Graphical abstract
Keywords: Prothrombin activation, Factor Xa, Molecular dynamics simulation
Abstract
Prothrombin is a key zymogen of the coagulation process and can be converted to thrombin by the prothrombinase complex, which consists of factor Xa (FXa), cofactor Va (FVa), and phospholipids. Prothrombin can be activated at two cleavage sites, R271 and R320, which generates two intermediates: prethrombin-2 via the initial cleavage at R271, and meizothrombin via the first cleavage at R320. Several mechanisms have been proposed to explain this activation preference, but the role of cleavage site sequences in prothrombin activation has not been thoroughly investigated. Here, we used an advanced sampling technique, parallel tempering metadynamics with a well-tempered ensemble (PTMetaD-WTE), to study the binding modes of prothrombin cleavage site sequences R266AIEGRTATSEY277 (denoted as Pep271) and S315YIDGRIVEGSD326 (denoted as Pep320) to the FXa catalytic triad. Our study indicates that there exist three binding modes for Pep271 to the FXa catalytic triad but only one binding mode for Pep320 to the FXa catalytic triad. Further molecular dynamics simulations revealed that due to the strong electrostatic interactions, especially the H-bond interactions and salt bridges formed between Pep320 and FXa, the binding mode in the Pep320-FXa system is more stable than the binding modes in the Pep271-FXa system. In view of experimental observations and our results that there exists only one binding mode for Pep320 to the FXa catalytic triad and especially R320 in Pep320 can stably bind to the FXa catalytic triad, we believe that the first cleavage at R320 is favored.
1. Introduction
Prothrombin is a multidomain vitamin K-dependent blood coagulation factor. Prothrombin activation plays key roles in fibrin clot formation and hemostasis [1], [2]. Prothrombin can be converted to thrombin by the activated form of coagulation factor X (FXa) at the two prothrombin cleavage sites, R271 and R320. Previous studies have shown that thrombin can be generated through two possible pathways (Fig. 1(a)) [3]. In one pathway, prothrombin is first cleaved at R320 and produces the enzymatically active intermediate meizothrombin. In another pathway, initial cleavage may occur at R271 and generate the intermediate prethrombin-2. It is well known that the rate and pathway of prothrombin activation mainly depend on the prothrombinase complex [4], [5], [6], which is assembled from FXa and cofactor Va (FVa) on a negatively charged phospholipid membrane in the presence of calcium ions. Compared with FXa alone, the prothrombinase complex can enhance the activity of FXa by five orders of magnitude [7].
Fig. 1.
(a) Schematic representation of the prothrombin activation process. Prothrombin consists of γ-carboxyglutamate (GLA), two kringles (K), and a serine protease domain comprising A and B chains. Prothrombin activation proceeds along two pathways: (1) first cleavage at R271, which leads to the formation of prethrombin-2, and (2) first cleavage at R320, which leads to the formation of meizothrombin. (b) The sequences R266AIEGRTATSEY277 (denoted as Pep271) around the prothrombin R271 cleavage site (in red) and S315YIDGRIVEGSD326 (denoted as Pep320) around the R320 site (in red). Pep271 is located in between the K2 domain and A chain of prothrombin. Pep320 is located in between the A and B chain of prothrombin. (c) and (d) PTMetaD simulation models. Pep271 and Pep320 were placed around the catalytic triad (H236, D282 and S379) of FXa, and the resulting systems were named as Pep271-FXa and Pep320-FXa, respectively. FXa is shown in white surface model and the catalytic triad of FXa in red surface model. Pep271 or Pep320 is indicated in green or cyan cartoon model. R271 or R320 is shown in green or cyan licorice model. Ions and water molecules are not shown for clarity. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Studies have been carried out to elucidate the prothrombin activation mechanisms associated with the two pathways. When the prothrombinase complex is assembled on synthetic phospholipid vesicles, the pathway corresponding to cleavage at the R320 site is favored [8], [9]. Matthew et al. [10] also found that the membrane of red blood cells helps prothrombin activation through the meizothrombin pathway. In contrast, Wood et al. [11] revealed that prothrombin activation on the activated platelet surface occurs through the prethrombin-2 pathway. Haynes et al. [12] suggested that the mechanism of prothrombin activation on the activated platelet surface most likely differs from that on the synthetic phospholipid vesicles and the platelet-associated prothrombinase structure may differ from that assembled on the synthetic phospholipid vesicles. The prothrombin-2 pathway is also preferred in the absence of FVa [4]. To date, it is still not clear which of the two pathways predominates in vivo. Previous studies have found that cofactor FVa in the prothrombinase complex plays a key role in the prothrombin activation process. Pozzi et al. [13] revealed that deletion of the linker connecting the two kringles of prothrombin mimics the action of cofactor FVa. FVa can compress the linker and morph prothrombin into a favorable conformation for FXa activation. Lechtenberg et al. [14] reported a crystal structure of the prothrombinase complex obtained from the venom of pseudonaja textilis. Their structure also supports the existence of a single substrate binding channel on FVa. Prothrombin and the intermediate meizothrombin can bind to this channel in two different orientations. However, the focus of these studies is on the role of cofactor FVa and the role of factor FXa in the prothrombin activation process has not been studied thoroughly. Very recently, Stojanovski et al. [15] investigated the role of sequences and positions of the cleavage sites in prothrombin activation. They swapped the prothrombin sequences at the two cleavage sites, i.e. with R310 ∼ E328 at the R271 site and D261 ∼ T279 at the R320 site. They found that at each cleavage site, the preferred pathway of prothrombin activation is dominated by both the sequence and its position. Inspired by their studies, it is of great interest to understand the interaction details of a specific sequence around each cleavage site of prothrombin with FXa at the atomic level. This will be helpful to understand the role of FXa in prothrombin activation and how the sequence at the cleavage site R271 or R320 affects the rate and pathway of prothrombin activation.
