcriptional activity and also at sites of recombination between homologous chromosomes (Baker et al. 2014; Lange et al. 2016). To distinguish between these states, we asked if AGO2 in meiotic cells preferentially localized to recombination hotspots. Only 15 meiotic AGO2 peaks overlapped previously defined recombination hotspots (Smagulova et al. 2016), compared with 280 overlaps with randomized AGO2 peaks. This finding reveals no significant association of AGO2 with recombination hotspots and indicates that AGO2 preferentially binds chromatin near sites of transcriptional activity in meiotic cells.
The finding that AGO2 binds both nuclear mRNAs (Fig. 2) and active chromatin suggested that its association with chromatin might be dependent on interaction with nascent transcripts. Because of the limited cell numbers available for spermatogenic cells, we addressed this question using mESCs. We treated mESC nuclei with an RNase cocktail to cleave any nascent transcripts and found that AGO2 binding did not change significantly in RNase-treated cells relative to control, implying that AGO2 interaction with chromatin does not require RNA transcripts (Supplemental Fig. S3F).
We then defined motifs enriched in AGO2 ChIP peaks in meiotic cells. Several enriched motifs matched binding sites for transcription factors expressed during male meiosis (Fig. 3F; Supplemental Fig. S3G), suggesting that one or more of these transcription factors may recruit AGO2 to target sites, similar to transcription factor–mediated recruitment of AGO2 to chromatin in other systems (Cernilogar et al. 2011; Moshkovich et al. 2011; Benhamed et al. 2012; Nazer et al. 2018). Notably, we also confirmed an interaction between AGO2 and the nucleosome remodeler SMARCA5 (also known as SNF2H) (Supplemental Fig. S1C,D), indicating that AGO2 may be recruited directly by the remodeling machinery, instead of or in addition to sequence-specific transcription factor binding.
AGO2 binds chromatin near hundreds of genes corresponding to AGO2-bound nuclear transcripts in meiotic cells
We next compared our ChIP and eCLIP data sets from meiotic cells to ask if there was a relationship between genomic sites and nuclear transcripts bound by AGO2. There was moderate enrichment for AGO2 chromatin binding at the specific genomic sites corresponding to AGO2 binding at mRNA transcripts in meiotic but not postmeiotic cells, determined by assessing AGO2 ChIP-seq signal intensity at eCLIP peaks (Fig. 3G; Supplemental Fig. S3H). The modest level of enrichment suggested variability in the exact placement of the ChIP peak relative to the CLIP peak when both occur at the same gene. Correspondingly, we consistently observed maximum ChIP signal near, but not directly overlapping, a CLIP peak (Fig. 3H; Supplemental Fig. S3I). Supporting this model, bodies of genes containing CLIP peaks were strongly enriched for ChIP peaks (odds ratio 7.75, P < 1015) (Fig. 3I). We identified 656 genes for which AGO2 binds chromatin at or near (<20 kb) the coding region and also binds the nuclear transcript (Fig. 3J; Supplemental Table S4). ChIP peaks associated with these genes were enriched for nine motifs matching transcription factors expressed in male meiotic germ cells (Fig. 3K), further refining a set of candidate protein partners that may direct AGO2 function on chromatin.
Nuclear AGO2 regulates the expression of spermatogenesis proteins
The association of AGO2 with hundreds of genes and transcripts during meiosis motivated us to ask whether loss of AGO2 affects expression of these nuclear targets and their encoded proteins. We deleted AGO2 in male germ cells using a Ddx4-Cre transgene (Gallardo et al. 2007; Hu et al. 2013) and a conditional Ago2 allele (O'Carroll et al. 2007) to eliminate AGO2 expression specifically in germ cells at birth. As expected, AGO2 protein expression was lost in adult spermatogenic cells (Supplemental Fig. S4A). We purified meiotic and postmeiotic germ cells from Ago2 cKO and control testes and evaluated changes in the germ cell proteome and transcriptome.
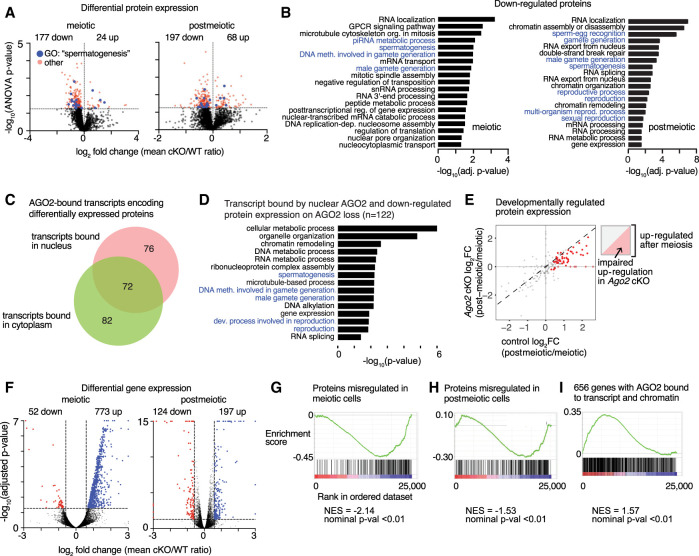
Despite the established function of AGO2 in reducing target protein expression, there was a strong bias toward protein down-regulation in both meiotic and postmeiotic Ago2 cKO germ cells (177/201 [88%] of meiotic and 197/265 [74%] of postmeiotic differentially expressed proteins) (Fig. 4A; Supplemental Fig. S4B; Supplemental Table S5). The sets of down-regulated proteins were significantly enriched for biological processes related to reproduction (Fig. 4B), whereas up-regulated proteins were not strongly enriched for any function (Supplemental Fig. S4C). These results imply that AGO2 plays a role in promoting spermatogenic protein expression, contrasting with its expected role in inhibiting translation.
Figure 4.
AGO2 regulates protein and transcript expression of male reproduction-related genes. (A) Volcano plots showing protein level changes in meiotic and postmeiotic male germ cells from Ago2 cKO animals. Statistically significant differentially expressed proteins (P ≤ 0.05) are shown by a dashed line and are highlighted in red. Differentially expressed proteins belonging to the GO category “spermatogenesis” (GO:0007283) are highlighted in blue. (B) Enriched GO terms associated with down-regulated proteins in Ago2 cKO meiotic and postmeiotic cells. GO terms related to male fertility are highlighted in blue. (C) Distribution of transcripts encoding differentially expressed proteins bound by AGO2 in nucleus, cytoplasm, or both. (D) Selected enriched GO terms for the set of genes with reduced protein expression in the Ago2 cKO and transcripts bound by AGO2 in meiotic or postmeiotic nuclei. (E) Changes in protein expression of meiotic AGO2 CLIP targets between meiotic and postmeiotic stages. Statistically significantly up-regulated proteins are in red. Cartoon plot illustrates that proteins in the upper right quadrant are up-regulated at postmeiotic stages, and up-regulation is attenuated in Ago2 cKO for proteins below the diagonal. (F) Volcano plots showing global transcriptional changes in Ago2 cKO male germ cells at meiotic (left) and postmeiotic (right) stages. Transcripts with log2 fold change ≤1.5 or ≥1.5 and adjusted P-value ≤ 0.05 are demarcated by dashed lines and highlighted in blue (up-regulated) or red (down-regulated). (G–I) Gene set enrichment analysis (GSEA) for differentially expressed transcripts relative to the set of proteins differentially expressed in meiotic cells (G), the set of proteins differentially expressed in postmeiotic cells (H), or genes for which AGO2 interacts with both chromatin and nuclear transcripts (I) (as shown in Fig. 3J).
