Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the pathogen responsible for the coronavirus disease 2019 (COVID-19) pandemic. Of particular interest for this topic are the signaling cascades that regulate cell survival and death, two opposite cell programs whose control is hijacked by viral infections. The AKT and the Unfolded Protein Response (UPR) pathways, which maintain cell homeostasis by regulating these two programs, have been shown to be deregulated during SARS-CoVs infection as well as in the development of cancer, one of the most important comorbidities in relation to COVID-19. Recent evidence revealed two way crosstalk mechanisms between the AKT and the UPR pathways, suggesting that they might constitute a unified homeostatic control system. Here, we review the role of the AKT and UPR pathways and their interaction in relation to SARS-CoV-2 infection as well as in tumor onset and progression. Feedback regulation between AKT and UPR pathways emerges as a master control mechanism of cell decision making in terms of survival or death and therefore represents a key potential target for developing treatments for both viral infection and cancer. In particular, drug repositioning, the investigation of existing drugs for new therapeutic purposes, could significantly reduce time and costs compared to de novo drug discovery.
Subject terms: Mechanisms of disease, Cancer, Infection, Cell signalling
Facts
The AKT and UPR pathways maintain cell homeostasis, regulating cell decision making mechanisms in terms of cell survival and death.
The AKT and UPR pathways are co-opted both during viral infection and cancer development.
The AKT and UPR pathways constitute a unified homeostatic control system, emerging as a master control mechanism both in physiological and pathological conditions.
The crosstalk between the AKT and UPR pathways represents a key potential target in the development of treatments for both viral infection and cancer.
Introduction: COVID-19, cancer and therapeutic targets
The coronavirus disease 2019 (COVID-19) outbreak was first reported in December 2019 in Wuhan, China [1]. Its causative agent -the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)- is an enveloped positive-sense RNA virus [2], belonging to the lineage B (Sarbecovirus) of the Betacoronavirus family. The genomic sequence of SARS-CoV-2 has 29,903 nucleotides in length [3], and it is closely related to SARS-CoV (79% genetic similarity) [4]. It presents 10–12 putative open reading frames (ORFs), including four specifically coding for its structural proteins: Spike (S), Envelope (E), Membrane (M), and Nucleocapsid (N). The Spike protein drives SARS-CoV-2 tropism and mediates cellular infection by binding to the angiotensin-converting enzyme (ACE-2) [5]. Since its first report in 2019, SARS-CoV-2 has rapidly spread worldwide, resulting in hundreds of millions of people infected and over 6 million deaths.
SARS-CoV-2 lethality is increased in people with different comorbidities, of which cancer, a group of diverse pathologies characterized by uncontrolled cell proliferation and dissemination, is one of the most prominent [6]. This has been related to the fact that many antitumor therapies generate immunosuppression [7]. Cancer patients are not only frequently immunosuppressed, but also suffer other side effects from antitumor treatments, having a higher risk of contracting infections, developing complications, and requiring intensive care, which is why they were considered a population at risk during the pandemic [8]. As an example, it has been reported that having received chemotherapy within 4 weeks prior to the onset of COVID-19 symptoms is associated with an increased risk of mortality [7, 8], indicating that the quality of the immune system can be a determining factor in the severity of the disease. Since there is evidence that antitumor treatments that compromise the immune system correlate with worse prognosis in COVID-19 patients, the development of alternative therapies that do not affect the immune system for cancer patients in the current context of pandemic is of great importance. In this sense, seeking pharmacological therapies associated with human proteins, which have a lower mutation rate than viral proteins, could be an effective strategy, complementary to the use of vaccines, which may present variability in the efficacy against different emerging variants of the virus. Furthermore, such a strategy could be relevant in the potential case of a future epidemic or pandemic caused by another strain or type of coronavirus. Of particular interest for this purpose are the AKT [9] and the Unfolded Protein Response (UPR) [10] pathways, which maintain cell homeostasis by regulating cell survival and cell death, two opposite cell programs whose control is hijacked by viral infections. As it will be reviewed below, these pathways have been shown to be deregulated during SARS-CoV infection as well as in the development of cancer. Feedback regulation between AKT and UPR pathways emerges as a master control mechanism of cell decision making in terms of survival or death and therefore represents a key potential target for developing treatments for cancer and viral infection (in particular, COVID-19). The repositioning of drugs, a strategy whereby new indications are found for existing drugs, could significantly reduce time and costs compared to de novo drug discovery [11].
AKT, cancer and disease
AKT (also known as protein kinase B or PKB) is a serine/threonine kinase member of the AGC family of protein kinases. In mammals, three cellular homologs of AKT (AKT1, AKT2, and AKT3) have been found, each transcribed from separate genes [12–14].
AKT contains an N-terminal pleckstrin homology (PH) domain that interacts with phosphatidylinositol (3,4,5)-triphosphate (PIP3), a central kinase (CAT) domain, and a C-terminal extension (EXT) that contains a hydrophobic motif (HM) with homology to other AGC kinases [15]. AKT plays a central role in growth, proliferation, differentiation, glucose uptake, metabolism, angiogenesis, protein translation, cell survival and apoptosis, and constitutes a vastly studied model of how signaling proteins transduce and process extracellular information into cell decisions and fates [9, 16].
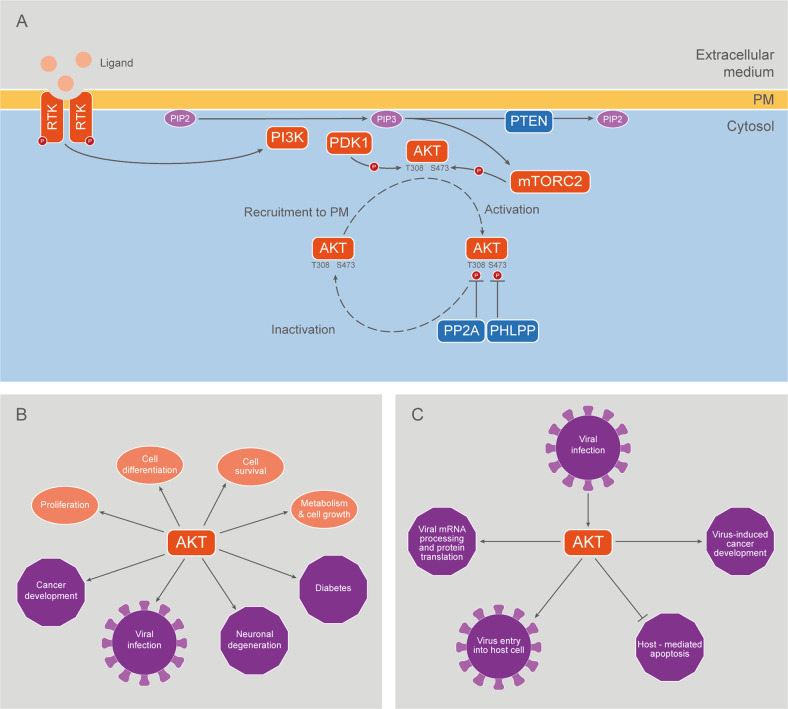
Traditional activation of AKT occurs when cognate ligands such as growth factors bind to tyrosine kinase receptors, resulting in their autophosphorylation, a process followed by the recruitment of small G proteins such as Ras (Fig. 1). One Ras effector is class I phosphatidylinositol-3-kinase (PI3K), which converts plasma membrane (PM) associated phosphatidylinositol-4,5-biphosphate (PIP2) to PIP3. AKT is then recruited to the PM by binding to PIP3 through its PH domain. This binding induces a conformational change that allows phosphorylation of AKT1 in T308 (T309 and T305 in AKT2 and AKT3, respectively) by Phosphoinositide-dependent kinase-1 (PDK1) [17] and in S473 (S474 and S472 in AKT2 and AKT3, respectively) by the mammalian target of rapamycin complex 2 (mTORC2) [18], resulting in AKT activation (Fig. 1). On the other hand, dephosphorylation of AKT leads to the termination of AKT activation: protein phosphatase 2 A (PP2A) downregulates AKT activity by inducing dephosphorylation of T308 from AKT1, while PH domain leucine-rich repeat protein phosphatase (PHLPP) suppresses AKT activity by dephosphorylating AKT1 in S473 [19, 20]. Also, dephosphorylation of PIP3 to PIP2, induced by phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase (PTEN), downregulates recruitment of AKT to PM and, consequently, results in reduced AKT activity [21] (Fig. 1).
Fig. 1. The AKT pathway.
A A traditional perspective of AKT activation assumes that PI3K, which converts PM-associated PIP2 to PIP3 in response to Growth Factors (GF) and other stimuli, leads to PIP3-mediated recruitment of cytosolic AKT to PM. The conformational change elicited by AKT PH domain binding to PIP3 allows phosphorylation of AKT by PDK1 and mTORC2. As a result, activated AKT concertedly regulates a great variety of targets and functions in the cytosol and the nucleus, among other subcellular compartments. B Examples of physiological and pathological processes in which AKT is implicated. C Key steps of viral infection regulated by AKT.
In the last few years, novel phosphorylation sites have been reported at the carboxy-terminus of AKT, which contribute to its activation [22], maturation, folding and stability [17, 23]. Additionally, AKT can be phosphorylated in several tyrosine residues, which seems to be a prerequisite to both T308 and S473 phosphorylations [24, 25]. Interestingly, in addition to phosphorylation, other AKT post-translational modifications (PTMs) have been described: O-glycosylation, ubiquitination, acetylation, oxidation and SUMOylation [16, 26]. It was also demonstrated that AKT is proline-hydroxylated [27] (Guo et al., 2016), methylated [28] and palmitoylated [29].
In addition to PM, some studies show that AKT can be also found in internal membranes such as those of the endoplasmic reticulum (ER), mitochondria, lysosomes, Golgi and early endosomes [29–34]. Once activated, AKT acts over multiple targets located in the nucleus, cytosol and different internal cell membranes. AKT is known to phosphorylate a large and diverse group of proteins containing the AKT consensus motif (RXRXXS*/T*B), where “S*/T*” represents the phosphorylatable serine or threonine, X is any residue, and B is a bulky hydrophobic residue [9]. Given the importance of the AKT pathway in all the aforementioned processes, it is consistent that its dysregulation has been associated with a wide variety of human diseases, including pathological cardiac hypertrophy, diabetes, neurodegeneration, vascular disorders, different types of cancer and viral infections [35–38] (Fig. 1).
The role of AKT in cancer development has been supported by numerous animal tumor models [39]. AKT was also shown to regulate hormone independence and tumor differentiation [40]. Additionally, AKT1, AKT2, and AKT3 isoforms are found to be overexpressed in several human cancers [35]. These results highlight the critical role of the PI3K/AKT pathway in cancer development.
In this context, AKT represents an attractive therapeutic target for cancer treatment and, as discussed below, for the treatment of specific viral infections. Given the variety of functions carried out by this kinase, only a fraction of them are relevant to tumor and viral progression. Thus, the choice of drug targets must take into account the adverse effects that may result from the inhibition of cellular processes regulated by AKT, such as those derived from its participation in other cell signaling pathways. Therefore, a deep understanding of the molecular mechanisms underlying the regulation of the activity of this protein kinase acquires special relevance [41]. In particular, inhibitors of the AKT pathway have been developed for therapeutic treatments and some of them are beginning to be used in clinical trials [42, 43].
UPR, cancer and disease
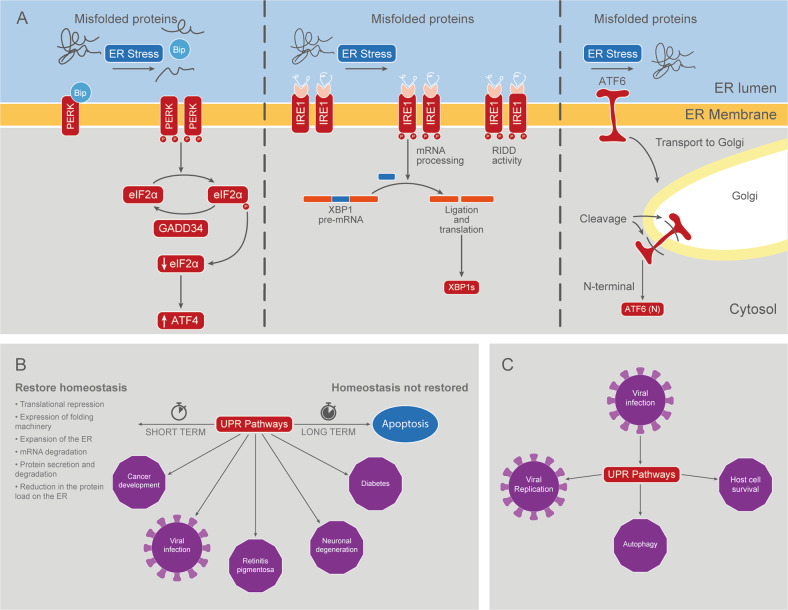
In eukaryotic cells, most of the secreted and transmembrane proteins fold and mature in the lumen of the endoplasmic reticulum (ER). The flow within this cell compartment is variable and can rapidly change in response to different contexts such as cell differentiation, diverse environmental conditions, and different physiological states of the cell. To handle this dynamic situation, cells developed a control system collectively known as the UPR, which is activated during different ER stress situations, such as the accumulation of misfolded proteins (Fig. 2).
Fig. 2. The UPR pathways.
A Activation of the UPR pathways. Three ER stress transducers have been identified: IRE1, ATF6 and PERK. These integral membrane proteins sense the protein folding status in the ER lumen and communicate this information to cytosolic target proteins that translocate to the nucleus to modulate gene expression. B Examples of physiological and pathological processes in which the UPR is implicated. C Key steps of viral infection regulated by the UPR.
The activation of the UPR mediates the expansion of ER membranes and the production of proteins that are part of the protein folding machinery and that reside in the ER lumen, such as chaperones and foldases. These processes help to increase the folding capacity of the cell. In parallel, this is accompanied by mechanisms that decrease the flow of proteins that transiently enter the ER. If the activity of the UPR persists, which means that cellular homeostasis cannot be restored, then the mechanisms mentioned above are turned off, and apoptosis is induced instead (Fig. 2). In this sense, the UPR consists of numerous feedback loops that establish and maintain the balance between survival and cell death [10].
