Abstract
Two truncated variants of AbrB, comprising either its first 53 (AbrBN53) or first 55 (AbrBN55) amino acid residues, were constructed and purified. Noncovalently linked homodimers of the truncated variants exhibited very weak DNA-binding activity. Cross-linking AbrBN55 dimers into tetramers and higher-order multimers (via disulfide bonding between penultimate cysteine residues) resulted in proteins having DNA-binding affinity comparable to and DNA-binding specificity identical to those of intact, wild-type AbrB. These results indicate that the DNA recognition and specificity determinants of AbrB binding lie solely within its N-terminal amino acid sequence.
In the life cycle of a bacterial cell, the transition between active growth and stationary phase is a time of profound changes in gene expression. For Bacillus subtilis and other spore-forming microbes, it is also the time when the exact environmental conditions must be sensed and then the decision made whether or not to enter the dedicated sporulation pathway. Numerous complex interconnected regulatory circuits govern the survival strategy that is ultimately chosen (9, 10).
The DNA-binding AbrB protein of B. subtilis is a global regulator of numerous genes that commence expression at the end of vegetative growth and the onset of stationary phase (10, 14). In solution, AbrB is a tetramer comprised of identical 94-amino-acid subunits (17). Over 30 different operons have been shown to be subject to AbrB-mediated regulatory effects that can be related to AbrB-binding sites within promoter regions (3, 8, 10, 11, 12, 16, 21; M. A. Strauch, unpublished data). However, the total number of operons subject to AbrB control is much larger than this, since AbrB regulates a number of other functions that affect transcription of separate regulons (4, 6, 14, 18; M. A. Strauch, unpublished). Over 40 different target regions of AbrB binding to chromosomally located sequences have been identified (3, 8, 11, 12, 16, 20, 21; M. A. Strauch, unpublished data). Comparison of these target regions reveals no apparent base sequence that can be defined as an AbrB consensus binding site. In vitro selection of optimal AbrB-binding targets (“aptomers”) reveals that there is a preferred consensus sequence that exhibits high-affinity binding by AbrB (20); yet, the presence of this consensus is scarce within chromosomally located AbrB-binding sites. These observations, coupled with recent nuclear magnetic resonance (NMR) studies (17), strengthen the notion first proposed a number of years ago (16) that AbrB exhibits a significant flexibility in its ability to recognize, and bind with high affinity, a subset of different base sequences, presumably via recognition of three-dimensional DNA structures.
Previous studies have suggested that most, if not all, of the DNA-binding determinants of AbrB lie within the first 55 amino acids of the N terminus (17, 19). To test this hypothesis, we constructed and purified truncated proteins corresponding to N-terminal portions of AbrB. We obtained DNA fragments (via PCR methods) using oligonucleotide primer pairs ABPC5 plus ABSTOP1 and ABPC5 plus ABSTOP2 (Fig. 1). These fragments were inserted into the expression vector pKQV4 (16) in order to place the truncated genes under isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible transcription originating from a pSPAC promoter. DNA sequencing confirmed that the desired constructions were obtained. The expression plasmids were introduced into Escherichia coli cells, and transformants were tested for IPTG-induced overexpression of polypeptides of the desired size (about 6,300 Da). All transformants tested exhibited induction of protein of about 6 kDa. One of each type was chosen as the vehicle for the production of proteins to be purified.
FIG. 1.
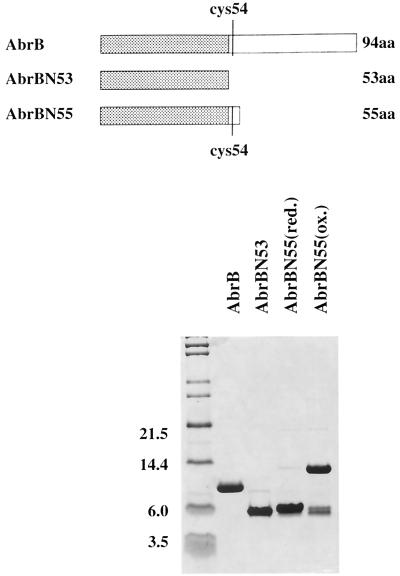
Construction of truncated abrB genes. (A) Sequence of the beginning of the abrB reading frame and the oligonucleotide (ABPC5) annealing to this region that was used in the PCR. (B) Sequence of the abrB gene encoding residues Leu45 to Val56 and the oligonucleotides annealing to this region that were used in PCR to construct the abrBN55 (ABSTOP1) and abrBN53 (ABSTOP2) truncations. For clarity, the sequences complementary to the oligonucleotides are shown along with the amino acid sequences that are present at the carboxyl terminals of the truncations. ∗, stop codons.