Comparison with our eCLIP data revealed that AGO2 binds transcripts corresponding to 230 of the 444 total misregulated proteins (52%), either in the nucleus, cytoplasm, or both (76, 82, and 72 genes, respectively) (Fig. 4C), implying that AGO2 binding is important for downstream protein expression. The set of 122 genes that are down-regulated at the protein level in the Ago2 cKO and whose transcripts are bound by nuclear AGO2 are enriched for functions in sperm development (Fig. 4D). To evaluate the significance of nuclear AGO2 transcript binding for developmental protein expression, we examined protein expression for the complete set of 1448 genes whose nuclear transcripts are bound by AGO2 in meiotic cells (Fig. 2C). Among the subset of nuclear AGO2 targets detectable at the protein level in our data (n = 190), 30% (57/190) were significantly up-regulated between the meiotic and postmeiotic stages (Supplemental Table S6). Developmental protein up-regulation was impaired in the absence of AGO2, and transcripts for half of these genes (49%) were bound by AGO2 specifically in the nucleus and not in the cytoplasm (Fig. 4E; Supplemental Fig. S4D), supporting the functional importance of nuclear AGO2 in promoting developmental regulation of proteins important for spermatogenesis. A luciferase reporter assay for intronic AGO2 binding sites identified by eCLIP further revealed a contribution of these sites to regulation of protein-level expression in a cell line derived from meiotic spermatogenic cells (GC2spd(ts)) (Hofmann et al. 1994), although the direction and strength of the effect dependent on the specific target site used (Supplemental Fig. S4E,F). Together, our data imply that nuclear AGO2 binds mRNA transcripts in the nucleus and selectively promotes protein-level expression of genes important for spermatogenesis.
Down-regulated proteins are associated with down-regulated transcripts in Ago2 mutants
We then asked whether the effect of Ago2 cKO on spermatogenic protein levels was explained by changes in the level of mRNA transcripts. We collected RNA-seq data from meiotic and postmeiotic cells from Ago2 cKO and wild-type control males. In meiotic cells, 94% (773/825) of differentially expressed genes (DEGs) were up-regulated, contrasting with protein-level effects (Fig. 4F; Supplemental Table S7), whereas postmeiotic DEGs had no bias toward up- or down-regulation. Meiotic up-regulated transcripts were enriched for Gene Ontology (GO) terms related to male germ cell development (Supplemental Fig. S4G), and there was no functional enrichment among postmeiotic or down-regulated meiotic DEGs (Supplemental Table S7).
We applied gene set enrichment analysis (GSEA) (Mootha et al. 2003; Subramanian et al. 2005) to evaluate the relationship between changes in mRNA and protein levels. Despite the strong bias toward up-regulation of transcripts in meiotic cells, changes in protein expression were associated with the minority of down-regulated transcripts, consistent with the trend toward down-regulation of proteins. There was an association between down-regulated transcripts in meiotic cells and changes in protein expression both before and after meiosis (Fig. 4G,H), implying that there is a developmental delay between altered mRNA levels and some protein-level effects. The set of all meiotic AGO2 ChIP-seq peaks was not significantly associated with DEGs, indicating that many differences in transcript level may be indirect effects of Ago2 loss. However, GSEA revealed enrichment for the set of 656 genes associated with nuclear AGO2 at both the chromatin and transcript levels (Fig. 3J) among DEGs (Fig. 4I; Supplemental Fig. S4H), supporting a model in which AGO2 associates with chromatin and stabilizes nascent transcripts at a subset of these regions.
Together, our results indicate that nuclear and cytoplasmic AGO2 regulates different sets of genes and has different regulatory effects on their targets. One-fifth (18%) of the genes whose expression at the protein level depends on AGO2 produce transcripts bound by AGO2 exclusively in the nucleus, and one-half (49%) of AGO2 target genes that are developmentally regulated between meiotic and postmeiotic stages are bound by AGO2 exclusively in the nucleus. These findings support separable contributions of nuclear and cytoplasmic AGO2 activities to protein expression in male germ cells.
AGO2 plays a limited role in splicing and nuclear export of bound transcripts in male germ cells
In addition to its association with chromatin, our IP-MS data indicate that AGO2 also interacts with the RNA splicing machinery and the nuclear pore complex (Fig. 1D; Supplemental Fig. S1C). We therefore evaluated a possible function for AGO2 in splicing and nuclear export in male germ cells. Using an established pipeline to evaluate splicing differences in our RNA-seq data sets (Shen et al. 2012, 2014), we found that a modest number of transcripts undergo altered splicing in Ago2 cKO germ cells (Supplemental Fig. S5A; Supplemental Table S8). Splicing differences are relatively small in magnitude (Supplemental Fig. S5B–D), and a minority (17%) of differentially spliced transcripts were directly bound by AGO2 in our eCLIP data. Differentially spliced transcripts were not enriched for reproductive or spermatogenic functions, and there was no bias toward retention of variant exons in the absence of AGO2, contrary to a previous report in HeLa cells (Ameyar-Zazoua et al. 2012).
AGO2 protein also interacted with subunits of the nuclear pore and with the nuclear export factor NXF1 (Fig. 1D; Supplemental Fig. S1C), leading us to test if AGO2 facilitates export of bound transcripts from the nucleus to cytoplasm. We selected two transcripts bound by nuclear AGO2 in germ cells (Ak7 and Hmgb2), along with two negative control unbound transcripts (Ift27 and Ppib), and evaluated nuclear/cytoplasmic transcript distribution semiquantitatively using single-molecule RNA FISH (Wang et al. 2012). We found a modestly increased ratio of nuclear to cytoplasmic transcripts for the AGO2-bound transcript Ak7, indicating increased nuclear retention (Supplemental Fig. S5E,F). Hmgb2 transcripts also showed a trend toward increased nuclear retention, although it was not statistically significant (P = 0.0949, Welch's t-test). The nuclear transcript fraction was unchanged for both control genes (Supplemental Fig. S5E,F). We conclude that although AGO2 is involved in regulating differential splicing and export of nuclear transcripts, neither function makes a strong contribution to gene expression or phenotype in male germ cells. These results suggest that nuclear AGO2 function is mediated primarily by its interaction with unspliced transcripts at chromatin.
Germline loss of AGO2 causes sperm head defects and alters offspring viability
Many of the genes bound by AGO2 at the chromatin and transcript levels in the nucleus and down-regulated at the protein level in the Ago2 cKO are required for sperm morphology and motility (Fig. 4D), leading us to predict that Ago2 cKO sperm show impaired development or function (Macleod 1971; Zhuang et al. 2014). We therefore tested for morphology and motility defects in Ago2 cKO sperm, after confirming down-regulation of a subset of spermatogenesis-associated DEPs by western blot (Fig. 5A).
Figure 5.