Three different signaling UPR pathways have been described (Fig. 2), the main players of which are ER transmembrane proteins: inositol-requiring protein-1 (IRE1), activating transcription factor-6 (ATF6) and protein kinase RNA (PKR)-like ER kinase (PERK).
PERK is linked through its luminal domain to the binding immunoglobulin protein (BiP), also known as the glucose-regulated protein 78 (GRP78), and its activation occurs when BiP dissociates to bind misfolded proteins. This leads to oligomerization and transphosphorylation of PERK, thus activating its tyrosine kinase domain. Active PERK phosphorylates the eukaryotic translation initiation factor α (eIF2α), inactivating it, and inhibiting CAP-dependent messenger RNA (mRNA) translation initiation (Fig. 2). In this way, it favors the re-establishment of cellular homeostasis in the short run by reducing the load of synthesized proteins [44]. However, the reduction in active eIF2α levels favors the translation of activating transcription factor 4 (ATF4). Once induced, the ATF4 transcription factor activates a cluster of genes involved in the homeostatic response. ATF4 also regulates the transcription of the C/EBP homologous protein (CHOP) mRNA, and together with CHOP they induce the expression of the growth arrest and DNA damage-inducible protein (GADD34), which finally dephosphorylates eIF2α, allowing for global protein translation to resume [45]. Then, if UPR activation is maintained, apoptosis is promoted through CHOP, a transcription factor involved in cell arrest and apoptosis [46].
IRE1 is also linked through its luminal domain to BiP. After oligomerization and phosphorylation of IRE1, its cytosolic domain RNAse is activated, catalyzing an unconventional cytosolic “splicing” of X-Box Binding Protein 1 (XBP) 1mRNA [47, 48] (Fig. 2). In humans, splicing of the 26 nucleotide fragment produces a shift in the reading frame that encodes a version of XBP1 with a different amino acid sequence and higher molecular weight, known as spliced XBP1 or XBP1s [48]. XBP1s is a transcription factor that induces the expression of genes that contain an UPR response element (UPRE) or a hormone-specific response element (HRE) in their promoters, i.e. genes linked to folding, glycosylation, the endoplasmic-reticulum-associated protein degradation (ERAD) pathway, lipid biogenesis and/or vesicular trafficking [49]. IRE1 can also bind to and degrade other mRNAs through a process called IRE1-dependent regulated messenger degradation (RIDD) [50, 51]. While low levels of RIDD activity facilitate homeostasis by degrading ER-associated mRNAs, prolonged RIDD activity degrades mRNA coding for proteins that contribute to cell survival, thus promoting apoptosis (Fig. 2).
Finally, ATF6 is a transcription factor initially synthesized as an ER transmembrane protein. During UPR, ATF6 migrates to the Golgi apparatus via vesicular transport (Fig. 2). There, it is cleaved by site 1 and site 2 proteases (S1P and S2P) that sequentially remove its luminal and transmembrane domain, respectively, in a process called regulated intramembrane proteolysis (RIP) [52]. The N-terminal (cytosolic) domain migrates from the Golgi to the nucleus where it binds to UPRE regions and activates target genes that play a role in the recovery of ER homeostasis, especially genes that code for chaperones such as GRP78 and GRP94 (Fig. 2). Historically, the ATF6 pathway has been associated with pro-survival functions, but in recent years, evidence has emerged that ATF6 is a major inducer of CHOP [53].
In summary, UPR mainly induces translational silencing, transcriptional activation of molecular folding machinery, mRNA degradation, and stimulates both protein secretion and degradation [10, 54]. These processes promote cell survival in the short term, but if homeostasis is not restored, cell cycle arrest and apoptosis are promoted instead [10] (Fig. 2).
Dysregulation of UPR is reported in various diseases, such as cancer, diabetes, viral infection and neurodegenerative diseases [10, 55]. When homeostasis cannot be reestablished, UPR can function as a trigger for apoptosis in cells that are beneficial to the body. On the other hand, it can also have a cytoprotective effect by killing cells that are dangerous for the rest of the body [10]. Examples of the former are retinitis pigmentosa and other inherited forms of blindness, in which retinal cells undergo apoptosis when misfolded mutant rhodopsins are produced [56]. Examples of the latter are cancer and viral infections. Although in some types of cancer the UPR may have a protective effect for the host by triggering apoptosis in tumor cells [57], for the most part UPR functions as an adaptive survival mechanism [58], favoring tumor growth by allowing cancer cells to survive unfavorable conditions [59]. Furthermore, there is recent evidence indicating that, in addition to promoting tumor progression, activation of the UPR may also limit the effectiveness of chemotherapy, contributing to the development of chemoresistance [60]. It is in this context that the UPR has become an attractive therapeutic target for many types of cancer in recent years. In the case of viral infections, viruses exploit the UPR by increasing the ER’s ability to assist in viral replication, and in many cases, they make use of late apoptosis to be released in vesicles without being detected by the immune system [61] (Fig. 2).
AKT, viral infection and SARS-CoV-2
AKT regulation during viral infection
Several viral proteins are capable of upregulating PI3K/AKT signaling by modulating this pathway at multiple points [38] (Supplementary Table 1 and Fig. 1). For example, the NS1 protein of Influenza A virus (IAV) binds to the p85β regulatory subunit of PI3K, increasing PI3K activity and resulting in the phosphorylation of AKT [62]. It has also been reported that NS1 directly interacts with phosphorylated AKT, enhancing AKT activity [63]. The ST protein of the simian virus 40 (SV40) binds to phosphatase PP2A, preventing AKT from being dephosphorylated and therefore inactivated [37, 64]. A similar mechanism has been proposed for the E7 protein of the Human Papillomavirus (HPV) [65].
Host organisms use apoptosis as an instrument to resist viral infections. Thus, activating the PI3K/AKT signaling pathway is a strategy used by some viruses to delay host cell apoptosis and extend viral replication, especially at the earliest stages of infection (Supplementary Table 1). As an example, NS1 and NS2 proteins of Respiratory Syncytial virus (RSV), which are expressed early in infection, induce the activation of NF-B and PI3K/AKT pathways [66]. Once AKT is activated, it phosphorylates MDM2 and leads to the degradation of p53, a pro-apoptotic protein [67]. Similarly, it has been shown that the activation of the PI3K/AKT pathway in cells infected with Sendai virus (SV) leads to the stabilization of XIAP, an anti-apoptotic protein that blocks caspase 9, preventing early apoptosis [68, 69]. Likewise, activation of the PI3K/AKT pathway in Newcastle Disease Virus (NDV) infected cells promotes an antiapoptotic response, as suggested by the fact that treatment with LY294002, a PI3K inhibitor, results in cleavage of PARP and caspase 3 at early stages of infection [70]. Vaccinia virus (VACV) and Cowpox virus (CPXV), two members of the Poxviridae family, also activate the PI3K/AKT pathway to avoid early apoptosis [71]. Furthermore, the PI3K/AKT pathway appears to play a key role in both VACV and CPVX mRNA expression, viral assembly and morphogenesis, which occurs at a late stage of viral infection [37, 38, 71, 72] (Supplementary Table 1).
It has also been reported that, in some cases, the AKT pathway has a role in early internalization events and endocytosis. Some viruses, like IAV, Hepatitis C virus (HCV), Ebola virus (EV), NDV and Herpes simplex virus (HSV), induce the activation of this pathway to promote entry of the virus into the host cell [68, 73–77]. Particularly, HSV makes use of an interesting strategy to enter the cell. As a consequence of the interaction of viral envelope glycoproteins with cell surface, a small amount of intracellular calcium is released, and phospholipid scramblase-1 (PLSCR1) is activated. PLSCR1 flips phosphatidylserines (PtdS) from the inner to the outer leaflet of the plasma membrane, which results in externalization of AKT. Therefore, externalized AKT interacts with viral glycoprotein B, which apparently allows AKT phosphorylation at S473 and T308 by yet undefined kinases [78, 79]. Activation of AKT pathway is required for the release of cytoplasmatic calcium that promotes the fusion of the plasma membrane and viral envelope along with the entry of viral capsids [80]. Finally, PtdS and AKT flip back to the inner leaflet of the plasma membrane in response to the interaction between PLSCR1 and viral glycoprotein L. As PtdS exposure is an apoptotic signal, HSV presumably prevents this by restoring plasma membrane architecture [79] (Supplementary Table 1).
Interestingly, some viruses co-opt host mRNA splicing machinery, either to expand their coding potential or to disrupt host mRNA splicing patterns, altering cellular mRNA processing to facilitate viral gene expression. As an example, HSV-1 protein ICP27 interacts with SR proteins, which are RNA-binding proteins essential for mRNA processing, especially splicing regulation. This interaction affects SR protein phosphorylation, resulting in the shutdown of host mRNAs splicing [81]. Additionally, the Dengue virus (DV) has been also shown to hijack U5 snRNP proteins and RBM10 to deregulate host cell splicing [82, 83]. The PI3K/AKT pathway has been widely shown to regulate mRNA processing and particularly splicing. In response to extracellular stimuli, AKT has been described to phosphorylate SR and other RNA-binding proteins such as hnRNP proteins. It has been reported that AKT can phosphorylate splicing proteins both in a direct manner and also indirectly, via SRPK1/2 [37, 84, 85]. The Human Immunodeficiency virus 1 (HIV-1) seems to promote PI3K signaling in favor of phosphorylating SR proteins in order to regulate viral mRNA splicing [37, 86]. Furthermore, the PI3K/AKT pathway plays a key role in the regulation of the HPV type 16 (HPV16) life cycle-dependent gene expression. AKT activity seems to benefit HPV16 early gene expression and suppress HPV16 late gene expression: inhibition of AKT activity leads to dephosphorylation of hnRNP L, which causes reduction in HPV16 early polyadenylation and activation of HPV16 late L1 mRNA splicing [87] (Supplementary Table 1).
The PI3K/AKT pathway can also be hijacked for viral protein expression. In the context of translation initiation, viral mRNAs can be translated in a cap-dependent or independent manner, which involves the use of internal ribosome entry sites (IRES). The AKT substrate SRSF1 (also known as SF2 or ASF) has been reported to increase the ratio between cap-dependent translation and IRES-dependent translation [88]. In particular, it has been shown that viruses like NDV and HPV16 induce PI3K/AKT/mTOR pathway activation in order to upregulate cap-dependent host machinery and facilitate viral mRNA translation [89, 90].
Regarding tumorigenesis, viral infections are associated with 10-15% of all human cancers worldwide [91]. Several viruses, including Hepatitis B virus (HBV) and HCV, Epstein-Barr virus (EBV), Kaposi’s sarcoma herpesvirus (KSHV) and HPV are oncogenic. The PI3K/AKT signaling pathway is frequently deregulated in many types of human cancers, and in particular, in virus-induced malignancies. Activation of this pathway produces an increase in cell proliferation, resistance to apoptosis, genetic instability and even changes in cytoskeletal dynamics that lead to malignant transformation of the infected cells [92, 93]. K1 and vGPCR proteins of KSHV induce AKT phosphorylation via PI3K activation, while K1 also induces PTEN inactivation. Constitutive activation of AKT enhances survival and proliferation of infected cells and has an essential role in KSHV carcinogenesis [76, 94, 95]. It has been demonstrated that LMP1 and LMP2A viral proteins are responsible for activating AKT in EBV-associated cancers [96]. HCV-infected hepatocellular carcinoma (HCC) patients show decreased levels of PTEN, which is associated with HCC pathogenesis and poor prognosis [93]. Similarly, HBx protein of HBV reduces PTEN activity and enhances carcinogenesis [97]. E6 and E7 oncoproteins of HPV, identified as those responsible for the maintenance of HPV-related oncogenesis, inhibit tumor suppressors p53 and Rb, hence preventing apoptosis. These proteins also induce the activation of the PI3K/AKT/mTOR pathway [89] and/or prevent its inactivation, as mentioned above [65] (Supplementary Table 1).
AKT and SARS-CoVs
Particularly, as a consequence of SARS-CoV infection, different signaling pathways are up- and downregulated. Activation of two opposing cellular programs has been demostrated: on the one hand, apoptosis to kill virus-infected cells and, on the other hand, cell survival through the production of antiviral cytokines. In particular, AKT has been shown to be a key player in the regulation of cell death and survival in cells infected by SARS-CoV [98] (Supplementary Table 1).
A pioneer study developed by Surjit and collaborators in 2004 reported that as a result of the expression of SARS-CoV N, the levels of phospho-AKT and Bcl-2 are downregulated in COS-1 monkey kidney cells, correlating with activation of caspases 3 and 7 [99]. Activation of apoptosis in these cells was independent of the p53 and Fas signaling cascade.
The same year, Morikawa and collegues showed that in confluent Vero E6 cells AKT is activated after SARS-CoV replication prior to cell death [100]. Phosphorylation in AKT S473 was detected about 8 h after infection, decreasing after 18 h. In contrast, no phosphorylation in AKT T308 was detected. In the following years, the same group reported that the AKT pathway is required for establishing persistent SARS-CoV infection in these cells and that proliferation of subconfluent Vero E6 cells ceased after SARS-CoV infection, a process accompanied by persistent AKT dephosphorylation [101, 102]. Apparently, SARS-CoV infection is made possible via the phosphorylation of AKT and JNK, both of which are induced by the viral protein N [103]. The authors concluded that four proteins, AKT, JNK, Bcl-2 and Bcl-xL are necessary for survival of persistently SARS-CoV-infected cells (Supplementary Table 1).
One year later, Chan et al. showed that the SARS-CoV M protein induces apoptosis through modulation of the AKT cell survival pathway and the release of mitochondrial cytochrome c in both HEK293T cells and transgenic Drosophila [104]. The same group reported 7 years later that the C-terminus of the M protein interacts with the PH domain of PDK1 and that this interaction disrupts the association between PDK1 and AKT, leading to downregulation of AKT activity [105]. This signaling cascade culminates in the activation of caspases 8 and 9.
After the COVID-19 outbreak, it was not surprising to find that AKT was among the most strongly regulated kinases following not only SARS-CoV but also SARS-CoV-2 infection [106–113].
