Abstract
Ewing sarcoma (ES) is an aggressive primary malignant bone tumor that predominantly affects children and young adults. Multimodal treatment approaches have markedly improved the survival of patients with localized ES. However, local recurrence and distant metastasis following curative therapies remain a main concern for patients with ES. Recent studies have suggested that slow-cycling cells (SCCs) are associated with tumor progression, local recurrence and distant metastasis in various types of cancers. According to the results of these studies, it was hypothesized that SCCs may play a critical role in tumor progression, chemoresistance and local/distal recurrence in patients with ES. The present study applied a label-retaining system using carboxyfluorescein diacetate succinimidyl ester (CFSE) to identify and isolate SCCs in ES cell lines. In addition, the properties of SCCs, including sphere formation ability, cell cycle distribution and chemoresistance, in comparison with non-SCCs were investigated. RNA sequencing also revealed several upregulated genes in SCCs as compared with non-SCCs; the identified genes not only inhibited cell cycle progression, but also promoted the malignant properties of SCCs. On the whole, the present study successfully identified SCCs in ES cells through a label-retaining system using CFSE. Moreover, to the best of our knowledge, the present study is the first to describe the characteristic properties of SCCs in ES. The findings of this study, if confirmed, may prove to be useful in elucidating the underlying molecular mechanisms and identifying effective therapeutic targets for ES.
Keywords: slow-cycling cells, cell cycle, Ewing sarcoma, recurrence, metastasis, carboxyfluorescein diacetate succinimidyl ester
Introduction
Ewing sarcoma (ES), the second most common malignant bone tumor, is an aggressive cancer that primarily affects children and young adults (1). ES arises predominantly in the pelvis, long bones and ribs (2,3). Recently, a multimodal treatment approach consisting of surgery and/or radiotherapy with intensive chemotherapy has markedly improved the survival of patients with localized ES. Thus, the current 5-year overall survival rate of patients with localized ES ranges from 65 to 75% (4,5). However, the rates of local recurrence and distant metastasis have been reported to be 6.2 to 7% and 12.9 to 31%, respectively (6,7), and the 5-year overall survival rate for patients with local recurrence and/or distant metastasis has been reported to be <25% (8-10). Hence, obtaining a better understanding of the mechanisms underlying local recurrence and distant metastasis following curative therapies is a time-sensitive matter of critical importance for patients with ES.
Human malignancies display intratumoral heterogeneity in phenotypic features, as regards cellular morphology, gene expression, metabolism, motility and angiogenic, proliferative, immunogenic, and metastatic potential (11). This phenotypic heterogeneity is considered to be one of the major causes of treatment failure when implementing state-of-the-art cancer therapies (12).
As one example, in heterogeneous tumor cell populations, there is generally a small subpopulation of slow-cycling cells (SCCs), defined as non-proliferating quiescent/dormant cells (13,14). Previous research has demonstrated the existence of SCCs, and has described their properties in various human cancer cell cultures and xenograft models. For example, a small subpopulation of SCCs was identified in a study evaluating melanoma cells, and these cells demonstrated chemoresistance (15). The high invasive ability of SCCs in melanoma has also been reported in prior research (16). Similarly, chemoresistance and the tumorigenic potential of SCCs have been observed in colon cancer and breast cancer cell lines (17), and SCCs in glioblastoma have demonstrated resistance to both chemotherapy and radiotherapy (18). However, to the best of our knowledge, SCCs have not been previously studied in ES, and their characteristics are unknown. Therefore, investigating the characteristics of SCCs in ES is a highly understudied topic of substantial clinical importance and has the potential to provide meaningful information for improving existing ES treatment regimens.
Several label-retaining systems have been reported to effectively identify SCCs/quiescent cells in both normal and cancer cell lines (13,16-21). Using these label-retaining systems, SCCs or quiescent cells can be distinguished from other cell populations by the slowed cell division of SCCs as compared to other cells. Moreover, among these systems, the use of carboxyfluorescein diacetate succinimidyl ester (CFSE) green fluorescent dye is a well-established method (13), and has been applied in studies investigating SCCs in several types of cancer (17,19-21).
More specifically, in the CFSE labeling system, once the cells are labeled with CFSE, the fluorescence in the cells is equally distributed between the daughter cells upon division. Since the fluorescence in the cells is gradually diluted by cell proliferation and only non-dividing cells or SCCs can retain strong fluorescent staining via CFSE for a long period of time, researchers can effectively distinguish SCCs (with strong fluorescence) from non-slow-cycling cells (non-SCCs) (with weak fluorescence).
According to previous studies (17,19-21), the present study hypothesized the existence of SCCs in ES. The present study therefore applied a label-retaining system based on CFSE to ES cell lines, and succeeded in identifying and isolating SCCs, in the first study of its kind conducted to date, to the best of our knowledge. Moreover, the distinctive properties of SCCs in ES were comprehensively described by evaluating their sphere formation ability, cell cycle distribution and chemoresistance. In addition, RNA sequencing revealed differentially expressed genes in SCCs as compared with non-SCCs, that not only inhibited cell cycle progression, but also supported malignant properties, including invasive capacity and metastatic potential.
Materials and methods
Cells and cell culture
In total, three ES cell lines were used in the present study. The SK-ES-1 (HTB-86) and A673 (CRL-1598) cells were purchased from the American Type Culture Collection (ATCC), and the TC71 (ACC 516) cells were obtained from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures GmbH. The SK-ES-1 cells were cultured in McCoy's 5A medium (MilliporeSigma) supplemented with 15% fetal bovine serum (FBS; MilliporeSigma). The A673 cells were cultured in Dulbecco's modified Eagle's medium (MilliporeSigma) supplemented with 15% FBS. The TC71 cells were cultured in Iscove's modified Dulbecco's medium (Thermo Fisher Scientific, Inc.) supplemented with 10% FBS. All cell culture media were supplemented with 100 U/ml penicillin (MilliporeSigma) and 100 µg/ml streptomycin (MilliporeSigma). The cells were maintained in a humidified atmosphere with 5% CO2 at 37°C.
Labeling of ES cells with CFSE
The CellTrace CFSE Cell Proliferation kit (cat. no. C34554; Thermo Fisher Scientific, Inc.) was used to label the ES cells. Briefly, both a cell suspension (1×106 cells/ml) and a solution of 4 µM CFSE was prepared in phosphate-buffered saline (PBS) supplemented with 1% FBS. Equal amounts of the cell suspension and CFSE solution were mixed, and the cells were incubated at a final concentration of 2 µM by incubation at 37°C for 10 min under dark conditions. The cells were quenched three times with cold PBS supplemented with 10% FBS. These cells were then used as CFSE-labeled cells for the experiments described below. CFSE fluorescence was confirmed using an all-in-one fluorescence microscope (BZ-X700; Keyence Corporation).