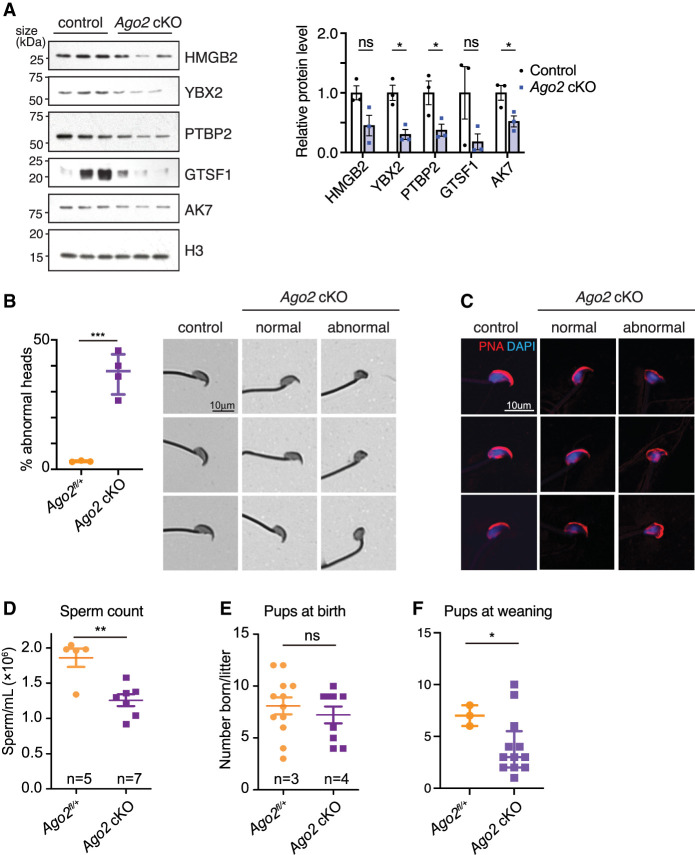
Loss of AGO2 impairs expression of proteins required for normal sperm production and results in oligospermia and abnormal sperm head morphology. (A) Western blot validation of spermiogenesis protein down-regulation. Bar chart shows levels of each protein relative to histone H3, quantified using densitometric analysis for three biological replicates per group. Error bars, SEM; (ns) not statistically significant, (*) P ≤ 0.05, Welch's t-test. (B, left) Percentage of Ago2 cKO spermatozoa with abnormal heads, representing n > 400 spermatozoa from each of three to four biological replicates. Points represent mean of each replicate, and bars show median and interquartile range of all replicates. (***) P < 0.001, Welch's t-test. (Right) Brightfield images of spermatozoa stained with Coomassie blue, showing representative cells collected from control and Ago2 cKO cauda epididymides. Both normal and abnormal examples of Ago2 cKO sperm are shown. (C) Representative spermatozoa stained with fluorophore-conjugated lectin-peanut agglutinin to visualize the acrosome (red) and DAPI to show DNA (blue). Both normal and abnormal examples of Ago2 cKO sperm are shown. (D) Epididymal sperm count in Ago2 cKO males and littermate controls. (E,F) Fertility in Ago2 cKO males and littermate controls, assessed as number of pups at birth (E) or at weaning (F). Error bars, SD; (*) P < 0.05, (**) P < 0.01, (***) P < 0.005, Welch's t-test.
We first tested for sperm morphology defects using Coomassie staining of cauda epididymal sperm and blinded scoring of sperm head shape. Ago2 cKO sperm had a significantly higher proportion of head morphology defects compared with the control (37% vs. 3%), predominantly in the form of small, rounded heads (Fig. 5B). Tail lengths were comparable between Ago2 cKO and control sperm (Supplemental Fig. S6A). Defects in formation of the acrosome, the organelle containing enzymes that break down the zona pellucida at fertilization, are the major cause of sperm head defects (Perrin et al. 2013), and lectin-peanut agglutinin staining of the acrosome showed that it was small and improperly formed in Ago2 cKO sperm (Fig. 5C). The set of head morphology defects observed in Ago2 cKO sperm parallels the defects expected following depletion of several of the proteins down-regulated in Ago2 cKO postmeiotic germ cells, including HMGB2, YBX2, and PTBP2 (Catena et al. 2006; Snyder et al. 2015; Hannigan et al. 2018; Lorès et al. 2018). Further, at least two of the transcription factors with binding motifs enriched among the set of dual AGO2 ChIP and eCLIP targets (NF1 and STAT4) (Fig. 3K) are known to regulate sperm head formation (Herrada and Wolgemuth 1997; Chohan et al. 2018). Phenotype enrichment analysis (Weng and Liao 2017) confirmed that down-regulated DEPs were statistically significantly (adjusted P-value ≤ 0.05) enriched for abnormal male germ cell morphology and male infertility. As expected based on the association between sperm head morphology defects and reduced sperm production, we observed a significant reduction in sperm count in Ago2 cKO males (Fig. 5D).
To test for effects on sperm motility, we assessed Ago2 cKO sperm using computer-assisted semen analysis (CASA). We found a 7% increase in the fraction of slow-moving sperm among Ago2 cKO cauda epididymal sperm after capacitation (functional sperm maturation), but no significant difference in overall progressive motility compared with the control (Supplemental Fig. S6B,C), leading us to conclude that loss of AGO2 has at most a minor effect on motility.
In agreement with a previous report (Hayashi et al. 2008), Ago2 cKO mice showed no obvious change in testis size, morphology, or testis-to-body weight ratio compared with the littermate controls, and spermatogenesis was grossly normal, with all spermatogenic cell types present in the seminiferous tubules (Supplemental Fig. S7A,B). There was no difference in numbers of apoptotic germ cells and no significant difference in the expression of transposable elements, which can cause spermatogenesis defects (Supplemental Fig. S7C,D; Webster et al. 2005; Carmell et al. 2007; Soper et al. 2008; Pastor et al. 2014). Loss of AGO2 was not strongly compensated by increased transcription of other family members (Supplemental Fig. S7E). In agreement with the normal fertility previously reported in Ago2 cKO male mice, Ago2 cKO males fathered the same number of pups as the littermate controls (Fig. 5E; Hayashi et al. 2008). This finding is consistent with the observation that male mice with sperm count reductions as large as 60% can still produce normal numbers of offspring under laboratory conditions (Kumar et al. 1997; Hudson et al. 1998; Schürmann et al. 2002; Santti et al. 2005; Zhang et al. 2006). However, although there was no difference in the number of neonates, we observed a reduction in the number of live pups at weaning (Fig. 5F), implying that altered AGO2 nuclear regulation during spermatogenesis may influence offspring physiology. Loss of Ago2 in the male germline therefore undermines sperm production and function despite the apparent fertility of Ago2 cKO males under laboratory conditions and may impair the ability of sperm to robustly withstand genetic and environmental perturbations.
Discussion
We report that AGO2 localizes to the nucleus of male germ cells and dynamically binds chromatin and nuclear transcripts of developmentally important genes during prophase of meiosis. Loss of AGO2 impairs expression of the encoded proteins, resulting in formation of morphologically abnormal sperm, decreased total sperm count, and impaired offspring viability. We propose a model (Fig. 6) in which AGO2 undergoes widespread, transient recruitment to active euchromatin, binds nearby nascent transcripts, and selectively promotes their stabilization and export from the nucleus. This function enhances expression of genes that contribute to spermatogenic development and contrasts with the canonical function of cytoplasmic AGO2 in down-regulating target gene expression.
Figure 6.