Endocytosis of SARS-CoV-2 after binding to the ACE2 receptor occurs via the clathrin-mediated pathway, which is regulated by the PI3K/AKT signaling cascade [114]. Moreover, the reduction of ACE2 at the cell surface after SARS-CoV-2 contributes to an increase of angiotensin II in serum, which in turn can trigger the activation of the PI3K/AKT signaling pathway after binding to the angiotensin II receptor type 1 (AT1R) [114].
Consistently, a proteo-transcriptomic analysis developed by Appelberg et al. revealed that AKT signaling is modulated in Huh7 cells infected with SARS-CoV-2, showing an activation 24 h after infection. Moreover, Li et al. showed that SARS-CoV-2 infection induces autophagy and apoptosis in human microvascular endothelial and bronchial epithelial cells by inhibiting the PI3K/AKT/mTOR pathway and increasing intracellular reactive oxygen species (ROS) levels [115]. Callahan et al. demonstrated that inhibiting the AKT pathway with the inhibitor GSK690693 markedly reduced gene expression of chemokines CXCL9, CXCL10 and CXCL11 in cells infected with SARS-CoV-2, controlling hyperinflammation [116]. Ren et al. further demonstrated that SARS-CoV-2 M and N proteins can induce caspase-dependent apoptosis by physically interacting with PDK1 and preventing PDK1-AKT interaction and inhibiting AKT signaling in Vero E6 and HepG2 cells [117]. Recent work from Malgotra and Sharma revealed that activation of the AKT pathway by SARS-CoV-2 contributes to the induction of glucose uptake via glucose transporters (GLUTs), leading to increased glycolysis and viral replication in host cells [118]. Finally, Pelzl et al. also found that antibody‐mediated procoagulant platelet formation in COVID‐19 is AKT dependent, suggesting that targeting this pathway might represent a promising strategy to reduce the risk for thrombosis in patients with severe COVID-19 [119] (Supplementary Table 1).
As discussed in the following sections, these results led to the conclusion that AKT could be an interesting target in patients infected with SARS-CoV-2 and other similar viruses [120–122].
UPR, viral infection and SARS-CoV-2
UPR regulation during viral infection
During the last two decades, a great deal of effort has been focused on establishing the link between ER stress, the unfolded protein response (UPR), and viral infection. Viruses induce ER stress since they require the host’s ER machinery to synthesize vast quantities of proteins so as to replicate successfully [123]. Moreover, several key stages of the viral life cycle take place in the ER, including protein synthesis and modification, genome replication and virus assembly [124].
In order to deal with ER stress and restore cellular homeostasis, the UPR is activated [125]. The three branches of the UPR function together as to decrease the load of unfolded or misfolded proteins in the ER by attenuating protein translation, expanding the ER folding capacity, increasing the ERAD, and by changing the protein secretory and intracellular trafficking routes [126]. If cells are not able to resolve ER stress during viral infection, they activate cell death pathways such as intrinsic mitochondrial apoptosis [127]. Nonetheless, several viruses are capable of hijacking the UPR to promote their survival and replication [61] (Supplementary Table 2 and Fig. 2).
Several past studies have focused on unraveling the ability of hijacking the UPR by certain viral pathogens, through the modulation of the different UPR branches. For instance, some viruses are capable of selectively activating the ATF6 branch of the UPR, inducing chaperone expression so as to synthesize viral proteins [128]. Previous research has shown that ATF6 induction is key for successful replication of the Lymphocytic Choriomeningitis virus (LCMV) [129], the African Swine Fever virus (ASFV) [130], the West Nile virus (WNV) [131], the HBV [132], and of the Zika virus (ZIKV) [133]. A recent study has also shown that ATF6 branch activation by HBx is essential for host cell survival in hepatoma cells, which could represent a key player in hepatocellular carcinoma development [134]. Moreover, the ATF6 branch was activated by the HCV core protein in a human hepatocellular carcinoma cell line. In this case, the HCV core protein induced autophagy through the activation of the ATF6 and PERK branches of the UPR (but not of the IRE1-XBP1 branch) [135]. Activation of autophagy via the induction of the ATF6 and PERK pathways was also reported in porcine cell lines infected with the Seneca Valley virus (SVV) [136] (Supplementary Table 2).
Activation of the IRE1-XBP1 branch of the UPR may also promote viral replication, since it increases the folding capacity in the ER and the synthesis of new ER membranes [137]. For instance, the exclusive activation of the IRE1-XBP1 branch leads to the replication of the IAV in human lung epithelial cells [138]. Moreover, in cells infected with the Japanese Encephalitis virus (JEV) [139, 140], the DV [140], the WNV [141] and the HBV, an activation of the IRE1-XBP1 branch is reported. For both the JEV and the DV, IRE1-XBP1 branch activation is associated with cell survival as evidenced by the fact that the knockdown of XBP1 brings along an enhanced cytopathic effect in a mice neuroblastoma cell line [140]. On the other hand, the Classical Swine Fever virus (CSFV) induces both the IRE1-XBP1 and PERK branches of the UPR, then triggering autophagy in order to enhance viral replication [142]. In addition, a recent study has revealed that Marburg virus (MARV) regulates IRE1-XBP1 in a time-dependent manner, upregulating the IRE1-XBP1 branch during the first 24 h post infection so as to promote viral replication [143]. Curiously, another study has shown that replication of Tick-borne Encephalitis flaviviruses (TBEV) in astrocytes was severely reduced when they were treated with IRE1 inhibitors before viral infection [144] (Supplementary Table 2).
Concerning the PERK branch of the UPR, ATF4 activity may play an important role in the re-establishment of cellular metabolism in virus-infected cells and in the resumption of protein synthesis. For instance, the upregulation of ATF4 has been shown to enhance the replication of HIV-1 [145]. Furthermore, both the human Cytomegalovirus (CMV) and the murine CMV induce ATF4 accumulation [146–148]. Interestingly, murine CMV prevented the expression of CHOP, a pro-apoptotic transcription factor downstream of ATF4, which could represent a viral strategy for promoting host cell survival [148]. Besides, a recent study has revealed that the Porcine Reproductive and Respiratory Syndrome virus (PRRSV) is capable of hijacking the ATF4 protein to viral replication complexes so as to promote its replication [149].
In opposition, other viruses do not induce the UPR since they have evolved strategies to suppress it. For instance, Herpes Simplex virus Type 1 (HSV-1) effectively suppresses the UPR at early stages of infection, with release of suppression at later stages as evidenced by an increased activity of eIF2α and ATF4, when virion assembly and liberation has been completed [150]. Furthermore, a recent study has revealed that the kinase activity of IRE1 was beneficial for replication of the HSV-1, while the RNase activity of IRE1 resulted pernicious in this process [151] (Supplementary Table 2).
UPR and SARS-CoVs
As described above, accumulating evidence suggests that ER stress and UPR activation are common outcomes during viral infection, and coronavirus infections are not the exception. The expression of ER-chaperones like GRP78 and GRP94 are clear indicators of ER stress. Different groups found that infection with SARS-CoV causes an increase in expression levels of GRP78 and GRP94 [152–154]. There are several aspects of coronavirus infection that may cause ER stress. Their replication occurs in the cytoplasm, strongly associated with the ER, and during this process great amounts of viral proteins are synthetized, some of which, are folded and modified in this compartment [155]. In fact, the S protein folding and maturation is mediated by ER chaperones [156]. If the accumulation of unfolded viral proteins saturates the folding capacity of the ER, this could induce ER stress and trigger the UPR. In addition, the ER-Golgi intermediate compartment (ERGIC), which is an extension of the ER, is where formation and budding of virions occur [157]. This process depletes the levels of ER membrane, which induces ER stress and activates the UPR [158, 159]. Furthermore, coronaviruses induce the formation of double membrane vesicles (DMVs) derived from the ER that also deplete membrane levels, and therefore trigger ER stress [160] (Supplementary Table 2).
In 2006, Chan et al. found that cells transfected with luciferase reporter constructs driven by GPR78 or GPR94 promoters were activated in cells infected with SARS-CoV or even merely overexpressing the S protein, thus confirming that infection with SARS-CoV induced ER stress through transcriptional activation of these chaperones [153]. Other SARS-CoV proteins such as E, M and NSP6 did not activate these promoters. S protein overexpression did not affect the activity of ATF4’s promoter, and only mildly affected CHOP´s promoter activity. When cells were co-transfected with a dominant-negative mutant of PERK or a dominant-negative mutant of eIF2α, transcription of GPR78 and GPR94 was blocked, showing that PERK activation and eIF2α phosphorylation were required for their induction. Chan et al. also found that eIF2α phosphorylation increased after infection with SARS-CoV or S protein overexpression. Finally, S protein overexpression did not significantly affect the IRE1 or ATF6 pathways. In conclusion, SARS-CoV S protein specifically activated PERK but did not affect IRE1 or ATF6. The authors suggested that this modulation of the UPR likely represents a viral strategy to combat the host’s response while facilitating viral replication, and that the effect of S was shown mainly in the transcriptional activation of chaperones, which would enhance the folding capacity of the ER. Although S activated the PERK/eif2α pathway, which blocks global protein synthesis of the cell, it had little influence over CHOP activation. CHOP activation would lead to apoptosis, which is undesirable for the virus when the infection is at an early stage. Low levels of IRE1 activation could also be beneficial for virus replication, since XBP1 upregulates chaperone expression, but high levels of activation could lead to apoptosis (Supplementary Table 2).
Three years later, Minakshi et al. showed that GRP78 and GRP94 promoters are activated when SARS-CoV 3a protein is overexpressed [161]. Of the three UPR sensors, only PERK was found to be activated in 3a expressing cells, based on the levels of eif2α phosphorylation and the inhibitory effects of a dominant negative version of eIF2α on the GRP78 promoter. 3a overexpression also increased the activity of the ATF4 and CHOP promoters. The authors concluded that this may be beneficial for viral replication, because while PERK activation leads to increased expression of ER chaperones, IRE1 and ATF6 activation would also activate ERAD, causing the degradation of viral proteins.
In 2011, DeDiego et al. found that infection with SARS-CoV lacking the E protein is attenuated in vivo [162]. To analyze the effects of the E protein, they infected cells either with SARS-CoV or a recombinant SARS-CoV lacking E protein (rSARS-CoV-ΔE) and analyzed differentially regulated genes. Most of these genes were involved in cytoplasmic, ER or mitochondrial stress, and their expression was higher in cells infected with rSARS-CoV-ΔE. In addition, levels of spliced XBP1 were higher in cells infected with rSARS-CoV-ΔE than SARS-CoV infected cells, but there was no significant activation of PERK or ATF6 pathways. These results suggest that ER stress in cells infected with SARS-CoV is downregulated when the E protein is expressed, which the authors suggest might work as a strategy for preventing premature cell death and facilitating efficient virus replication (Supplementary Table 2).
ER stress and UPR have already been associated with SARS-CoV-2 infection in previous works [163–165]. Tang´s group found in 2021 that open reading frame 8 (ORF8) protein induces ER stress and UPR: cells transfected with either of the two different genotypes of ORF8 protein (ORF8L and ORF8S) had an increase in luciferase activity of reporter constructs driven by the promoters of GRP78 or GRP94, and mRNA levels of these two chaperones were upregulated too [166]. They also observed a reduction in full-length ATF6 levels, and a 35–40% increase in cleaved ATF6 levels, indicating that the ATF6 branch of the UPR was activated. There was also an increase in XBP1 splicing, and in the amount of ERdj4 protein, a downstream target of spliced XBP1, indicating an upregulation of the IRE1 branch. However, there was no induction of the PERK branch.
As stated before, both SARS-CoV and SARS-CoV2 infection upregulate GRP78 in the infected host cells. Interestingly, Ahmed´s group suggested that GRP78, which is the main responsible for directing the misfolded proteins either for refolding or degradation, could be an interesting target for viral infections [167]. While under normal conditions GRP78 is found in the ER, it has been shown that under stress conditions this protein can translocate into the cytoplasm and on the cell membrane, where it is known as cell surface GRP78 (CS-GRP78) [168, 169]. On the other hand, there is evidence that CS-GRP78 facilitates pathogenic entry to the cell in many different types of viral infection. Moreover, bioinformatic studies suggest that CS-GRP78 can interact with the spike protein of SARS-CoV-2 and improve virus attachment and host cell entry [170]. The authors suggest that lowering the concentration of CS-GRP78 could reduce the number of internalized viral particles.
In the last months, evidence of activation of the UPR during SARS-CoV-2 infection has been rapidly accumulating. In 2021, Balakrishnan and Lai overexpressed the S protein in HEK293T cells, and found augmented ER stress (evidenced by a higher level of GRP78), a significant increase in the levels of phosphorylated eIF2α (suggesting PERK branch activation) as well as induction of autophagy and cell death [171].
That same year, Rosa-Fernandes et al. provided evidence that SARS-CoV-2 hijacks the glycosylation biosynthetic, ER-stress and UPR machineries for viral replication, using a spatio-temporal mass spectrometry-based quantitative approach comprised of proteome, membranome and N-deglycoproteome of SARS-CoV-2 infected Vero cells [172]. They also performed immunoblotting for ER-stress and UPR markers, and found increased phosphorylation levels of PERK and eIF2α, together with increased protein levels of ATF4 2 h after viral infection. They also observed higher levels of ATF6 and phosphorylated IRE1α 48 h after infection. Finally, they re-analyzed transcriptome data obtained from lung autopsies of patients who died as a result of COVID-19: differentially regulated transcripts were involved in several processes linked to ER stress, such as cell death, chaperone-mediated folding, ‘de novo’ protein folding, protein localization to ER and programmed cell death [172].
In 2021, Chuan-min´s group found that expression of SARS-CoV-2 protein ORF3a in Hela cells increased GRP78, ATF4, CHOP, cleaved ATF6 and XBP1s protein levels, indicating the induction of the three UPR signal pathways [173]. They also reported that transfection of both ATF6 and IRE1 siRNA inhibited ORF3a-induced autophagy, while transfection of PERK siRNA did not, indicating that ORF3a promotes the induction of autophagy via the ATF6 and IRE1 pathways [173].
