Summary
Mitochondrial damage-associated molecular patterns (mitoDAMPs) are released from cells dying uncontrolled, non-apoptotic deaths, usually secondary to disease or trauma. Here, we describe preparation of mitoDAMPs from mouse liver, but this protocol can be adapted for preparation of mitoDAMPs from other species and tissues. Tissues are dissociated and then processed to isolate mitochondria. Mitochondria are then sonicated and mitoDAMPs are collected by ultracentrifugation. This procedure produces μg quantities of mitoDAMPs and facilitates research to understand their impacts in health and disease.
For complete details on the use and execution of this protocol, please refer to Westhaver et al. (2022).
Subject areas: Cell Biology, Cell isolation, Cell separation/fractionation, Cell-based Assays, Immunology, Metabolomics
Graphical abstract
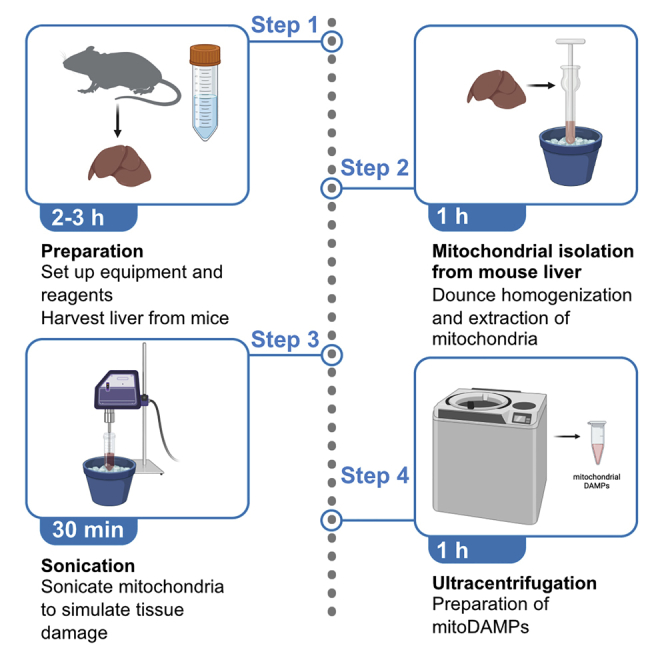
Highlights
-
•
Isolation protocol for mitochondrial damage-associated molecular patterns (mitoDAMPs)
-
•
Optimized isolation from mouse liver with optional substitution with cultured cells
-
•
We describe tissue harvest, mitochondria isolation, and simulated tissue damage
-
•
Applicable to study of diseases involving tissue damage, cancer, and necrotic cell death
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Mitochondrial damage-associated molecular patterns (mitoDAMPs) are released from cells dying uncontrolled, non-apoptotic deaths, usually secondary to disease or trauma. Here, we describe preparation of mitoDAMPs from mouse liver, but this protocol can be adapted for preparation of mitoDAMPs from other species and tissues. Tissues are dissociated and then processed to isolate mitochondria. Mitochondria are then sonicated and mitoDAMPs are collected by ultracentrifugation. This procedure produces μg quantities of mitoDAMPs and facilitates research to understand their impacts in health and disease.
Before you begin
This protocol describes isolation of mitochondrial DAMPs from mouse liver, and modified from Zhang et al. (2010). (Zhang et al., 2010; Westhaver et al., 2022).
We first homogenize liver tissues, then use a commercially available kit to isolate mitochondria from cells, which are then dissociated to create mitoDAMPs. We describe the use of a commercial mitochondria isolation kit from Thermo Fisher Scientific, which relies on differential centrifugation, and we find this to be simple and cost-effective. However, mitochondrial isolation via comparable commercial products or traditional sucrose-gradient protocols (see troubleshooting section, below) are suitable alternatives (Clayton and Shadel, 2014).