Definition of SCCs and the sorting of cells into SCCs and non-SCCs
CFSE fluorescence in ES cells was preliminarily evaluated in both sphere and adherent cultures, and it was confirmed that the CFSE fluorescence intensity was more heterogeneous in the sphere cultures (Fig. S1). These findings indicated that cell division was more variable in spheres than in adherent cultures; therefore, sphere cultures were used in all the experiments in the present study. Since the cells retaining a strong CFSE fluorescence decreased in a culture period of >5 days in the preliminary study (data not shown), the cells were collected at day 5 after seeding for the experiments.
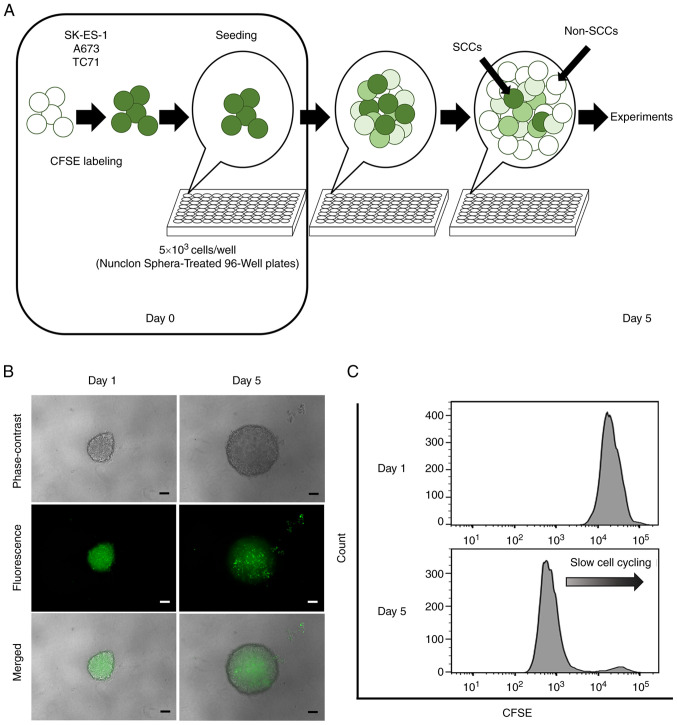
CFSE-labeled cells (5×103 cells/well) were seeded into Nunclon Sphera 96-well, Nunclon Sphera-treated, U-shaped-bottom Microplates (cat. no. 174929; Thermo Fisher Scientific, Inc.). The plates were centrifuged at 228 × g for 3 min at room temperature. Following 5 days of culture, the CFSE-labeled cells formed a single sphere in each well (Fig. 1A). The spheres were confirmed under a fluorescence microscope (BZ-X700; Keyence Corporation).
Figure 1.
Identification of SCCs by a label-retaining system using CFSE. (A) Schematic diagram using a CFSE label-retaining system. (B) Phase-contrast, fluorescence and merge images of single spheres derived from the SK-ES-1 cell line at 1 and 5 days after CFSE labeling. Scale bars, 100 µm. (C) Flow cytometric analyses of CFSE fluorescence in spheres derived from the SK-ES-1 cell line at 1 and 5 days after CFSE labeling. CFSE, carboxyfluorescein diacetate succinimidyl ester; SCCs, slow-cycling cells.
For the flow cytometric analysis, each sphere was collected and dissociated using Accumax (cat. no. AM105; Nacalai Tesque, Inc.) to prepare single-cell suspensions, and the CFSE fluorescence of each cell was evaluated using a BD LSRFortessa cell analyzer (BD Biosciences). In the present study, cells retaining a strong CFSE fluorescence (i.e., in the top 10%) were defined as SCCs and other cells as non-SCCs on day 5 after seeding. For sorting, single cells obtained from each sphere were resuspended in Hank's Balanced Salt Solution (HBSS; cat. no. 14175079; Thermo Fisher Scientific, Inc.) containing 1 mM UltraPure 0.5 M ethylenediaminetetraacetic acid (EDTA; pH 8.0; cat. no. 15575020; Thermo Fisher Scientific, Inc.), and 25 mM 4-[2-hydroxyethyl]-1-piperazineethanesulfonic acid (HEPES; cat. no. H3375; MillporeSigma), 2% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin. The sorting of cells into SCCs and non-SCCs was conducted using a BD FACS Aria III cell sorter (BD Biosciences) with the exclusion of doublets and dead cells.
Sphere formation assay
The sorted SCCs (1×105 cells/well) and non-SCCs (1×105 cells/well) from the SK-ES-1 cells were seeded into Costar 24-well Clear Flat Bottom Ultra-low Attachment Multiple Well Plates (Corning, Inc.) in serum-free McCoy's 5A medium containing 10 ng/ml basic fibroblast growth factor (cat. no. 062-06661; FUJIFILM Wako Pure Chemical Corporation), 10 µg/ml human insulin (cat. no. 0105; Cell Science & Technology Institute, Inc.), 100 µg/ml human transferrin (cat. no. 10652202; MillporeSigma) and 100 µg/ml bovine serum albumin (cat. no. 019-21272; Nacalai Tesque, Inc.) and were incubated at 37°C for 6 days in order to form spheres, as previously described (22). Formed spheres ≥50 µm in size in each well were manually counted under a fluorescence microscope (BZ-X700; Keyence Corporation) by two examiners blinded to the sorting conditions.
Cell cycle analysis
Single cells (1×106 cells) obtained from spheres were fixed with cold 70% ethanol for 30 min on ice under dark conditions and washed twice with PBS. The cells were then centrifuged at 130 × g for 5 min at room temperature and resuspended in 0.5 ml propidium iodide (PI, in PI/RNase Staining Buffer; cat. no. 550825; BD Biosciences). Following incubation for 15 min on ice under dark conditions, the DNA content of the samples was analyzed using PI and flow cytometry (BD LSRFortessa cell analyzer; BD Biosciences). Data were analyzed using FlowJo analysis software (version 10.7.2; BD Biosciences), and the cell cycle distribution was determined by applying the Dean-Jett-Fox cell-cycle modeling algorithm to the PI fluorescence intensity profile of the cells.