The truncated proteins (AbrBN53 and AbrBN55) were purified using a series of steps including ammonium sulfate fractionation and DEAE and heparin agarose column chromatography (details available upon request). While assaying fractions (via sodium dodecyl sulfate-polyacrylamide gel electrophoresis) during the latter purification steps of AbrBN55 (but not AbrBN53), we observed a progressive increase in the amount of a protein with a molecular mass of approximately 13 kDa that cofractionated with the 6.3-kDa AbrBN55 protein. We suspected that this 13-kDA species represented a disulfide-linked form of AbrBN55 that resulted from partial oxidation occurring during the preparation of the samples. To ascertain if this were indeed the case, we subjected a portion of our purified preparation to increased amounts of reducing agent (β-mercaptoethanol), followed by alkylation of cysteine residues using iodoacetamide in order to break any disulfide bonds and block their reformation. This treatment confirmed that the 13-kDa species was a dimeric form of AbrBN55 that resulted from disulfide bond formation between the lone cysteine residues (Cys54) present on the monomers. Figure 2 illustrates the purity and nature of the AbrBN53, reduced AbrBN55 [AbrBN55(red)], and oxidized AbrBN55 [AbrBN55(ox)] preparations we obtained.
FIG. 2.
Purified proteins used in DNA-binding assays. At the top is a cartoon depicting the nature of the monomeric subunits of the proteins. The purity of the proteins used and the comparative difference between the reduced and oxidized (disulfide-linked) preparations of AbrBN55 are shown at the bottom by a sodium dodecyl sulfate-polyacrylamide gel electrophoresis fractionation. Note that the truncated proteins used in this study, as well as wild-type AbrB (16), have the sequence MKST… at their amino terminus (N terminus). However, in the original report announcing the sequence of the abrB gene (7), the reading frame was presumed to start at a Met codon located 6 bp upstream (and in frame) from the codon that specifies the Met residue at the N terminus of the pure proteins. Most of the previous literature and database sequence depositories have followed the original convention indicating that the N terminus of AbrB is MFMKST… While we cannot rule out the possibility that in vivo translation starts at the upstream Met codon, with subsequent processing of the protein to produce the observed N terminus, we believe the true in vivo start to be at the downstream Met codon. In any event, since our pure proteins had MKST… at their N terminus, we were compelled to number the residues of the protein accordingly. Unfortunately, if reference is made to the previous literature, a discrepancy of two residues will be observed in the numbering of the amino acids, e.g., the lone Cys residue in the monomer is Cys54 in this paper but was Cys56 previously. Molecular masses (in kilodaltons) are shown.
Gel retardation assays were used to test the binding activities of the various preparations towards various DNA fragments containing known AbrB recognition sites. In an effort to obtain an AbrBN55 preparation containing as high a proportion of disulfide-linked forms as possible, we subjected the protein to various conditions that could be expected to increase the oxidation reaction between sulfhydryls. We found that treatment with Cu2+ ions (1) significantly increased disulfide-linked forms: for some preparations, we estimated that as much as 90% of the protein was in a disulfide-linked form (Fig. 2).
For each DNA target tested, we observed that the AbrBN53 and AbrBN55(red) proteins had very little binding affinity. However, the disulfide-linked AbrBN55(ox) protein displayed a DNA-binding affinity equivalent to, or very comparable to, that shown by the wild-type protein. Table 1 illustrates the data for binding to three different targets. [The values reported for the AbrBN55(ox) preparations are likely to be underestimates of the actual affinities. We did not attempt to correct the values based upon the percentage of the preparation that was in a disulfide form—usually 80 to 90%; see above.] From this, we concluded that the N-terminal portion of the AbrB protein contains the majority of or all of the determinants responsible for at least some type of DNA-binding activity. However, stable DNA binding requires that at least two of the AbrBN55 domains be covalently linked (in this case, via a disulfide bond between the Cys54 residues). We also cross-linked AbrBN55 monomers by using bis-maleimidohexane (BMH), which would form a link between Cys54 residues containing a 16-Å spacer segment. BMH-linked AbrBN55 domains did not exhibit DNA-binding activity (data not shown), possibly implying that a very specific spatial relationship between AbrBN55 domains is required for binding recognition or stability, or both. Not surprisingly, alkylation (via iodoacetamide) and carboxyethylation (via iodoacetic acid) of Cys54 in AbrBN55 preparations inhibited any appreciable binding activity [data not shown; comparable to values given in Table 1 for AbrBN53 and AbrBN55(red)].