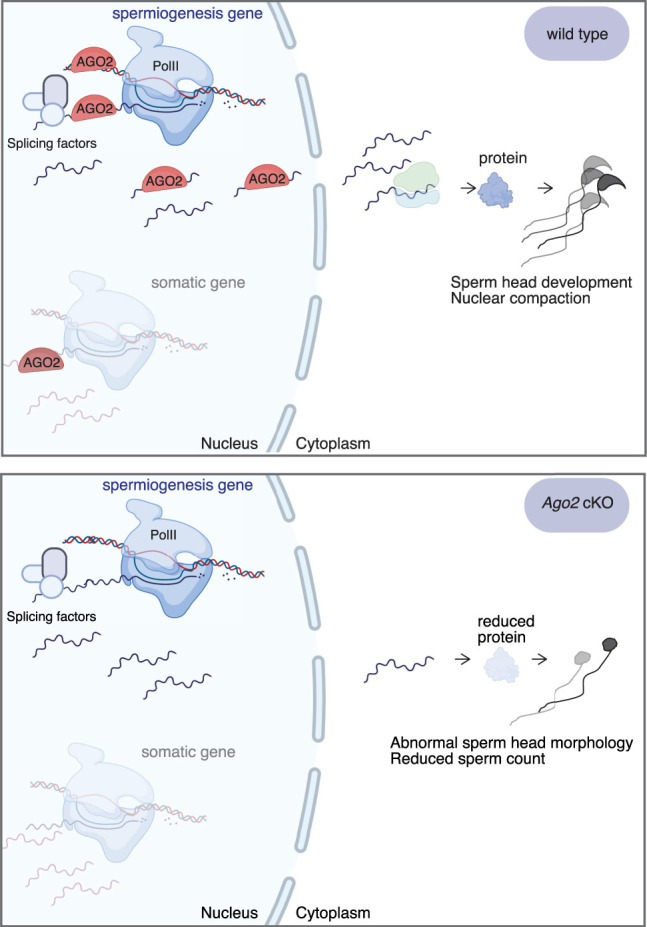
Model for RISC-independent nuclear AGO2 function in germ cells during meiotic prophase. AGO2 interacts with chromatin and binds nearby nascent transcripts. At genes important for spermatogenic development, AGO2 maintains its interaction with the nuclear transcript and helps to stabilize and shuttle these transcripts to the cytoplasm, eventually resulting in increased protein expression and promoting sperm production. Diagrams created with BioRender (https://biorender.com).
In human cell lines and Xenopus oocytes, cytoplasmic AGO2 can also have a positive effect on protein levels by acting in complex with fragile X mental retardation syndrome–related protein 1a (FXR1a) to promote translation in a miRNA-dependent manner (Vasudevan et al. 2007; Mortensen et al. 2011; Bukhari et al. 2016). This function could contribute to AGO2-dependent enhancement of protein expression for the subset of mRNA targets bound by AGO2 in both the cytoplasm and the nucleus of male germ cells. However, because nuclear AGO2 binds a large fraction of the transcripts encoding down-regulated proteins, including many not bound in the cytoplasm, we suggest that its positive effect on protein expression is mediated primarily by its nuclear role. Future experiments in which AGO2 is excluded from the nucleus, but unaltered in the cytoplasm, will be required to definitively rule out cytoplasmic AGO2 function as an explanation for the sperm phenotype observed in the Ago2 cKO.
Additional work will also be needed to fully define the molecular mechanism by which AGO2 promotes protein expression of its nuclear targets. Loss of AGO2 did not have a strong effect on splicing in male germ cells, in contrast to previous reports (Ameyar-Zazoua et al. 2012; Tarallo et al. 2017; Nazer et al. 2018; Chu et al. 2021). We found that AGO2 may play a role in export of transcripts from the nucleus in germ cells. A nuclear export function for AGO2 has not previously been described and merits deeper investigation in the future. We found several strongly enriched DNA sequence motifs in AGO2 ChIP peaks, implying that AGO2 may partner with specific transcription factors or transcription factor complexes at chromatin. Based on its interaction with the chromatin remodeler SMARCA5, AGO2 could also be recruited via direct interaction with the chromatin remodeling machinery, with sequence-specific transcription factors modulating its function on chromatin and interaction with target transcripts. Alternatively, sequence specificity could be mediated by RNA-dependent interactions, analogous to the role of noncoding RNAs in recruitment and stabilization of chromatin regulators at heterochromatin (Johnson et al. 2017; Velazquez Camacho et al. 2017). Although AGO2–chromatin interactions appear to be resistant to RNase digestion in mESCs (Supplemental Fig. S3F), RNAs tightly associated with AGO2 in the miRNA-binding pocket could be protected from RNase or could play a role in recruitment but be dispensable for subsequent stabilization of binding at chromatin.
Our data indicate that nuclear AGO2 activity in the male germ line occurs primarily during meiotic prophase, a cellular state unique to germ cells. Meiotic prophase can be broken down into four substages, each of which is associated with drastically different chromosome conformations and chromatin activities. Our “meiotic” cell population is primarily made up of cells in the pachytene stage of meiotic prophase; although technically challenging, it will be informative to further dissect the substages of meiotic prophase and to associate AGO2 nuclear function more precisely with the events of homologous recombination, transposable element repression, and sex chromosome silencing occurring during this time.
Canonically, cytoplasmic AGO2 associates with miRNAs to buffer gene expression against perturbations to normal cell function (Hornstein and Shomron 2006; Ebert and Sharp 2012; Strovas et al. 2014). The nuclear function of AGO2 function we identify here may contribute to maintaining robust germline gene expression in the context of environmental stressors, such as temperature or chemical exposure, and internal stressors, such as aging or inflammatory activity. If so, we predict that the relatively mild spermatogenesis phenotype found in Ago2 cKO mice would be enhanced under stress conditions. Alternatively, the mild phenotype could be explained by partial compensation of AGO2 loss by other members of the AGO family. Simultaneous depletion of multiple AGO family members in the male germline will help to answer this important question.
Finally, a critical question is whether AGO2 nuclear function is specific to the male germ line or whether it also occurs during developmental transitions in other cell types. On one hand, germ cells have unique requirements for gene regulation. AGO2 may participate in protection of gene expression programs during the massive chromosome rearrangements that occur during meiosis, when most large-scale chromatin compartments are lost (Alavattam et al. 2019). On the other hand, chromatin reorganization during meiosis may reveal general regulatory functions that are relevant but not immediately apparent in other cell types. Supporting a more generalizable role for nuclear AGO2 is the fact that many of its gene and transcript targets are involved in somatic, rather than spermatogenic, development. The majority of nuclear transcripts bound by AGO2 were not detectable at the protein level in germ cells and do not have known functions in spermatogenesis, potentially indicating relatively broad, non-tissue-specific regulation of immature nuclear mRNAs by AGO2 (Fig. 2D). In addition, 16 of the nuclear AGO2 protein partners we identify here were also found to interact with AGO2 in nuclei of mouse neurons (Ravid et al. 2020). Nuclear AGO2 may target a common set of genes across multiple cell types, with tissue-specific functions imposed by transcription factor partners or additional downstream regulatory mechanisms. Future work will help to untangle these possibilities and further elucidate the biological and molecular significance of nuclear AGO2 function within and beyond the male germ line.
Methods
Antibodies
ChIP and eCLIP were performed using a mouse monoclonal antibody (Millipore 04-642) that has been previously validated for ChIP-seq in human cells (Woolnough et al. 2015). A full list of antibodies is provided in the Supplemental Methods.
Cell culture
mESCs were grown under standard serum conditions and passaged 1:10 every 2–3 d. Details are provided in the Supplemental Methods.
Mouse strains and animal care
All mice were maintained on a C57BL/6J genetic background. Ago2flox/flox mice (B6.129P2(129S4)-Ago2tm1.1Tara/J) (O'Carroll et al. 2007) were obtained from Jackson Laboratory (strain 016520). Ago2flox/flox mice were mated with mice carrying one copy of the Ddx4-Cre transgene (B6-Ddx4tm1.1(cre/mOrange)Dcp) (Gallardo et al. 2007; Hu et al. 2013) in which Cre is expressed specifically in germ cells beginning at embryonic day 15.5, to generate Ago2 cKO males (Ddx4-Cre/+;Ago2flox/Δ). Primers used for genotyping are listed in Supplemental Table S9. These studies were approved by the Yale University institutional animal care and use committee under protocol 2020-20169. All mice used in these studies were maintained and euthanized according to the principles and procedures described in the National Institutes of Health guide for the care and use of laboratory animals.
Testis collection and cell dissociation
Seminiferous tubules were isolated by enzymatic digestion as described previously (Bryant et al. 2013). Male germ cells were obtained from seminiferous tubules using a second enzymatic digestion (0.25% trypsin [EDTA-free, Sigma-Aldrich], 2 mg/mL DNase I; Stem Cell Technologies 07900). Additional details are provided in the Supplemental Methods.