To clarify the role of FXa in prothrombin activation, we focus on studying the binding modes of R271 and R320 cleavage site sequences to the catalytic triad of FXa, which comprises H236, D282 and S379. It is known that molecular dynamics (MD) simulations can be very helpful for studying biological processes at the molecular level [16], [17], [18], [19]. However, MD simulations still suffer from high computational costs, which often leads to insufficient sampling for large systems. This limitation can be circumvented by using a combination of MD simulation and an enhanced sampling approach, such as parallel tempering metadynamics with a well-tempered ensemble (PTMetaD-WTE) [18], [20]. PTMetaD-WTE with well-selected collective variables (CVs) enables enhanced sampling of a large system and helps reconstruct the free energy surface defined by the CVs. In this study, two short peptides Pep271 and Pep320 around the R271 and R320 cleavage sites were selected, respectively (Fig. 1 (b)). PTMetaD-WTE simulations were first carried out to construct the free energy surfaces for Pep271 and Pep320 binding to FXa, respectively (Fig. 1(c) and (d)), which were further used to study the binding modes in the Pep271-FXa and Pep320-FXa complexes. Thereafter, long-time unbiased MD simulations were performed to study the stability of the complexes. Finally, we analyzed in detail the processes of Pep271 and Pep320 binding to FXa and clarified the role of cleavage site sequences in prothrombin activation.
2. Method
2.1. Simulation setup
All the simulations were performed with GROMACS 2021.3 [21] patched with plumed 2.7.2 [22]. The Amber99SB-ILDN force field [23] was used. The time step was 2 fs for all the simulations. The FXa structure used in this work is 1XKA. It has been shown by Shim et al.[24] that the all atom model built on this structure, prothrombin, and the FVa/FXa complex from Pseudonaja textilis can stabilize the orientation of prothrombin and locate the active site of FXa in the vicinity of prothrombin cleavage positions. The protonation state of FXa was checked by using the PDB2PQR webservice tool [25]. Two short peptides, with the sequences R266AIEGRTATSEY277 (denoted as Pep271) around the prothrombin R271 cleavage site and S315YIDGRIVEGSD326 (denoted as Pep320) around the R320 site (Fig. 1 (b)) were selected based on the all-atom human ternary model of Shim et al [24]. These two peptides were selected with the residues within 2.5 nm of the FXa catalytic triad, which are sufficiently large for including the residues most accessible to the interactions with FXa. Pep271 and Pep320 were placed around the catalytic triad (H236, D282 and S379) of FXa (see Fig. 1(c) and (d)), and the resulting systems were named as Pep271-FXa and Pep320-FXa, respectively. Each system was further placed in a cubic box with the size of 6.5 × 6.5 × 6.5 nm3, and the shortest distance between the solute and a surface of the box was about 8 Å. For the Pep271-FXa system, 7680 TIP3P water molecules and 4 counter ions (Cl-) were added to the simulation box. For the Pep320-FXa system, 7685 TIP3P water molecules and 2 counter ions (Cl-) were added to the simulation box. For all the simulations, the periodic boundary conditions (PBCs) were used. The cut-offs for the van der Waals (vdW) and short-range electrostatic interactions were set to 1.2 nm. The particle mesh Ewald method (PME) [26] was used to recover the long-range electrostatic interaction. The Lorentz-Berthelot (LB) rule [27] was used to model the Lennard-Jones (LJ) interactions between different types of atoms. All bonds were constrained with the LINCS algorithm [28]. The temperature was maintained via the velocity rescaling method [29]. For all the simulations, the NVT ensemble was used. Since our focus is on the binding modes of R271 and R320 cleavage site sequences to the catalytic triad (H236, D282 and S379) of FXa, the FXa heavy atoms within 1.5 nm of the FXa catalytic triad were not restrained and other FXa heavy atoms were restrained with a harmonic potential with the force constant set to 1000 kJ mol−1 nm−2 during the simulations.
