Abstract
Genomic footprints of pathogens shed by infected individuals can be traced in environmental samples, which can serve as a noninvasive method of infectious disease surveillance. The research evaluates the efficacy of environmental monitoring of SARS-CoV-2 RNA in air, surface swabs and wastewater to predict COVID-19 cases. Using a prospective experimental design, air, surface swabs, and wastewater samples were collected from a college dormitory housing roughly 500 students from March to May 2021 at the University of Miami, Coral Gables, FL. Students were randomly screened for COVID-19 during the study period. SARS-CoV-2 concentration in environmental samples was quantified using Volcano 2nd Generation-qPCR. Descriptive analyses were conducted to examine the associations between time-lagged SARS-CoV-2 in environmental samples and COVID-19 cases. SARS-CoV-2 was detected in air, surface swab and wastewater samples on 52 (63.4 %), 40 (50.0 %) and 57 (68.6 %) days, respectively. On 19 (24 %) of 78 days SARS-CoV-2 was detected in all three sample types. COVID-19 cases were reported on 11 days during the study period and SARS-CoV-2 was also detected two days before the case diagnosis on all 11 (100 %), 9 (81.8 %) and 8 (72.7 %) days in air, surface swab and wastewater samples, respectively. SARS-CoV-2 detection in environmental samples was an indicator of the presence of local COVID-19 cases and a 3-day lead indicator for a potential outbreak at the dormitory building scale. Proactive environmental surveillance of SARS-CoV-2 or other pathogens in multiple environmental media has potential to guide targeted measures to contain and/or mitigate infectious disease outbreaks within communities.
Keywords: SARS-CoV-2; Environmental surveillance; COVID-19; Air, surface and wastewater; COVID-19 epidemiology
Graphical abstract
1. Introduction
The COVID-19 pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in 485 million cases and 6.14 million deaths worldwide as of April 1, 2022 (WHO, 2021). As infected individuals travel across space and time, they shed the virus through bodily fluids and exhalation (in the form of air droplets). As a result, the virus makes its genomic footprint in the environment, including in air, on surfaces and in wastewater. The virus RNA signal may persist in these environments for hours to days, especially indoors, depending on the surface type and meteorological conditions (Biryukov et al., 2020; van Doremalen et al., 2020; Setti et al., 2020).
Environmental monitoring of pathogens is non-invasive, cost-effective and can be conducted in public spaces. Thus, it can provide early indications of community level infection and early warnings of disease outbreaks more effectively than human surveillance methods (i.e., testing and tracing infected individuals). Given the multimodal transmission of SARS-CoV-2, researchers have been monitoring the virus in wastewater (Babady et al., 2020; Kaplan et al., 2020; Peccia et al., 2020), on surfaces (Parker et al., 2020) and in the air (Cheng et al., 2020; Zhang et al., 2022). Each medium has its unique advantages and disadvantages. For example, wastewater monitoring can localize the source of infection to a building, a neighborhood, and/or a community. Research shows that the concentrations of SARS-CoV-2 found in wastewater samples correspond with COVID-19 case incidence rate (Betancourt et al., 2021; Harris-Lovett et al., 2021; Scott et al., 2021; Sharkey et al., 2021; Gibas et al., 2021; Zhan et al., 2022). Although there is some consistency in the association between SARS-CoV-2 in the wastewater samples and COVID-19 cases, this association can vary with respect to scale (ranging from building to community sewage plant) and time-lag between COVID-19 case reporting and SARS-CoV-2 detected in the wastewater sample (Sharkey et al., 2021). A recent study showed that the four-day lagged SARS-CoV-2 concentration in the wastewater samples had the strongest association with COVID-19 cases at a university campus in 2020–21 and this association varied across different parts of the campus (Sharkey et al., 2021). While wastewater surveillance provides an early indication of community level infection, it is challenging to trace potential location(s) of cases in the absence of building specific monitoring of wastewater. Moreover, the virus concentration in the wastewater can be influenced by dilution, distance and time between the source and sample site, time of sampling, and area of the sewershed.
SARS-CoV-2 in air, surface and wastewater samples has been detected (Setti et al., 2020; Chia et al., 2020; Pan et al., 2021). However, limited data are available on their collective and relative comparison and efficacy in COVID-19 case prediction rate (see Table S1 in the supplement online material (SOM) for relevant literature). This paper aims to address this research gap by using the data from a controlled experimental design. We compare time- and building-matched concentrations of SARS-CoV-2 in daily air, surface and wastewater samples in a student dormitory (of about 500 students) for three months in Spring 2021 at the University of Miami, Coral Gables, FL, USA campus (UM). Regular screening of the student population for COVID-19 by building allowed for a direct comparison of environmental detection of SARS-CoV-2.