Spermatogenic cell isolation
STA-PUT was performed as described previously (Bellvé 1993). For flow cytometry isolation, round spermatids were sorted based on 1C DNA content and size, and differentiating spermatogonia (“premeiotic cells”) were isolated using an anti-KIT antibody directly conjugated to PE (eBioscience 12-1171-82). Additional STA-PUT and flow cytometry details are provided in the Supplemental Methods.
Morphological assessment of spermatozoa
Cauda epididymides were dissected from adult (8–16 wk old) male mice and allowed to swim out into PBS in a 12-well culture dish for 15 min and then were fixed and smeared onto a slide for staining and visual inspection. Additional details are provided in the Supplemental Methods.
Sperm motility assessment
Cauda epididymal sperm were collected by swim-out in HEPES buffered saline (HS) medium and analyzed by CASA (Sperm Class Analyzer version 6.3, Microptic). Over 200 motile sperm were analyzed for each trial. Three and two biological replicates were performed for the control and Ago2 cKO, respectively. Additional details are provided in the Supplemental Methods.
Nuclear/cytoplasmic fractionation
A pellet of 0.5–2 × 107 fresh spermatogenic cells was resuspended in 0.5–1 mLCell lysis buffer 1 (50 mM Tris-HCl at pH 7.4, 100 mM NaCl, 0.1% NP-40, with freshly added protease inhibitor cocktail) and incubated for 20 min on ice with inversion every 4 min. Cell pellets were then spun down at 1200g for 5 min at 4°C, and supernatants were kept as cytoplasmic lysate. Pellets were washed once with 1 mL of cell lysis buffer 2 (50 mM Tris-HCl at pH 7.4, 100 mM NaCl, 0.2% NP-40) and once with 1 mL of cell lysis buffer 3 (50 mM Tris-HCl at pH 7.4, 100 mM NaCl, 0.5% NP-40) and were incubated for 5 min on ice each time. Finally, the nuclear pellets were spun down at 1200g for 5 min at 4°C; supernatants were discarded; and pellets were resuspended with 500 μL of 1 mL of iCLIP lysis buffer (50 mM Tris-HCl at pH 7.4, 100 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate) and incubated for 5 min on ice. Lysates were centrifuged at 15000g for 15 min at 4°C, and the supernatants were collected as nuclear lysate.
Immunoblotting
Immunoblotting was performed using standard methods. Details are provided in the Supplemental Methods.
Coimmunoprecipitation of AGO2 protein interactors
Coimmunoprecipitation was performed using standard methods. Details are provided in the Supplemental Methods.
RNA isolation
RNA was isolated in TRIzol, followed by washing and elution using spin columns from the RNeasy MinElute kit (Qiagen 74104). The quality of the eluted RNA was determined by measuring 28S/18S ribosomal ratios and RNA integrity number (RIN) using an Agilent Bioanalyzer. Additional details are provided in the Supplemental Methods.
Real-time quantitative PCR
RT-qPCR was performed on an Applied Biosystems QuantStudio 3 real-time PCR system using Power SYBR Green PCR master mix (Applied Biosystems 4367659). Primer sequences are provided in Supplemental Table S9, and additional details are provided in the Supplemental Methods.
Luciferase assays
GC-2spd(ts) cells (ATCC CRL-2196) were transfected with pmirGLO dual-luciferase miRNA target expression vectors (Promega E1330) harboring sequences corresponding to targets of Ago2 binding at Ak7, Spag17, or Tdrd9 mRNA. The inserted sequences are centered on the eCLIP peak (12–30 bp) and extended in both directions for a final insert length of 400 bp (Supplemental Table S10). Firefly luciferase signals were normalized to Renilla luciferase measurements according to the manufacturer's instructions. Additional details are provided in the Supplemental Methods.
eCLIP
AGO2 eCLIP in spermatogenic cell fractions and whole-testis tissue was performed as described previously (Van Nostrand et al. 2016; Brugiolo et al. 2017; Biancon et al. 2022). Details are provided in the Supplemental Methods.
Chromatin immunoprecipitation
ChIP was performed as described previously (Lesch et al. 2013, 2016). Details are provided in the Supplemental Methods.
RNase treatment
mESC nuclei were treated with RNase according to a protocol adapted from Sigova et al. (2015). Additional details are provided in the Supplemental Methods.
Mass spectrometry analysis for AGO2 protein interactors
Nuclear and cytoplasmic fractions from wild-type round spermatids purified by STA-PUT were isolated as described above and incubated with anti-AGO2 antibody (Millipore 04-642) or normal mouse IgG as a negative control. Immunoprecipitated samples were then separated by SDS-PAGE, and the gel was stained with Coomassie blue. All bands except immunoglobulin heavy-chain and light-chain were excised from the gel and subjected to mass spectrometry protein identification at the Keck MS & Proteomics Resource at Yale University. The data were processed, and protein identification was searched using Proteome Discoverer (v. 2.2, Thermo Fisher Scientific) and Mascot search algorithm (v. 2.6, Matrix Science). A protein was considered identified when Mascot listed it as significant at a false-discovery rate of 1% and more than two unique peptides matched the same protein. Additional details are provided in the Supplemental Methods.
Quantitative mass spectrometry analysis of protein expression
Pachytene spermatocytes or round spermatids were isolated by STA-PUT as described above, and pellets containing 1–3 × 106 pachytene spermatocytes or 0.4–1 × 107 round spermatids were washed in PBS with protease inhibitors before sonicating in 400 µL of RIPA buffer and then processed for label-free quantitative mass spectrometry at the Keck MS & Proteomics Resource. The LC-MS/MS data were processed with Progenesis QI software (Nonlinear Dynamics, version 2.4) with protein identification performed using an in-house Mascot search engine (v2.6). A normalization factor for each run was calculated to account for differences in sample load between injections and for differences in ionization. Mascot search results from the MS/MS spectra were input into Progenesis QI software, where peptides were synched with the corresponding quantified features and their corresponding abundances. Protein abundances (requiring at least two unique peptides) were then calculated from the sum of all unique normalized peptide ions for a specific protein on each run. Additional details are provided in the Supplemental Methods.
High-throughput DNA sequencing
Details of quality control, library preparation, and sequencing are provided in the Supplemental Methods.
ChIP-seq data analysis
ChIP-seq data were quality-filtered with the fastq_quality_filter tool from FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit), aligned to the mouse genome build mm10 using Bowtie 2 (Langmead and Salzberg 2012), and used to call peaks with MACS2 at q = 0.05 (Zhang et al. 2008). Shuffled controls were generated using the shuffle function in BEDTools (Quinlan and Hall 2010). Additional details and specific parameters are provided in the Supplemental Methods.
RNA-seq data analysis
RNA-seq data were quality-filtered with fastq_quality_filter and quantified using kallisto v0.45.0 (Bray et al. 2016). DEGs were called using DESeq2 with batch correction (Love et al. 2014). Splicing was assessed using rMATS with default parameters (Shen et al. 2014), and downstream analysis was conducted using the maser package in R (https://github.com/DiogoVeiga/maser). Additional details and specific parameters are provided in the Supplemental Methods.
eCLIP data analysis
AGO2 IP and size-matched input control data were processed as previously described (Van Nostrand et al. 2016). Libraries were demultiplexed; adapters were trimmed using cutadapt v1.18 (Martin 2011); and libraries were filtered for repeat elements. Filtered libraries were aligned to mm10 using using STAR v.2.6.1d (Dobin et al. 2013). PCR duplicates were removed based on UMIs. Peaks were called using the tag2peak.pl script from a CLIP tool kit (CTK) (Shah et al. 2017) with an adjusted P-value threshold of 0.01 and normalized to the mm10 background model. eCLIP targets were confirmed by an additional independent peak calling pipeline (Biancon et al. 2022). Additional details and specific parameters are provided in the Supplemental Methods.