2.2. Metadynamics and molecular dynamics (MD) simulations
In this work, parallel tempering metadynamics (PTMetaD) [30] with a well-tempered ensemble (WTE) [31] (PTMetaD-WTE) was used to study the binding modes of Pep271 and Pep320 to the FXa catalytic triad in the Pep271-FXa and Pep320-FXa systems, respectively. Compared with metadynamics and PTMetaD, PTMetaD-WTE performs better in terms of sampling efficiency and can reduce the number of replicas used in a PTMetaD simulation. The details of PTMetaD-WTE setup are according to the work of Barducci et al. [18]. Briefly, 16 replicas distributed in the temperature range of 290–656 K were used for each system. The temperatures of the replicas follow the distribution suggested in ref [32]. According to the WTE approach, the potential energy of a system was selected as a CV and was firstly converged for all the 16 replicas in 20 ns in a PTMetaD simulation. This WTE approach can significantly enhance replica diffusion in the temperature space, resulting in an average exchange acceptance probability of ≈ 0.2 and reducing the number of replicas. The bias was then applied over two types of CVs: (1) the distance between the backbone Cα atom of R271 and the side chain oxygen atom of S379 in the FXa catalytic triad (denoted as D271) for the Pep271-FXa system or that between R320 and S379 (denoted as D320) for the Pep320-FXa system; (2) the contact number between Pep271 or Pep320 and any atoms within 0.6 nm of S379 in FXa (denoted as N271 or N320). These CVs were selected according to the catalytic mechanism of serine proteases [33], where the peptide substrate binds to the surface of a serine protease and the scissile bond is inserted into the active site of the enzyme, with the carbonyl carbon of this bond positioned near the nucleophilic serine. In this case, S379 on the FXa catalytic triad attacks the Cα atom of R271 or R320 of prothrombin to further promote activation. The length of each PTMetaD-WTE simulation was 200 ns, which yields a total simulation time of 3.2 μs for each system. From the PTMetaD-WTE simulation results (Fig. 2(c) and (d)), we found that Pep271 has three main binding modes to the FXa catalytic triad, and Pep320 has only one binding mode to the FXa catalytic triad. For each of these binding modes, a 1000 ns unbiased MD simulation with the temperature set at 300 K was performed to study its stability.
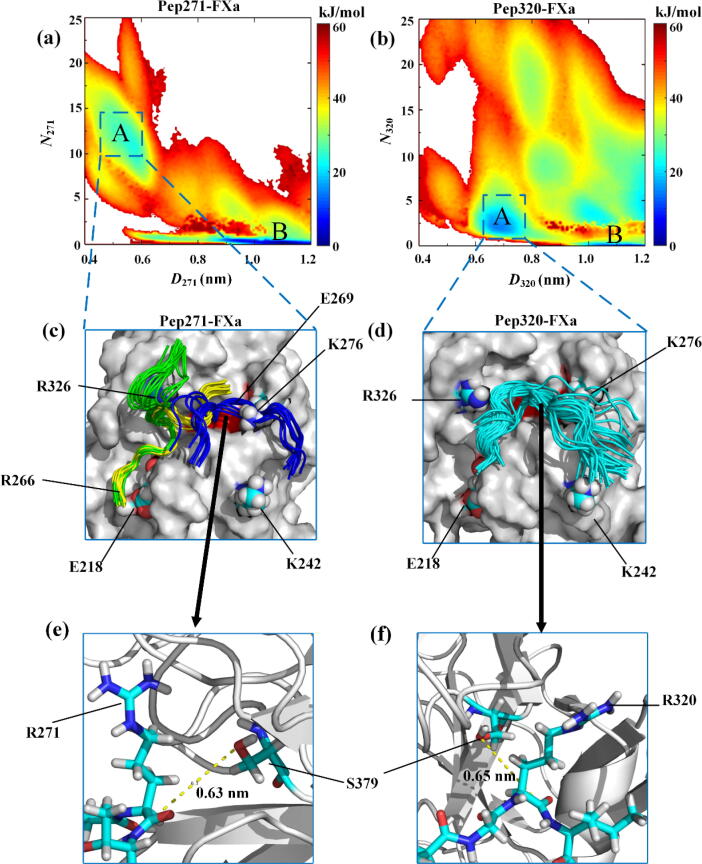
Fig. 2.