TABLE 1.
Binding affinity of AbrBN55 to target DNA sites
| Targetb |
Kd (nM)a
|
||
|---|---|---|---|
| AbrBN55(red) | AbrBN55(ox) | AbrB(wt) | |
| Spo0E | >50,000 | 600 | 300 |
| BS18 | 10,000 | 80 | 30 |
| C47 | 4,000 | 40 | 50 |
Relative apparent dissociation constants (see text). The Kd values are expressed as the concentration of protein required to produce half maximal binding to the input DNA (20).
The Spo0E target was an 110-bp fragment isolated from pMAS3011 (13), which contains the 39-bp AbrB footprint region sequence from the spo0E promoter; BS18 and C47 targets are 97-bp fragments containing sequences selected in vitro for high-affinity binding to wild-type AbrB (20). The DNase I footprint of AbrB upon C47 is approximately twice as long as the footprint on BS18 (45 versus 25 bp; see reference 20). Gel retardations were performed at 20°C essentially as has been described previously (16) except that the binding buffer lacked a reducing agent, such as β-mercaptoethanol or dithiothreitol, when necessary. Radiolabeled DNA target sequences were prepared as described previously (16). Apparent dissociation constants (Kd) were derived using data obtained from the quantification of gel retardation assays using a Molecular Dynamics PhosphorImager. wt, wild type.
AbrBN53 has been found to exist as a dimer in solution by using a number of techniques, including NMR (17). The dimerization results from a number of noncovalent interactions between β-sheet regions of the monomers (17). Using fast-performance liquid chromatography sizing techniques (data not shown), we observed that AbrBN53 and AbrBN55(red) preparations behaved as dimers whereas AbrBN55(ox) preparations, while having a small proportion of dimers, consisted mainly of tetramers. Some higher-order multimeric or concatenated forms were also present in these preparations (minor peaks having the expected elution behavior for hexameric and octameric forms were observed). The exact nature of these higher-order forms is not known, but presumably they must reflect both additional disulfide bonding and noncovalent interactions between various multimers. We have not yet been able to obtain a preparation of tetramers that is entirely free from these larger forms. However, in no instances have monomeric forms of any of the truncated proteins ever been observed, nor have monomeric forms of the full-length wild-type protein ever been observed under nondenaturing conditions. It has recently been shown that an active form of wild-type AbrB is a tetramer (17). Given that an active form of the wild-type protein is a tetramer, we feel that the active form of AbrBN55(ox) is probably also tetrameric; however, for comparison purposes, we have elected to report the Kd values in Table 1 based upon the molarity of total individual polypeptide chains present.
By their nature, the gel retardation experiments measured only the total affinity of binding of AbrBN55(ox) to the DNA targets employed. The question thus arose as to whether or not the protein exhibited the same specificity of binding to defined regions on the DNA targets as did the wild-type protein. Therefore, we conducted DNase I footprinting assays to ascertain the exact regions on the DNA targets that were interacting with AbrBN55(ox).