Drug treatment of CFSE-labeled ES cells
Drug treatment assays were performed in the sphere culture of CFSE-labeled SK-ES-1 cells, not using sorted SCCs and non-SCCs. The assays were performed in a similar environment as the in vivo conditions, and using a sphere culture known to be possible to create the in vivo conditions (23). In addition, the process of the cell sorting may affect the condition of the cells during the drug treatment assays, and the affect should be avoided. CFSE-labeled SK-ES-1 cells (5×103 cells/well) were seeded in each well of Nunclon Sphera-Treated 96-Well Plates (Thermo Fisher Scientific, Inc.) and cultured for 3 days in order to form single spheres. The medium was then replaced with medium containing no drug (i.e., the control medium), 30 nM doxorubicin (Dox; cat. no. D1515; MilliporeSigma), or 5 nM vincristine (Vin; cat. no. V8879; MilliporeSigma) followed by incubation for 2 days at 37°C. To evaluate the CFSE fluorescence intensity following drug treatment, Z-stack images of the spheres were captured under a fluorescence microscope (BZ-X700; Keyence Corporation). The area with CFSE fluorescence (%) in each sphere was quantitatively calculated using a hybrid cell count application available in the BZ-X Analyzer software (BZ-H4A; Keyence Corporation).
The apoptotic analysis of drug-treated cells was performed using flow cytometry with dual staining with APC Annexin V (Annexin V; cat. no. 550475; BD Biosciences) and PI (cat. no. 556463; BD Biosciences). Briefly, single cells obtained from each sphere after 2 days of drug treatment were washed twice with cold PBS and resuspended in 1X Annexin V binding buffer (cat. no. 556454; BD Biosciences) at a concentration of 2×106 cells/ml. The cell solution (2×105 cells in 100 µl) was then transferred into a 1.5-ml tube, and 5 µl Annexin V and 2 µl PI were added. The tubes were vortexed gently and incubated for 15 min at room temperature under dark conditions. Following this, 400 µl 1X Annexin V binding buffer was added to each tube, and the samples were analyzed using a flow cytometer (BD LSRFortessa cell analyzer; BD Biosciences). Data were analyzed using FlowJo analysis software (version 10.7.2; BD Biosciences).
RNA sequencing
For RNA sequencing, total RNA was extracted from the cells using TRIzol® reagent according to the manufacturer's instructions (cat. no. 15596026; Thermo Fisher Scientific, Inc.). After sorting the SK-ES-1 cells into SCCs and non-SCCs, 2×105 cells from each cell fraction were lysed and homogenized in 1 ml TRIzol® reagent in a tube. The samples were incubated for 5 min at room temperature to permit the complete dissociation of the nucleoprotein complexes. Following this, 200 µl chloroform (cat. no. 038-02606; FUJIFILM Wako Pure Chemical Corporation) were added, and the tubes were shaken vigorously for 15 sec. This was followed by incubation at room temperature for 3 min. The samples were transferred to phasemaker tubes (Thermo Fisher Scientific, Inc.) and centrifuged at 12,000 × g for 15 min at 4°C. The aqueous phase in the phasemaker tube was transferred to a fresh tube and mixed with 500 µl isopropyl alcohol. Following 10 min of incubation at room temperature, the mixture was washed once with 75% ethanol and centrifuged at 7,500 × g for 5 min at 4°C. The supernatant liquid was discarded, and the RNA pellets were air-dried and dissolved in distilled water. For RNA quality control, the concentration and purity were examined using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Inc.). Total RNA samples from SCCs and non-SCCs were submitted to Macrogen for library preparation using the TruSeq Stranded mRNA LT Sample Prep kit (Illumina, Inc.). Paired-end RNA sequencing was performed using the Illumina NovaSeq6000 System (Illumina, Inc.). Reads were aligned to human transcriptome (hg38) reference sequences using the Strand NGS software program (Strand Life Sciences). The aligned reads were normalized to transcripts per million (TPM) and the normalized counts were set to 1. TPM values (log2) were used to compare gene expression levels between SCCs and non-SCCs. Gene expression data were visualized using scatter plots, and pathway analyses were conducted using WikiPathways (https://wikipathways.org/) within the Strand NGS software program (https://www.strand-ngs.com/).
Statistical analysis
Statistical analyses were conducted using EZR statistical software (Saitama Medical Centre, Jichi Medical University; http://www.jichi.ac.jp/saitama-sct/SaitamaHP.files/statmedEN.html; Kanda, 2012). All values are presented as the mean ± standard error of the mean. Comparisons between two groups were performed using unpaired t-tests. Comparisons between multiple groups were determined using one-way analysis of variance (ANOVA) followed by post-hoc testing using the Tukey procedure (i.e., Tukey's honest significant difference test). A two-sided P-value <0.05 was considered to indicate a statistically significant difference.
Results
Identification of SCCs in ES
The formation of a single sphere with CFSE fluorescence in each well was confirmed under a fluorescence microscope at 1 day after seeding (Fig. 1B). Microscopic evaluation and flow cytometric analyses performed on day 5 revealed that the detected CFSE fluorescence gradually weakened in the majority of the cells, and that only a small number of cells retained strong CFSE fluorescence (Fig. 1B and C). Cells that retained a strong fluorescence (top 10%) as of day 5 were defined as SCCs, and other cells were categorized as non-SCCs.
Sphere formation ability of SCCs as compared to non-SCCs
Previous research has reported that sphere formation ability is one of the characteristics of SCCs in cancer cell lines (18), and that this ability is also associated with tumorigenicity in ES (24). Therefore, the present study examined the sphere formation ability of SCCs within ES cells in comparison with that of non-SCCs. Sphere formation assays demonstrated that the number of spheres was significantly higher in SCCs than in non-SCCs (P<0.05; Fig. 2A and B). These results indicated that the sphere formation ability was enhanced in SCCs within ES.
Figure 2.
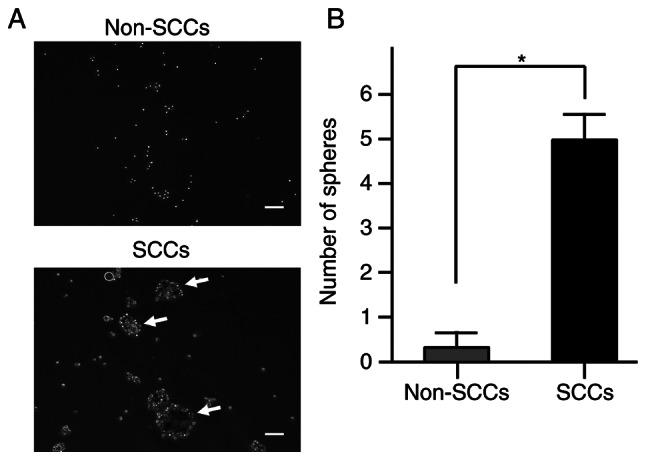
Sphere formation ability of SCCs and non-SCCs in the SK-ES-1 cell line. (A) Representative images of sphere formation assays in sorted SCCs and non-SCCs in the SK-ES-1 cell line. Scale bars, 50 µm. Formed spheres ≥50 µm in size (shown using white arrows) were counted under microscopy. (B) Comparative number of spheres between SCCs and non-SCCs. The error bars indicate the standard error of the mean. *P<0.05. SCCs, slow-cycling cells.