2. Methods and materials
2.1. Experimental strategy
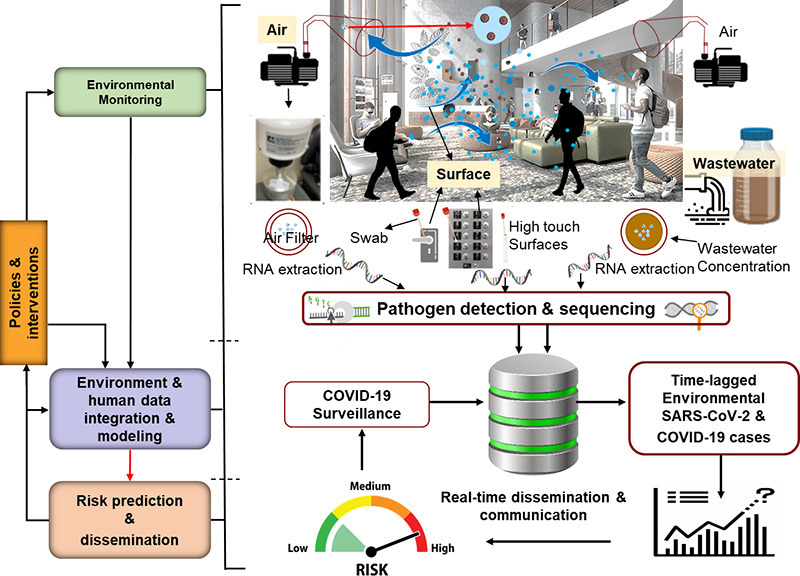
We used a prospective control design for this research. We selected a campus site (Y-leg of Lakeside Village [student dormitory], YLV) with corresponding wastewater and high traffic access points. YLV houses about 500 students and is serviced by two interconnected lobbies, C and D. This dorm is secure and can be accessed by students by swiping their university issued identification cards. Based on the swipe registry data, >5000 individuals (mostly students) entered YLV in a seven-day period. The wastewater from the YLV drains to a designated manhole (K). Since there are two access points to YLV, we conducted daily (active) air sampling and surface swab sampling in both lobbies. Thus, 24 h air samples and swab samples of the same high touch surfaces, namely elevator buttons, door handles and door push bars, corresponded to daily wastewater samples from manhole K (Fig. S1). Random screening of students residing in the dormitory also occurred 2–3 times/week during the study period: 2nd March 2, 2021 to 28th May 28, 2021. Methods of sample collection and analyses of these samples are detailed below. Daily air, surface swab and wastewater samples were collected between 8:00 AM and 9:00 AM Monday through Friday and 10:00 AM to 11:00 AM during weekends.
2.2. Sample collection
2.2.1. Wastewater
Samples were collected from one manhole located at the Lakeside Village of the University of Miami corresponding to the undergraduate student dorm selected for this study (Fig. S1). Daily sampling occurred from 2nd March 2021 to 25th May 2021. For the first two weeks, only grab samples were collected. After two weeks, an autosampler (ISCO 6712) was installed and composite samples were collected along with the grab samples. Composite samples consisted of a mixture of equal volume hourly samples drawn in a 5-liter sterile bottle during a 24-h cycle.
Before sample collection on a given day, the 5-liter bottle with the composite sample was retrieved from the autosampler and replaced with a new sterile bottle. The wastewater collected in this bottle was mixed thoroughly and dispensed into a 500 mL plastic cup using a stainless strainer with 2 mm openings to capture large solid particles in the wastewater. The sensor end of a pre-calibrated sonde (Xylem YSI Pro DSS) was immersed in the sample to measure sample characteristics, such as temperature, pH, salinity, dissolve oxygen, and turbidity (see Table S2 for summary statistics wastewater characteristics). This sample was then transferred to a sterile 250 mL labeled plastic bottle. This sample was stored in an ice cooler and transported to our laboratory for processing on the same day. All instruments were rinsed with water, dried, sprayed with 70 % isopropyl alcohol and again dried with a lint-free Kim wipe.
2.2.2. Air pollution monitoring pumps with flowmeters (LMP AIR)
Were installed in the Lobby C and Lobby D of the dorm selected for air sampling (Fig. S2). The sampling set-up was customized so that the air was drawn to the air filter from both elevators. A flow of 1.42/m3 air was drawn by the blower from two inlets. Using an L-connection, the air was branched (from the main stream of air flow) to the impactor, which was connected to a flow meter, and a vacuum pump drew air through the flowmeter at a set flow rate. Each day, a new sterile polycarbonate membrane filter (Isopore, 47 mm polycarbonate filter with 0.4 μm: a standard method for sampling aerosols) with a 47 mm supporting cellulose pad was deployed in the impactor. The pump was turned on and the initial flow was set between 11 L/min ± 1 L/min. Start date, time and initial flow rate were noted. The sample was collected the next day, generally after 24 h ± 2 h. We also deployed a real-time air pollution sensor that monitored airborne particle matter (PM) of three different sizes ≤1 μm, ≤ 2.5 μm and ≤ 10 μm (PM1, PM2.5 and PM10, respectively), total volatile organic compound (VOCs), temperature and relative humidity (see Table S3 for the summary statistics of variables). These data were also noted at the beginning and end of the sampling period each day. The sensor also recorded these data every 3 min. After 24 h, the flow was noted, pump was turned off, and the filter was retrieved from the impactor. The filter was rolled (cylindrically) and transferred to a 5 mL DNA/RNAase free conical tube. The impactor was cleaned with 70 % isopropyl alcohol, wiped with a lint free Kim wipe, and a new set of PC membrane filters and a 47 mm supporting cellulose pad was deployed for the next 24 h. The retrieved samples from both Lobby C and Lobby D of the dorms were stored in an ice cooler, which were then transported to our laboratory for processing.
2.2.3. Surface swab samples
Were collected daily from the same high touch surfaces including elevator buttons, door handles and door push bars using sterile polyester swabs with polystyrene handles. Following the collection, the swabbed surfaces were wiped with alcohol wipes for sterilization. The swab tips were stored in 1.5 mL conical tubes in a cooler with ice packs and transported to our laboratory for processing.
2.2.4. Disinfection practices
Were prioritized throughout the process with oversight from the University's Health and Safety Office. Safety protocols were followed in the field, and all study team members used proper PPE, such as using disposable lab coats, disposable gloves, and reusable goggles. Masks and face shields were worn as part of COVID-19 social distancing protocols.