Metagene analysis
Metagene analysis was performed using deepTools version 3.5.1 (Ramírez et al. 2016). Read distributions were compiled around peak centers. Raw H3K9me3 ChIP-seq data from meiotic cells (pachytene spermatocytes) were obtained from the NCBI Gene Expression Omnibus (GEO: https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE137742 (Liu et al. 2019) and processed as described above. Raw ATAC-seq data from meiotic cells (pachytene spermatocytes) were obtained from GEO accession number GSE102954 (Maezawa et al. 2018) and processed as described above for ChIP-seq data. Locations of mouse recombination hotspots were obtained from DMC1 immunoprecipitation and sequencing-based detection of ssDNA (SSDS) data from GEO accession number GSE75419 (Smagulova et al. 2016) and intersected with meiotic AGO2 peaks using the intersect function from BEDTools (Quinlan and Hall 2010). Shuffled peaks were generated using the shuffle function from BEDTools.
Motif analysis
Motif analysis was performed using the findMotifsGenome.pl script in HOMER with default parameters to screen a library of known motifs (Heinz et al. 2010).
Functional enrichment analysis
GO enrichment analysis was performed using PANTHER GO annotations (https://doi.org/10.5281/zenodo.4495804 released: February 1, 2021) at the GO Consortium website (Mi et al. 2017; http://www.geneontology.org). GSEA was conducted as previously described using version 4.1.0 with default parameters (Mootha et al. 2003; Subramanian et al. 2005).
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE181264. The mass spectrometry proteomics data have been submitted to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2019) partner repository (http:// proteomecentral.proteomexchange.org/cgi/GetDataset) under the data set identifier PXD027935.
Supplementary Material
Acknowledgments
We thank P. Reddi for the gift of the ACRV1 antibody, J. Hwang for help with STA-PUT, Y. Kong for assistance with eCLIP analysis, S. Kataruka and S. Reilly for advice on luciferase assays, and S. Nicoli for critical reading of the manuscript. We thank Florine Collin from the MS & Proteomics Resource at Yale University for assistance with the proteomics sample preparation and data collection. Proteomics data were collected at the MS & Proteomics Resource at Yale University with mass spectrometers supported by the National Institutes of Health (NIH) Scientific Interest Groups (S10OD02365101A1, S10OD019967, and S10OD018034) and the Yale School of Medicine. We appreciate help from the Yale Center for Genome Analysis for high-throughput sequencing. This study was funded in part by the Edward P. Evans Foundation, the NIH/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) R01DK102792, and the NIH/NIDDK R01DK124788 (to S.H.). G.B. was supported by NIH/NIDDK Cooperative Centers of Excellence in Hematology (CCEH) U24DK126127. T.T. was supported by a pilot grant from the Yale CCEH (NIH/NIDDK U54DK106857) and by the Associazione Italiana per la Ricerca sul Cancro under MFAG 2020 (ID. 24883 project). We acknowledge start-up funds from Yale University School of Medicine and funding from the NIH/Eunice Kennedy Shriver National Institute of Child Health and Human Development to J.-J.C. (R01HD096745) and to B.J.L. (R01HD098128) and support from the Searle Scholars Program (SSP-2019-108) and Pew Biomedical Scholars Program (Pew Charitable Trusts, 00035285) to B.J.L. B.J.L. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund.
Author contributions: Conceptualization was by K.N.G., B.W.W., H.L., and B.J.L. Validation was by K.N.G. and B.W.W. Formal analysis was done by K.N.G., B.W.W., H.L., G.B., T.T., T.T.L., and B.J.L. Investigation was done by K.N.G., B.W.W., H.L., H.W., G.B., C.B.K., J.K., and T.T.L. Resources were provided by T.T.L., S.H., J.-J.C., and B.J.L. Writing the original draft was done by K.N.G., B.W.W., and H.L. Reviewing and editing were done by G.B., A.L.C., J.-J.C., and B.J.L. Visualization was by K.N.G., B.W.W., H.L., and B.J.L. Funding acquisition was by T.T.L., S.H., J.-J.C., and B.J.L. Supervision was by B.J.L.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276578.122.
Competing interest statement
S.H. is a consultant for Forma Therapeutics.
References
- Alavattam KG, Maezawa S, Sakashita A, Khoury H, Barski A, Kaplan N, Namekawa SH. 2019. Attenuated chromatin compartmentalization in meiosis and its maturation in sperm development. Nat Struct Mol Biol 26: 175–184. 10.1038/s41594-019-0189-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alisch RS, Jin P, Epstein M, Caspary T, Warren ST. 2007. Argonaute2 is essential for mammalian gastrulation and proper mesoderm formation. PLoS Genet 3: e227. 10.1371/journal.pgen.0030227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau J-C, et al. 2012. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol 19: 998–1004. 10.1038/nsmb.2373 [DOI] [PubMed] [Google Scholar]
- Baker CL, Walker M, Kajita S, Petkov PM, Paigen K. 2014. PRDM9 binding organizes hotspot nucleosomes and limits Holliday junction migration. Genome Res 24: 724–732. 10.1101/gr.170167.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellvé AR. 1993. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol 225: 84–113. 10.1016/0076-6879(93)25009-Q [DOI] [PubMed] [Google Scholar]
- Bellvé AR, Millette CF, Bhatnagar YM, O'Brien DA. 1977. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J Histochem Cytochem 25: 480–494. 10.1177/25.7.893996 [DOI] [PubMed] [Google Scholar]
- Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. 2012. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat Cell Biol 14: 266–275. 10.1038/ncb2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biancon G, Joshi P, Zimmer JT, Hunck T, Gao Y, Lessard MD, Courchaine E, Barentine AES, Machyna M, Botti V, et al. 2022. Precision analysis of mutant U2AF1 activity reveals deployment of stress granules in myeloid malignancies. Mol Cell 82: 1107–1122.e7. 10.1016/j.molcel.2022.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun RE. 2001. Packaging paternal chromosomes with protamine. Nat Genet 28: 10–12. 10.1038/ng0501-10 [DOI] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Brugiolo M, Botti V, Liu N, Müller-McNicoll M, Neugebauer KM. 2017. Fractionation iCLIP detects persistent SR protein binding to conserved, retained introns in chromatin, nucleoplasm and cytoplasm. Nucleic Acids Res 45: 10452–10465. 10.1093/nar/gkx671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant JM, Meyer-Ficca ML, Dang VM, Berger SL, Meyer RG. 2013. Separation of spermatogenic cell types using STA-PUT velocity sedimentation. J Vis Exp (80): e50648. 10.3791/50648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari SIA, Truesdell SS, Lee S, Kollu S, Classon A, Boukhali M, Jain E, Mortensen RD, Yanagiya A, Sadreyev RI, et al. 2016. A specialized mechanism of translation mediated by FXR1a-associated microRNP in cellular quiescence. Mol Cell 61: 760–773. 10.1016/j.molcel.2016.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJG, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514. 10.1016/j.devcel.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Catena R, Ronfani L, Sassone-Corsi P, Davidson I. 2006. Changes in intranuclear chromatin architecture induce bipolar nuclear localization of histone variant H1T2 in male haploid spermatids. Dev Biol 296: 231–238. 10.1016/j.ydbio.2006.04.458 [DOI] [PubMed] [Google Scholar]
- Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H, et al. 2011. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 480: 391–395. 10.1038/nature10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. 2009. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature 460: 479–486. 10.1038/nature08170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chohan H, Esfandiarei M, Arman D, Raamsdonk CDV, van Breemen C, Friedman JM, Jett KA. 2018. Neurofibromin haploinsufficiency results in altered spermatogenesis in a mouse model of neurofibromatosis type 1. PLoS One 13: e0208835. 10.1371/journal.pone.0208835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Yokota S, Liu J, Kilikevicius A, Johnson KC, Corey DR. 2021. Argonaute binding within human nuclear RNA and its impact on alternative splicing. RNA 27: 991–1003. 10.1261/rna.078707.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. 2012. Roles for microRNAs in conferring robustness to biological processes. Cell 149: 515–524. 10.1016/j.cell.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571. 10.1038/ng.368 [DOI] [PubMed] [Google Scholar]
- Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. 2014. RNAi factors are present and active in human cell nuclei. Cell Rep 6: 211–221. 10.1016/j.celrep.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo T, Shirley L, John GB, Castrillon DH. 2007. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45: 413–417. 10.1002/dvg.20310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. 2006. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell 124: 957–971. 10.1016/j.cell.2006.01.036 [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp A-C, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannigan MM, Fujioka H, Brett-Morris A, Mears JA, Licatalosi DD. 2018. PTBP2-dependent alternative splicing regulates protein transport and mitochondria morphology in post-meiotic germ cells. bioRxiv 10.1101/497461 [DOI]
- Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, Tang F, Hajkova P, Lao K, O'Carroll D, Das PP, Tarakhovsky A, Miska EA, et al. 2008. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One 3: e1738. 10.1371/journal.pone.0001738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrada G, Wolgemuth DJ. 1997. The mouse transcription factor Stat4 is expressed in haploid male germ cells and is present in the perinuclear theca of spermatozoa. J Cell Sci 110: 1543–1553. 10.1242/jcs.110.14.1543 [DOI] [PubMed] [Google Scholar]
- Hofmann MC, Hess RA, Goldberg E, Millán JL. 1994. Immortalized germ cells undergo meiosis in vitro. Proc Natl Acad Sci 91: 5533–5537. 10.1073/pnas.91.12.5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein E, Shomron N. 2006. Canalization of development by microRNAs. Nat Genet 38: S20–S24. 10.1038/ng1803 [DOI] [PubMed] [Google Scholar]
- Hu YC, de Rooij DG, Page DC. 2013. Tumor suppressor gene Rb is required for self-renewal of spermatogonial stem cells in mice. Proc Natl Acad Sci 110: 12685–12690. 10.1073/pnas.1311548110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang V, Li L-C. 2014. Demystifying the nuclear function of Argonaute proteins. RNA Biol 11: 18–24. 10.4161/rna.27604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson DF, Fowler KJ, Earle E, Saffery R, Kalitsis P, Trowell H, Hill J, Wreford NG, de Kretser DM, Cancilla MR, et al. 1998. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J Cell Biol 141: 309–319. 10.1083/jcb.141.2.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, Zamore PD. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060. 10.1126/science.1073827 [DOI] [PubMed] [Google Scholar]
- Johnson WL, Yewdell WT, Bell JC, McNulty SM, Duda Z, O'Neill RJ, Sullivan BA, Straight AF. 2017. RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. eLife 6: e25299. 10.7554/eLife.25299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Tang F, O'Carroll D, Lao K, Surani MA. 2009. Essential role for Argonaute2 protein in mouse oogenesis. Epigenetics Chromatin 2: 9. 10.1186/1756-8935-2-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BS, Jung JS, Jang JH, Kang KS, Kang SK. 2011. Nuclear Argonaute 2 regulates adipose tissue-derived stem cell survival through direct control of miR10b and selenoprotein N1 expression. Aging Cell 10: 277–291. 10.1111/j.1474-9726.2011.00670.x [DOI] [PubMed] [Google Scholar]
- Kumar TR, Wang Y, Lu N, Matzuk MM. 1997. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat Genet 15: 201–204. 10.1038/ng0297-201 [DOI] [PubMed] [Google Scholar]
- Lambrot R, Siklenka K, Lafleur C, Kimmins S. 2019. The genomic distribution of histone H3K4me2 in spermatogonia is highly conserved in sperm. Biol Reprod 100: 1661–1672. 10.1093/biolre/ioz055 [DOI] [PubMed] [Google Scholar]
- Lange J, Yamada S, Tischfield SE, Pan J, Kim S, Zhu X, Socci ND, Jasin M, Keeney S. 2016. The landscape of mouse meiotic double-strand break formation, processing, and repair. Cell 167: 695–708.e16. 10.1016/j.cell.2016.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R, Feinbaum R, Ambros V. 2004. A short history of a short RNA. Cell 116: S89–S92. 10.1016/S0092-8674(04)00035-2 [DOI] [PubMed] [Google Scholar]
- Lesch BJ, Dokshin GA, Young RA, McCarrey JR, Page DC. 2013. A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proc Natl Acad Sci 110: 16061–16066. 10.1073/pnas.1315204110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch BJ, Silber SJ, McCarrey JR, Page DC. 2016. Parallel evolution of male germline epigenetic poising and somatic development in animals. Nat Genet 48: 888–894. 10.1038/ng.3591 [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773. 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang Y, Yin J, Gao Y, Li Y, Bai D, He W, Li X, Zhang P, Li R, et al. 2019. Distinct H3K9me3 and DNA methylation modifications during mouse spermatogenesis. J Biol Chem 294: 18714–18725. 10.1074/jbc.RA119.010496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorès P, Coutton C, El Khouri E, Stouvenel L, Givelet M, Thomas L, Rode B, Schmitt A, Louis B, Sakheli Z, et al. 2018. Homozygous missense mutation L673P in adenylate kinase 7 (AK7) leads to primary male infertility and multiple morphological anomalies of the flagella but not to primary ciliary dyskinesia. Hum Mol Genet 27: 1196–1211. 10.1093/hmg/ddy034 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatouk DM, Loveland KL, McManus MT, Moore K, Harfe BD. 2008. Dicer1 is required for differentiation of the mouse male Germline. Biol Reprod 79: 696–703. 10.1095/biolreprod.108.067827 [DOI] [PubMed] [Google Scholar]
- Macleod J. 1971. Human male infertility. Obstet Gynecol Surv 26: 335–351. 10.1097/00006254-197105000-00001 [DOI] [PubMed] [Google Scholar]
- Maezawa S, Yukawa M, Alavattam KG, Barski A, Namekawa SH. 2018. Dynamic reorganization of open chromatin underlies diverse transcriptomes during spermatogenesis. Nucleic Acids Res 46: 593–608. 10.1093/nar/gkx1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. 2017. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45: D183–D189. 10.1093/nar/gkw1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modzelewski AJ, Holmes RJ, Hilz S, Grimson A, Cohen PE. 2012. AGO4 regulates entry into meiosis and influences silencing of sex chromosomes in the male mouse germline. Dev Cell 23: 251–264. 10.1016/j.devcel.2012.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Mortensen RD, Serra M, Steitz JA, Vasudevan S. 2011. Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA–protein complexes (microRNPs). Proc Natl Acad Sci 108: 8281–8286. 10.1073/pnas.1105401108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. 2011. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev 25: 1686–1701. 10.1101/gad.16651211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima R, Sato T, Ogawa T, Okano H, Noce T. 2017. A noncoding RNA containing a SINE-B1 motif associates with meiotic metaphase chromatin and has an indispensable function during spermatogenesis. PLoS One 12: e0179585. 10.1371/journal.pone.0179585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazer E, Dale RK, Chinen M, Radmanesh B, Lei EP. 2018. Argonaute2 and LaminB modulate gene expression by controlling chromatin topology. PLoS Genet 14: e1007276. 10.1371/journal.pgen.1007276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A. 2007. A slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev 21: 1999–2004. 10.1101/gad.1565607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Stroud H, Nee K, Liu W, Pezic D, Manakov S, Lee SA, Moissiard G, Zamudio N, Bourc'his D, et al. 2014. MORC1 represses transposable elements in the mouse male germline. Nat Commun 5: 5795. 10.1038/ncomms6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47: D442–D450. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin A, Coat C, Nguyen MH, Talagas M, Morel F, Amice J, De Braekeleer M. 2013. Molecular cytogenetic and genetic aspects of globozoospermia: a review. Andrologia 45: 1–9. 10.1111/j.1439-0272.2012.01308.x [DOI] [PubMed] [Google Scholar]