Proportion of cells in the G0/G1 phase in SCCs vs. non-SCCs
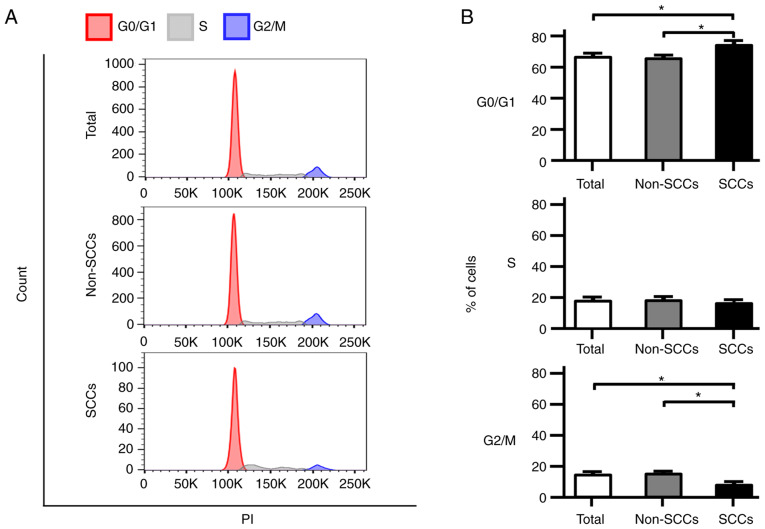
Cell cycle analyses were performed using flow cytometry, and the phase distribution of the cell cycle was compared among total cells, non-SCCs and SCCs in the SK-ES-1 cell line. Notably, the proportion of cells in the G0/G1 phase was significantly higher in the SCCs than in both total cells and non-SCCs (P<0.05; Fig. 3A and B). In addition, the number of cells in the G2/M phase in SCCs was significantly lower than that in both total cells and non-SCCs (P<0.05). Analyses were also performed using the A673 and TC71 ES cell lines; similarly, an increased number of cells in the G0/G1 phase and a decreased number of cells in the G2/M phase were observed in both cell lines for SCCs as compared with non-SCCs (Fig. S2A and B).
Figure 3.
Cell cycle distribution of total cells, non-SCCs and SCCs in the SK-ES-1 cell line. (A) Representative cell cycle distributions for the SK-ES-1 cell line are shown using flow cytometry after PI staining. The G0/G1 phase is shown in black, the S phase is shown in light gray, and the G2/M phase is shown in dark gray. (B) Comparative proportions of cells in each cell cycle phase among total cells, non-SCCs and SCCs in the SK-ES-1 cell line. The error bars indicate the standard error of the mean. *P<0.05. PI, propidium iodide; SCCs, slow-cycling cells.
Resistance to doxorubicin and vincristine in SCCs vs. non-SCCs
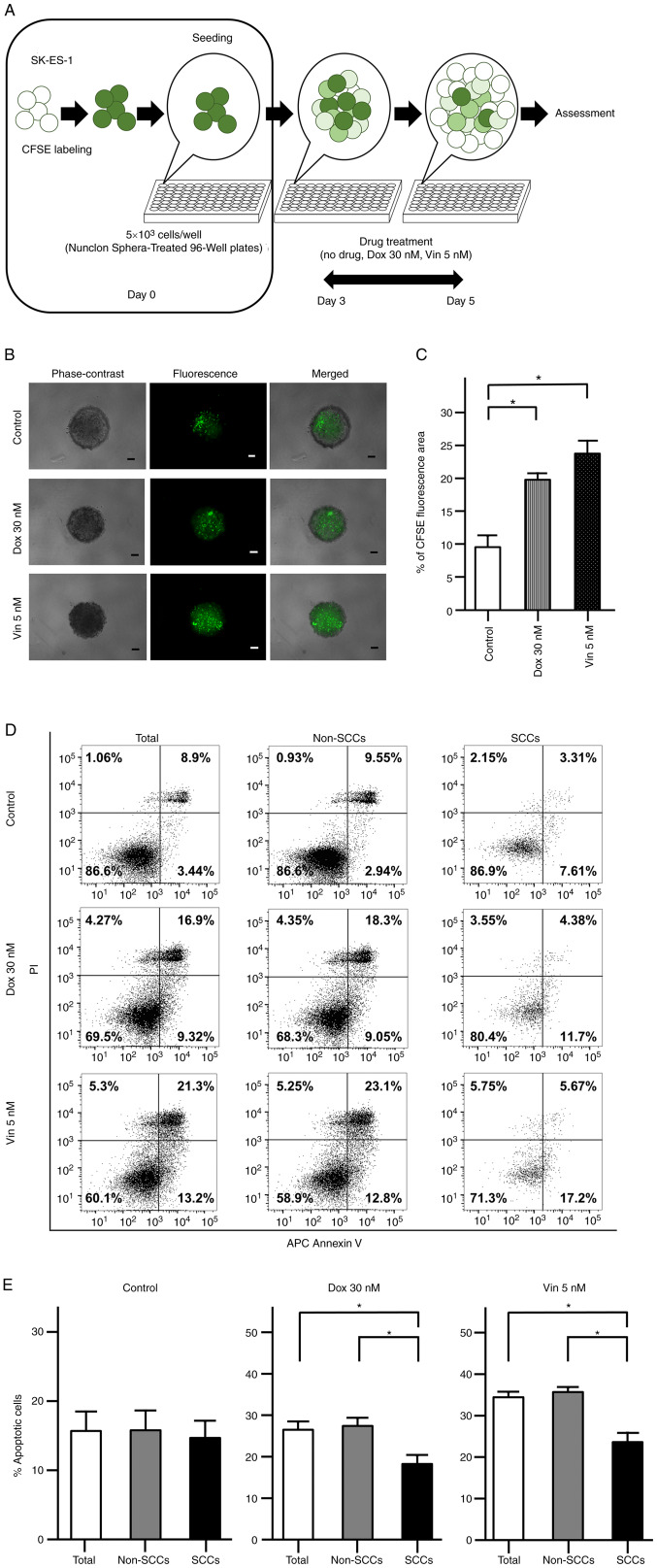
As previously reported, SCCs exhibit chemoresistance within several types of cancer (17,18). The present study thus assessed the chemoresistance of SCCs found in ES to Dox and Vin using spheres formed by CFSE-labeled cells (Fig. 4A). Following 2 days of drug treatment, CFSE fluorescence was observed in the evaluated spheres under a fluorescence microscope (Fig. 4B). The area with CFSE fluorescence was significantly greater in the spheres treated with Dox and Vin than in the spheres subjected to the control treatment (no drugs) (P<0.05; Fig. 4C).