2.3. Sample processing
2.3.1. Bacteria by culture
All wastewater samples were analyzed for the concentration of fecal indicator bacteria (FIB) to determine the concentration of human fecal input needed to standardize the concentration of SARS-CoV-2 with respect to estimated human bodily excretion. The wastewater sample was mixed, and an aliquot of 10 mL was removed and added in a sterile 15 mL centrifuge tube. 0.5 mL of the 10 mL (untreated) wastewater sample was added to 50 mL sterile phosphate buffered saline (PBS) to dilute the sample (100:1 dilution). After mixing the wastewater sample in the 50 mL PBS, 0.1 mL and 1 mL aliquots of the diluted samples were added in the 20 mL PBS in a sterile funnel and filtered through 47 mm grided membrane filters. Membrane filters were placed on the mFC agar plate and incubated for 24 h ± 2 h at 45 °C (Method 9222, APHA 2005). Agar plates were photographed and FIB colonies were counted using ImageJ (NIH, n.d.) (see Table S2 for a summary of fecal coliform in wastewater samples).
2.3.2. RNA concentration - electronegative filtration
The remaining 250 mL wastewater sample was used for the RNA concentration of SARS-CoV-2 using electronegative filtration (Sharkey et al., 2021; APHA, 2005). A magnetic stir bar was added to the sample to assist with maintaining homogeneity once placed on a magnetic plate. Heat inactivated (at 56 °C for 15 min) 35 μL of beta coronavirus OC43 with known number of copies, was added to 250 mL of wastewater sample, which served as a positive control for RNA extraction and its quantitation. To assist with adhesion to the filter, 2.35 mL of magnesium chloride was added per sample then drop by drop 10 % HCl (acid) was added to acidify the sample to a target pH value between 4.5 and 3.5. The number of HCl droplets added to the sample and the final pH value of the homogenized sample were noted. 40 mL ± 10 mL (depending on the presence of solid material dissolved in the water) was filtered through an electronegative membrane filter (0.45 μm pore size, 47 mm diameter). The filter was rolled conically and placed in an RNase/DNase free 5 mL conical tube. One mL of the DNA/RNA shield (Zymo) was added to the 5 mL tube. The tube was vortexed and then centrifuged. The concentrate was mixed by repeat pipetting and a 250 μL aliquot was used for subsequent RNA extraction.
2.3.3. RNA concentration - air filter samples
Although assessing overall concentration of other airborne viruses can be used to quantitate RNA recovery, it was beyond the scope of this work given these samples were not subject to genomic sequencing. Therefore, we added heat inactivated (56 °C for 15 min) 35 μL of beta coronavirus OC43 with a known concentration between 105 and 106 genomic copies (gc)/L as droplets to the filters while still in the 5 mL the conical tube. Moreover, it has been used as a standard method of RNA recovery in wastewater samples (APHA, 2005; Babler et al., 2022). 500 μL of DNA/RNA shield was then added around the top of the filters to collect any particles on the filter. This step was repeated, and air filter was submerged in 1 mL of DNA/RNA shield. The tube was vortexed and centrifuged, and the concentrate was mixed by pipetting and a 250 μL aliquot of the concentrated was used for RNA extraction.
2.3.4. RNA concentration - surface swab samples
Heat inactivated (56 °C for 15 min) 35 μL of beta coronavirus OC43 with a known concentration between 105 and 106 gc/L was added as droplets to the swabs while still in the 1.5 mL conical tube. One mL of DNA/RNA shield was then added around in the tube and processed the same way as for the air filters as described above.
2.3.5. SARS-CoV-2 RNA extraction process
The nucleic acid extraction process consisted of three processes: isolation, purification, and concentration (Fig. S3). We used a QuickRNA-Viral 96 Kit from Zymo Research Inc. for RNA extraction from all three sample types following their R1040/R1041 protocol. We added 500 μL of Viral RNA binding buffer to the concentrates of 250 μL wastewater, air filter, and surface swab samples previously prepared in 1 mL DNA/RNA shield. This reagent facilitates particle lysis and binding of RNA from other biological liquids, such as urine, in the wastewater. The samples were then centrifuged and transferred to columns and collection tubes. Next, 500 μL of wash buffer was added to the column before centrifuging. This was repeated twice. This “wash” removes proteins, salts, and other contaminants from the sample. Five hundred μL of 100 % ethanol was then added to the samples before centrifuging, which allowed the RNA to precipitate since nucleic acids are insoluble in ethanol. Finally, we added 15 μL of RNase-free water to the columns. After waiting for a minute, and the columns were centrifuged and RNA from the columns were collected in the 1.5 mL RNase free conical tubes for follow up qPCR analysis.
2.3.6. RNA quantification
The qPCR method utilized here was Volcano Second Generation (V2G) as described in Sharkey et al. (2021); aliquots of purified RNA were used in singleplex reactions to quantitate the SARS-CoV-2 nucleocapsid gene. The nucleocapsid gene utilized was a modified version of the N3 gene as described in Babler et al. (Sharkey et al., 2021), which was found to improve specificity of V2G amplifications and reduce detection limits to 70 gc/L (Sharkey et al., 2021). An advantage of the V2G-qPCR method over the more mainstream RT-qPCR is that it can read both RNA and DNA templates which eliminates the prior cDNA synthesis step (Blatter et al., 2013). According to research, the coronavirus nucleocapsid protein assists in the replication and transcription of viral RNA and interferes with cell-cycle processes of host cells, and as a result, plays a critical role in SARS-CoV-2 pathogenesis. The nucleocapsid proteins of many coronaviruses are immunogenic and expressed abundantly during infection (Cong et al., 2020). Detection, quantification, and analysis of its presence in wastewater, air filter, and surface swab samples have allowed for relatively accurate prediction of SARS-CoV-2 cases in a student dormitory and has correlated with reported cases at the University of Miami.