As described in Westhaver et al. (2022), mitoDAMP preparations were characterized via liquid chromatography-mass spectrometry; a comprehensive list of the >550 unique proteins identified within mouse mitoDAMPs is available here. As expected, most proteins identified exist exclusively within mitochondria and are associated with mitochondrial processes (Westhaver et al., 2022). This LCMS data were included as supplementary data in our manuscript and can be used to compare your results for quality assurance. We encourage users to validate the presence of their specific proteins of interest to ensure that they are intact after isolation. If desired, the mitoDAMP preparation could be further fractionated into DNA, protein or any other fraction of interest, using commercially available reagents or standard protocols in cell biology. We have used this protocol to prepare mitoDAMPs from mouse, cow, and sheep liver. The initial steps of this protocol can be adjusted for isolation of mitoDAMPs from cultured cells rather than hepatic tissue; we have used it to isolate mitoDAMPs from the human hepatoblastoma cell line, Hep-G2 (see troubleshooting section, below for details). This protocol is scalable as appropriate for downstream applications and the concentration of mitoDAMPs can be standardized for use in experiments via a simple Bradford protein quantification assay (please see key resources table for details).
Isolated mitoDAMPs can be stored at −80°C until use in both in vitro and in vivo experiments. We recommend that the preparation of mitoDAMPs be performed under sterile conditions, and in a biological safety cabinet. We further recommend testing preparations for endotoxin via routine limulus amebocyte lysate assay (please see key resources table for details).
Institutional permissions
Ensure that you have received the appropriate institutional approvals prior to working with live vertebrate tissues. We perform this protocol using treatment naive 8–12-week-old male 25–30 gram C57BL/6NCrl mice. All experimental protocols described below were conducted with approval from the Dalhousie University Committee on Laboratory Animals (18-021), according to guidelines set by the Canadian Council on Animal Care.
Equipment and reagent preparation
Timing: 2 h
-
1.Sterilize equipment, including Dounce homogenizer, tweezers, and scissors.
-
a.Sterile Dounce homogenizer, tweezers, and scissors i.e., via autoclaving.
-
b.For components for which autoclaving is not possible (i.e., sonicator probe), clean with 70% EtOH.
-
a.
-
2.
Prepare Mitochondrial Isolation Kit reagents according to manufacturer’s instructions; details can be found in materials and equipment, below.
-
3.
Ensure that all Mitochondrial Isolation Kit reagents, Halt protease inhibitor, HBSS, and PBS (or ultrapure water for reconstitution of BupH™ Phosphate Buffered Saline) have been placed on ice. Pre-chill the Dounce homogenizer on ice.
-
4.
Set benchtop microcentrifuge and ultracentrifuge to 2°C–8°C.
Optional: Prepare and label tubes for aliquots of isolated mitoDAMPs for storage at −80°C. We find that 1.5 mL microfuge tubes with ∼100 microliters (μL) of mitoDAMPs each works well for our in vitro experiments with lymphocytes.
Collection of liver tissue from mice
Timing: 30 min
-
5.Retrieve mouse liver according to institutional guidelines and after ethics approval.
-
a.Remove gallbladder from liver using a scalpel, scissors, or razorblade (Figures 1A and 1B).
-
a.
Note: We generally begin with 3–6 mouse livers; see expected outcomes section for anticipated yield.
Note: Cultured cells can be used in place of mouse liver; we have also performed this protocol using the Hep-G2 human hepatocellular carcinoma cell line. See troubleshooting section, below, for details.
Note: Perfusion of liver with PBS prior to removal from mouse may be employed if desired.
-
6.
Place liver in 50 mL conical tube on ice and proceed to step-by-step method details, below.
Optional: Mouse liver can be stored at −80°C and used for future mitochondrial isolation, depending on downstream application (i.e., if intact, functional protein or nucleic acid is required, use only fresh liver).
CRITICAL: Reagents and samples must be maintained on ice to maintain mitochondrial integrity and increase mitochondrial yield. Work quickly and avoid prolonged delays in protocol completion.
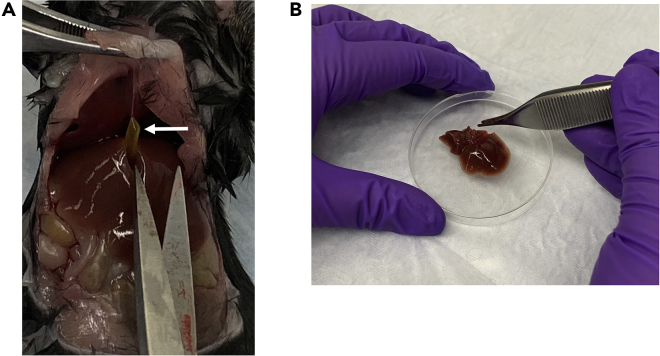
Figure 1.