Figure 4.
Chemoresistance of CFSE-labeled SK-ES-1 cells. (A) Schematic diagram evaluating the chemoresistance of CFSE-labeled SK-ES-1 cells. (B) Representative phase-contrast, fluorescence and merged images of spheres derived from CFSE-labeled SK-ES-1 cells. Scale bars, 100 µm. (C) Percentage of the CFSE fluorescence area following 2 days of drug treatment. The error bars indicate the standard error of the mean. *P<0.05. (D) Apoptosis assays by dual staining with APC Annexin V and PI using flow cytometry. The percentage of the cells was calculated after 2 days of drug treatment in each fraction: Left lower panel, APC Annexin V-negative/PI-negative; right lower panel, APC Annexin V-positive/PI-negative; left upper panel, APC Annexin V-negative/PI-positive; right upper panel, APC Annexin V-positive/PI-positive. (E) Percentage of apoptotic cells after 2 days of drug treatment in total cells, non-SCCs and SCCs in the SK-ES-1 cell line. The error bars indicate the standard error of the mean. *P<0.05. CFSE, carboxyfluorescein diacetate succinimidyl ester; Dox, doxorubicin; PI, propidium iodide; SCCs, slow-cycling cells; Vin, vincristine.
The apoptotic cells were further examined following drug treatment using flow cytometry with dual staining with Annexin V and PI (Fig. 4D). In the control group, there were no significant differences in the percentages of Annexin V-positive cells among the total cells, non-SCCs and SCCs. OF note, the percentages of Annexin V-positive cells were significantly decreased in the SCCs as compared with both the total cells and non-SCCs when the spheres were treated with Dox or Vin (P<0.05; Fig. 4E).
Upregulated expression of various genes and involved pathways in RNA sequencing
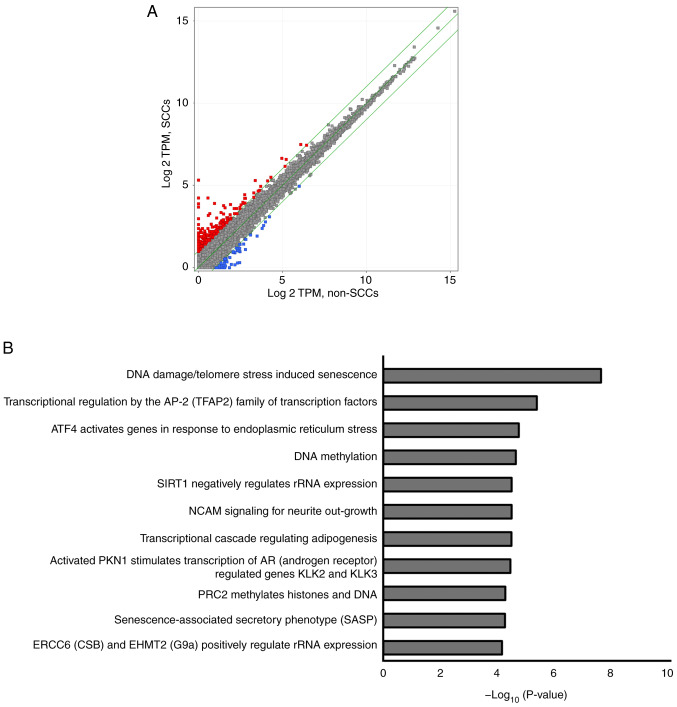
RNA sequencing was performed using the SK-ES-1 cells to evaluate differences in gene expression patterns between SCCs and non-SCCs. As illustrated in Fig. 5A, 255 genes were upregulated by >2-fold in the SCCs as compared to the non-SCCs (red dots), whereas 58 genes were downregulated by ≤2-fold in SCCs as compared to the non-SCCs (blue dots).
Figure 5.
RNA sequencing of SCCs and non-SCCs. (A) Scatter plots illustrating >2-fold or higher upregulated genes (shown using red dots) and ≤2-fold downregulated genes (shown using blue dots). (B) List of 11 pathways with an associated P-value <10−5. SCCs, slow-cycling cells.
A total of 11 pathways were detected using pathway analysis, including 'DNA damage/telomere stress induced senescence', 'Transcriptional regulation by the AP-2 (TFAP2) family of transcription factors', and 'ATF4 activates genes in response to endoplasmic reticulum stress'. Each detected pathway was associated with a P-value <10−5, and these pathways were associated with the aforementioned 255 upregulated genes found in SCCs (Fig. 5B). Genes involved in each pathway included cyclin-dependent kinase inhibitor 1A (CDKN1A), activating transcription factor 3 (ATF3) and DNA damage-inducible transcript 3 (DDIT3) (Table I). Pathway analyses of the aforementioned downregulated genes did not reveal any statistically significant pathways (data not shown).
Table I.
Genes involved in the 11 pathways detected using pathway analyses.
| Pathway | Genes |
|---|---|
| DNA damage/telomere stress Induced Senescence | CDKN1A, HIST1H2BC, HIST1H2AC, HIST1H1E, HIST1H2BJ, HIST2H2BE |
| Transcriptional regulation by the AP-2 (TFAP2) family of transcription factors | CDKN1A, APOE, KCTD15 |
| ATF4 activates genes in response to endoplasmic reticulum stress | ATF3, DDIT3 |
| DNA methylation | HIST1H2BC, HIST1H2AC, HIST1H2BJ, HIST2H2BE |
| SIRT1 negatively regulates rRNA expression | HIST1H2BC, HIST1H2AC, HIST1H2BJ |
| NCAM signaling for neurite out-growth | 3x4Hyp-5Hyl-COL6A2, NCAN, SPTBN4 |
| Transcriptional cascade regulating adipogenesis | KLF2, EGR2, DDIT3 |
| Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 | HIST1H2BC, HIST1H2AC, HIST1H2BJ, HIST2H2BE |
| PRC2 methylates histones and DNA | HIST1H2BC, HIST1H2AC, HIST1H2BJ, HIST2H2BE |
| Senescence-associated secretory phenotype (SASP) | CDKN1A, HIST1H2BC, HIST1H2AC, HIST1H2BJ, HIST2H2BE |
| ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression | HIST1H2BC, HIST1H2AC, HIST1H2BJ, HIST2H2BE |
Discussion
In recent years, SCCs have been studied and are presently considered to be associated with local recurrence and/or distant metastasis in various types of cancer (13-21). However, to the best of our knowledge, SCCs have not been investigated in ES to date, and the present study was the first to successfully identify and isolate SCCs from ES cell lines. As specific markers for SCCs are currently unknown, a label-retaining system was employed using CFSE; this system is relatively easy to manage and has been widely used in the research field in previous studies evaluating SCCs (17,19-21). Moreover, the distinctive properties of SCCs in ES were elucidated, such as a higher sphere formation ability, an increased cell proportion in the G0/G1 phase, and chemoresistance to doxorubicin and vincristine. These results are consistent with the properties of SCCs previously reported for other types of cancer (13-15,17-21).