2.3.7. qPCR process
Twenty μL of HIV-1 RNA spike was added to all samples before the qPCR process to assess PCR inhibition. Inhibition was assessed by an assessment of delta Ct values which were all less than a value of 2. A master mix protocol, created in-house based on the number of samples being run, and involved the following volumes and reagents for SARS-CoV-2 detection:
| qPCR master mix reagents | Volume per reaction |
|---|---|
| RNase-free water | 17.7 μL |
| 5× Volcano (2G) Buffer | 6.6 μL |
| 10 mM dNTPs | 0.6 μL |
| 5 units/μL anti-Taq antibody | 0.15 μL |
| 5 units/μL Volcano (2G) Polymerase | 0.3 μL |
| 20 μM CV3b/f primer | 0.75 μL |
| 20 μM CV3c/r primer | 0.75 μL |
| 100 μM CV3 probe | 0.075 μL |
| 400× Rox | 0.075 μL |
Utilizing a 96-well plate (BioRad Hard Shell #HSP9601), 27 μL of master mix was added to individual wells before the RNA of samples, or any controls were inserted. Three μL was utilized for all inputs to the plate which included sample RNA for “unknown” wells, nuclease-free water for no template, or negative, controls (NTCs), and Twist positive standards. To utilize a standard curve, 3 μL of known synthetic SARS-CoV-2 concentration ranging from 101 to 105 cp/uL were also added. The plate was sealed by firmly pressing on an adhesive Microseal B (BioRad #MSB1001), and briefly centrifuged prior to being loaded in the machine. A BioRad CFX-Connect instrument was used for real time results of SARS-CoV-2 detection.
This analysis was repeated for the viral recovery control OC43 with a known concentration between 105 and 106 gc/L following its addition to all sample types (air, swab, wastewater). Utilizing a recovery control allowed for a downstream percent recovery to be calculated per sample as a surrogate. Average percent recoveries were 15.5 %, 28.3 % and 19.0 % for air filters, surface swabs and wastewater, respectively. For OC43 detection by qPCR, a similar master mix was created in-house based on the number of samples being run, except for the target-specific primers and probe in which 20 μM OC43 f/r primers and a 100 μM OC43 probe were utilized prior to analysis with qPCR.
Similarly to before, 27 μL of the master mix was added to individual wells of a 96-well plate (BioRad Hard Shell #HSP9601) and then 3 μL of sample was added to the wells i. A 3 μL volume was also utilized of nuclease-free water for NTCs as well as for positive Twist control standards. The standards included for OC43 analysis ranged from 101 to 105 cp/μL consisted of pre-quantified PCR-amplified OC43 product. The plate was sealed by firmly pressing on an adhesive Microseal B (BioRad #MSB1001), and briefly centrifuged prior to its placement in the machine. A BioRad CFX-Connect instrument was also used for real time results of OC43 detection.
2.4. COVID-19 surveillance
Of the students residing at the YLV dormitory consisted primarily of random screenings 2–3 days/week using nasal swabs which were analyzed by RT-PCR. Some additional tests may have been from symptomatic students or from students who were believed to have been exposed as a results of contact tracing. These anonymous data on the total number of tests (performed) and COVID-19 cases by date were acquired from the University administration.
2.5. Analysis
Statistical analyses included descriptive analyses of aggregated and disaggregated data. Data of air and surface swab samples from both lobbies were aggregated to compare them with the data from the wastewater samples. We computed time-lagged SARS-CoV-2 concentrations in all three types of environmental samples to assess whether the virus in the wastewater samples was detected prior to the COVID-19 case diagnosis. For example, if a case was diagnosed on a given day, we computed SARS-CoV-2 concentration in the environmental samples for seven days prior (to the day of diagnosis) separately. We also used moving averages to smooth SARS-CoV-2 concentrations in environmental samples. χ2 tests were performed to assess statistical differences across groups and a p-value of 0.05 or below was considered as significant.
3. Results
A total of 445 air, surface swab and wastewater samples were collected from 2nd March 2 to 25th May 2021. Of these, 165 air samples and 166 surface swab samples were collected from two lobbies of YLV at the UM campus. A total of 114 daily wastewater samples were collected from manhole K. On 24 days, we collected both grab and 24 h composite wastewater samples. On 4 of these 24 days when SARS-CoV-2 was detected in the composite samples it was below the limit of detection (LOD) in the grab samples. On 3 of the 24 days SARS-CoV-2 was detected in the grab samples but it was below the LOD in the composite samples on these days. On the days when both grab and composite samples were collected, the mean concentration of SARS-CoV-2 in samples with above LOD (n = 27) was slightly higher in the composite samples as compared to grab (918 gc/L versus 597 gc/L on average). But this difference was statistically insignificant.
The concentration and frequency of SARS-CoV-2 detection in the air and surface samples collected from each of the two lobbies of YLV did not vary significantly. We detected SARS-CoV-2 in 50 (30 %) of the 165 air samples. The average SARS-CoV-2 concentration in the air sample was 14.8 gc/m3. Of the 166 surface swab samples, SARS-CoV-2 was detected in 33 (20 %) of them. The average concentration of SARS-CoV-2 in these samples was 16.5 gc/m2 surface area. Of the 114 grab and composite wastewater samples SARS-CoV-2 was detected in 57 (or 50 %) of the samples (on 43 days) and the average concentration in these samples was 1390 gc/L (Table 1 ). The greater detection in wastewater was likely due to the fact that when students were tested positive, they were subject to isolation where food was brought to their rooms. Thus they were not expected to use common areas such as elevators and lobbies. However, they continued to contribute to the wastewater throughout their illness when isolated in their rooms.