In 2022, Galli´s group demonstrated that SARS‐CoV‐2 infection in VERO‐E6 cells stimulated the expression of the ER stress signaling protein IRE‐1α [163], and that IRE‐1α activation in SARS‐CoV‐2 infected cells is associated with the inflammatory response (NF‐kB activation and pro‐inflammatory cytokine induction), which are key pathogenic events in COVID‐19. They also show that treatment with the antiviral Nelfinavir significantly reduced IRE‐1α activity of the infected cell, and restored homeostatic processes of the host cell. They conclude UPR signaling and ER stress are main aspects of the SARS‐CoV‐2‐host interaction, apparently contributing to viral replication and to its inflammatory complications (Supplementary Table 2).
The cross-talk between AKT and UPR pathways
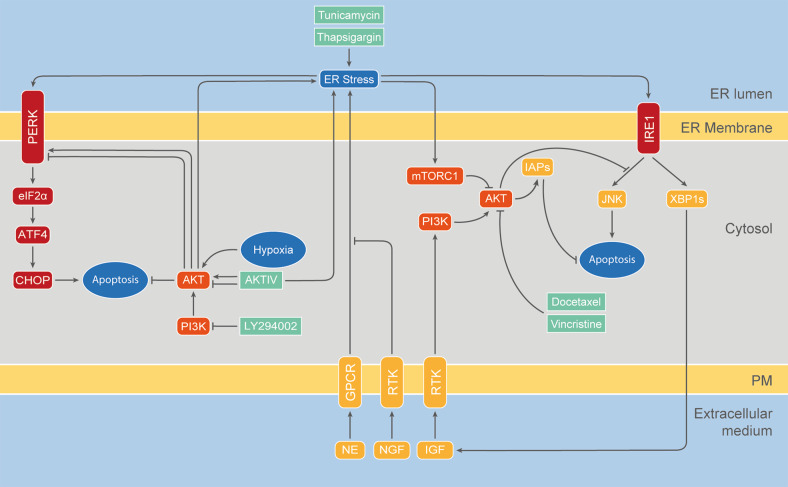
Even though AKT and UPR pathways maintain homeostasis regulating similar fundamental biological processes, they have been studied independently from each other for a long time. It was not until 2004 that the Exton group found a link between these pathways. First, they showed that PI3K and AKT are activated in different human cancer cell lines in response to ER stressors such as thapsigargin and tunicamycin. This activation was associated with the induction of two members of the Inhibitor of Apoptosis (IAP) family of caspase suppressors, cIAP-2 and XIAP [174]. Interestingly, this work also showed that either a PI3K inhibitor or a dominant-negative AKT version reversed cIAP-2 and XIAP induction in response to ER stress and sensitized cells to cell death. Knockdown of cIAP-2 and XIAP by RNA interference was sufficient to sensitize cells to ER stress-induced death, one of the first demonstrations that activation of endogenous AKT/IAPs plays a critical role in controlling cell survival by resisting ER stress-induced cell death signaling (Fig. 3).
Fig. 3. Crosstalk between the AKT and the UPR pathways.
The main interactions between both signaling pathways are shown, together with the main biological processes regulated by these pathways. Pharmacological compounds targeting these feedback loops with a potential therapeutic role in SARS-CoV-2 infection are listed below.
Two years later, in 2006, Mao et al., reported that the proapoptotic effect of norepinephrine (NE) in PC-12 cells is associated with both the activation of UPR and the inhibition of the PI3K/AKT pathway [175]. While stimulation with Nerve Growth Factor (NGF) activated PI3K/AKT and prevented ER stress-induced cell death, different PI3K inhibitors abolished this protective effect.
In 2007, Hosoi et al. discovered that PI3K/AKT is also regulated by ER stress in glial cells [33]. Interestingly, they found a dual effect: while AKT activation was upregulated by short-term exposure to ER stress, it was decreased during long-term exposure to ER stress. This was the first study to report AKT recruitment to the ER by immunohistochemistry and they also found increased AKT presence in the microsomal fraction during ER stress.
The same year, Hu et al. found that stable overexpression of XBP1s is sufficient to upregulate IGF expression and AKT phosphorylation in a zebrafish embryonic cell line (ZF4) [176]. Similarly, Zhang´s group found that XBP1 mediates the activation of AKT under ER stress in human melanoma cells [177, 178]. Remarkably, this process allowed tumor-derived cells to acquire resistance to chemotherapeutic drugs, such as docetaxel and vincristine, a phenomenon blocked by knockdown of XBP1 [177].
In 2010, Ishigaki et al. provided another piece of evidence in favor of the link between the UPR pathways and AKT [179]. Particularly, they found an induction of AKT1 during UPR, which depended on the activation of the PERK–eIF2α pathway, the upregulation of the Antagonizing Transcription Factor (AATF) and the Signal Transducer and Activator of Transcription 3 (STAT3). The activation of this signaling pathway mediated an antiapoptotic effect in rat insulinoma cells exposed to ER stress, which depended on the presence of AKT1. The same year, Qin et al., reported that ER stress-induced cell death in MEF cells is mediated by AKT inhibition, since the latter directly leads to downregulation of mTOR and enhancement of autophagy [180]. Similarly, Deldicque et al. reported that ER stress induces anabolic resistance in muscle cells in a manner that is dependent on the downregulation of AKT and consequent inhibition of mTORC1 [181]. On the other hand, Kato et al. described that mTORC1 suppressed AKT1 activity and enhanced the IRE1-JNK pathway during ER stress in the rat renal tubular epithelial cell line NRK-52E, resulting in the induction of apoptosis [182]. In summary, all these different groups have shown that the pro-apoptotic effects triggered by ER stress are alleviated by UPR-mediated activation of the AKT survival pathway, constituting an incoherent feed-forward loop.
Additionally, two different works, in 2011 and 2013, reported regulation of the UPR pathways by AKT for the first time. On the one hand, using pharmacological compounds such as the PI3K inhibitor LY294002, Mounir et al. found that AKT phosphorylates PERK at Thr799 leading to its inhibition in mammalian and Drosophila cells as well as in mouse mammary gland tumors [183]. On the other hand, Blaustein et al. reported that AKT phosphorylates PERK at a still unknown residue in response to hypoxia and to the antiproliferative and antiviral drug AKT-IV, leading to PERK–eIF2α activation and cell death [32]. In agreement with a positive role of AKT in PERK activation, Winnay et al. recently reported that PI3K/AKT inhibition leads to dephosphorylation of PERK at Thr980, a process associated with its inactivation [184]. The authors also observed that PI3K/AKT blockade leads to IRE1 inactivation, concluding that inhibition of the PI3K/AKT pathway blocks UPR and reduces sensitivity to ER stress-dependent apoptosis. The results of Mounir et al., Blaustein et al. and Winney et al. combined suggest that AKT might regulate the UPR by affecting PERK phosphorylation at different residues, eliciting different and even opposite responses depending on the cell context.
All together, these observations reveal the existence of a two way crosstalk between UPR and AKT pathways and suggest that they might constitute a unified homeostatic control system (Fig. 3). Feedback regulation between AKT and UPR pathways emerges as a master control mechanism of cell decision-making in terms of survival or death, showing the remarkable flexibility of signaling pathways which can direct cells to opposing fates depending on the dynamics of their activation. Recent work confirms the relevance of the crosstalk between the AKT and the UPR pathways [185–190].
Discussion: AKT and UPR signaling as possible targets for therapies against COVID-19 and cancer
Particularly, as we have shown along this review, AKT and UPR pathways are hijacked and deregulated during virus and particularly coronavirus infection, as well as during cancer development. Cancer is one of the most common comorbidities in relation with COVID-19. Particularly, SARS-CoV-2 replication in host cells depends on altered glucose metabolism. This metabolism is similar to the Warburg effect well studied in cancer, for which AKT and UPR pathways play a key role [191, 192]. The Warburg effect appears to be involved not only in several steps of cancer progression but also of COVID-19 infection. Interestingly, Mukhopadhyay et al. highlighted the potential of AKT-related miRNAs in the development of diagnostics, biomarkers, and novel targets for SARS-CoV-2 associated with lung cancer [193]. Interestingly, two other well-known COVID-19 comorbidities, obesity and diabetes, have been linked with a cross-talk between AKT and UPR pathways [194, 195]. Therefore, the search for potential therapeutic drugs that target AKT and/or UPR pathways appears as a promising strategy in the fight against COVID-19, particularly for patients with comorbidities such as cancer, obesity and/or diabetes.
Interestingly, AKT has been recommended as a viable target to treat COVID-19 patients [114, 196, 197] (Fig. 3). On the one hand, AKT signaling has been associated with disease progression [198]. On the other hand, it has been suggested that AKT inhibition would increase the number of regulatory T cells, suppressing inflammation, promoting vascular regeneration and wound resolution [114, 120]. Consistently, it has been proposed that targeting AKT can inhibit entry and replication of SARS-CoV-2, modulate the immune response, suppress the cytokine storm, and protect against thrombosis associated with severe COVID-19 cases [119, 197, 199].
Leonardi and Proenca have also proposed a link between AKT and Fas (CD95) to quell aberrant T cell differentiation and apoptosis in COVID-19 [122]. Interestingly, Diacerein, a drug derived from anthraquinone whose active metabolite, rhein, elicits antiviral activity, inhibited the interaction between SARS-CoV protein S and ACE2, through the downregulation of AKT, among others, suggesting that rhein is a potential therapeutic agent for the treatment of SARS-CoV-2 infection [200]. Recently, green tea polyphenol catechins have been suggested as a potential therapy to treat or prevent SARS-CoV-2 infection [201]. Catechins, which display anti-inflammatory and antiviral activities, have been shown to inhibit SARS-CoV replication, potentiate adaptive immunity and attenuate acute lung injury in mice through the inhibition of the PI3K/AKT pathway [201]. Shirato et al. showed that Asparagus officinalis stem standardized extract (EAS) attenuates SARS-CoV-2 spike protein S1 subunit (S1)-induced IL-6 and IL-1β production by suppressing p44/42 MAPK and AKT signaling in murine primary macrophages, suggesting that EAS may be beneficial in controlling excessive inflammation in COVID-19 patients [202]. Moreover, Yang et al. have confirmed the relationship between AKT and pulmonary fibrosis caused by COVID-19, showing that D-limonene has a potential therapeutic value in SARS-CoV-2-triggered pulmonary fibrosis by inhibiting the AKT pathway [121]. Recently, Wang et al also discovered that high-cannabidiol cannabis extracts attenuated ACE2 and transmembrane protease serine 2 (TMPRSS2) expression and the induction of inflammatory proteins COX2, IL-6, and IL-8 through the AKT pathway [203]. Finally, hesperetin, a molecule with antioxidant, anti-inflammatory, and antiviral properties which is mainly found in citrus honey, has recently been suggested by Khezri et al. as a therapeutic treatment against SARS-CoV-2 infection in a probable connection with the PI3K/AKT pathway [204].
Remarkably, an antiviral drug screen performed by García Jr. et al. identified different AKT inhibitors (dactolisib, AZD2014 and torin2) as potent blockers of SARS-CoV-2 replication [205]. Moreover, Sun et al. found an AKT inhibitor, capivasertib, which restricted the entry of SARS-CoV-2 into cells under non-cytotoxic concentrations [206]. Consistently, work by Stukalov et al. found that the AKT inhibitor ipatasertib exhibited high antiviral activity against SARS-CoV-2 [111]. Finally, inhibition of AKT with the pharmacological compound MK-2206 showed a significant reduction in virus production as well [207].
Similarly, the UPR has also been recently proposed as a therapeutic target for COVID-19 (Fig. 3). As it has been pointed out, replication of coronaviruses induces UPR in the infected cells. Interfering with this response may provide new therapeutic targets and antiviral agents against COVID-19 [173, 208]. It is also known that dysregulation of the vascular barrier function contributes to the irreversible outcomes of SARS. Since the UPR modulates lung endothelial permeability, it has been suggested that UPR manipulation by pharmacologic intervention can serve to oppose the devastating outcomes of COVID-19 [209]. The fact that COVID-19 tends to affect men more severely than women has also been suggested to have a link with the UPR: estrogen production would serve to alleviate ER stress shielding women from COVID-19 complications [210]. Consistently, different authors have reported the potential utility of drugs, such as Gene-Eden-VIR/Novirin, targeting the UPR as COVID-19 therapeutic treatments [211–213]. Interestingly, Ahmed´s group suggests that GRP78, which is the main responsible for directing the misfolded proteins either for refolding or degradation, could be an interesting target for both viral infections and cancer [167]. CS-GRP78 is found in many types of aggressive cancers. In non-small-cell lung cancer (NSCLC) and glioblastoma multiforme (GBM), for example, overexpression of CS-GRP78 is one of the responsibles for radio-resistance [214], while administration of anti-GRP78 delayed tumor growth and enhanced the efficacy of the radiation treatment. Other groups also found that targeting CS-GRP78 improves treatment efficacy [215, 216]. Moreover, crosstalk between GRP78 and AKT has been reported several times [217]. Reducing the concentration of CS-GRP78 could both reduce the number of internalized viral particles and the cancer-associated resistance, making this protein an attractive target for patients with cancer, COVID-19, or both [170]. Finally, autophagy, a process strongly regulated both by AKT and UPR pathways, as it has been described above, has been also suggested as a potential target for anti-COVID-19 therapies [218, 219].
In a context in which the use of antitumor therapies such as chemotherapy or radiotherapy generate immunosuppression [7, 8], the search for cancer therapies that do not compromise patients immunologically is urgent. Drug repositioning, the investigation of existing drugs for new therapeutic purposes, could be a therapeutic alternative for cancer and/or COVID-19 patients [11, 205, 206]. Drug repositioning could significantly reduce time and costs compared to de novo drug discovery. For instance, Nelfinavir, an antiretroviral drug used to treat HIV, has been shown to regulate AKT as well as the UPR and has been recently proposed both in cancer and in COVID-19 therapy [220–222]. Similarly, the broad spectrum corticosteroid Dexamethasone has been described to modify both, AKT as well as UPR activities and has been used in cancer and COVID-19 therapy [223–225].