The present study revealed that SCCs in ES exhibited a higher sphere-formation ability than non-SCCs. Previously, Zeng et al (18) reported a high sphere-forming ability of SCCs in glioblastoma cell lines, and Wahl et al (24) reported that single cells derived from spheres exhibited a high tumorigenicity in ES cell lines. These studies strongly suggest that SCCs in ES have a high tumorigenicity associated with a high sphere-formation ability. In addition, the sphere formation ability has previously been used to evaluate anchorage-independent growth (25), a critical step in metastasis (26). Thus, the high sphere formation ability evidenced in the present study may also reflect the metastatic potential of SCCs in ES.
SCCs/quiescent cells are considered to be in a particular phase of the cell cycle (16,17,27-29). However, the cell cycle phases of SCCs remain controversial. Bleau et al (27) found that SCCs were enriched in the G0/G1 phase in a sphere culture of a non-small cell lung cancer cell line, and Carcereri de Prati et al (28) reported that breast cancer cells entered the quiescent state in the G0/G1 phase under hypoxic conditions. By contrast, a high proportion of SCCs in the G2/M phase has been reported in melanoma (16) and colon cancer cell lines (17); Gao et al (29) demonstrated that SCCs were enriched in the S phase in ovarian cancer cells derived from a patient. In the present study, it was revealed that SCCs in ES cell lines were enriched in the G0/G1 phase, and these findings indicated an extended G0/G1 phase in ES SCCs. Moreover, as regards the cell cycle, RNA sequencing detected CDKN1A as one of the upregulated genes in SCCs in ES cells; this gene is involved in three of the detected pathways (Table I). Moreover, CDKN1A encodes p21, which is a widely known inhibitor of the G1/S transition (30,31). Therefore, CDKN1A may play a crucial role in the extension of the G0/G1 phase in ES SCCs.
Previous studies have indicated the chemoresistance of SCCs in various types of cancer, including glioblastoma (32), and colon (17) and breast cancer (17). In addition, Moore et al (17) demonstrated that SCCs in colon and breast cancer cells could re-enter the cell cycle and actively proliferate following the removal of anticancer drugs. In the present study, a statistically significant increase was observed in the percentage of the CFSE fluorescence area in ES spheres, and a statistically significant decrease in the apoptotic activity of ES SCCs under treatment with both Dox and Vin (i.e., the drugs used in conventional chemotherapeutic regimens for ES) (33).
The main mechanism underlying the anticancer effects of Dox is the cessation of the DNA replication process (34), which occurs at the S phase. Thus, SCCs in ES likely evade the effects of Dox by extending the G0/G1 phase. Moreover, SCCs, which are enriched in the G0/G1 phase, may be less susceptible to Vin as this drug functions as an inhibitor in the M phase (35). The findings of the present study strongly suggest that resistance to Dox and Vin in SCCs within ES is caused by the enrichment of cells in the G0/G1 phase, and that the effects of anticancer agents may be improved by promoting re-entry into the cell cycle.
Using RNA sequencing, the present study identified a pathway 'ATF4 activates genes in response to endoplasmic reticulum stress', by the pathway analysis of upregulated genes in SCCs. Endoplasmic reticulum stress has previously been reported to play a critical role in cell cycle arrest and chemoresistance in leukemia and squamous carcinoma (36,37). Moreover, ATF3 and DDIT3 are upregulated in SCCs and are each involved in this pathway. Li et al (38) previously reported that ATF3 knockdown impaired the invasion of lung cancer cells. Bandyopadhyay et al (39) revealed that ATF3 overexpression promoted invasiveness in vitro, as well as that the nuclear expression of ATF3 was positively associated with metastases in prostate cancer. As regards the cell cycle, both ATF3 and DDIT3 are known to block the G1/S transition (40-42), thereby potentially resulting in the enrichment of cells in the G0/G1 phase of SCCs in ES. Therefore, ATF3 and DDIT3 may be involved in mechanisms relevant to SCCs and, pending confirmation, may be attractive therapeutic targets specific to SCCs within ES.
Several limitations of the present study should be acknowledged. First, all the experiments were performed in vitro without any in vivo experimentation. As SCCs comprise a very small population of total cells, it was difficult to collect a sufficient number of SCCs for in vivo experiments. Second, the present study did not attempt to confirm the existence of cells with a molecular signature similar to that of the SCCs identified in the present study in tumor tissues from patients with ES. It is thus recommended that future research investigations build on these efforts in order to elucidate these questions more comprehensively.
In conclusion, in the present study, SCCs were identified in ES cell lines using a label-retaining system with CFSE. It was found that SCCs in ES exhibited an enhanced sphere formation ability, a distinctive cell cycle distribution in the G0/G1 phase, and chemoresistance to Dox and Vin. To the best of our knowledge, the present study was the first to identify SCCs in ES and demonstrate their characteristics. Moreover, RNA sequencing identified 255 genes upregulated by >2-fold in SCC, and 11 pathways were detected by pathway analysis using the 255 genes. The genes and the pathways may be used to develop biomarkers or as therapeutic targets for SCCs and/or Ewing sarcoma. Although additional studies are required to elucidate the characteristics of SCCs in ES, exploring SCCs using the employed label-retaining system with CFSE may be useful in revealing molecular mechanisms and identifying effective therapeutic targets in ES.
Supplementary Data
Acknowledgments
The authors would like to thank Ms. Minako Nagata, Ms. Maya Yasuda and Ms. Kyoko Tanaka (Department of Orthopaedic Surgery, Kobe University Graduate School of Medicine, Kobe, Japan) for their expert technical assistance.