(a) The free energy surface (FES) for the Pep271-FXa system, and (b) the FES for the Pep320-FXa system. Two major basins on each FES were identified, which were named as states A and B, respectively. (c) and (d) The Pep271 and Pep320 conformational ensembles corresponding to the intermediate state A in the Pep271-FXa and Pep320-FXa systems, respectively. The Pep271 ensemble is shown in green, yellow, and blue, which correspond to the three binding modes, respectively. The Pep320 ensemble is shown in cyan. (e) and (f) Details of the interactions of R271 or R320 with S379, where the Cα atom of R271 or R320 is close to the oxygen atom on the S379 side chain. The arrows highlight the Pep271 binding mode (in blue) in Fig. 2(c), and the Pep320 binding mode (in cyan) in Fig. 2(d), respectively. R271, R320, and S379 are shown in licorice model. R326, K276, R266, K242, and E218 are shown in van der Waals model. Nitrogen is in blue, hydrogen in white, oxygen in red, and carbon in cyan. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Results
3.1. Free energy surfaces for Pep271 and Pep320 to the FXa catalytic triad
Change of the free energy at 300 K along D271 for the Pep271-FXa system and that along D320 for the Pep320-FXa system are shown in Figure S1. As we can see, for each system the free energy fluctuates slightly after ∼ 200 ns of the PTMetaD-WTE simulations and the relative free energy between the major minima remains essentially unchanged. In Fig. 2(a) and (b), we show the free energy surface (FES) at 300 K in the space defined by the selected CVs for the two systems, respectively, obtained after 200 ns of the simulations. For the Pep271-FXa system, two main basins can be found on the FES, which were named as states A and B, respectively. State B is located in the region with D271 ranging from 0.9 ∼ 1.2 nm and N271 close to 0, and corresponds to the global free-energy minimum. In this state, none of the Pep271 atoms are close to S379 of FXa, as indicated by the fact that the CV N271 is close to 0. State A is located in the region with 0.4 < D271 < 0.6 nm and 7 < N271 < 17. In this state, the Cα atom of R271 is in contact with the oxygen atom on the S379 side chain. Compared with the FES for the Pep271-FXa system, the FES for the Pep320-FXa system also exhibits two major basins, which were also named as states A and B, respectively. State B corresponds to the global free-energy minimum and is located in the region similar to that on the FES for the Pep271-FXa system. In this state, none of the Pep320 atoms are close to S379 of FXa. The intermediate state A is located in the region with 0.6 < D320 < 0.8 nm and 2 < N320 < 7. In this state, the Cα atom of R320 is close to the oxygen atom on the S379 side chain, which is similar to that of R271 in the intermediate state A for the Pep271-FXa system. However, compared with the intermediate state A in the Pep271-FXa system, the intermediate state A in the Pep320-FXa system is found to have a deeper free energy basin. This reflects that the binding of R320 to S379 of FXa in the Pep320-FXa system is more stable than that of R271 to S379 in the Pep271-FXa system.
3.2. Binding modes of Pep271 or Pep320 to the FXa catalytic triad
To study the binding modes of Pep271 or Pep320 to the FXa catalytic triad in the intermediate state A, we analyzed the conformational ensemble of Pep271 or Pep320 around the FXa catalytic triad in the state (see Fig. 2(c) and (d), respectively). Three major binding modes have been found for the binding of Pep271 to the FXa catalytic triad, which are shown in green, yellow, and blue, respectively (Fig. 2(c)). However, only one major binding mode (in cyan) has been found for the binding of Pep320 to the FXa catalytic triad. Figure S2 shows the contribution of each residue to the binding free energy in the intermediate state A for each of the Pep271-FXa and Pep320-FXa systems, obtained using the g_mmpbsa tool [34]. As we can see from the figure, residues R326, E218, K276, and K242 on FXa are the key residues for the binding of Pep271 or Pep320 to the FXa catalytic triad. These residues are charged and can form strong electrostatic interactions with Pep271 or Pep320.