On the other hand, pharmacological treatment can complement the use of vaccines, taking into account the challenges in production, affordability, allocation and global access to them [226]. Moreover, the efficacy of vaccines, designed on the basis of specific viral nucleotide or protein sequences, is challenged by the emergence of new SARS-CoV-2 variants, due to its high mutation rate [227]. The fact that some people, especially elderly or those with comorbidities such as cancer, are susceptible to severe manifestations of COVID-19 or even to death despite a complete vaccination schedule, makes the search for drugs to complement vaccination essential [228]. In other cases, such as immunosuppressed patients who cannot develop an immunological response to vaccines, this need is even more evident. In this sense, searching for pharmacological therapies associated with human proteins, with a lower mutation rate than viral proteins, could be an effective and complementary strategy to the use of vaccines. Furthermore, it could be relevant in the potential case of future epidemics or pandemics.
Supplementary information
Acknowledgements
We like to show our most sincere gratitude to Karin E. Giacomuzzi for her creative and inspired graphic design work. We also wish to thank Alejandro Colman-Lerner, Anabella Srebrow, Alejandro Amoroso, Ricardo Biondi and Susana Silberstein for encouraging discussions that greatly helped to write this manuscript. M.B. is a member of CONICET and was supported by the National Agency for Science and Technology, Argentina. M.G.C. was supported by a postdoctoral fellowship from CONICET. A.V., A.A. and G.M.S. were supported by doctoral fellowships from CONICET. M.S and M.C were supported by undergraduate research fellowships from the UBA.
Author contributions
M.B. conceptualized, designed and wrote the manuscript. M.S., A.V., A.A. and M.G.C. designed the supplementary tables and wrote the manuscript. G.M.S. designed the figures and wrote the manuscript. M.C. revised and edited the manuscript. All contributing authors revised the figures. All authors read and approved the final paper.
Data availability
All data are included in this article and its supplementary materials.
Competing interests
The authors declare no competing interests.
Footnotes
Edited by Professor Hans-Uwe Simon
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Mariana Suaya, Gonzalo Manuel Sánchez, Antonella Vila, Analía Amante, Mercedes García Carrillo.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-022-05250-5.
References
- 1.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–3. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16:1–22. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim D, Lee J-Y, Yang J-S, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–21. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–74. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–9. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 6.Chavez-MacGregor M, Lei X, Zhao H, Scheet P, Giordano SH. Evaluation of COVID-19 mortality and adverse outcomes in US patients with or without cancer. JAMA Oncol. 2022;8:69–78. doi: 10.1001/jamaoncol.2021.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang W, Guan W, Chen R, Wang W, Li J, Xu K, et al. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21:335–7. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian J, Yuan X, Xiao J, Zhong Q, Yang C, Liu B et al. Clinical characteristics and risk factors associated with COVID-19 disease severity in patients with cancer in Wuhan, China: a multicentre, retrospective, cohort study. Lancet Oncol. 2020. 10.1016/S1470-2045(20)30309-0. [DOI] [PMC free article] [PubMed]
- 9.Manning BD, Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–6. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 11.Altay O, Mohammadi E, Lam S, Turkez H, Boren J, Nielsen J, et al. Current status of COVID-19 therapies and drug repositioning applications. iScience. 2020;23:101303. doi: 10.1016/j.isci.2020.101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones PF, Jakubowicz T, Pitossi FJ, Maurer F, Hemmings BA. Molecular cloning and identification of a serine/threonine protein kinase of the second-messenger subfamily. Proc Natl Acad Sci USA. 1991;88:4171–5. doi: 10.1073/pnas.88.10.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones PF, Jakubowicz T, Hemmings BA. Molecular cloning of a second form of rac protein kinase. Cell Regul. 1991;2:1001–9. doi: 10.1091/mbc.2.12.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brodbeck D, Cron P, Hemmings BA. A human protein kinase Bγ with regulatory phosphorylation sites in the activation loop and in the C-terminal hydrophobic domain. J Biol Chem. 1999;274:9133–6. doi: 10.1074/jbc.274.14.9133. [DOI] [PubMed] [Google Scholar]
- 15.Kumar CC, Madison V. AKT crystal structure and AKT-specific inhibitors. Oncogene. 2005;24:7493–501. doi: 10.1038/sj.onc.1209087. [DOI] [PubMed] [Google Scholar]
- 16.Blaustein M AKT. Encyclopedia of signaling molecules. Switzerland: Springer; 2018.
- 17.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, et al. Mechanism of activation of protein kinase B by insulin and IGF‐1. EMBO J. 1996;15:6541–51. doi: 10.1002/j.1460-2075.1996.tb01045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 19.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Andjelković M, Jakubowicz T, Cron P, Ming X-F, Han J-W, Hemmings BA. Activation and phosphorylation of a pleckstrin homology domain containing protein kinase (RAC-PK/PKB) promoted by serum and protein phosphatase inhibitors. Proc Natl Acad Sci USA. 1996;93:5699–704. doi: 10.1073/pnas.93.12.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3, 4, 5-trisphosphate. J Biol Chem. 1998;273:13375–8. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 22.Liu P, Begley M, Michowski W, Inuzuka H, Ginzberg M, Gao D, et al. Cell-cycle-regulated activation of Akt kinase by phosphorylation at its carboxyl terminus. Nature. 2014;508:541–5. doi: 10.1038/nature13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Facchinetti V, Ouyang W, Wei H, Soto N, Lazorchak A, Gould C, et al. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 2008;27:1932–43. doi: 10.1038/emboj.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conus NM, Hannan KM, Cristiano BE, Hemmings BA, Pearson RB. Direct identification of tyrosine 474 as a regulatory phosphorylation site for the Akt protein kinase* 210. J Biol Chem. 2002;277:38021–8. doi: 10.1074/jbc.M203387200. [DOI] [PubMed] [Google Scholar]
- 25.Mahajan K, Coppola D, Challa S, Fang B, Chen YA, Zhu W, et al. Ack1 mediated AKT/PKB tyrosine 176 phosphorylation regulates its activation. PLoS One. 2010;5:e9646. doi: 10.1371/journal.pone.0009646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Risso G, Blaustein M, Pozzi B, Mammi P, Srebrow A. Akt/PKB: one kinase, many modifications. Biochemical J. 2015;468:203–14. doi: 10.1042/BJ20150041. [DOI] [PubMed] [Google Scholar]
- 27.Guo J, Chakraborty AA, Liu P, Gan W, Zheng X, Inuzuka H, et al. pVHL suppresses kinase activity of Akt in a proline-hydroxylation–dependent manner. Science. 2016;353:929–32. doi: 10.1126/science.aad5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshioka Y, Suzuki T, Matsuo Y, Nakakido M, Tsurita G, Simone C, et al. SMYD3-mediated lysine methylation in the PH domain is critical for activation of AKT1. Oncotarget. 2016;7:75023–37. doi: 10.18632/oncotarget.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blaustein M, Piegari E, Martínez Calejman C, Vila A, Amante A, Manese MV, et al. Akt is S-palmitoylated: a new layer of regulation for Akt. Front cell Develop Biol. 2021;9:229. doi: 10.3389/fcell.2021.626404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S-L, Wang Z-G, Hu Y, Xin Y, Singaram I, Gorai S, et al. Quantitative lipid imaging reveals a new signaling function of phosphatidylinositol-3,4-bisphophate: isoform- and site-specific activation of Akt. Mol Cell. 2018;71:1092–1104.e5. doi: 10.1016/j.molcel.2018.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arias E, Koga H, Diaz A, Mocholi E, Patel B, Cuervo AM. Lysosomal mTORC2/PHLPP1/Akt regulate chaperone-mediated autophagy. Mol Cell. 2015;59:270–84. doi: 10.1016/j.molcel.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blaustein M, Pérez-Munizaga D, Sánchez MA, Urrutia C, Grande A, Risso G, et al. Modulation of the Akt pathway reveals a novel link with PERK/eIF2α, which is relevant during hypoxia. PLoS One. 2013;8:e69668. doi: 10.1371/journal.pone.0069668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosoi T, Hyoda K, Okuma Y, Nomura Y, Ozawa K. Akt up-and down-regulation in response to endoplasmic reticulum stress. Brain Res. 2007;1152:27–31. doi: 10.1016/j.brainres.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 34.Santi SA, Lee H. The Akt isoforms are present at distinct subcellular locations. Am J Physiol Cell Physiol. 2010;298:C580–C591. doi: 10.1152/ajpcell.00375.2009. [DOI] [PubMed] [Google Scholar]
- 35.Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–64. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]
- 36.Cheung M, Testa JR. Diverse mechanisms of AKT pathway activation in human malignancy. Curr Cancer Drug Targets. 2013;13:234–44. doi: 10.2174/1568009611313030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunn EF, Connor JH. HijAkt: The PI3K/Akt pathway in virus replication and pathogenesis. Prog Mol Biol Transl Sci. 2012;106:223–50. doi: 10.1016/B978-0-12-396456-4.00002-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diehl N, Schaal H. Make yourself at home: viral hijacking of the PI3K/Akt signaling pathway. Viruses. 2013;5:3192–212. doi: 10.3390/v5123192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen M-L, Xu P-Z, Peng X, Chen WS, Guzman G, Yang X, et al. The deficiency of Akt1 is sufficient to suppress tumor development in Pten + /− mice. Genes Dev. 2006;20:1569–74. doi: 10.1101/gad.1395006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riggio M, Polo ML, Blaustein M, Colman-Lerner A, Lüthy I, Lanari C, et al. PI3K/AKT pathway regulates phosphorylation of steroid receptors, hormone independence and tumor differentiation in breast cancer. Carcinogenesis. 2012;33:509–18. doi: 10.1093/carcin/bgr303. [DOI] [PubMed] [Google Scholar]
- 41.Pons-Tostivint E, Thibault B, Guillermet-Guibert J. Targeting PI3K signaling in combination cancer therapy. Trends Cancer. 2017;3:454–69. doi: 10.1016/j.trecan.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 42.de Bono JS, De Giorgi U, Rodrigues DN, Massard C, Bracarda S, Font A, et al. Randomized phase II study evaluating Akt blockade with ipatasertib, in combination with abiraterone, in patients with metastatic prostate cancer with and without PTEN loss. Clin Cancer Res. 2019;25:928–36. doi: 10.1158/1078-0432.CCR-18-0981. [DOI] [PubMed] [Google Scholar]
- 43.Xing Y, Lin NU, Maurer MA, Chen H, Mahvash A, Sahin A, et al. Phase II trial of AKT inhibitor MK-2206 in patients with advanced breast cancer who have tumors with PIK3CA or AKT mutations, and/or PTEN loss/PTEN mutation. Breast Cancer Res. 2019;21:1–12. doi: 10.1186/s13058-019-1154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 45.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–77. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han J, Back SH, Hur J, Lin Y-H, Gildersleeve R, Shan J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15:481–90. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–9. doi: 10.1016/S0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 48.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–91. doi: 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 49.Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: assembling the IRE1α interactome. Mol Cell. 2009;35:551–61. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–31. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–7. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins: involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273:33741–9. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Guo Y, Tang J, Jiang J, Chen Z. New insights into the roles of CHOP-induced apoptosis in ER stress. Acta Biochim Biophys Sin. 2014;46:629–40. doi: 10.1093/abbs/gmu048. [DOI] [PubMed] [Google Scholar]
- 54.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–58. doi: 10.1016/S0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 55.Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. J Cell Biol. 2012;197:857–67. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Athanasiou D, Aguila M, Bellingham J, Kanuga N, Adamson P, Cheetham ME. The role of the ER stress-response protein PERK in rhodopsin retinitis pigmentosa. Hum Mol Genet. 2017;26:4896–905. doi: 10.1093/hmg/ddx370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Storm M, Sheng X, Arnoldussen YJ, Saatcioglu F. Prostate cancer and the unfolded protein response. Oncotarget. 2016;7:54051. doi: 10.18632/oncotarget.9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Auf G, Jabouille A, Guérit S, Pineau R, Delugin M, Bouchecareilh M, et al. Inositol-requiring enzyme 1α is a key regulator of angiogenesis and invasion in malignant glioma. Proc Natl Acad Sci USA. 2010;107:15553–8. doi: 10.1073/pnas.0914072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng H, Takahashi H, Li X, Hara T, Masuda S, Guan Y, et al. Overexpression of GRP78 and GRP94 are markers for aggressive behavior and poor prognosis in gastric carcinomas. Hum Pathol. 2008;39:1042–9. doi: 10.1016/j.humpath.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 60.Urra H, Dufey E, Avril T, Chevet E, Hetz C. Endoplasmic reticulum stress and the hallmarks of cancer. Trends Cancer. 2016;2:252–62. doi: 10.1016/j.trecan.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 61.Chan S-W. The unfolded protein response in virus infections. Front Microbiol. 2014;5:518. doi: 10.3389/fmicb.2014.00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, Anderson DH, Liu Q, Zhou Y. Mechanism of influenza A virus NS1 protein interaction with the p85β, but not the p85α, subunit of phosphatidylinositol 3-kinase (PI3K) and up-regulation of PI3K activity. J Biol Chem. 2008;283:23397–409. doi: 10.1074/jbc.M802737200. [DOI] [PubMed] [Google Scholar]