Removal of mouse liver and excision of gall bladder
(A) Gallbladder shown in-situ; arrow indicates gallbladder.
(B) Mouse liver following removal.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Source of mitoDAMPs – livers or cell lines, as desired | ||
| Chemicals, peptides, and recombinant proteins | ||
| Pierce Chromogenic Endotoxin Quant Kit | Thermo Fisher Scientific | Cat # A39552 |
| Halt protease inhibitor cocktail | Thermo Fisher Scientific | Cat # 78430 |
| Hanks Balanced Salt Solution (HBSS) | Thermo Fisher Scientific | Cat # 14170112 |
| Critical commercial assays | ||
| Mitochondrial Isolation Kit for Tissue | Thermo Fisher Scientific | Cat # 89874 |
| Mitochondrial Isolation Kit for Cultured Cells | Thermo Fisher Scientific | Cat # 89801 |
| Bio-Rad Protein assay kit II with bovine serum albumin standard | Bio-Rad | Cat # 5000002 |
| Experimental models: Cell lines | ||
| Hep-G2 human hepatocellular carcinoma (male) | ATCC | Cat # HB-8065; RRID: CVCL_0027 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6NCrl, 8–12-week-old | Charles River Laboratories | https://www.criver.com/products-services/find-model/c57bl6-mouse?region=24 |
| Other | ||
| Ultrasonic Processor Sound-Abating Enclosure | Cole-Parmer | Cat # UZ-04712-50 |
| Sonic Dismembrator Fisherbrand™ Model 120 (referred to as “Sonicator” in this protocol) | Thermo Fisher Scientific | Cat # FB120110 |
| Avanti J30I Ultracentrifuge | Beckman Coulter | Cat # J-30I |
| DWK Life Sciences Kimble™ Kontes™ Dounce Tissue Grinder 15 mL using small/looser fitting pestle | Fisher Scientific (of Thermo Fisher Scientific) | Cat # K885300-0015 |
| 2 mL conical microfuge tubes (i.e., Eppendorf Safe-Lock Tubes, 2.0 mL) | Eppendorf | Cat # 022363352 |
| 15 or 50 mL conical sterile polypropylene centrifuge tubes | Thermo Fisher Scientific | Cat # 339650 or 339652 |
Materials and equipment
Note: We perform this protocol using naïve 8-12-week-old male C57BL/6NCrl mice. Mice are permitted to acclimate to facility at least 1 week prior to use. Mice are not fasted or otherwise subjected to experimental procedures. For the purposes of this protocol, 1 mouse liver is assumed to weigh approximately 1.2 g as wet tissue. Adjust protocol according to empirically determined mass of mouse liver tissue.
HBSS/protease inhibitor solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 1× HBSS | N/A | 0.33 mL per 1.2 g mouse liver (equivalent to 0.275 mL/g of wet tissue) |
| 1× Halt protease inhibitor cocktail | 1:100 | 10 μL/mL of HBSS |
| Total | N/A | 0.337 mL per 1.2 g mouse liver (equivalent to 0.275 mL/g of wet tissue) |
Prepare HBSS with protease inhibitor fresh for each mitoDAMP isolation and maintain on ice; do not store.
PBS (Mitochondrial Isolation Kit for Tissue; prepared according to manufacturer’s instructions)
| Reagent | Final concentration | Amount |
|---|---|---|
| BupH™ Phosphate Buffered Saline | 0.1 M sodium phosphate 0.15 M sodium chloride |
1 package (included in kit) |
| Sterile ultrapure water | N/A | 500 mL |
| Total | N/A | 500 mL |
Sterile filter and maintain at 4°C for long term storage.
Alternatives: Sterile, commercial PBS may also be used.
Reagent A/BSA/protease inhibitor solution.
Mitochondrial Isolation Kit for Tissue; prepared according to manufacturer’s instructions
| Reagent | Final concentration | Amount |
|---|---|---|
| Mitochondrial Isolation Reagent A | N/A | 4.8 mL per 1.2 g mouse liver (equivalent to 4 mL/g of wet tissue) |
| 1× Halt protease inhibitor cocktail | 1:100 | 10 μL/mL of Mitochondrial Isolation Reagent A |
| BSA | 4 mg/mL | 4 mg/mL of Mitochondrial Isolation Reagent A |
| Total | N/A | 4.85 mL per 1.2 g mouse liver |
Prepare immediately before use and maintain on ice; do not store.