For the intermediate state A in the Pep271-FXa system, we find that there exist three binding modes for Pep271 to the FXa catalytic triad. In two of the binding modes the positively charged R266 of Pep271 interacts with the negatively charged E218 of FXa (see Fig. 2(c)) and in one binding mode only the negatively charged E269 of Pep271 interacts with the positively charged K276 of FXa. For the intermediate state A in the Pep320-FXa system, we find only one binding mode. In this binding mode three negatively charged residues D318, E323, and D326 of Pep320 form strong electrostatic interactions with the positively charged residues R326, K276, and K242 on FXa, respectively (see Fig. 2(d) and S3(a)). Thus, we believe that it is the strong electrostatic interaction between Pep320 and the FXa catalytic triad that stabilizes the intermediate state A. As reflected by the FESs for the two systems, the binding modes in the Pep271-FXa system are less stable than that in the Pep320-FXa system.
3.3. Binding stability of R271 or R320 to the FXa catalytic triad
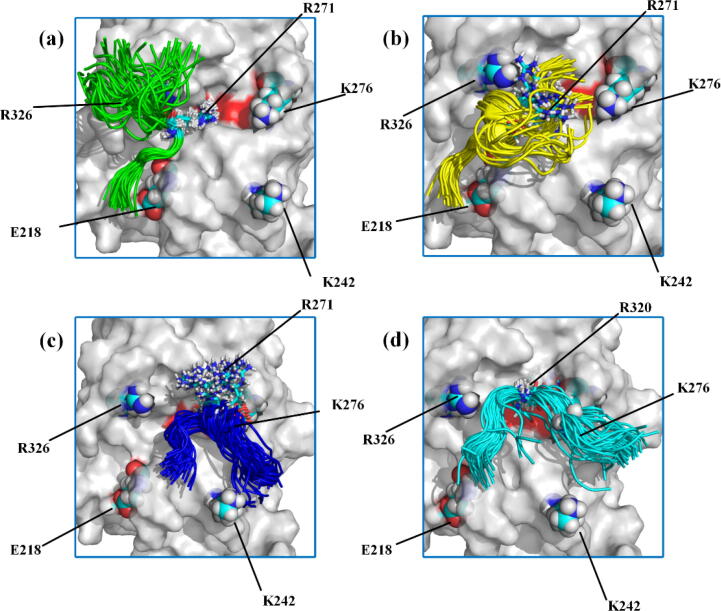
The binding of R271 or R320 to the FXa catalytic triad is important for prothrombin activation by FXa [33]. We therefore carried out conventional unbiased MD simulations to study the binding stability of R271 or R320 to the FXa catalytic triad in the four binding modes shown in Fig. 2, which were named as Pep271-FXa-BM1 (in green), Pep271-FXa-BM2 (in yellow), Pep271-FXa-BM3 (in blue), and Pep320-FXa-BM (in cyan), respectively. Four 1000 ns MD simulations, with each simulation corresponding a binding mode, were performed. For each simulation, the initial structure was taken randomly from the conformations for the corresponding binding mode (Fig. 2(c) and (d)). The evolution of D271 or D320 with the simulation time for each system (see Fig. 3) was used to estimate the binding stability of R271 or R320 to the FXa catalytic triad because D271 or D320 is the distance between the backbone Cα atom of R271 or R320 and the side chain oxygen atom of S379 in the FXa catalytic triad. For Pep320-FXa-BM, D320 (in cyan) reaches a steady value very quickly in the very beginning of the simulation and has essentially a minor change during the 1000 ns simulation. As we can see from the figure, during the simulation, D320 is stabilized at ∼ 0.7 nm, which agrees well with the location of the intermediate state A (0.6 < D320 < 0.8 nm) identified from the FES for the Pep320-FXa system (Fig. 2(b)). The D271 value for Pep271-FXa-BM1 (in green) fluctuates slightly around 0.8 nm during the 1000 ns simulation. However, for the other two binding modes, i.e. Pep271-FXa-BM2 and Pep271-FXa-BM3, the D271 values (in yellow and blue) show significant changes during the 1000 ns simulations. The D271 value for Pep271-FXa-BM3 (in blue) went rapidly up to ∼ 1.0 nm and reached ∼ 1.2 nm at the end of the simulation. The D271 value for Pep271-FXa-BM2 (in yellow) maintains at ∼ 0.8 nm in the first 500 ns of the simulation, but finally reached ∼ 1.2 nm at the end of the simulation. This observation reflects that the binding of R271 to the catalytic triad is not stable in these two binding modes. The Pep271 or Pep320 conformational ensembles during the 1000 ns MD simulations are shown in Fig. 4. One can also find that R271 in Pep271-FXa-BM1 (in green) and R320 in Pep320-FXa-BM (in cyan) have small fluctuations, which indicates that the binding of R271 or R320 to FXa is stable in these two binding modes. However, since R271 has a large fluctuation in Pep271-FXa-BM2 (in yellow) or Pep271-FXa-BM3 (in blue), the stable binding of R271 to FXa cannot be observed in these two binding modes, which are consistent with the results shown in Fig. 3.
Fig. 3.
D271or D320 as a function of the simulation time for the systems corresponding to the four binding modes shown in Fig. 2, which are Pep271-FXa-BM1 (in green), Pep271-FXa-BM2 (in yellow), Pep271-FXa-BM3 (in blue), and Pep320-FXa-BM (in cyan), respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Cartoon representation of the Pep271 or Pep320 conformational ensembles around the catalytic triad of FXa (in red) during the 1000 ns MD simulations for (a) Pep271-FXa-BM1 (in green), (b) Pep271-FXa-BM2 (in yellow), (c) Pep271-FXa-BM3 (in blue), and (d) Pep320-FXa-BM (in cyan), respectively. R271 and R320 are shown in licorice model. R326, K276, R266, K242, and E218 are shown in van der Waals model. The Pep271 ensemble is shown in green, yellow, and blue, which correspond to the three binding modes, respectively. The Pep320 ensemble is shown in cyan. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
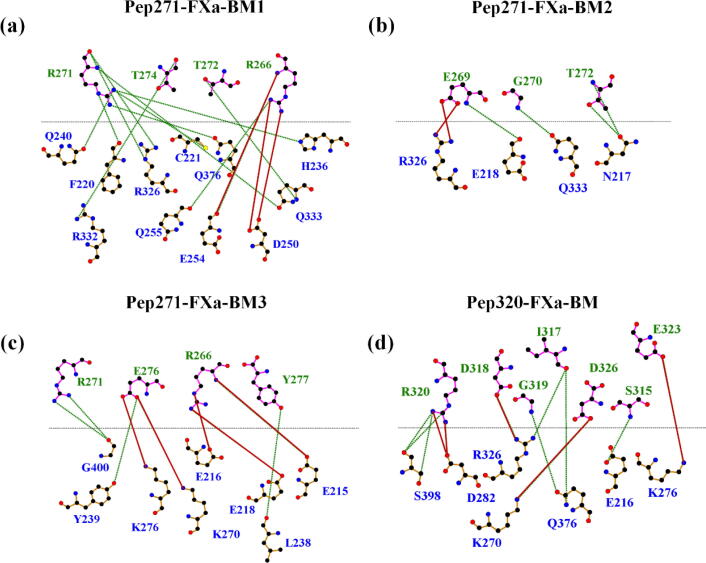