- 63.Matsuda M, Suizu F, Hirata N, Miyazaki T, Obuse C, Noguchi M. Characterization of the interaction of influenza virus NS1 with Akt. Biochem Biophys Res Commun. 2010;395:312–7. doi: 10.1016/j.bbrc.2010.03.166. [DOI] [PubMed] [Google Scholar]
- 64.Yuan H, Veldman T, Rundell K, Schlegel R. Simian virus 40 small tumor antigen activates AKT and telomerase and induces anchorage-independent growth of human epithelial cells. J Virol. 2002;76:10685–91. doi: 10.1128/JVI.76.21.10685-10691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pim D, Massimi P, Dilworth SM, Banks L. Activation of the protein kinase B pathway by the HPV-16 E7 oncoprotein occurs through a mechanism involving interaction with PP2A. Oncogene. 2005;24:7830–8. doi: 10.1038/sj.onc.1208935. [DOI] [PubMed] [Google Scholar]
- 66.Bitko V, Shulyayeva O, Mazumder B, Musiyenko A, Ramaswamy M, Look DC, et al. Nonstructural proteins of respiratory syncytial virus suppress premature apoptosis by an NF-κB-dependent, interferon-independent mechanism and facilitate virus growth. J Virol. 2007;81:1786–95. doi: 10.1128/JVI.01420-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Groskreutz DJ, Monick MM, Yarovinsky TO, Powers LS, Quelle DE, Varga SM, et al. Respiratory syncytial virus decreases p53 protein to prolong survival of airway epithelial cells. J Immunol. 2007;179:2741–7. doi: 10.4049/jimmunol.179.5.2741. [DOI] [PubMed] [Google Scholar]
- 68.Blanco J, Cameirao C, López MC, Muñoz-Barroso I. Phosphatidylinositol-3-kinase-Akt pathway in negative-stranded RNA virus infection: a minireview. Arch Virol. 2020;165:2165–76. [DOI] [PubMed]
- 69.White CL, Chattopadhyay S, Sen GC. Phosphatidylinositol 3-kinase signaling delays Sendai virus-induced apoptosis by preventing XIAP degradation. J Virol. 2011;85:5224–7. doi: 10.1128/JVI.00053-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kang Y, Yuan R, Zhao X, Xiang B, Gao S, Gao P, et al. Transient activation of the PI3K/Akt pathway promotes Newcastle disease virus replication and enhances anti-apoptotic signaling responses. Oncotarget. 2017;8:23551–63. doi: 10.18632/oncotarget.15796. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Soares JA, Leite FG, Andrade LG, Torres AA, De Sousa LP, Barcelos LS, et al. Activation of the PI3K/Akt pathway early during vaccinia and cowpox virus infections is required for both host survival and viral replication. J Virol. 2009;83:6883–99. doi: 10.1128/JVI.00245-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McNulty S, Bornmann W, Schriewer J, Werner C, Smith SK, Olson VA, et al. Multiple phosphatidylinositol 3-kinases regulate vaccinia virus morphogenesis. PLoS One. 2010;5:e10884. doi: 10.1371/journal.pone.0010884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tiwari V, Shukla D. Phosphoinositide 3 kinase signalling may affect multiple steps during herpes simplex virus type-1 entry. J Gen Virol. 2010;91:3002. doi: 10.1099/vir.0.024166-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Z, Tian Y, Machida K, Lai MM, Luo G, Foung SK, et al. Transient activation of the PI3K-AKT pathway by hepatitis C virus to enhance viral entry. J Biol Chem. 2012;287:41922–30. doi: 10.1074/jbc.M112.414789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ehrhardt C, Marjuki H, Wolff T, Nürnberg B, Planz O, Pleschka S, et al. Bivalent role of the phosphatidylinositol‐3‐kinase (PI3K) during influenza virus infection and host cell defence. Cell Microbiol. 2006;8:1336–48. doi: 10.1111/j.1462-5822.2006.00713.x. [DOI] [PubMed] [Google Scholar]
- 76.Liu X, Cohen JI. The role of PI3K/Akt in human herpesvirus infection: from the bench to the bedside. Virology. 2015;479:568–77. doi: 10.1016/j.virol.2015.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saeed MF, Kolokoltsov AA, Freiberg AN, Holbrook MR, Davey RA. Phosphoinositide-3 kinase-Akt pathway controls cellular entry of Ebola virus. PLoS Pathog. 2008;4:e1000141. doi: 10.1371/journal.ppat.1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheshenko N, Trepanier JB, González PA, Eugenin EA, Jacobs WR, Jr, Herold BC. Herpes simplex virus type 2 glycoprotein H interacts with integrin αvβ3 to facilitate viral entry and calcium signaling in human genital tract epithelial cells. J Virol. 2014;88:10026–38. doi: 10.1128/JVI.00725-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheshenko N, Pierce C, Herold BC. Herpes simplex viruses activate phospholipid scramblase to redistribute phosphatidylserines and Akt to the outer leaflet of the plasma membrane and promote viral entry. PLoS Pathog. 2018;14:e1006766. doi: 10.1371/journal.ppat.1006766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cheshenko N, Trepanier JB, Stefanidou M, Buckley N, Gonzalez P, Jacobs W, et al. HSV activates Akt to trigger calcium release and promote viral entry: novel candidate target for treatment and suppression. FASEB J. 2013;27:2584–99. doi: 10.1096/fj.12-220285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sciabica KS, Dai QJ, Sandri‐Goldin RM. ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J. 2003;22:1608–19. doi: 10.1093/emboj/cdg166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pozzi B, Bragado L, Mammi P, Torti MF, Gaioli N, Gebhard LG, et al. Dengue virus targets RBM10 deregulating host cell splicing and innate immune response. Nucleic acids Res. 2020;48:6824–38. doi: 10.1093/nar/gkaa340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Maio FA, Risso G, Iglesias NG, Shah P, Pozzi B, Gebhard LG, et al. The dengue virus NS5 protein intrudes in the cellular spliceosome and modulates splicing. PLoS Pathog. 2016;12:e1005841. doi: 10.1371/journal.ppat.1005841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Blaustein M, Pelisch F, Tanos T, Munoz MJ, Wengier D, Quadrana L, et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat Struct Mol Biol. 2005;12:1037–44. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- 85.Zhou Z, Qiu J, Liu W, Zhou Y, Plocinik RM, Li H, et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell. 2012;47:422–33. doi: 10.1016/j.molcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hillebrand F, Erkelenz S, Diehl N, Widera M, Noffke J, Avota E, et al. The PI3K pathway acting on alternative HIV-1 pre-mRNA splicing. J Gen Virol. 2014;95:1809–15. doi: 10.1099/vir.0.064618-0. [DOI] [PubMed] [Google Scholar]
- 87.Kajitani N, Glahder J, Wu C, Yu H, Nilsson K, Schwartz S. hnRNP L controls HPV16 RNA polyadenylation and splicing in an Akt kinase-dependent manner. Nucleic Acids Res. 2017;45:9654–78. doi: 10.1093/nar/gkx606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blaustein M, Quadrana L, Risso G, Mata Mde L, Pelisch F, Srebrow A. SF2/ASF regulates proteomic diversity by affecting the balance between translation initiation mechanisms. J Cell Biochem. 2009;107:826–33. doi: 10.1002/jcb.22181. [DOI] [PubMed] [Google Scholar]
- 89.Zhang L, Wu J, Ling MT, Zhao L, Zhao K-N. The role of the PI3K/Akt/mTOR signalling pathway in human cancers induced by infection with human papillomaviruses. Mol Cancer. 2015;14:1–13. doi: 10.1186/s12943-015-0361-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhan Y, Yu S, Yang S, Qiu X, Meng C, Tan L, et al. Newcastle disease virus infection activates PI3K/Akt/mTOR and p38 MAPK/Mnk1 pathways to benefit viral mRNA translation via interaction of the viral NP protein and host eIF4E. PLoS Pathog. 2020;16:e1008610. doi: 10.1371/journal.ppat.1008610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10:878–89. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ji W-T, Liu HJ. PI3K-Akt signaling and viral infection. Recent Pat Biotechnol. 2008;2:218–26. doi: 10.2174/187220808786241042. [DOI] [PubMed] [Google Scholar]
- 93.Cheng D, Zhang L, Yang G, Zhao L, Peng F, Tian Y, et al. Hepatitis C virus NS 5A drives a PTEN‐PI 3K/Akt feedback loop to support cell survival. Liver Int. 2015;35:1682–91. doi: 10.1111/liv.12733. [DOI] [PubMed] [Google Scholar]
- 94.Sodhi A, Montaner S, Patel V, Gómez-Román JJ, Li Y, Sausville EA, et al. Akt plays a central role in sarcomagenesis induced by Kaposi’s sarcoma herpesvirus-encoded G protein-coupled receptor. Proc Natl Acad Sci USA. 2004;101:4821–6. doi: 10.1073/pnas.0400835101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tomlinson CC, Damania B. The K1 protein of Kaposi’s sarcoma-associated herpesvirus activates the Akt signaling pathway. J Virol. 2004;78:1918–27. doi: 10.1128/JVI.78.4.1918-1927.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen J. Roles of the PI3K/Akt pathway in Epstein-Barr virus-induced cancers and therapeutic implications. World J Virol. 2012;1:154. doi: 10.5501/wjv.v1.i6.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chung T-W, Lee Y-C, Ko J-H, Kim C-H, Hepatitis B. Virus X protein modulates the expression of PTEN by inhibiting the function of p53, a transcriptional activator in liver cells. Cancer Res. 2003;63:3453–8. [PubMed] [Google Scholar]
- 98.Mizutani T. Signal transduction in SARS-CoV-infected cells. Ann N. Y Acad Sci. 2007;1102:86–95. doi: 10.1196/annals.1408.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Surjit M, Liu B, Jameel S, Chow VTK, Lal SK. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochemical J. 2004;383:13–18. doi: 10.1042/BJ20040984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. Importance of Akt signaling pathway for apoptosis in SARS-CoV-infected Vero E6 cells. Virology. 2004;327:169–74. doi: 10.1016/j.virol.2004.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. JNK and PI3k/Akt signaling pathways are required for establishing persistent SARS-CoV infection in Vero E6 cells. Biochim Biophys Acta. 2005;1741:4–10. doi: 10.1016/j.bbadis.2005.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mizutani T, Fukushi S, Iizuka D, Inanami O, Kuwabara M, Takashima H, et al. Inhibition of cell proliferation by SARS-CoV infection in Vero E6 cells. FEMS Immunol Med Microbiol. 2006;46:236–43. doi: 10.1111/j.1574-695X.2005.00028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mizutani T, Fukushi S, Ishii K, Sasaki Y, Kenri T, Saijo M, et al. Mechanisms of establishment of persistent SARS-CoV-infected cells. Biochemical Biophysical Res Commun. 2006;347:261–5. doi: 10.1016/j.bbrc.2006.06.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chan C-M, Ma C-W, Chan W-Y, Chan HYE. The SARS-Coronavirus membrane protein induces apoptosis through modulating the Akt survival pathway. Arch Biochem Biophysics. 2007;459:197–207. doi: 10.1016/j.abb.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tsoi H, Li L, Chen ZS, Lau K-F, Tsui SKW, Chan HYE. The SARS-coronavirus membrane protein induces apoptosis via interfering with PDK1–PKB/Akt signalling. Biochemical J. 2014;464:439–47. doi: 10.1042/BJ20131461. [DOI] [PubMed] [Google Scholar]
- 106.Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Correa Marrero M, et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712.e19. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Parthasarathi KT, Munjal NS, Dey G, Kumar A, Pandey A, Balakrishnan L, et al. A pathway map of signaling events triggered upon SARS-CoV infection. J cell Commun Signal. 2021;15:595–600. doi: 10.1007/s12079-021-00642-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ahsan N, Rao RSP, Wilson RS, Punyamurtula U, Salvato F, Petersen M, et al. Mass spectrometry‐based proteomic platforms for better understanding of SARS‐CoV‐2 induced pathogenesis and potential diagnostic approaches. Proteomics. 2021;21:2000279. doi: 10.1002/pmic.202000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ping H, Zhang K, Wang Y, Tong X, Chen Z, Cai C, et al. Cell death and pathological findings of the spleen in COVID-19 patients. Pathol Res Pract. 2021;227:153610. doi: 10.1016/j.prp.2021.153610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rex D, Dagamajalu S, Kandasamy RK, Raju R, Prasad T. SARS-CoV-2 signaling pathway map: a functional landscape of molecular mechanisms in COVID-19. J Cell Commun Signal. 2021;15:601–8. doi: 10.1007/s12079-021-00632-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stukalov A, Girault V, Grass V, Karayel O, Bergant V, Urban C, et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature. 2021;594:246–52. doi: 10.1038/s41586-021-03493-4. [DOI] [PubMed] [Google Scholar]
- 112.Nicoletti AS, Visacri MB, da Ronda CRDSC, Vasconcelos PEDNS, Quintanilha JCF, de Souza RN et al. Differentially expressed plasmatic microRNAs in Brazilian patients with coronavirus disease 2019 (COVID-19): preliminary results. Mol Biol Rep. 2022;49:1–13. [DOI] [PMC free article] [PubMed]
- 113.Chatterjee B, Thakur SS. SARS-CoV-2 infection triggers phosphorylation: potential target for Anti-COVID-19 therapeutics. Front Immunol. 2022;13:829474. [DOI] [PMC free article] [PubMed]