Reagent C/protease inhibitor solution.
Mitochondrial Isolation Kit for Tissue; prepared according to manufacturer’s instructions
| Reagent | Final concentration | Amount |
|---|---|---|
| Mitochondrial Isolation Reagent C | N/A | 4.8 mL per 1.2 g mouse liver (equivalent to 4 mL/g of wet tissue) |
| 1× Halt protease inhibitor cocktail | 1:100 | 10 μL/mL of Mitochondrial Isolation Reagent C |
| Total | N/A | 4.85 mL per 1.2 g mouse liver |
Prepare immediately before use and maintain on ice; do not store.
Wash Buffer/protease inhibitor solution.
Mitochondrial Isolation Kit for Tissue; prepared according to manufacturer’s instructions
| Reagent | Final concentration | Amount |
|---|---|---|
| Mitochondrial Isolation Reagent C | N/A | 1.5 mL per 1.2 g mouse liver (equivalent to 1.25 mL/g of wet tissue) |
| Sterile ultrapure water | N/A | 1.5 mL per 1.2 g mouse liver (equivalent to 1.25 mL/g of wet tissue) |
| 1× Halt protease inhibitor cocktail | 1:100 | 10 μL/mL of Wash Buffer |
| Total | N/A | 3.03 mL per 1.2 g mouse liver |
Prepare immediately before use and maintain on ice; do not store.
CRITICAL: Reagents and samples must be maintained on ice to maintain mitochondrial integrity and increase mitochondrial yield.
Step-by-step method details
Mitochondrial isolation from mouse liver tissue
Timing: 1 h
This section follows the manufacturer’s instructions for the Mitochondrial Isolation Kit for Tissue, Option A (soft tissues), Protocol 2: Dounce Homogenization for Soft Tissues. We start with 3–6 mouse livers, and process 1–2 pooled livers simultaneously in a Dounce homogenizer with a liquid volume capacity of 15 mL.
-
1.
Prepare Mitochondria Isolation Kit Reagents and PBS according to materials and equipment section, above. Prepare immediately prior to use and maintain on ice.
-
2.
Wash each mouse liver in 20–40 mL PBS to remove excess blood and debris from tissue. Swirl tissue in PBS in 50 mL conical tubes using sterile tweezers, or cap tube and gently invert to wash (Figure 2).
Note: We perform two successive washes in 50 mL conical tubes from “dirty” to “clean”. Sterile culture dishes could also be used.
-
3.
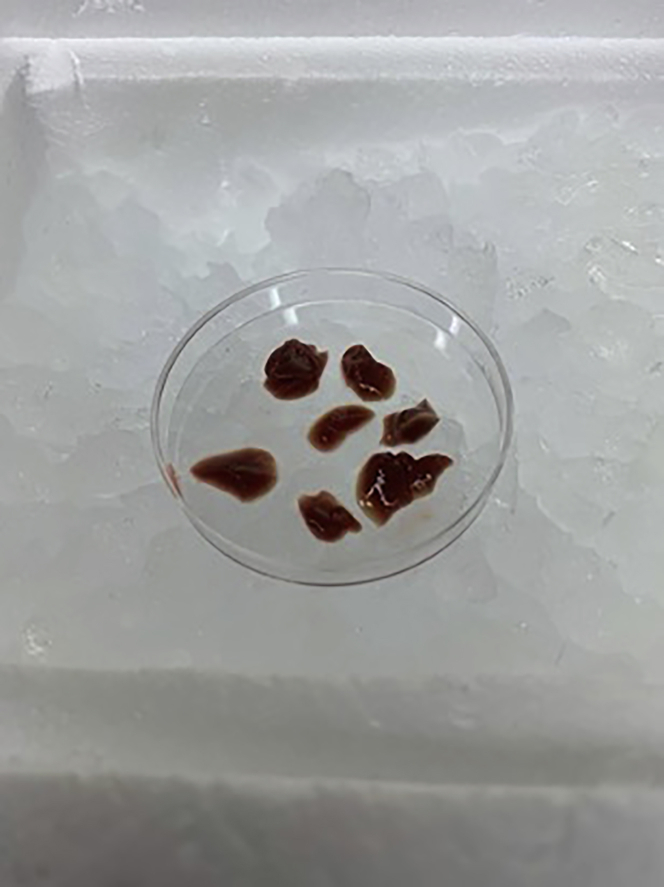
Use scissors or razor blade to cut liver tissue into smaller pieces (∼ 6–8 50–200 mg pieces) (Figure 3); this will allow for easier Dounce homogenization.