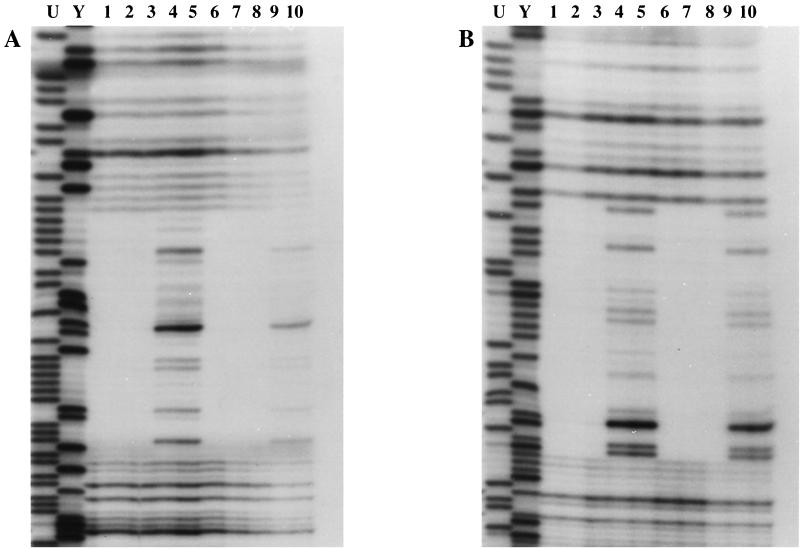
For every DNA target examined, the DNase I footprint of AbrBN55(ox) was identical to the footprint given by the wild-type protein. Figure 3 illustrates a representative result. Even at very high protein/DNA ratios, only a well-defined footprint identical to the wild-type binding region was observed. There was no evidence of any type of altered recognition or relaxed specificity exhibited by AbrBN55(ox) relative to those of intact, native AbrB. These results show that the recognition and specificity determinants of AbrB lie solely within its N-terminal amino acid sequence.
FIG. 3.
AbrBN55(ox) and AbrB display identical binding recognition and specificity. The results of DNase I protection assays (footprinting) of protein binding to the BS18 aptomer sequence are shown. The target DNA used was an 105-bp EcoRI-HindIII fragment labeled either on one strand at its EcoRI end (A) or on the opposite strand at the HindIII end (B). Lanes 1 to 3 contained 1, 0.6, and 0.2 mg of AbrBN55(ox)/ml, respectively; lanes 6 to 8 contained 1.5, 0.9, and 0.4 mg of AbrB/ml, respectively; and lanes 4, 5, 9, and 10 contained no binding protein. The Maxam-Gilbert purine (lane U) and pyrimidine (lane Y) sequencing ladders for each fragment are shown for reference. DNase I footprinting assays were performed at 20°C essentially as described previously (16). Radiolabeled DNAs were prepared as described previously (16).
The preponderance of the evidence presented here and elsewhere (17, 19) argues that high-affinity DNA binding requires a multimeric association between intact full-length AbrB monomers (as well as between truncated N-terminal domains of the protein). In this regard, we should emphasize a correction to some of our previous conclusions concerning the multimeric nature of AbrB: earlier studies had been interpreted as indicating that AbrB is hexameric in structure (16, 19), but recent, more refined and detailed analysis indicates that AbrB is actually a tetramer in solution (17; M. A. Strauch, unpublished data).
The NMR structure of AbrBN53 has recently been solved (17). In solution, AbrBN53 exists as a stable noncovalently linked dimer. There is a single α-helical region present in each monomer (residues 19 to 26). In the dimer, the two α-helices are aligned lengthwise, forming the base of a cleft that is predicted to accommodate DNA. Molecular modeling and several lines of empirical evidence indicate that this cleft is the site of binding to DNA with the amino acid residues of the α-helix being critical for interaction (17, 19; M. A. Strauch, unpublished data). Yet, the dimers of AbrBN53 or of AbrBN55 (i.e., the noncovalently linked, reduced form) have a very weak ability to form stable protein-DNA complexes (Table 1). Thus, noncovalent dimerization of N-terminal DNA-binding domains is not sufficient for stable interaction with a DNA target: further multimerization appears to be required. Unfortunately, it is not yet clear why this is structurally necessary, and work is in progress to address this point.