- Policarpi C, Crepaldi L, Brookes E, Nitarska J, French SM, Coatti A, Riccio A. 2017. Enhancer SINEs link Pol III to Pol II transcription in neurons. Cell Rep 21: 2879–2894. 10.1016/j.celrep.2017.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid R, Siany A, Rivkin N, Eitan C, Marmor-Kollet H, Yanowski E, Savidor A, Levin Y, Rot G, Meister G, et al. 2020. A nuclear role for ARGONAUTE-2 in regulation of neuronal alternative polyadenylation. bioRxiv 10.1101/2020.12.15.422806 [DOI]
- Romero Y, Meikar O, Papaioannou MD, Conne B, Grey C, Weier M, Pralong F, Massy BD, Kaessmann H, Vassalli J-D, et al. 2011. Dicer1 depletion in male germ cells leads to infertility due to cumulative meiotic and spermiogenic defects. PLoS One 6: e25241. 10.1371/journal.pone.0025241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santti H, Mikkonen L, Anand A, Hirvonen-Santti S, Toppari J, Panhuysen M, Vauti F, Perera M, Corte G, Wurst W, et al. 2005. Disruption of the murine PIASx gene results in reduced testis weight. J Mol Endocrinol 34: 645–654. 10.1677/jme.1.01666 [DOI] [PubMed] [Google Scholar]
- Schürmann A, Koling S, Jacobs S, Saftig P, Krauβ S, Wennemuth G, Kluge R, Joost H-G. 2002. Reduced sperm count and normal fertility in male mice with targeted disruption of the ADP-ribosylation factor-like 4 (Arl4) gene. Mol Cell Biol 22: 2761–2768. 10.1128/MCB.22.8.2761-2768.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A, Qian Y, Weyn-Vanhentenryck SM, Zhang C. 2017. CLIP Tool Kit (CTK): a flexible and robust pipeline to analyze CLIP sequencing data. Bioinformatics 33: 566–567. 10.1093/bioinformatics/btw653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Park JW, Huang J, Dittmar KA, Lu Z, Zhou Q, Carstens RP, Xing Y. 2012. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res 40: e61. 10.1093/nar/gkr1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Park JW, Lu Z, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y. 2014. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci 111: E5593–E5601. 10.1073/pnas.1419161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, Jangi M, Giallourakis CC, Sharp PA, Young RA. 2015. Transcription factor trapping by RNA in gene regulatory elements. Science 350: 978–981. 10.1126/science.aad3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Brick K, Pu Y, Camerini-Otero RD, Petukhova GV. 2016. The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes Dev 30: 266–280. 10.1101/gad.270009.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder E, Soundararajan R, Sharma M, Dearth A, Smith B, Braun RE. 2015. Compound heterozygosity for Y box proteins causes sterility due to loss of translational repression. PLoS Genet 11: e1005690. 10.1371/journal.pgen.1005690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper SFC, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. 2008. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell 15: 285–297. 10.1016/j.devcel.2008.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthès P, Kokkinaki M, Nef S, Gnirke A, et al. 2013. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep 3: 2179–2190. 10.1016/j.celrep.2013.05.031 [DOI] [PubMed] [Google Scholar]
- Strovas TJ, Rosenberg AB, Kuypers BE, Muscat RA, Seelig G. 2014. MicroRNA-based single-gene circuits buffer protein synthesis rates against perturbations. ACS Synth Biol 3: 324–331. 10.1021/sb4001867 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliaferro JM, Aspden JL, Bradley T, Marwha D, Blanchette M, Rio DC. 2013. Two new and distinct roles for Drosophila Argonaute-2 in the nucleus: alternative pre-mRNA splicing and transcriptional repression. Genes Dev 27: 378–389. 10.1101/gad.210708.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarallo R, Giurato G, Bruno G, Ravo M, Rizzo F, Salvati A, Ricciardi L, Marchese G, Cordella A, Rocco T, et al. 2017. The nuclear receptor ERβ engages AGO2 in regulation of gene transcription, RNA splicing and RISC loading. Genome Biol 18: 189. 10.1186/s13059-017-1321-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro K, Teissier A, Kobayashi N, Nakanishi A, Sasaki T, Yan K, Tarabykin V, Vigier L, Sumiyama K, Hirakawa M, et al. 2011. A mammalian conserved element derived from SINE displays enhancer properties recapitulating Satb2 expression in early-born callosal projection neurons. PLoS One 6: e28497. 10.1371/journal.pone.0028497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. 2016. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13: 508–514. 10.1038/nmeth.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. 2007. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 318: 1931–1934. 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- Velazquez Camacho O, Galan C, Swist-Rosowska K, Ching R, Gamalinda M, Karabiber F, De La Rosa-Velazquez I, Engist B, Koschorz B, Shukeir N, et al. 2017. Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. eLife 6: e25293. 10.7554/eLife.25293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, Moazed D. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676. 10.1126/science.1093686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang L-C, Bui S, Nielson A, Wu X, Vo H-T, Ma X-J, Luo Y. 2012. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14: 22–29. 10.1016/j.jmoldx.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster KE, O'Bryan MK, Fletcher S, Crewther PE, Aapola U, Craig J, Harrison DK, Aung H, Phutikanit N, Lyle R, et al. 2005. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc Natl Acad Sci 102: 4068–4073. 10.1073/pnas.0500702102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang Y-G, Qi Y. 2012. A role for small RNAs in DNA double-strand break repair. Cell 149: 101–112. 10.1016/j.cell.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Weng M-P, Liao B-Y. 2017. modPhEA: model organism Phenotype Enrichment Analysis of eukaryotic gene sets. Bioinformatics 33: 3505–3507. 10.1093/bioinformatics/btx426 [DOI] [PubMed] [Google Scholar]
- Woolnough JL, Atwood BL, Giles KE. 2015. Argonaute 2 binds directly to tRNA genes and promotes gene repression in cis. Mol Cell Biol 35: 2278–2294. 10.1128/MCB.00076-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Song R, Ortogero N, Zheng H, Evanoff R, Small CL, Griswold MD, Namekawa SH, Royo H, Turner JM, et al. 2012. The RNase III enzyme DROSHA is essential for microRNA production and spermatogenesis. J Biol Chem 287: 25173–25190. 10.1074/jbc.M112.362053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yeh S, Chen Y-T, Wu C-C, Chuang K-H, Lin H-Y, Wang R-S, Chang Y-J, Mendis-Handagama C, Hu L, et al. 2006. Oligozoospermia with normal fertility in male mice lacking the androgen receptor in testis peritubular myoid cells. Proc Natl Acad Sci 103: 17718–17723. 10.1073/pnas.0608556103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang T, Hess RA, Kolla V, Higashi M, Raabe TD, Brodeur GM. 2014. CHD5 is required for spermiogenesis and chromatin condensation. Mech Dev 131: 35–46. 10.1016/j.mod.2013.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone Methylation. Science 299: 716–719. 10.1126/science.1079695 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.