-
4.
Transfer liver pieces to Dounce homogenizer and add appropriate volume of Reagent A/BSA/protease inhibitor solution (Figure 4) according to materials and equipment section, above, and volume capacity of the Dounce Homogenizer.
CRITICAL: Over-filling Dounce homogenizer will result in ejection of fluid and loss of tissue from homogenizer during Dounce strokes.
-
5.
Perform Dounce homogenization on ice.
Note: We perform 75–100 strokes per 1–2 mouse livers, although this may vary slightly depending on Dounce homogenizer used. Continue with Dounce homogenization until liver is completely dissociated and no visible pieces of intact liver tissue remains (Figure 5).
-
6.
Discontinue once no visible pieces remain to avoid overprocessing.
CRITICAL: Avoid excessive homogenization (i.e., more homogenization than is necessary to remove intact liver tissue, or greater than 130 strokes) as this may compromise mitochondrial integrity and affect downstream applications.
-
7.
Transfer homogenized mixture of liver tissue and Reagent A/BSA/protease inhibitor solution to 2 mL microfuge tubes, adding 0.8 mL to each tube.
-
8.
Add 0.8 mL Reagent C/protease inhibitor solution to each tube, cap, and invert gently to mix (Figure 6). Do not vortex.
-
9.
Use benchtop microcentrifuge (pre-chilled to 4°C) to centrifuge at 700 × g for 10 min at 4°C.
-
10.
Remove supernatant, being careful not to disturb the pelleted material, and transfer supernatant to new 2 mL microfuge tube. Discard pelleted material (Figure 7).
-
11.
Use benchtop microcentrifuge to centrifuge at 3,000 × g for 15 min at 4°C.
Note: The manufacturer’s protocol allows for the optional centrifugation at either 3,000 × g or 12,000 × g. We centrifuge at 3,000 × g. The resulting pellet contains mitochondria from the heavy mitochondrial fraction (Islinger et al., 2010).
-
12.
Remove supernatant and discard; the pelleted material contains isolated mitochondria.
Note: Keep the supernatant containing the cytosolic cell fraction for future analysis, if desired, for downstream applications.
-
13.
Perform surface wash by adding 500 mL of Wash Buffer/protease inhibitor solution to the mitochondrial pellet; it is not necessary to resuspend the pellet (Figure 8).
-
14.
Use benchtop microcentrifuge to centrifuge at 12, 000 × g for 5 min at 4°C.
-
15.
Discard supernatant.
Note: Sometimes a layer of white fatty substance may appear on surface of supernatant. This is normal, but care should be taken to discard this layer along with the rest of the supernatant: see (Figure 12) in troubleshooting Section, below for details.
-
16.
Resuspend mitochondrial pellets in HBSS/protease inhibitor solution and combine in a single 15- or 50-mL conical tube (Figure 9). Volume of HBSS/protease inhibitor used to resuspend pellets will depend on quantity of mouse livers used in isolation protocol (generally 337 μL/liver). See materials and equipment Section and troubleshooting section for details.
Note: Mitochondrial pellet in HBSS/protease inhibitor solution will be viscous. Take caution when pipetting.
-
17.
Maintain mitochondrial pellet in HBSS/protease inhibitor solution on ice. Proceed directly to sonication, below.
Figure 2.
Washing of liver in cold PBS, using sterile tweezers
Figure 3.
Mouse liver cut into smaller pieces to facilitate Dounce homogenization
Figure 4.
Liver pieces and Reagent A/BSA solution with protease inhibitor in Dounce Homogenizer
Figure 5.
Liver tissue that has been completely dissociated following Dounce homogenization
Figure 6.