Table 1.
SARS-CoV-2 detection and concentration in air, surface swab and wastewater samples.
| Environmental Matrix | Number of samples collected | Number of SARS-CoV-2 positive samples (% total) | loge (SARS-CoV-2 gc) (95 % Confidence Interval (CI; n) |
|---|---|---|---|
| Air (gc/m3) | 165 | 50 (30.3) | 2.7 (2.4–3.0; 50) |
| Surface (gc/m2) | 166 | 33 (19.9) | 2.8 (2.4–3.2; 33) |
| Wastewater (gc/L) | 114 | 57 (50.0) | 6.5 (6.2–6.8; 57) |
| Total | 445 | 140 (31.5) | 4.3 (3.9–4.6; 140) |
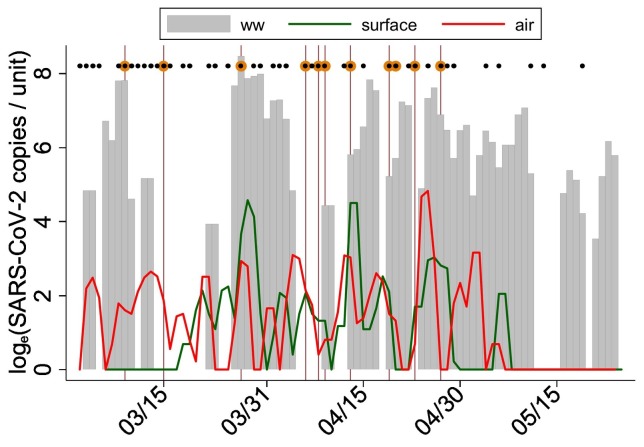
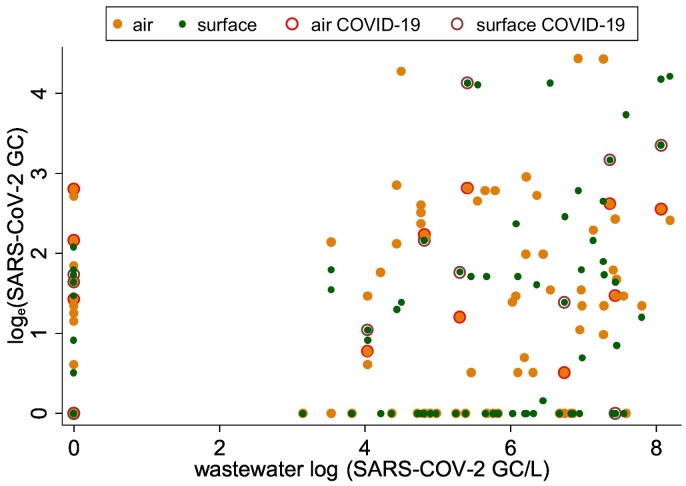
Given the major focus of our project has been on wastewater detection, air and surface swab samples were aggregated by day to compare against the wastewater samples. Some consistency was observed in the concentration and detection of SARS-CoV-2 across air, surface and wastewater samples (Fig. 1 ; Table 2 ). On 36 (or 46 %) of 78 days there was complete agreement in the detection of SARS-CoV-2 in all air, surface, and wastewater samples. However, on many of the days, SARS-CoV-2 was detected in the wastewater samples but not in air and surface swab samples, and vice-versa (Fig. 2 ). For example, SARS-CoV-2 was not detected in wastewater samples on 24 days, but 17 and 14 of these 24 days the virus was detected in the air and surface swab samples, respectively (Table 2). The differences in the frequency distribution of the virus detection across different sample types was marginally significant (χ2 = 3.06; p ~ 0.058). When 1-day lagged moving average of SARS-CoV-2 was detected in wastewater samples on 43 days, it was also detected on 33 and 26 days in air and surface samples, respectively. The frequency distribution of the virus detection across air and surface samples when it was also detected in wastewater samples did not show statistically significant differences (χ2 = 3.02; p ~ 0.082) (Table 2), suggesting some agreement in the detection of SARS-CoV-2 across three different types of environmental samples.
Fig. 1.
Concentration of SARS-CoV-2 in wastewater, surface and air samples and COVID-19 cases in a UM dormitory, March–May 2021. Maroon vertical lines indicate dates when COVID-19 cases were reported; on two days, 4/19/2021 and 4/23/201 represented by a single line two COVID-19 cases were reported, lines on these days represent two cases each. Black dots indicate days when COVID-19 monitoring occurred in the dormitory and dark orange symbols indicate days when COVID-19 case(s) were detected.
Table 2.
Number of days SARS-CoV-2 detection in air, surface swab and wastewater samples, March to May 2021 (% days in parenthesis).
| SARS-CoV-2 detection | Surface swab sample detection |
|||
|---|---|---|---|---|
| No | Yes | Total | ||
| Wastewater sample detection (No) | ||||
| Air sample detection | No | 5 (20.8) | 2 (8.3) | 7 (29.2) |
| Yes | 5 (20.8) | 12 (50.0) | 17 (70.8) | |
| Total | 10 (41.7) | 14 (58.3) | 24 (100) | |
| Wastewater sample detection (Yes) | ||||
| Air sample detection | No | 14 (25.9) | 7 (13.0) | 21 (38.9) |
| Yes | 14 (25.9) | 19 (35.2) | 33 (61.1) | |
| Total | 28 (51.9) | 26 (48.1) | 54 (100) | |
Fig. 2.