In a binding mode, the binding stability of R271 or R320 to the FXa catalytic triad is closely related to the interactions between the FXa catalytic triad and Pep271 or Pep320. In Figure S4, we show the average electrostatic and van der Waals interaction energies between the FXa catalytic triad and Pep271 or Pep320 in each binding mode. From the figure, we can see that the electrostatic interaction dominates the interactions of Pep271 or Pep320 with FXa. The electrostatic interaction between Pep320 and the FXa catalytic triad in the binding mode of the Pep320-FXa system is stronger than those between Pep271 and FXa in the binding modes of the Pep271-FXa system. We further analyzed the H-bonds and salt bridges formed between Pep271 or Pep320 and FXa by using the Ligplot software [35] (Fig. 5). The maximum distance for the formation of H-bonds in the Ligplot software is defined as 3.9 Å. Only three or four residues of Pep271 can form H-bonds with FXa in each of the binding modes of the Pep271-FXa system. However, seven residues of Pep320 can form H-bonds with FXa in the binding mode of the Pep320-FXa system. Moreover, the charged residues of Pep320 (e.g., D318, R320, D323, and E326) can form salt bridges with the charged residues of FXa (e.g., K270, K276, D282, and R326). Due to the strong electrostatic interactions, especially the H-bond interactions and salt bridges formed between Pep320 and FXa, the binding mode in the Pep320-FXa system is more stable than any of the three binding modes found in the Pep271-FXa system, which also contributes significantly to the stability of R320 in the FXa catalytic triad. One could also find that among all the three binding modes identified in the Pep271-FXa system, R271 in the Pep271-FXa-BM1 binding mode forms the largest number of H-bonds with FXa. This can be the reason why the stable binding of R271 to FXa was observed only in this binding mode. However, R271 cannot stably bind to the FXa catalytic triad in the other two binding modes, i.e. Pep271-FXa-BM2 and Pep271-FXa-BM3, since R271 cannot form H-bonds with FXa in Pep271-FXa-BM2 and can only form two H-bonds with FXa in Pep271-FXa-BM3. Note that in most force fields, H-bond interactions are described as electrostatic interactions. Therefore, we can accordingly find that the electrostatic interactions between Pep271 and FXa in Pep271-FXa-BM2 and Pep271-FXa-BM3 are much weaker than in the binding mode in the Pep320-FXa system (see Figure S4).
Fig. 5.
Ligplot hydrogen bonds (H-bonds) and salt bridges analyses. The H-bonds and salt bridges between the residues of Pep271 or Pep320 (shown above the black line) and those of FXa (shown below the black line) were analyzed for (a) Pep271-FXa-BM1, (b) Pep271-FXa-BM2, (c) Pep271-FXa-BM3, and (d) Pep320-FXa-BM, respectively. H-bonds are shown as green lines, and salt bridges are shown as red lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
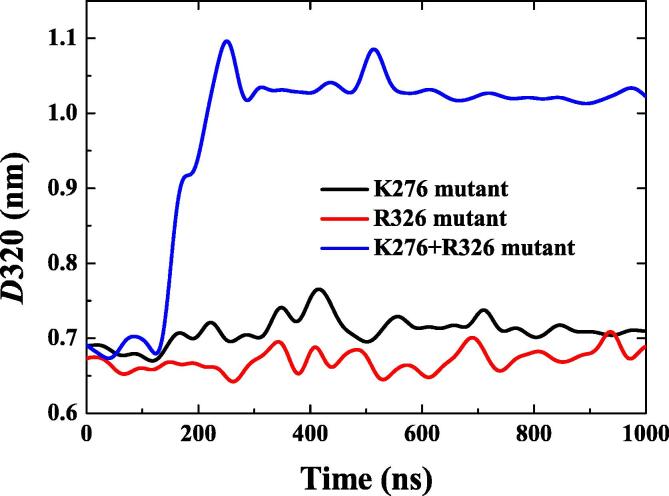
To know if specific substitutions of one or more of the charged residues of FXa enhance or disrupt the interactions between Pep320 and FXa, we used the current in silico approach to evaluate the effect of substitutions. We mutated the positively charged residues R326 or K276, or both, into alanine. In Fig. 6, it can be found that when R326 or K276 was mutated to alanine, the D320 value has hardly changed during the 1000 ns simulation, which means that substitution of just one key residue could not disrupt the intermediate state. However, when both R326 and K276 were mutated into alanine, R320 is disrupted from the FXa catalytic triad.
Fig. 6.
Effects of K276 and R326 mutations on D320 in the Pep320-FXa system.
4. Discussion
In this study, an enhanced sampling technique, called PTMetaD-WTE, was used to characterize the binding modes of Pep271 or Pep320 to the FXa catalytic triad. We found that there exist three binding modes for the binding of Pep271 to the FXa catalytic triad in the Pep271-FXa system but only one binding mode for Pep320 in the Pep320-FXa system. These binding modes correspond to the intermediate states on the free energy surfaces (FESs) of the two systems, respectively. Further MD simulations suggest that, due to the strong electrostatic interactions, especially the H-bond and salt bridges formed between Pep320 and FXa, the binding mode in the Pep320-FXa system is more stable than any of the three binding modes found in the Pep271-FXa system. This is in line with the previous studies that the electrostatic recognition is a major factor in protease substrate recognition in the serine protease family [36], [37]. It has also been found that the association rates between the serine protease and its substrate are largely governed by electrostatic interactions [38]. In the binding mode of the Pep320-FXa system, R320 can stably bind to the FXa catalytic triad.