Liver homogenate in 2 mL tube, following addition of Reagent C with protease inhibitor
Figure 7.
Liver homogenate and Reagent C solution, following centrifugation at 700 × g for 10 min
Figure 8.
Mitochondrial pellet following surface wash with Wash Buffer and subsequent removal of supernatant
Figure 12.
Lipid layer on surface the liver homogenate following centrifugation
Arrow indicates white lipid layer.
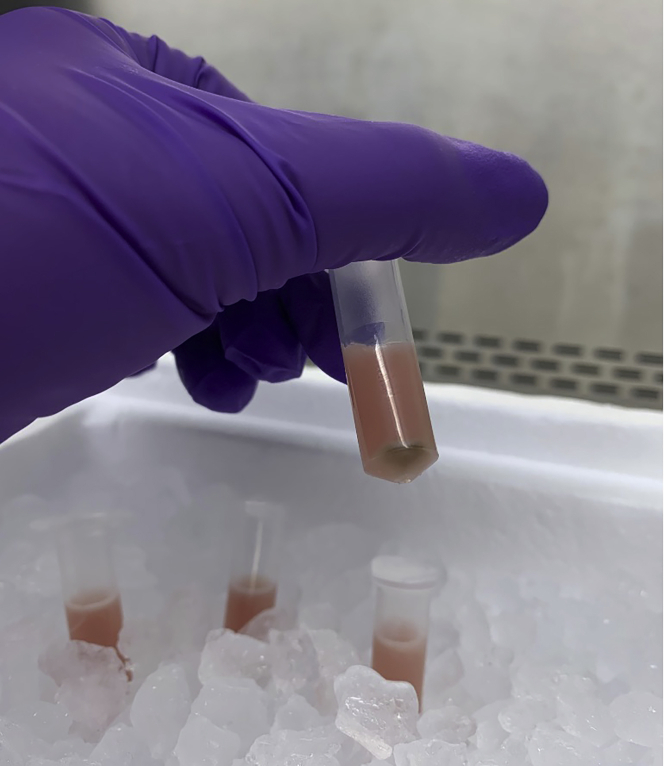
Figure 9.
Isolated mitochondria resuspended in HBSS with protease inhibitor
Sonication
Timing: 30 min
This section describes the sonication of isolated mitochondria to simulate tissue damage, as described (Zhang et al., 2010). Where possible, care should be taken to maintain sterility by working in a biological safety cabinet.
CRITICAL: Sonication generates heat and must therefore be performed on ice to protect mitochondrial protein from denaturation.
-
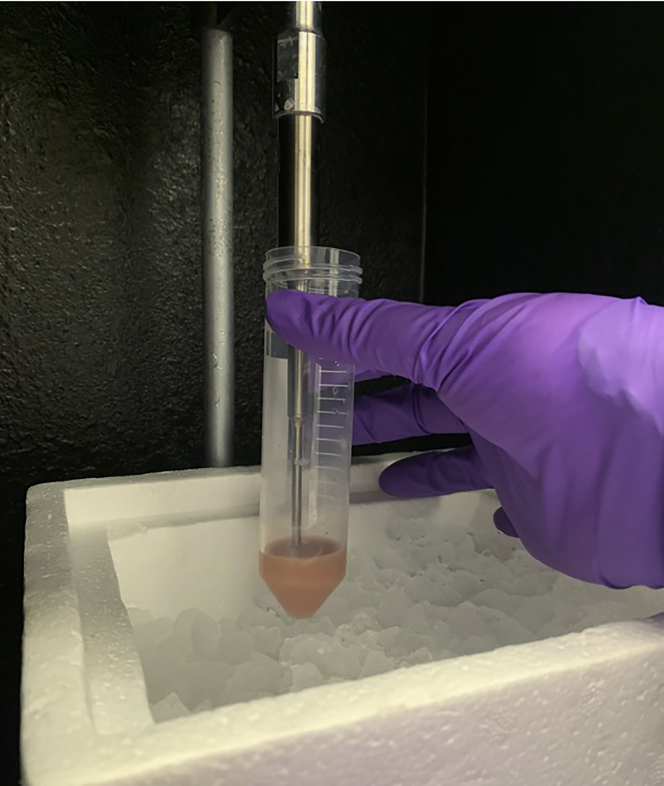
18.