One-day lagged moving average of SARS-CoV-2 in air and surface samples (on y-axis) with respect to SARS-CoV-2 in wastewater samples (on x-axis), March–May 2021 (wastewater concentration on x-axis). Dark orange symbol shows SARS-CoV-2 values in air and wastewater samples, respectively; red circle around orange indicates COVID-19 case detection; green symbol shows SARS-CoV-2 values on surface and in wastewater samples; brown circle around green circle indicates COVID-19 case detection on days these samples were collected.
Human surveillance in the residential dormitory corresponded to testing 2.3 students/day, on average, during 44 of the 85 days during the study period. COVID-19 cases were detected on 11 of these 44 days of testing: 1 student tested positive during 9 of the days, and 2 students tested positive during the other 2 days. Considering that human surveillance was conducted on only 44 of the 85 days during the study, the comparison of COVID-19 cases and environmental SARS-CoV-2 was restricted to days of COVID-19 screening or over which averages were computed. On the 11 days when COVID-19 cases were detected, SARS-CoV-2 was detected in air, surface swab and wastewater samples on 6, 6, and 7 of these 11 days respectively. Conversely, COVID-19 case(s) were detected in the building air, surface swab, and wastewater samples but were negative for SARS-CoV-2 on 5, 5, and 4 days. The efficacy of 1-day lag SARS-CoV-2 (detection) in air, surface swab and wastewater samples to predict COVID-19 cases was 90.9, 72.7 % and 63.6 %, which improved to 100 % for air, 81.8 % for surface swab and 90.1 % for wastewater SARS-CoV-2 three days before COVID-19 case reporting (Table S4 in SOM).
We also computed 1, 2 and 3 day lagged moving average of the SARS-CoV-2 concentration in all collected samples. The days when SARS-CoV-2 was below the LOD, zero values were assigned. Thus, the number of days that SARS-CoV-2 was above LOD increased for lagged analysis due to the averaging effects from days with values above detection. One-day lagged moving averages of SARS-CoV-2 were above detection limit on 52 (63.4 %), 40 (50.0 %) and 57 (68.6 %) of 83, 82, and 80 days in air, surface swab and wastewater samples, respectively (data not shown). The efficacy of SARS-CoV-2 detection in the 2-day lagged moving average of air, surface swab and wastewater samples to predict days with COVID-19 case(s) was 100 %, 81.8 % and 72.7 %, respectively (Table 3 ). For the 3-day lagged moving average the efficacy of air, surface swab, and wastewater samples to predict COVID-19 cases was 100 %, 81.8 %, and 90.9 %.
Table 3.
Lagged moving average of the SARS-CoV-2 concentration for days with positive COVID-19 cases and percentage of days that the time-lagged concentrations were able to predict days with positive COVID-19 cases and negative COVID-19 cases.
| Date | Number of COVID-19 cases | Air SARS-CoV-2 GC/m3 |
Surface SARS-CoV-2 GC/m2 |
Wastewater SARS-CoV-2 GC/L |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lag-1 | Lag-2 | Lag-3 | Lag-1 | Lag-2 | Lag-3 | Lag-1 | Lag-2 | Lag-3 | ||
| 3/9/2021 | 1 | 4.0 | 3.3 | 2.5 | ND | ND | ND | 2475 | 1700 | 1481 |
| 3/15/2021 | 1 | 5.5 | 7.7 | 9.4 | ND | ND | ND | ND | ND | 88 |
| 3/27/2021 | 1 | 17.8 | 11.8 | 8.9 | 38.3 | 27.5 | 23.4 | 4775 | 3183 | 2388 |
| 4/6/2021 | 1 | 7.5 | 15.5 | 14.4 | 7.0 | 4.7 | 3.8 | ND | ND | ND |
| 4/8/2021 | 1 | 0.5 | 3.2 | 4.0 | 2.8 | 4.2 | 4.9 | ND | ND | ND |
| 4/9/2021 | 1 | 1.3 | 1.2 | 3.0 | 2.8 | 1.8 | 3.1 | 83 | 56 | 42 |
| 4/13/2021 | 1 | 19.8 | 15.7 | 11.8 | 89.5 | 61.2 | 45.9 | 333 | 222 | 167 |
| 4/19/2021 | 2 | 3.5 | 8.3 | 8.0 | 7.3 | 7.7 | 5.8 | 183 | 122 | 1033 |
| 4/20/2021 | 1 | 2.8 | 2.3 | 6.3 | ND | 4.8 | 5.8 | 300 | 200 | 150 |
| 4/23/2021 | 2 | 1.0 | 0.7 | 0.5 | 4.5 | 3.0 | 2.3 | ND | 844 | 692 |
| 4/27/2021 | 1 | ND | 12.7 | 62.0 | 15.8 | 22.7 | 17.0 | 975 | 1583 | 1254 |
| % days COVID-19+ | 90.9 | 100 | 100 | 72.7 | 81.8 | 81.8 | 63.6 | 72.7 | 72.7 | |
| % days COVID-19- | 63.6 | 72.7 | 84.8 | 51.5 | 51.5 | 51.5 | 66.7 | 72.7 | 84.8 | |
ND = below limit of detection.