- 114.Khezri MR. PI3K/AKT signaling pathway: a possible target for adjuvant therapy in COVID-19. Hum Cell. 2021;34:700–1. doi: 10.1007/s13577-021-00484-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li F, Li J, Wang P-H, Yang N, Huang J, Ou J, et al. SARS-CoV-2 spike promotes inflammation and apoptosis through autophagy by ROS-suppressed PI3K/AKT/mTOR signaling. Biochim Biophysica Acta. 2021;1867:166260. doi: 10.1016/j.bbadis.2021.166260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Callahan V, Hawks S, Crawford MA, Lehman CW, Morrison HA, Ivester HM, et al. The pro-inflammatory chemokines CXCL9, CXCL10 and CXCL11 are upregulated following SARS-CoV-2 infection in an AKT-dependent manner. Viruses. 2021;13:1062. doi: 10.3390/v13061062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ren Y, Wang A, Fang Y, Shu T, Wu D, Wang C et al. SARS-CoV-2 membrane glycoprotein M triggers apoptosis with the assistance of nucleocapsid protein N in cells. Front Cell Infect Microbiol. 2021;11:627. [DOI] [PMC free article] [PubMed]
- 118.Malgotra V, Sharma V. 2-Deoxy-d-glucose inhibits replication of novel coronavirus (SARS-CoV-2) with adverse effects on host cell metabolism. Clin trials. 2021;7:10. [Google Scholar]
- 119.Pelzl L, Singh A, Funk J, Witzemann A, Marini I, Zlamal J, et al. Antibody‐mediated procoagulant platelet formation in COVID‐19 is AKT dependent. J Thrombosis Haemost. 2022;20:387–98. doi: 10.1111/jth.15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Somanath PR. Is targeting Akt a viable option to treat advanced-stage COVID-19 patients? Am J Physiol Lung Cell Mol Physiol. 2020;319:L45–L47. doi: 10.1152/ajplung.00124.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang F, Chen R, Li W, Zhu H, Chen X, Hou Z, et al. D-Limonene is a potential monoterpene to inhibit PI3K/Akt/IKK-α/NF-κB p65 signaling pathway in coronavirus dsease 2019 pulmonary fibrosis. Front Med. 2021;8:140. doi: 10.3389/fmed.2021.591830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Leonardi AJ, Proenca RB. Akt-Fas to quell aberrant T cell differentiation and apoptosis in Covid-19. Front Immunol. 2020. 10.3389/fimmu.2020.600405. [DOI] [PMC free article] [PubMed]
- 123.Li S, Kong L, Yu X. The expanding roles of endoplasmic reticulum stress in virus replication and pathogenesis. Crit Rev Microbiol. 2015;41:150–64. doi: 10.3109/1040841X.2013.813899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Inoue T, Tsai B. How viruses use the endoplasmic reticulum for entry, replication, and assembly. Cold Spring Harb Perspect Biol. 2013;5:a013250. doi: 10.1101/cshperspect.a013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Smith JA. A new paradigm: innate immune sensing of viruses via the unfolded protein response. Front Microbiol. 2014;5:222. doi: 10.3389/fmicb.2014.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kohli E, Causse S, Baverel V, Dubrez L, Borges-Bonan N, Demidov O, et al. Endoplasmic reticulum chaperones in viral infection: therapeutic perspectives. Microbiol Mol Biol Rev. 2021;85:e00035–21. doi: 10.1128/MMBR.00035-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Choi J, Song C-H. Insights into the role of endoplasmic reticulum stress in infectious diseases. Front Immunol. 2020;10:3147. doi: 10.3389/fimmu.2019.03147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhao D, Yang J, Han K, Liu Q, Wang H, Liu Y, et al. The unfolded protein response induced by Tembusu virus infection. BMC Vet Res. 2019;15:1–13. doi: 10.1186/s12917-019-1781-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pasqual G, Burri DJ, Pasquato A, de la Torre JC, Kunz S. Role of the host cell’s unfolded protein response in arenavirus infection. J Virol. 2011;85:1662–70. doi: 10.1128/JVI.01782-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Galindo I, Hernaez B, Munoz-Moreno R, Cuesta-Geijo M, Dalmau-Mena I, Alonso C. The ATF6 branch of unfolded protein response and apoptosis are activated to promote African swine fever virus infection. Cell Death Dis. 2012;3:e341–e341. doi: 10.1038/cddis.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ambrose RL, Mackenzie JM. ATF6 signaling is required for efficient West Nile virus replication by promoting cell survival and inhibition of innate immune responses. J Virol. 2013;87:2206–14. doi: 10.1128/JVI.02097-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li B, Gao B, Ye L, Han X, Wang W, Kong L, et al. Hepatitis B virus X protein (HBx) activates ATF6 and IRE1-XBP1 pathways of unfolded protein response. Virus Res. 2007;124:44–49. doi: 10.1016/j.virusres.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 133.Tan Z, Zhang W, Sun J, Fu Z, Ke X, Zheng C, et al. ZIKV infection activates the IRE1-XBP1 and ATF6 pathways of unfolded protein response in neural cells. J Neuroinflammation. 2018;15:1–16. doi: 10.1186/s12974-018-1311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li J, He J, Fu Y, Hu X, Sun L-Q, Huang Y, et al. Hepatitis B virus X protein inhibits apoptosis by modulating endoplasmic reticulum stress response. Oncotarget. 2017;8:96027. doi: 10.18632/oncotarget.21630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang J, Kang R, Huang H, Xi X, Wang B, Wang J, et al. Hepatitis C virus core protein activates autophagy through EIF2AK3 and ATF6 UPR pathway-mediated MAP1LC3B and ATG12 expression. Autophagy. 2014;10:766–84. doi: 10.4161/auto.27954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hou L, Dong J, Zhu S, Yuan F, Wei L, Wang J, et al. Seneca valley virus activates autophagy through the PERK and ATF6 UPR pathways. Virology. 2019;537:254–63. doi: 10.1016/j.virol.2019.08.029. [DOI] [PubMed] [Google Scholar]
- 137.Ron D, Hampton RY. Membrane biogenesis and the unfolded protein response. J Cell Biol. 2004;167:23–25. doi: 10.1083/jcb.200408117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hassan IH, Zhang MS, Powers LS, Shao JQ, Baltrusaitis J, Rutkowski DT, et al. Influenza A viral replication is blocked by inhibition of the inositol-requiring enzyme 1 (IRE1) stress pathway. J Biol Chem. 2012;287:4679–89. doi: 10.1074/jbc.M111.284695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Su H-L, Liao C-L, Lin Y-L. Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J Virol. 2002;76:4162–71. doi: 10.1128/JVI.76.9.4162-4171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yu C-Y, Hsu Y-W, Liao C-L, Lin Y-L. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virol. 2006;80:11868–80. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Medigeshi GR, Lancaster AM, Hirsch AJ, Briese T, Lipkin WI, DeFilippis V, et al. West Nile virus infection activates the unfolded protein response, leading to CHOP induction and apoptosis. J Virol. 2007;81:10849–60. doi: 10.1128/JVI.01151-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhu E, Wu H, Chen W, Qin Y, Liu J, Fan S, et al. Classical swine fever virus employs the PERK-and IRE1-dependent autophagy for viral replication in cultured cells. Virulence. 2021;12:130–49. doi: 10.1080/21505594.2020.1845040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rohde C, Becker S, Krähling V. Marburg virus regulates the IRE1/XBP1-dependent unfolded protein response to ensure efficient viral replication. Emerg Microbes Infect. 2019;8:1300–13. doi: 10.1080/22221751.2019.1659552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Breitkopf VJ, Dobler G, Claus P, Naim HY, Steffen I. IRE1-mediated unfolded protein response promotes the replication of tick-Borne flaviviruses in a virus and cell-type dependent manner. Viruses. 2021;13:2164. doi: 10.3390/v13112164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Caselli E, Benedetti S, Gentili V, Grigolato J, Di Luca D. Activating transcription factor 4 (ATF4) promotes HIV type 1 activation. AIDS Res Hum Retrovir. 2012;28:907–12. doi: 10.1089/aid.2011.0252. [DOI] [PubMed] [Google Scholar]
- 146.Isler JA, Skalet AH, Alwine JC. Human cytomegalovirus infection activates and regulates the unfolded protein response. J Virol. 2005;79:6890–9. doi: 10.1128/JVI.79.11.6890-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xuan B, Qian Z, Torigoi E, Yu D. Human cytomegalovirus protein pUL38 induces ATF4 expression, inhibits persistent JNK phosphorylation, and suppresses endoplasmic reticulum stress-induced cell death. J Virol. 2009;83:3463–74. doi: 10.1128/JVI.02307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Qian Z, Xuan B, Chapa TJ, Gualberto N, Yu D. Murine cytomegalovirus targets transcription factor ATF4 to exploit the unfolded-protein response. J Virol. 2012;86:6712–23. doi: 10.1128/JVI.00200-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gao P, Chai Y, Song J, Liu T, Chen P, Zhou L, et al. Reprogramming the unfolded protein response for replication by porcine reproductive and respiratory syndrome virus. PLoS Pathog. 2019;15:e1008169. doi: 10.1371/journal.ppat.1008169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Burnett HF, Audas TE, Liang G, Lu RR. Herpes simplex virus-1 disarms the unfolded protein response in the early stages of infection. Cell Stress Chaperones. 2012;17:473–83. doi: 10.1007/s12192-012-0324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Su A, Wang H, Li Y, Wang X, Chen D, Wu Z. Opposite roles of RNase and kinase activities of inositol-requiring enzyme 1 (IRE1) on HSV-1 replication. Viruses. 2017;9:235. doi: 10.3390/v9090235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tang BS, Chan K, Cheng VC, Woo PC, Lau SK, Lam CC, et al. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by severe acute respiratory syndrome coronavirus and human coronavirus 229E. J Virol. 2005;79:6180–93. doi: 10.1128/JVI.79.10.6180-6193.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chan C-P, Siu K-L, Chin K-T, Yuen K-Y, Zheng B, Jin D-Y. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2006;80:9279–87. doi: 10.1128/JVI.00659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yeung Y-S, Yip C-W, Hon C-C, Chow KY, Ma IC, Zeng F, et al. Transcriptional profiling of Vero E6 cells over-expressing SARS-CoV S2 subunit: insights on viral regulation of apoptosis and proliferation. Virology. 2008;371:32–43. doi: 10.1016/j.virol.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fung TS, Liu DX. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13:405–30. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fukushi M, Yoshinaka Y, Matsuoka Y, Hatakeyama S, Ishizaka Y, Kirikae T, et al. Monitoring of S protein maturation in the endoplasmic reticulum by calnexin is important for the infectivity of severe acute respiratory syndrome coronavirus. J Virol. 2012;86:11745–53. doi: 10.1128/JVI.01250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Stertz S, Reichelt M, Spiegel M, Kuri T, Martínez-Sobrido L, García-Sastre A, et al. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–15. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1 a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.van der Sanden MH, Houweling M, van Golde LM, Vaandrager AB. Inhibition of phosphatidylcholine synthesis induces expression of the endoplasmic reticulum stress and apoptosis-related protein CCAAT/enhancer-binding protein-homologous protein (CHOP/GADD153) Biochem J. 2003;369:643–50. doi: 10.1042/bj20020285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Minakshi R, Padhan K, Rani M, Khan N, Ahmad F, Jameel S. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PloS One. 2009;4:e8342. doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.DeDiego ML, Nieto-Torres JL, Jiménez-Guardeño JM, Regla-Nava JA, Alvarez E, Oliveros JC, et al. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog. 2011;7:e1002315. doi: 10.1371/journal.ppat.1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bartolini D, Stabile AM, Vacca C, Pistilli A, Rende M, Gioiello A, et al. Endoplasmic reticulum stress and NF‐kB activation in SARS‐CoV‐2 infected cells and their response to antiviral therapy. IUBMB life. 2022;74:93–100. doi: 10.1002/iub.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Boga JA, Coto-Montes A. ER stress and autophagy induced by SARS-CoV-2: The targets for melatonin treatment. Melatonin Res. 2020;3:346–61. doi: 10.32794/mr11250067. [DOI] [Google Scholar]
- 165.Siri M, Dastghaib S, Zamani M, Rahmani-Kukia N, Geraylow KR, Fakher S, et al. Autophagy, unfolded protein response, and neuropilin-1 cross-talk in SARS-CoV-2 infection: what can Be learned from other coronaviruses. Int J Mol Sci. 2021;22:5992. doi: 10.3390/ijms22115992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rashid F, Dzakah EE, Wang H, Tang S. The ORF8 protein of SARS-CoV-2 induced endoplasmic reticulum stress and mediated immune evasion by antagonizing production of interferon beta. Virus Res. 2021;296:198350. doi: 10.1016/j.virusres.2021.198350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Elfiky AA, Baghdady AM, Ali SA, Ahmed MI. GRP78 targeting: hitting two birds with a stone. Life Sci. 2020;260:118317. [DOI] [PMC free article] [PubMed]
- 168.Ibrahim IM, Abdelmalek DH, Elfiky AA. GRP78: A cell’s response to stress. Life Sci. 2019;226:156–63. doi: 10.1016/j.lfs.2019.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tsai Y-L, Lee AS Cell surface GRP78: anchoring and translocation mechanisms and therapeutic potential in cancer. In: Pizzo SV, editor. Cell surface GRP78, a new paradigm in signal transduction biology. Elsevier; 2018. p. 41–62.
- 170.Ibrahim IM, Abdelmalek DH, Elshahat ME, Elfiky AA. COVID-19 spike-host cell receptor GRP78 binding site prediction. J Infect. 2020;80:554–62. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Balakrishnan B, Lai K. Modulation of SARS-CoV-2 spike-induced Unfolded Protein Response (UPR) in HEK293T cells by selected small chemical molecules. bioRxiv. 2021;80:9279–87.
- 172.Rosa-Fernandes L, Lazari LC, da Silva JM, de Morais Gomes V, Machado RRG, dos Santos AF et al. SARS-CoV-2 activates ER stress and Unfolded protein response. bioRxiv. 2021. 10.1101/2021.06.21.449284.