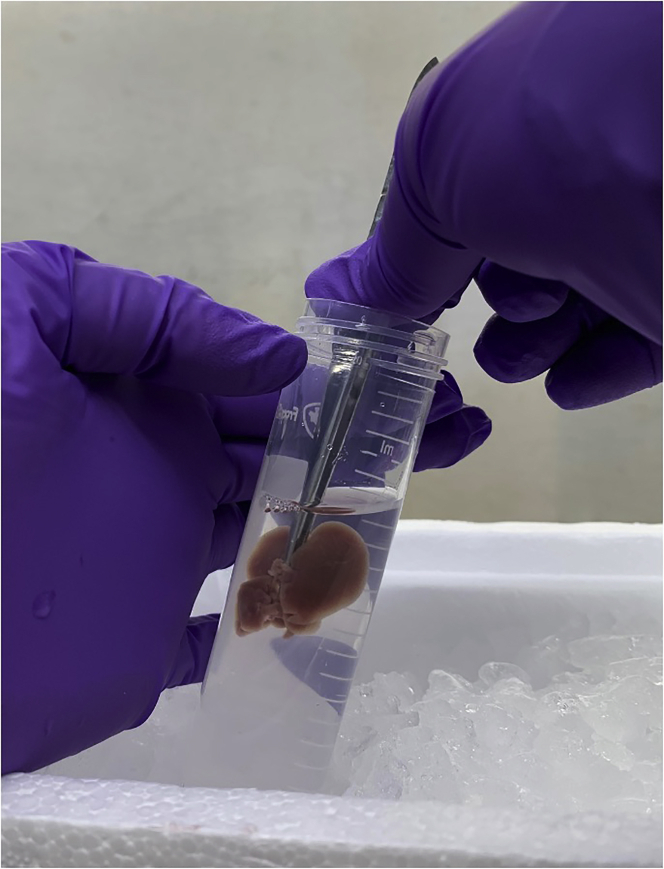
Sonicate mitochondria solution on ice at 100% amplitude in 30 s intervals, with 30 s of rest between sonication pulses, for a total of 10 min of sonication (Figure 10).
Note: Sonication at 100% amplitude may result in disruption of labile proteins – reduce sonication amplitude or duration if preservation of these proteins is desired for your experiment.
-
19.
Following sonication, transfer mitochondria solution to a tube(s) that is suitable for ultracentrifugation at 100,000 × g. Please consult centrifuge-specific manufacturer regarding recommendations for suitable tubes.
-
20.
Maintain tube containing mitochondrial solution on ice and proceed directly to ultracentrifugation.
Figure 10.
Sonication of isolated mitochondria on ice
Ultracentrifugation
Timing: 1 h of processing, 24 h of cold storage at −80°C
-
21.
Carefully balance tube(s) containing mitochondria solution for ultracentrifugation.
Note: We weigh both sample and balance tubes and adaptors on a digital scale to ensure there is <1 mg difference in weight.
CRITICAL: Care must be taken to precisely balance tube/adaptors/etc. prior to ultracentrifugation; please follow equipment and institution specific standard operating protocols.
-
22.
Centrifuge at 12,000 × g for 10 min at 4°C.
Note: Confirm that the centrifuge was properly balanced during step 21, and carefully re-balance tube(s) before proceeding to step 22.
-
23.
Centrifuge at 100,000 × g for 30 min at 4°C.