When using SARS-CoV-2 in multiple environmental samples together, COVID-19 prediction improved. For example, with 1 day-lagged SARS-CoV-2 in wastewater and surface samples the COVID-19 case prediction efficiency was 91 %. However, when using 1-day lagged SARS-CoV-2 in wastewater and air samples, the COVID-19 case prediction efficiency was 100 %. When using 0, 1, 2, 3 and 4 day-lagged SARS-CoV-2 in any of the three air, surface swab and wastewater samples on a given day, the efficacy to predict COVID-19 case(s) was 90.9 %, 90.9 %, 100 %, 90.9 % and 100 % respectively. However, when the 1-day lagged moving average of SARS-CoV-2 in any of the three types of samples was used, COVID-19 case(s) prediction rate from environmental SARS-CoV-2 increased to 100 %, suggesting if the virus is not detected in one or two environmental media (e.g. in air or on surface), it can be detected in third environmental media (e.g. in wastewater).
4. Discussion
SARS-CoV-2 detection in air, surface swab and wastewater samples in a building with clinically confirmed COVID-19 cases suggests that the genomic footprints of the virus, shed by infected individuals, can be traced in the environment. Our analysis further suggests that SARS-CoV-2 detection in environmental samples (air, surface swab and wastewater) predicted all clinically diagnosed COVID-19 cases in the selected dormitory depending upon the lag time chosen. Some of these findings are consistent with the emerging literature, which suggests association between COVID-19 case prediction with the aid of SARS-CoV-2 in wastewater samples (Betancourt et al., 2021; Gibas et al., 2021). Daily samples from multiple environmental media were evaluated in this research, which provide a novel insight into variations in COVID-19 case prediction with respect to changes in time-lagged SARS-CoV-2 detection in air, surface swab and wastewater samples. The 3-day lagged SARS-CoV-2 in the wastewater samples showed the strongest association with COVID-19 case detection, which is consistent with previous research. Sharkey et al., 2021 describe that the 4-day lagged SARS-CoV-2 in wastewater had the strongest association with COVID-19 cases (Sharkey et al., 2021). This current study, for the first-time, shows association between COVID-19 cases and time-lagged SARS-CoV-2 detection in three different types of samples collected daily. In fact, the strongest association between COVID-19 diagnosis and SARS-CoV-2 in air and surface swabs was observed three days before COVID-19 case reporting. Thus, SARS-CoV-2 in these environmental samples can provide early warnings of an outbreak even at the scale of a building. A unique finding of this research is that if one environmental sample type was falsely negative (for SARS-CoV-2) it was detected in another sample type, suggesting that sampling air, surface swab and wastewater on the same day can improve the efficacy of environmental surveillance of infectious diseases. Among three types of environmental samples, air samples were most efficient in COVID-19 case prediction. One day-lagged SARS-CoV-2 in air and wastewater samples or in air and surface swabs predicted all COVID-19 cases.
Our research has public health and policy implications. The detection of the virus in environmental samples several days prior to clinical diagnosis can guide timely interventions to reduce pathogen transmission. Unlike sentinel human screening, environmental monitoring is non-invasive and less costly (Hassard et al., 2021; Renninger et al., 2021). Moreover, asymptomatic individuals do not necessarily seek care and hence can be missed by clinical screening (Yao et al., 2021). Environmental samples can help identify location- and time-specific cluster(s) of both symptomatic and asymptomatic cases several days prior to their clinical diagnosis. For example, strategic (active) air, surface and wastewater sampling in places such as airports, schools and malls can provide insight to the spread and potential outbreak of the disease at multiple scales, and trace sources of disease transmission. Air sampling has a unique advantage to capture airborne pathogens where people may not use toilet or may avoid touching surface(s) such automatic door opening, but they will be breathing. Thus, deploying high volume air samplers at strategic locations, such as at entrances and waiting areas in public places can capture airborne pathogens they shed. However, air sampling is more costly and labor intensive than swab and wastewater sampling. Moreover, wastewater sampling is being increasingly implemented worldwide for the surveillance of emerging and remerging pathogens associated with different primary modes of transmission, such as monkey pox and polio.
Results of this research must be interpreted with caution due to the following limitations. First, some of the samples analyzed could be false negatives due to low concentrations of virus quantified from the sample or due to inhibition during sample preparation and analysis. Second, SARS-CoV-2 recovery from the samples can be subject to bias due to a low recovery rate from air, surface swab and wastewater samples. Thus, the plausibility of false negatives cannot be discounted. Third, COVID-19 case data can also be subject to bias because of the limited testing of students. For example, SARS-CoV-2 was detected in the environmental samples on many days, but COVID-19 cases were not reported. Specifically, students who may quarantine on campus can shed SARS-CoV-2 for many days without subsequent daily testing. Routine daily testing of students was not implemented, especially during weekends. Finally, SARS-CoV-2 concentrations in environmental samples were not adjusted for potential confounders, such as local meteorological conditions, ventilation and airborne particulate matter which have been shown to impact SARS-CoV-2 concentrations in air and on surface swabs (Renninger et al., 2021; Wathore et al., 2020). Despite these limitations this research sheds light on the relevance of proactive environmental surveillance for emerging disease-causing pathogens and their management.
5. Conclusion
SARS-CoV-2 RNA was detected in all three environmental samples: air, surface swabs and wastewater. The relative efficiency of predicting COVID-19 cases improved to 100 % when multiple environmental media were monitored (air plus wastewater or air plus surface swabs). SARS-CoV-2 was also detected in environmental samples when COVID-19 cases were not reported, indicating underreporting of COVID-19 cases. Thus, environmental monitoring of SARS-CoV-2 serves as effective method of community surveillance of the COVID-19 disease. Environmental monitoring in public places, such as airports, school, metro-stations and shopping malls has potential for the surveillance of other infectious diseases.
CRediT authorship contribution statement
Conceptualization: NK, HS.
Methodology: NK, HS, MS.
Investigation: NK, HS, SK, SA, JC, MS, AM, WL, JTJ, EK, NS, RK, BS, SW.