- 173.Su W, Yu X, Zhou C. SARS-CoV-2 ORF3a induces incomplete autophagy via the unfolded protein response. Viruses. 2021;13:2467. doi: 10.3390/v13122467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hu P, Han Z, Couvillon AD, Exton JH. Critical role of endogenous Akt/IAPs and MEK1/ERK pathways in counteracting endoplasmic reticulum stress-induced cell death. J Biol Chem. 2004;279:49420–9. doi: 10.1074/jbc.M407700200. [DOI] [PubMed] [Google Scholar]
- 175.Mao W, Iwai C, Keng PC, Vulapalli R, Liang C. Norepinephrine-induced oxidative stress causes PC-12 cell apoptosis by both endoplasmic reticulum stress and mitochondrial intrinsic pathway: inhibition of phosphatidylinositol 3-kinase survival pathway. Am J Physiol-Cell Physiol. 2006;290:C1373–C1384. doi: 10.1152/ajpcell.00369.2005. [DOI] [PubMed] [Google Scholar]
- 176.Hu M-C, Gong H-Y, Lin G-H, Hu S-Y, Chen MH-C, Huang S-J, et al. XBP-1, a key regulator of unfolded protein response, activates transcription of IGF1 and Akt phosphorylation in zebrafish embryonic cell line. Biochem Biophys Res Commun. 2007;359:778–83. doi: 10.1016/j.bbrc.2007.05.183. [DOI] [PubMed] [Google Scholar]
- 177.Jiang CC, Yang F, Thorne RF, Zhu BK, Hersey P, Zhang XD. Human melanoma cells under endoplasmic reticulum stress acquire resistance to microtubule-targeting drugs through XBP-1-mediated activation of Akt. Neoplasia. 2009;11:436. doi: 10.1593/neo.09208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dong L, Jiang CC, Thorne RF, Croft A, Yang F, Liu H, et al. Ets-1 mediates upregulation of Mcl-1 downstream of XBP-1 in human melanoma cells upon ER stress. Oncogene. 2011;30:3716–26. doi: 10.1038/onc.2011.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ishigaki S, Fonseca SG, Oslowski CM, Jurczyk A, Shearstone JR, Zhu LJ, et al. AATF mediates an antiapoptotic effect of the unfolded protein response through transcriptional regulation of AKT1. Cell Death Differ. 2010;17:774–86. doi: 10.1038/cdd.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Qin L, Wang Z, Tao L, Wang Y. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy. 2010;6:239–47. doi: 10.4161/auto.6.2.11062. [DOI] [PubMed] [Google Scholar]
- 181.Deldicque L, Bertrand L, Patton A, Francaux M, Baar K. ER stress induces anabolic resistance in muscle cells through PKB-induced blockade of mTORC1. PLoS One. 2011;6:e20993. doi: 10.1371/journal.pone.0020993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kato H, Nakajima S, Saito Y, Takahashi S, Katoh R, Kitamura M. mTORC1 serves ER stress-triggered apoptosis via selective activation of the IRE1–JNK pathway. Cell Death Differ. 2012;19:310–20. doi: 10.1038/cdd.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mounir Z, Krishnamoorthy JL, Wang S, Papadopoulou B, Campbell S, Muller WJ, et al. Akt determines cell fate through inhibition of the PERK-eIF2α phosphorylation pathway. Sci Signal. 2011;4:ra62. doi: 10.1126/scisignal.2001630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Winnay JN, Solheim MH, Sakaguchi M, Njølstad PR, Kahn CR. Inhibition of the PI 3-kinase pathway disrupts the unfolded protein response and reduces sensitivity to ER stress-dependent apoptosis. FASEB J. 2020;34:12521–32. [DOI] [PMC free article] [PubMed]
- 185.Wang Y, He Z, Yang Q, Zhou G. XBP 1 inhibits mesangial cell apoptosis in response to oxidative stress via the PTEN/AKT pathway in diabetic nephropathy. FEBS Open Bio. 2019;9:1249–58. doi: 10.1002/2211-5463.12655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 186.Zhang J, Zhong W, Liu Y, Chen W, Lu Y, Zeng Z, et al. Extracellular HSP90α interacts with ER stress to promote fibroblasts activation through PI3K/AKT pathway in pulmonary fibrosis. Front Pharmacol. 2021;12:708462. doi: 10.3389/fphar.2021.708462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sanchez-Alvarez M, Del Pozo MA, Bakal C. AKT-mTOR signaling modulates the dynamics of IRE1 RNAse activity by regulating ER-mitochondria contacts. Sci Rep. 2017;7:1–15. doi: 10.1038/s41598-017-16662-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wang S, Duan J, Liao J, Wang Y, Xiao X, Li L, et al. LncRNA H19 inhibits ER stress induced apoptosis and improves diabetic cardiomyopathy by regulating PI3K/AKT/mTOR axis. Aging (Albany NY) 2022;14:6809. doi: 10.18632/aging.204256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Fleifel AM, Soubh AA, Abdallah DM, Ahmed KA, El-Abhar HS. Preferential effect of Montelukast on Dapagliflozin: Modulation of IRS-1/AKT/GLUT4 and ER stress response elements improves insulin sensitivity in soleus muscle of a type-2 diabetic rat model. Life Sci. 2022;307:120865. doi: 10.1016/j.lfs.2022.120865. [DOI] [PubMed] [Google Scholar]
- 190.Shao Z, Wu P, Wang X, Jin M, Liu S, Ma X, et al. Tetramethylpyrazine protects against early brain injury and inhibits the PERK/Akt pathway in a rat model of subarachnoid hemorrhage. Neurochem Res. 2018;43:1650–9. doi: 10.1007/s11064-018-2581-0. [DOI] [PubMed] [Google Scholar]
- 191.Icard P, Lincet H, Wu Z, Coquerel A, Forgez P, Alifano M, et al. The key role of Warburg effect in SARS-CoV-2 replication and associated inflammatory response. Biochimie. 2021;180:169–77. doi: 10.1016/j.biochi.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Scriven P, Brown NJ, Pockley AG, Wyld L. The unfolded protein response and cancer: a brighter future unfolding? J Mol Med. 2007;85:331–41. doi: 10.1007/s00109-006-0150-5. [DOI] [PubMed] [Google Scholar]
- 193.Mukhopadhyay D, AlSawaftah N, Husseini GA. Identification of novel microRNAs targeting SARS-CoV-2 through the regulation of TMPRSS2/PI3K/AKT/PTEN alignment in lung cancer: an in Silico. Anal ACS Pharm Transl Sci. 2021;4:1075–8. doi: 10.1021/acsptsci.1c00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Winnay JN, Boucher J, Mori MA, Ueki K, Kahn CR. A regulatory subunit of phosphoinositide 3-kinase increases the nuclear accumulation of X-box–binding protein-1 to modulate the unfolded protein response. Nat Med. 2010;16:438–45. doi: 10.1038/nm.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Özcan U, Cao Q, Yilmaz E, Lee A-H, Iwakoshi NN, Özdelen E, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–61. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 196.Khezri MR, Varzandeh R, Ghasemnejad-Berenji M. The probable role and therapeutic potential of the PI3K/AKT signaling pathway in SARS-CoV-2 induced coagulopathy. Cell Mol Biol Lett. 2022;27:1–10. doi: 10.1186/s11658-022-00308-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Basile MS, Cavalli E, McCubrey J, Hernández-Bello J, Muñoz-Valle JF, Fagone P et al. The PI3K/Akt/mTOR pathway: a potential pharmacological target in COVID-19. Drug Discov Today. 2021;27:848–56. [DOI] [PMC free article] [PubMed]
- 198.Fagone P, Ciurleo R, Lombardo SD, Iacobello C, Palermo CI, Shoenfeld Y, et al. Transcriptional landscape of SARS-CoV-2 infection dismantles pathogenic pathways activated by the virus, proposes unique sex-specific differences and predicts tailored therapeutic strategies. Autoimmun Rev. 2020;19:102571. doi: 10.1016/j.autrev.2020.102571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Abu-Eid R, Ward FJ. Targeting the PI3K/Akt/mTOR pathway: a therapeutic strategy in COVID-19 patients. Immunol Lett. 2021;240:1–8. doi: 10.1016/j.imlet.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.de Oliveira PG, Termini L, Durigon EL, Lepique AP, Sposito AC, Boccardo E. Diacerein: A potential multi-target therapeutic drug for COVID-19. Med Hypotheses. 2020;144:109920. doi: 10.1016/j.mehy.2020.109920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yang C-C, Wu C-J, Chien C-Y, Chien C-T. Green tea polyphenol catechins inhibit coronavirus replication and potentiate the adaptive immunity and autophagy-dependent protective mechanism to improve acute lung injury in mice. Antioxidants. 2021;10:928. doi: 10.3390/antiox10060928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Shirato K, Takanari J, Kizaki T. Standardized extract of asparagus officinalis stem attenuates SARS-CoV-2 spike protein-induced IL-6 and IL-1β production by suppressing p44/42 MAPK and Akt phosphorylation in murine primary macrophages. Molecules. 2021;26:6189. doi: 10.3390/molecules26206189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wang B, Li D, Fiselier A, Kovalchuk I, Kovalchuk O. New AKT-dependent mechanisms of anti-COVID-19 action of high-CBD Cannabis sativa extracts. Cell Death Discov. 2022;8:1–12. doi: 10.1038/s41420-022-00876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Khezri MR, Ghasemnejad‐Berenji M, Moloodsouri D. Hesperetin and the PI3K/AKT pathway: could their interaction play a role in the entry and replication of the SARS‐CoV‐2? J Food Biochem. 2022;46:e14212. [DOI] [PMC free article] [PubMed]
- 205.Garcia G, Jr, Sharma A, Ramaiah A, Sen C, Purkayastha A, Kohn DB, et al. Antiviral drug screen identifies DNA-damage response inhibitor as potent blocker of SARS-CoV-2 replication. Cell Rep. 2021;35:108940. doi: 10.1016/j.celrep.2021.108940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sun F, Mu C, Kwok HF, Xu J, Wu Y, Liu W, et al. Capivasertib restricts SARS-CoV-2 cellular entry: a potential clinical application for COVID-19. Int J Biol Sci. 2021;17:2348. doi: 10.7150/ijbs.57810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Appelberg S, Gupta S, Svensson Akusjärvi S, Ambikan AT, Mikaeloff F, Saccon E, et al. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg Microbes Infect. 2020;9:1748–60. doi: 10.1080/22221751.2020.1799723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sureda A, Alizadeh J, Nabavi SF, Berindan-Neagoe I, Cismaru CA, Jeandet P, et al. Endoplasmic reticulum as a potential therapeutic target for covid-19 infection management? Eur J Pharmacol. 2020;882:173288. doi: 10.1016/j.ejphar.2020.173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Barabutis N. Unfolded protein response in the COVID-19 context. Aging Health Res. 2020;1:100001. [DOI] [PMC free article] [PubMed]
- 210.Shabbir S, Hafeez A, Rafiq MA, Khan MJ. Estrogen shields women from COVID-19 complications by reducing ER stress. Med Hypotheses. 2020;143:110148. doi: 10.1016/j.mehy.2020.110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Banerjee A, Czinn SJ, Reiter RJ, Blanchard TG. Crosstalk between endoplasmic reticulum stress and anti-viral activities: a novel therapeutic target for COVID-19. Life Sci. 2020;255:117842. doi: 10.1016/j.lfs.2020.117842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Polansky H, Lori G. Coronavirus disease 2019 (COVID-19): first indication of efficacy of Gene-Eden-VIR/Novirin in SARS-CoV-2 infection. Int J Antimicrob Agents. 2020;55:105971. doi: 10.1016/j.ijantimicag.2020.105971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Nabirotchkin S, Peluffo AE, Bouaziz J, Cohen D. Focusing on the unfolded protein response and autophagy related pathways to reposition common approved drugs against COVID-19. 2020. 10.20944/preprints202003.0302.v1.
- 214.Dadey DY, Kapoor V, Hoye K, Khudanyan A, Collins A, Thotala D, et al. Antibody targeting GRP78 enhances the efficacy of radiation therapy in human glioblastoma and non–small cell lung cancer cell lines and tumor models. Clin Cancer Res. 2017;23:2556–64. doi: 10.1158/1078-0432.CCR-16-1935. [DOI] [PubMed] [Google Scholar]
- 215.Pfaffenbach KT, Lee AS. The critical role of GRP78 in physiologic and pathologic stress. Curr Opin Cell Biol. 2011;23:150–6. doi: 10.1016/j.ceb.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhao T, Du J, Zeng H. Interplay between endoplasmic reticulum stress and non-coding RNAs in cancer. J Hematol Oncol. 2020;13:1–20. doi: 10.1186/s13045-020-01002-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Liu R, Li X, Gao W, Zhou Y, Wey S, Mitra SK, et al. Monoclonal antibody against cell surface GRP78 as a novel agent in suppressing PI3K/AKT signaling, tumor growth, and metastasis. Clin Cancer Res. 2013;19:6802–11. doi: 10.1158/1078-0432.CCR-13-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Miller K, McGrath ME, Hu Z, Ariannejad S, Weston S, Frieman M, et al. Coronavirus interactions with the cellular autophagy machinery. Autophagy. 2020;16:2131–9. doi: 10.1080/15548627.2020.1817280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Pereira GJDS, Leão AHFF, Erustes AG, Morais IBM, Vrechi TAM, Zamarioli LDS, et al. Pharmacological modulators of autophagy as a potential strategy for the treatment of COVID-19. Int J Mol Sci. 2021;22:4067. doi: 10.3390/ijms22084067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Gills JJ, LoPiccolo J, Tsurutani J, Shoemaker RH, Best CJ, Abu-Asab MS, et al. Nelfinavir, A lead HIV protease inhibitor, is a broad-spectrum, anticancer agent that induces endoplasmic reticulum stress, autophagy, and apoptosis in vitro and in vivo. Clin Cancer Res. 2007;13:5183–94. doi: 10.1158/1078-0432.CCR-07-0161. [DOI] [PubMed] [Google Scholar]
- 221.Gupta AK, Li B, Cerniglia GJ, Ahmed MS, Hahn SM, Maity A. The HIV protease inhibitor nelfinavir downregulates Akt phosphorylation by inhibiting proteasomal activity and inducing the unfolded protein response. Neoplasia. 2007;9:271–8. doi: 10.1593/neo.07124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ohashi H, Watashi K, Saso W, Shionoya K, Iwanami S, Hirokawa T, et al. Potential anti-COVID-19 agents, cepharanthine and nelfinavir, and their usage for combination treatment. iScience. 2021;24:102367. doi: 10.1016/j.isci.2021.102367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Han S, Bui NT, Ho MT, Kim YM, Cho M, Shin DB. Dexamethasone inhibits TGF-β1–induced cell migration by regulating the ERK and AKT pathways in human colon cancer cells via CYR61. Cancer Res Treat. 2016;48:1141. doi: 10.4143/crt.2015.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Johnson RM, Vinetz JM. Dexamethasone in the management of covid-19. bmj 2020. 10.1136/bmj.m2648. [DOI] [PubMed]
- 225.Sudsaward S, Khunchai S, Thepmalee C, Othman A, Limjindaporn T, Yenchitsomanus P, et al. Endoplasmic reticulum stress, unfolded protein response and autophagy contribute to resistance to glucocorticoid treatment in human acute lymphoblastic leukaemia cells. Int J Oncol. 2020;57:835–44. doi: 10.3892/ijo.2020.5089. [DOI] [PubMed] [Google Scholar]
- 226.Wouters OJ, Shadlen KC, Salcher-Konrad M, Pollard AJ, Larson HJ, Teerawattananon Y et al. Challenges in ensuring global access to COVID-19 vaccines: production, affordability, allocation, and deployment. Lancet. 2021. 10.1016/S0140-6736(21)00306-8. [DOI] [PMC free article] [PubMed]
- 227.Abdool Karim SS, de Oliveira T. New SARS-CoV-2 variants—clinical, public health, and vaccine implications. N. Engl J Med. 2021;384:1866–8. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Schmidt A, Labaki C, Hsu C-Y, Bakouny Z, Balanchivadze N, Berg S, et al. COVID-19 vaccination and breakthrough infections in patients with cancer. Ann Oncol. 2022;33:340–6. doi: 10.1016/j.annonc.2021.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in this article and its supplementary materials.