Given that multimerization of AbrBN55 dimers into tetramers (i.e., via disulfide linkage) can result in in vitro recognition and specificity identical to and binding affinity comparable to those of native AbrB, what is the function of the C-terminal domain of the latter? That the C-terminal domain is important for the functioning of the native protein is known, since mutations affecting various residues in this region of the protein result in an AbrB phenotype in vivo and altered activity in vitro (19; M. A. Strauch, unpublished data). Additionally, the truncated abrBN53 and abrBN55 genes produce an AbrB phenotype in vivo (Q. Qian and M. A. Strauch, unpublished data). Although we cannot rule out that the C-terminal portion of AbrB may provide some DNA contacts that increase binding stability, we do know that isolated C-terminal truncations (residues K31 to K94) exhibit no inherent DNA-binding activity (S. Rao and M. A. Strauch, unpublished data). At present, we believe that the C-terminal domain is a type of higher-order multimerization element serving to arrange or stabilize the N-terminal DNA-binding domains into a configuration necessary for optimal interaction with DNA. Relevant to this are our observations that purified mutant proteins containing lesions in the C-terminal domain exhibit alterations in multimeric form or multimeric stability (19; D. K. Strickland and M. A. Strauch, unpublished data).
Although multimerization of AbrBN55 to produce an active DNA-binding protein in vitro can be achieved via disulfide bonding between the penultimate Cys54 residues on the truncated domains, multimerization mediated by the C-terminal domains of intact wild-type AbrB does not appear to involve disulfide bridges (Z. E. Phillips and M. A. Strauch, unpublished data). Yet, mutation or modification of Cys54 in full-length AbrB leads to altered multimerization and DNA-binding properties (7, 15, 19; Z. E. Phillips and M. A. Strauch, unpublished data). Thus, the exact role played by Cys54 in the native protein structure is enigmatic and is being actively investigated.
The N-terminal domain of AbrB represents a heretofore-uncharacterized example of a DNA-binding structural motif (17). Recent genome sequencing projects of Bacillus and Clostridium species have uncovered numerous additional genes (17 at the time of this writing) encoding proteins having N-terminal domains exhibiting a high degree of amino acid identity to the sequence of the first 50 residues of AbrB. B. subtilis possess two such additional AbrB homologs, which are known as Abh (5; M. A. Strauch, unpublished data) and SpoVT (2). Based on available evidence, both Abh and SpoVT are DNA-binding regulatory proteins affecting the transcription of genes that are expressed during and after entry of the cell into stationary phase and sporulation. Despite the high degree of sequence identity in their presumptive DNA-binding domains, AbrB, Abh, and SpoVT appear to control quite different regulons (although the extent and overlap between these regulons has not yet been fully determined). Undoubtedly, the specific amino acid differences in the N-terminal domains play significant roles in DNA sequence recognition. Also striking is the fact that these three proteins show no significant amino acid similarity in their C-terminal domains. Considering the role of the C-terminal domain in mediating multimer formation that produces active AbrB protein, it seems likely that the C-terminal domains of Abh and SpoVT are also involved in multimerization. Perhaps the divergent C-terminal sequences of these proteins ensures against the formation of nonfunctional proteins containing mixed subunits. We are presently exploring this possibility.
Acknowledgments
We thank Deborah L. McCann for assistance in preparation of some of the figures.
We also thank the Stein Endowment Fund (The Scripps Research Institute) for partial defrayment of the cost of some oligonucleotides used in this study. This work was supported in part by grant GM46700, National Institutes of Health.
REFERENCES
- 1.Ahmed A K, Schaffer S W, Wetlaufer D B. Nonenzymatic reactivation of reduced bovine pancreatic ribonuclease by air oxidation and by glutathione oxidoreduction buffers. J Biol Chem. 1975;250:8477–8482. [PubMed] [Google Scholar]
- 2.Bagyan I, Hobot J, Cutting S. A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J Bacteriol. 1996;178:4500–4507. doi: 10.1128/jb.178.15.4500-4507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher S H, Strauch M A, Atkinson M R, Wray L V., Jr Modulation of Bacillus subtilis catabolite repression by transition state regulatory protein AbrB. J Bacteriol. 1994;176:1903–1912. doi: 10.1128/jb.176.7.1903-1912.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kallio P T, Fagelson J E, Hoch J A, Strauch M A. The transition state regulator Hpr of Bacillus subtilis is a DNA-binding protein. J Biol Chem. 1991;266:13411–13417. [PubMed] [Google Scholar]