Funding Statement
The present study was supported by JSPS KAKENHI grant no. 22K15527.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The RNA sequencing data have been deposited in NCBI GEO under accession no. GSE204671.
Authors' contributions
SY and SF wrote the manuscript, collected and assembled the data, and performed the data analysis and interpretation. HH, NF, RS, TT, TMiyamoto, YM, KK, YH, SH, TMatsumoto and TMatsuhita collected and/or assembled the data. MKA performed the data analysis and interpretation. TK, TaA, RK and ToA designed, collected and/or assembled the data, performed the data analysis and interpretation, and gave the final approval of the manuscript. SY, TK and SF confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Balamuth NJ, Womer RB. Ewing's sarcoma. Lancet Oncol. 2010;11:184–192. doi: 10.1016/S1470-2045(09)70286-4. [DOI] [PubMed] [Google Scholar]
- 2.Damron TA, Ward WG, Stewart A. Osteosarcoma, chondrosarcoma, and Ewing's sarcoma: National cancer data base report. Clin Orthop Relat Res. 2007;459:40–47. doi: 10.1097/BLO.0b013e318059b8c9. [DOI] [PubMed] [Google Scholar]
- 3.Potratz J, Dirksen U, Jürgens H, Craft A. Ewing sarcoma: Clinical state-of-the-art. Pediatr Hematol Oncol. 2012;29:1–11. doi: 10.3109/08880018.2011.622034. [DOI] [PubMed] [Google Scholar]
- 4.Gaspar N, Hawkins DS, Dirksen U, Lewis IJ, Ferrari S, Le Deley MC, Kovar H, Grimer R, Whelan J, Claude L, et al. Ewing sarcoma: Current management and future approaches through collaboration. J Clin Oncol. 2015;33:3036–3046. doi: 10.1200/JCO.2014.59.5256. [DOI] [PubMed] [Google Scholar]
- 5.Rodríguez-Galindo C, Liu T, Krasin MJ, Wu J, Billups CA, Daw NC, Spunt SL, Rao BN, Santana VM, Navid F. Analysis of prognostic factors in Ewing sarcoma family of tumors: Review of St. Jude children's research hospital studies. Cancer. 2007;110:375–384. doi: 10.1002/cncr.22821. [DOI] [PubMed] [Google Scholar]
- 6.Paulussen M, Ahrens S, Dunst J, Winkelmann W, Exner GU, Kotz R, Amann G, Dockhorn-Dworniczak B, Harms D, Müller-Weihrich S, et al. Localized Ewing tumor of bone: Final results of the cooperative Ewing's sarcoma study CESS 86. J Clin Oncol. 2001;19:1818–1829. doi: 10.1200/JCO.2001.19.6.1818. [DOI] [PubMed] [Google Scholar]
- 7.Granowetter L, Womer R, Devidas M, Krailo M, Wang C, Bernstein M, Marina N, Leavey P, Gebhardt M, Healey J, et al. Dose-intensified compared with standard chemotherapy for nonmetastatic Ewing sarcoma family of tumors: A children's oncology group study. J Clin Oncol. 2009;27:2536–2541. doi: 10.1200/JCO.2008.19.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bacci G, Ferrari S, Longhi A, Donati D, De Paolis M, Forni C, Versari M, Setola E, Briccoli A, Barbieri E. Therapy and survival after recurrence of Ewing's tumors: The Rizzoli experience in 195 patients treated with adjuvant and neoadjuvant chemotherapy from 1979 to 1997. Ann Oncol. 2003;14:1654–1659. doi: 10.1093/annonc/mdg457. [DOI] [PubMed] [Google Scholar]
- 9.Barker LM, Pendergrass TW, Sanders JE, Hawkins DS. Survival after recurrence of Ewing's sarcoma family of tumors. J Clin Oncol. 2005;23:4354–4362. doi: 10.1200/JCO.2005.05.105. [DOI] [PubMed] [Google Scholar]
- 10.Stahl M, Ranft A, Paulussen M, Bölling T, Vieth V, Bielack S, Görtitz I, Braun-Munzinger G, Hardes J, Jürgens H, Dirksen U. Risk of recurrence and survival after relapse in patients with Ewing sarcoma. Pediatr Blood Cancer. 2011;57:549–553. doi: 10.1002/pbc.23040. [DOI] [PubMed] [Google Scholar]
- 11.Marusyk A, Polyak K. Tumor heterogeneity: Causes and consequences. Biochim Biophys Acta. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen S, Vagner S, Robert C. Persistent cancer cells: The deadly survivors. Cell. 2020;183:860–874. doi: 10.1016/j.cell.2020.10.027. [DOI] [PubMed] [Google Scholar]
- 13.Basu S, Dong Y, Kumar R, Jeter C, Tang DG. Slow-cycling (dormant) cancer cells in therapy resistance, cancer relapse and metastasis. Semin Cancer Biol. 2022;78:90–103. doi: 10.1016/j.semcancer.2021.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis JE, Jr, Kirk J, Ji Y, Tang DG. Tumor dormancy and slow-cycling cancer cells. Adv Exp Med Biol. 2019;1164:199–206. doi: 10.1007/978-3-030-22254-3_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perego M, Maurer M, Wang JX, Shaffer S, Müller AC, Parapatics K, Li L, Hristova D, Shin S, Keeney F, et al. A slow-cycling subpopulation of melanoma cells with highly invasive properties. Oncogene. 2018;37:302–312. doi: 10.1038/onc.2017.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore N, Houghton J, Lyle S. Slow-cycling therapy-resistant cancer cells. Stem Cells Dev. 2012;21:1822–1830. doi: 10.1089/scd.2011.0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng L, Zhao Y, Ouyang T, Zhao T, Zhang S, Chen J, Yu J, Lei T. Label-retaining assay enriches tumor-initiating cells in glioblastoma spheres cultivated in serum-free medium. Oncol Lett. 2016;12:815–824. doi: 10.3892/ol.2016.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebinger S, Özdemir EZ, Ziegenhain C, Tiedt S, Castro Alves C, Grunert M, Dworzak M, Lutz C, Turati VA, Enver T, et al. Characterization of rare, dormant, and therapy-resistant cells in acute lymphoblastic leukemia. Cancer Cell. 2016;30:849–862. doi: 10.1016/j.ccell.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho J, Min HY, Pei H, Wei X, Sim JY, Park SH, Hwang SJ, Lee HJ, Hong S, Shin YK, Lee HY. The ATF6-EGF pathway mediates the awakening of slow-cycling chemoresistant cells and tumor recurrence by stimulating tumor angiogenesis. Cancers (Basal) 2020;12:1772. doi: 10.3390/cancers12071772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho J, Min HY, Lee HJ, Hyun SY, Sim JY, Noh M, Hwang SJ, Park SH, Boo HJ, Lee HJ, et al. RGS2-mediated translational control mediates cancer cell dormancy and tumor relapse. J Clin Invest. 