We have noticed that prothrombin activation is closely related to the formation of the prothrombinase complex. Previous studies have shown that when the prothrombinase complex is assembled on synthetic phospholipid vesicles, the meizothrombin pathway (first cleavage at R320) is favored [8], [9]. Several studies have focused on the role of FVa in prothrombin activation [13], [14]. These studies suggest that with the assistance of FVa, prothrombin most likely has a favorable conformation for the FXa activation through the meizothrombin pathway (first cleavage at R320). The thrombin generating model proposed by Shim et al. [34] shows that R271 and R320 of prothrombin are close to the FXa catalytic triad in order to facilitate the prothrombin activation. They also showed that the sequence of prothrombin cleavage site R271 or R320 forms a large loop. The process of prothrombin activation by FXa can be divided into two steps, which are step 1, the initial binding of prothrombin to FXa, and step 2, the Pep271 or Pep320 loop region binding to the catalytic site of FXa. Recently, Stojanovski et al. [15] have addressed the role of sequences at the cleavage sites in prothrombin activation. They found that the sequence at the cleavage site R271 or R320 of prothrombin can play important roles in prothrombin activation. Therefore, we believe that the binding modes of Pep271 or Pep320 most likely appear in the second step of prothrombin activation. However, the interaction details of a specific sequence around each cleavage site of prothrombin with FXa at the atomic level are still unclear. As a complement to previous results, our study shows that there exists only one binding mode for Pep320 binding to the FXa catalytic triad and R320 can stably bind to the FXa catalytic triad, which means that this binding mode favors the first cleavage of prothrombin by the FXa catalytic triad at R320. It is also well known that the binding of a substrate to the active site is the first step and most likely a decisive factor in the catalytic process of an enzyme. Therefore, we believe that our study of the binding modes of Pep271 or Pep320 to the FXa catalytic triad can help us reveal the prothrombin activation mechanism.
CRediT authorship contribution statement
Jiachen Li: Conceptualization, Methodology, Software, Data curation, Writing – original draft. Qi Wang: Supervision, Funding acquisition. Yaoquan Tu: Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported by National Natural Science Foundation of China (21673206). Jiachen Li acknowledges the China Scholarship Council for financial support. We thank Swedish National Infrastructure for Computing (SNIC) for providing computational resources at PDC and NSC for the projects SNIC 2021-5-457 and SNIC 2021-3-22.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.09.030.
Contributor Information
Qi Wang, Email: qiwang@zju.edu.cn.
Yaoquan Tu, Email: yaoquan@kth.se.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Davie E.W., Fujikawa K., Kisiel W. The coagulation cascade: initiation, maintenance, and regulation. Biochemistry. 1991;30(43):10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 2.Wolberg A.S., Campbell R.A. Thrombin generation, fibrin clot formation and hemostasis. Transfus Apher Sci. 2008;38(1):15–23. doi: 10.1016/j.transci.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krishnaswamy S. Exosite-driven substrate specificity and function in coagulation. J Thromb Haemost. 2005;3(1):54–67. doi: 10.1111/j.1538-7836.2004.01021.x. [DOI] [PubMed] [Google Scholar]
- 4.Krishnaswamy S., Church W.R., Nesheim M.E., Mann K.G. Activation of human prothrombin by human prothrombinase. Influence of factor Va on the reaction mechanism. J Biol Chem. 1987;262(7):3291–3299. [PubMed] [Google Scholar]
- 5.Brufatto N., Nesheim M.E. Analysis of the kinetics of prothrombin activation and evidence that two equilibrating forms of prothrombinase are involved in the process. J Biol Chem. 2003;278(9):6755–6764. doi: 10.1074/jbc.M206413200. [DOI] [PubMed] [Google Scholar]
- 6.Mann K.G. Thrombin formation. Chest. 2003;124(3):4S–10S. doi: 10.1378/chest.124.3_suppl.4s. [DOI] [PubMed] [Google Scholar]
- 7.Nesheim M.E., Taswell J.B., Mann K.G. The contribution of bovine Factor V and Factor Va to the activity of prothrombinase. J Biol Chem. 1979;254(21):10952–10962. [PubMed] [Google Scholar]
- 8.Lentz B.R. Exposure of platelet membrane phosphatidylserine regulates blood coagulation. Prog Lipid Res. 2003;42(5):423–438. doi: 10.1016/s0163-7827(03)00025-0. [DOI] [PubMed] [Google Scholar]
- 9.Tans G., Janssen-Claessen T., Hemker H.C., Zwaal R.F., Rosing J. Meizothrombin formation during factor Xa-catalyzed prothrombin activation. Formation in a purified system and in plasma. J Biol Chem. 1991;266(32):21864–21873. [PubMed] [Google Scholar]