- 5.Kobayashi K, Shoji K, Shimizu T, Nakano K, Sato T, Kobayashi Y. Analysis of a suppressor mutation ssb (kinC) of sur0B20 (spo0A) mutation in Bacillus subtilis reveals that kinC encodes a histidine protein kinase. J Bacteriol. 1995;177:176–182. doi: 10.1128/jb.177.1.176-182.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perego M, Hoch J A. Sequence analysis and regulation of the hpr locus, a regulatory gene for protease production and sporulation in Bacillus subtilis. J Bacteriol. 1988;170:2560–2567. doi: 10.1128/jb.170.6.2560-2567.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perego M, Spiegelman G B, Hoch J A. Structure of the gene for the transition state regulator abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol. 1988;2:689–696. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 8.Robertson J B, Gocht M, Marahiel M A, Zuber P. AbrB, a regulator of gene expression in Bacillus, interacts with the transcription initiation regions of a sporulation gene and an antibiotic biosynthesis gene. Proc Natl Acad Sci USA. 1989;86:8457–8461. doi: 10.1073/pnas.86.21.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonenshein A L. Endospore-forming bacteria: an overview. In: Brun Y V, Shimkets L J, editors. Prokaryotic development. Washington, D.C.: American Society for Microbiology; 2000. pp. 133–150. [Google Scholar]
- 10.Strauch M A. Regulation of Bacillus subtilis gene expression during the transition from exponential growth to stationary phase. Prog Nucleic Acid Res Mol Biol. 1993;46:121–153. doi: 10.1016/s0079-6603(08)61020-x. [DOI] [PubMed] [Google Scholar]
- 11.Strauch M A. Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promoter. J Bacteriol. 1995;177:6999–7002. doi: 10.1128/jb.177.23.6999-7002.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strauch M A. AbrB modulates expression and catabolite repression of a Bacillus subtilis ribose transport operon. J Bacteriol. 1995;177:6727–6731. doi: 10.1128/jb.177.23.6727-6731.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strauch M A. Dissection of the Bacillus subtilis spo0E binding site for the global regulator AbrB reveals smaller recognition elements. Mol Gen Genet. 1996;250:742–749. doi: 10.1007/BF02172986. [DOI] [PubMed] [Google Scholar]
- 14.Strauch M A, Hoch J A. Transition-state regulators: sentinels of Bacillus subtilis post-exponential gene expression. Mol Microbiol. 1993;7:337–342. doi: 10.1111/j.1365-2958.1993.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 15.Strauch M A, Perego M, Burbulys D, Hoch J A. The transition state transcription regulator AbrB of Bacillus subtilis is autoregulated during vegetative growth. Mol Microbiol. 1989;3:1203–1209. doi: 10.1111/j.1365-2958.1989.tb00270.x. [DOI] [PubMed] [Google Scholar]
- 16.Strauch M A, Spiegelman G B, Perego M, Johnson W C, Burbulys D, Hoch J A. The transition state transcription regulator AbrB of Bacillus subtilis is a DNA binding protein. EMBO J. 1989;8:1615–1621. doi: 10.1002/j.1460-2075.1989.tb03546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaughn J L, Skelton N, Feher V, Naylor S, Strauch M A, Cavanagh J. Novel DNA recognition topology and gene regulation model for transition-state regulators. Nat Struct Biol. 2000;7:1139–1146. doi: 10.1038/81999. [DOI] [PubMed] [Google Scholar]
- 18.Weir J, Predich M, Dubnau E, Nair G, Smith I. Regulation of spo0H, a gene coding for the Bacillus subtilis ςH factor. J Bacteriol. 1991;173:521–529. doi: 10.1128/jb.173.2.521-529.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu K, Clark D, Strauch M A. Analysis of abrB mutations, mutant proteins, and why abrB does not utilize a perfect consensus in the −35 region of its sigma A promoter. J Biol Chem. 1996;271:2621–2626. doi: 10.1074/jbc.271.5.2621. [DOI] [PubMed] [Google Scholar]
- 20.Xu K, Strauch M A. In vitro selection of optimal AbrB-binding sites: comparison to known in vivo sites indicates flexibility in AbrB binding and recognition of three-dimensional DNA structures. Mol Microbiol. 1996;19:145–158. doi: 10.1046/j.1365-2958.1996.358882.x. [DOI] [PubMed] [Google Scholar]
- 21.Xu K, Strauch M A. Identification, sequence, and expression of the gene encoding γ-glutamyltranspeptidase in Bacillus subtilis. J Bacteriol. 1996;178:4319–4322. doi: 10.1128/jb.178.14.4319-4322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]