Visualization: NK.
Funding acquisition: HS, SS, CM.
Project administration: GG, HS, NK, SA, SK, DV.
Supervision: NK, HS.
Writing – original draft: NK, SK, SA.
Writing – review & editing: HS, NK, TB, SA.
Funding
This work in part was supported by the following agencies: National Institute of Health grant R01EY026174 (NK), National Institute of Health grant U01DA053941 (HS, SS, CM).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.159188.
Appendix A. Supplementary data
Supplementary material
Data availability
Environmental sample data can be made available upon a reasonable requested to the corresponding author. Human subject data on COVID-19 will not be available due to confidentiality reasons.
References
- APHA . In: Standard Methods for the Examination of Water & Wastewater, Centennial Edition. 21st edition. Eaton A.D., et al., editors. American Public Health Association; Washington DC: 2005. [Google Scholar]
- Babady N.E., et al. Performance of severe acute respiratory syndrome coronavirus 2 real-time RT-PCR tests on oral rinses and saliva samples. J. Mol. Diagn. 2020;23:3–9. doi: 10.1016/j.jmoldx.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babler K.M., et al. Comparison of electronegative filtration to magnetic bead-based concentration and V2G-qPCR to RT-qPCR for quantifying viral SARS-CoV-2 RNA from wastewater. ACS Es&T Water. 2022;23(1):1–3. doi: 10.1021/acsestwater.2c00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.Q., et al. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biryukov J., et al. Increasing temperature and relative humidity accelerates inactivation of SARS-CoV-2 on surfaces. mSphere. 2020;5(4) doi: 10.1128/mSphere.00441-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatter N., et al. Structure and function of an RNA-reading thermostable DNA polymerase. Angew. Chem. Int. Ed. Engl. 2013;52(45):11935–11939. doi: 10.1002/anie.201306655. [DOI] [PubMed] [Google Scholar]
- Cheng V.C., et al. Air and environmental sampling for SARS-CoV-2 around hospitalized patients with coronavirus disease 2019 (COVID-19) Infect. Control Hosp. Epidemiol. 2020;41(11):1258–1265. doi: 10.1017/ice.2020.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia P.Y., et al. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat. Commun. 2020;11(1):2800. doi: 10.1038/s41467-020-16670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 2020;94(4) doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382(16):1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas C., et al. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci. Total Environ. 2021;782 doi: 10.1016/j.scitotenv.2021.146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Lovett S., et al. Wastewater surveillance for SARS-CoV-2 on college campuses: initial efforts, lessons learned, and research needs. Int. J. Environ. Res. Public Health. 2021;18(9) doi: 10.3390/ijerph18094455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassard F., et al. Innovation in wastewater near-source tracking for rapid identification of COVID-19 in schools. Lancet Microbe. 2021;2(1):e4–e5. doi: 10.1016/S2666-5247(20)30193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan E.H., et al. Aligning SARS-CoV-2 indicators via an epidemic model: application to hospital admissions and RNA detection in sewage sludge. Health Care Manag. Sci. 2020 doi: 10.1007/s10729-020-09525-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH ImageJ 1.46r. 2013 [cited 2014 10/10/2014] http://imagej.nih.gov/ij/docs/index.html Available from:
- Pan J., et al. SARS-CoV-2 on surfaces and HVAC filters in dormitory rooms. Environ. Sci. Technol. Lett. 2021 doi: 10.1021/acs.estlett.1c00892. (forthcoming) [DOI] [Google Scholar]
- Parker C.W., et al. End-to-end protocol for the detection of SARS-CoV-2 fromBuilt Environments. mSystems. 2020;5(5) doi: 10.1128/mSystems.00771-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38(10):1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renninger N., et al. Indoor dust as a matrix for surveillance of COVID-19. mSystems. 2021;6(2) doi: 10.1128/mSystems.01350-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L.C., et al. Targeted wastewater surveillance of SARS-CoV-2 on a university campus for COVID-19 outbreak detection and mitigation. Environ. Res. 2021;200 doi: 10.1016/j.envres.2021.111374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setti L., et al. Airborne transmission route of COVID-19: why 2 meters/6 feet of inter-personal distance could not be enough. Int. J. Environ. Res. Public Health. 2020;17(8) doi: 10.3390/ijerph17082932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey M.E., et al. Lessons learned from SARS-CoV-2 measurements in wastewater. Sci. Total Environ. 2021;798 doi: 10.1016/j.scitotenv.2021.149177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathore R., et al. Understanding air and water borne transmission and survival of coronavirus: insights and way forward for SARS-CoV-2. Sci. Total Environ. 2020;749 doi: 10.1016/j.scitotenv.2020.141486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; 2021. WHO Coronavirus (COVID-19) Dashboard. [Google Scholar]
- Yao L., et al. Detection of coronavirus in environmental surveillance and risk monitoring for pandemic control. Chem. Soc. Rev. 2021;50(6):3656–3676. doi: 10.1039/d0cs00595a. [DOI] [PubMed] [Google Scholar]
- Zhan Q., et al. Relationships between SARS-CoV-2 in wastewater and COVID-19 clinical cases and hospitalizations, with and without normalization against indicators of human waste. ACS ES&T Water. 2022 doi: 10.1021/acsestwater.2c00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., et al. Monitoring SARS-CoV-2 in air and on surfaces and estimating infection risk in buildings and buses on a university campus. J. Expo. Sci. Environ. Epidemiol. 2022;32:751–758. doi: 10.1038/s41370-022-00442-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Environmental sample data can be made available upon a reasonable requested to the corresponding author. Human subject data on COVID-19 will not be available due to confidentiality reasons.