- 10.Whelihan M.F., Zachary V., Orfeo T., Mann K.G. Prothrombin activation in blood coagulation: the erythrocyte contribution to thrombin generation. Blood. 2012;120(18):3837–3845. doi: 10.1182/blood-2012-05-427856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood J.P., Silveira J.R., Maille N.M., Haynes L.M., Tracy P.B. Prothrombin activation on the activated platelet surface optimizes expression of procoagulant activity. Blood. 2011;117(5):1710–1718. doi: 10.1182/blood-2010-09-311035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haynes L.M., Bouchard B.A., Tracy P.B., Mann K.G. Prothrombin activation by platelet-associated prothrombinase proceeds through the prethrombin-2 pathway via a concerted mechanism. J Biol Chem. 2012;287(46):38647–38655. doi: 10.1074/jbc.M112.407791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pozzi N., Chen Z., Pelc L.A., Shropshire D.B., Di Cera E. The linker connecting the two kringles plays a key role in prothrombin activation. Proc Natl Acad Sci. 2014;111(21):7630–7635. doi: 10.1073/pnas.1403779111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lechtenberg B.C., Murray-Rust T.A., Johnson D.J., Adams T.E., Krishnaswamy S., Camire R.M., et al. Crystal structure of the prothrombinase complex from the venom of Pseudonaja textilis. Blood. 2013;122(16):2777–2783. doi: 10.1182/blood-2013-06-511733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stojanovski B.M., Di Cera E. Role of sequence and position of the cleavage sites in prothrombin activation. J Biol Chem. 2021;297(2) doi: 10.1016/j.jbc.2021.100955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adcock S.A., McCammon J.A. Molecular dynamics: survey of methods for simulating the activity of proteins. Chem Rev. 2006;106(5):1589–1615. doi: 10.1021/cr040426m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernardi R.C., Melo M.C., Schulten K. Enhanced sampling techniques in molecular dynamics simulations of biological systems. Biochim Biophys Acta-Gen Subj. 2015;1850(5):872–877. doi: 10.1016/j.bbagen.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barducci A., Bonomi M., Prakash M.K., Parrinello M. Free-energy landscape of protein oligomerization from atomistic simulations. Proc Natl Acad Sci. 2013;110(49):E4708–E4713. doi: 10.1073/pnas.1320077110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C., Codina N., Tang J., Yu H., Chakroun N., Kozielski F., et al. Comparison of the pH-and thermally-induced fluctuations of a therapeutic antibody Fab fragment by molecular dynamics simulation. Comput Struct Biotechnol J. 2021;19:2726–2741. doi: 10.1016/j.csbj.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bussi G., Gervasio F.L., Laio A., Parrinello M. Free-energy landscape for β hairpin folding from combined parallel tempering and metadynamics. J Am Chem Soc. 2006;128(41):13435–13441. doi: 10.1021/ja062463w. [DOI] [PubMed] [Google Scholar]
- 21.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 22.Bonomi M., Branduardi D., Bussi G., Camilloni C., Provasi D., Raiteri P., et al. PLUMED: A portable plugin for free-energy calculations with molecular dynamics. Comput Phys Commun. 2009;180(10):1961–1972. [Google Scholar]
- 23.Lindorff Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shim J., Lee C.J., Wu S., Pedersen L.G. A model for the unique role of factor Va A2 domain extension in the human ternary thrombin-generating complex. Biophys Chem. 2015;199:46–50. doi: 10.1016/j.bpc.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Jurrus E., Engel D., Star K., Monson K., Brandi J., Felberg L.E., et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018;27(1):112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darden T., York D., Pedersen L. Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. J Chem Phys. 1993;98(12):10089–10092. [Google Scholar]
- 27.Hirschfelder JO, Curtiss CF, Bird RB. Molecular theory of gases and liquids. Molecular theory of gases and liquids 1964.
- 28.Hess B., Bekker H., Berendsen H.J., Fraaije J.G. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18(12):1463–1472. [Google Scholar]
- 29.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126(1):14101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 30.Hansmann U.H. Parallel tempering algorithm for conformational studies of biological molecules. Chem Phys Lett. 1997;281(1–3):140–150. [Google Scholar]
- 31.Bonomi M., Parrinello M. Enhanced sampling in the well-tempered ensemble. Phys Rev Lett. 2010;104(19) doi: 10.1103/PhysRevLett.104.190601. [DOI] [PubMed] [Google Scholar]
- 32.Prakash M.K., Barducci A., Parrinello M. Replica temperatures for uniform exchange and efficient roundtrip times in explicit solvent parallel tempering simulations. J Chem Theory Comput. 2011;7(7):2025–2027. doi: 10.1021/ct200208h. [DOI] [PubMed] [Google Scholar]
- 33.Polgár L. The catalytic triad of serine peptidases. Cell Mol Life Sci. 2005;62(19):2161–2172. doi: 10.1007/s00018-005-5160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumari R., Kumar R., Open S.D.D.C., Lynn A., g_mmpbsa A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 35.Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 36.Batra J., Szabó A., Caulfield T.R., Soares A.S., Sahin-Tóth M., Radisky E.S. Long-range electrostatic complementarity governs substrate recognition by human chymotrypsin C, a key regulator of digestive enzyme activation. J Biol Chem. 2013;288(14):9848–9859. doi: 10.1074/jbc.M113.457382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waldner B.J., Kraml J., Kahler U., Spinn A., Schauperl M., Podewitz M., et al. Electrostatic recognition in substrate binding to serine proteases. J Mol Recognit. 2018;31(10) doi: 10.1002/jmr.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schreiber G., Haran G., Zhou H. Fundamental aspects of protein-protein association kinetics. Chem Rev. 2009;109(3):839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.