2021;131:e136779. doi: 10.1172/JCI136779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oshima N, Yamada Y, Nagayama S, Kawada K, Hasegawa S, Okabe H, Sakai Y, Aoi T. Induction of cancer stem cell properties in colon cancer cells by defined factors. PLoS One. 2014;9:e101735. doi: 10.1371/journal.pone.0101735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nath S, Devi GR. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol Ther. 2016;163:94–108. doi: 10.1016/j.pharmthera.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wahl J, Bogatyreva L, Boukamp P, Rojewski M, van Valen F, Fiedler J, Hipp N, Debatin KM, Beltinger C. Ewing's sarcoma cells with CD57-associated increase of tumorigenicity and with neural crest-like differentiation capacity. Int J Cancer. 2010;127:1295–1307. doi: 10.1002/ijc.25163. [DOI] [PubMed] [Google Scholar]
- 25.Song IS, Jeong YJ, Jeong SH, Kim JE, Han J, Kim TH, Jang SW. Modulation of mitochondrial ERβ expression inhibits triple-negative breast cancer tumor progression by activating mitochondrial function. Cell Physiol Biochem. 2019;52:468–485. doi: 10.33594/000000034. [DOI] [PubMed] [Google Scholar]
- 26.Simpson CD, Anyiwe K, Schimmer AD. Anoikis resistance and tumor metastasis. Cancer Lett. 2008;272:177–185. doi: 10.1016/j.canlet.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 27.Bleau AM, Zandueta C, Redrado M, Martínez-Canarias S, Larzábal L, Montuenga LM, Calvo A, Lecanda F. Sphere-derived tumor cells exhibit impaired metastasis by a host-mediated quiescent phenotype. Oncotarget. 2015;6:27288–27303. doi: 10.18632/oncotarget.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carcereri de Prati A, Butturini E, Rigo A, Oppici E, Rossin M, Boriero D, Mariotto S. Metastatic breast cancer cells enter into dormant state and express cancer stem cells phenotype under chronic hypoxia. J Cell Biochem. 2017;118:3237–3248. doi: 10.1002/jcb.25972. [DOI] [PubMed] [Google Scholar]
- 29.Gao MQ, Choi YP, Kang S, Youn JH, Cho NH. CD24+ cells from hierarchically organized ovarian cancer are enriched in cancer stem cells. Oncogene. 2010;29:2672–2680. doi: 10.1038/onc.2010.35. [DOI] [PubMed] [Google Scholar]
- 30.Abass T, Dutta A. p21 in cancer: Intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karimian A, Ahmadi Y, Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Hoang-Minh LB, Siebzehnrubl FA, Yang C, Suzuki-Hatano S, Dajac K, Loche T, Andrews N, Schmoll Massari M, Patel J, Amin K, et al. Infiltrative and drug-resistant slow-cycling cells support metabolic heterogeneity in glioblastoma. EMBO J. 2018;37:e98772. doi: 10.15252/embj.201798772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Mater D, Wagner L. Management of recurrent Ewing sarcoma: Challenges and approaches. Onco Targets Ther. 2019;12:2279–2288. doi: 10.2147/OTT.S170585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rivankar S. An overview of doxorubicin formulations in cancer therapy. J Cancer Res Ther. 2014;10:853–858. doi: 10.4103/0973-1482.139267. [DOI] [PubMed] [Google Scholar]
- 35.Martino E, Casamassima G, Castiglione S, Cellupica E, Pantalone S, Papagni F, Rui M, Siciliano AM, Collina S. Vinca alkaloids and analogues as anti-cancer agents: Looking back, peering ahead. Bioorg Med Chem Lett. 2018;28:2816–2826. doi: 10.1016/j.bmcl.2018.06.044. [DOI] [PubMed] [Google Scholar]
- 36.Liu BQ, Gao YY, Niu XF, Xie JS, Meng X, Guan Y, Wang HQ. Implication of unfolded protein response in resveratrol-induced inhibition of K562 cell proliferation. Biochem Biophys Res Commun. 2010;391:778–782. doi: 10.1016/j.bbrc.2009.11.137. [DOI] [PubMed] [Google Scholar]
- 37.Ranganathan AC, Zhang L, Adam AP, Aguirre-Ghiso JA. Functional coupling of p38-induced up-regulation of BiP and activation of RNA-dependent protein kinase-like endoplasmic reticulum kinase to drug resistance of dormant carcinoma cells. Cancer Res. 2006;66:1702–1711. doi: 10.1158/0008-5472.CAN-05-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Zhou X, Li Y, Zu L, Pan H, Liu B, Shen W, Fan Y, Zhou Q. Activating transcription factor 3 promotes malignance of lung cancer cells in vitro. Thorac Cancer. 2017;8:181–191. doi: 10.1111/1759-7714.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bandyopadhyay S, Wang Y, Zhan R, Pai SK, Watabe M, Iiizumi M, Furuta E, Mohinta S, Liu W, Hirota S, et al. The tumor metastasis suppressor gene Drg-1 down-regulates the expression of activating transcription factor 3 in prostate cancer. Cancer Res. 2006;66:11983–11990. doi: 10.1158/0008-5472.CAN-06-0943. [DOI] [PubMed] [Google Scholar]
- 40.Fan F, Jin S, Amundson SA, Tong T, Fan W, Zhao H, Zhu X, Mazzacurati L, Li X, Petrik KL, et al. ATF3 induction following DNA damage is regulated by distinct signaling pathways and over-expression of ATF3 protein suppresses cells growth. Oncogene. 2002;21:7488–7496. doi: 10.1038/sj.onc.1205896. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Zang S, Cheng H, Li J, Huang A. Overexpression of activating transcription factor 3 exerts suppressive effects in HepG2 cells. Mol Med Rep. 2019;19:869–876. doi: 10.3892/mmr.2018.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barone MV, Crozat A, Tabaee A, Philipson L, Ron D. CHOP (GADD153) and its oncogenic variant, TLS-CHOP, have opposing effects on the induction of G1/S arrest. Genes Dev. 1994;8:453–464. doi: 10.1101/gad.8.4.453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The RNA sequencing data have been deposited in NCBI GEO under accession no. GSE204671.