-
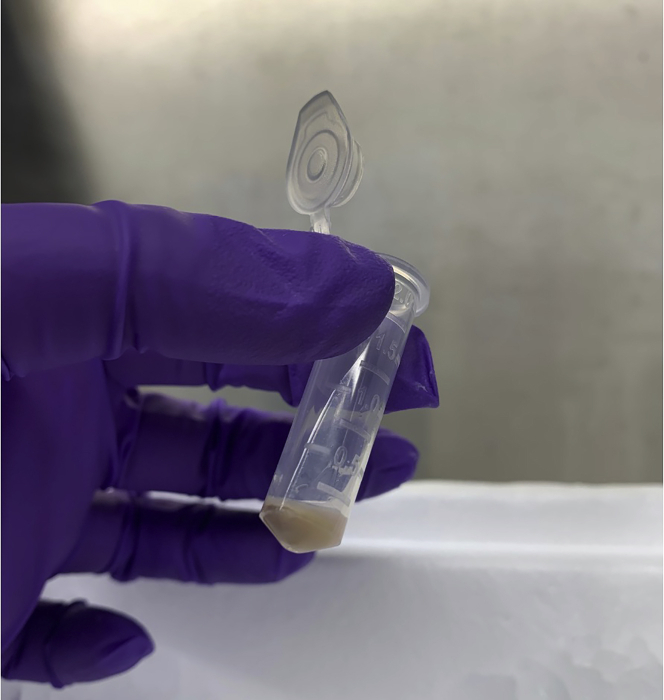
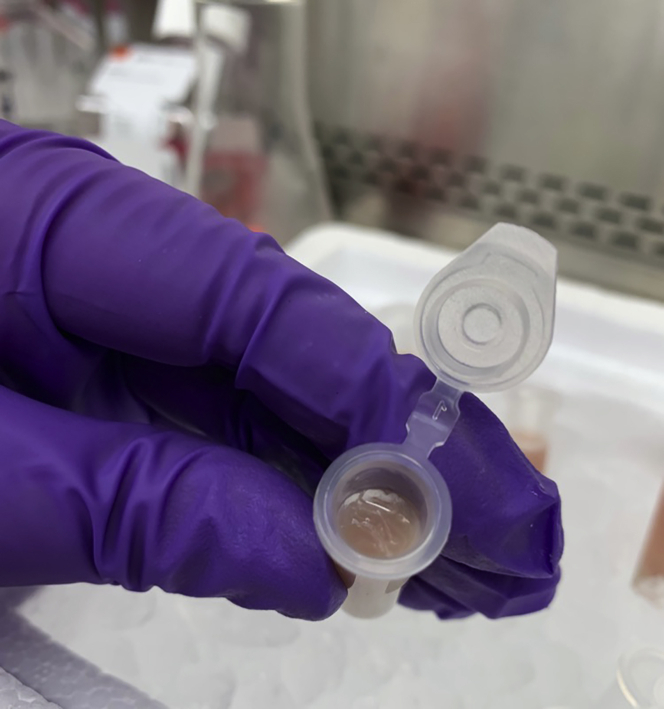
24.
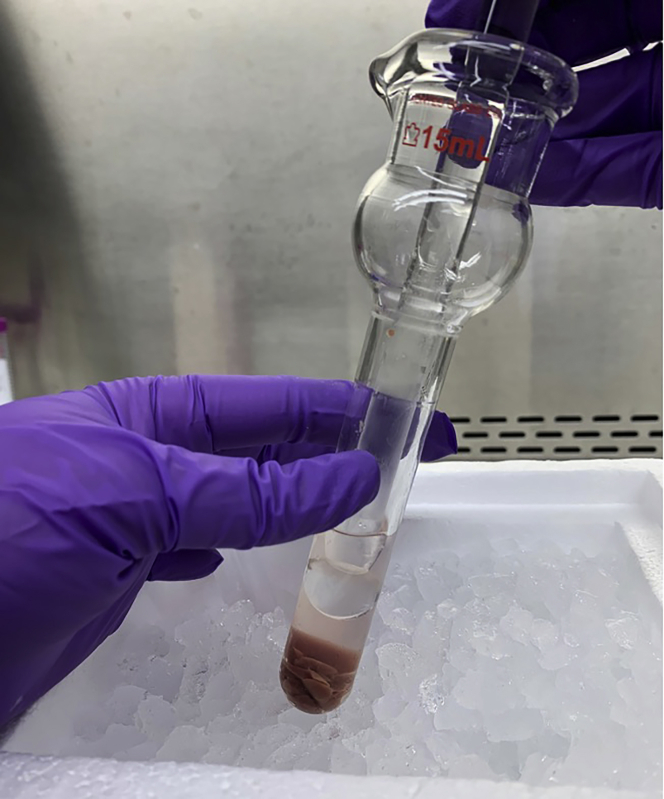
Remove supernatant (this is the isolated mitoDAMP fraction) and aliquot, if desired, into microfuge tubes (Figure 11). Store mitoDAMP preparation at −80°C until use.
-
25.
After aliquots have been at -80 for at least 24 h, a standard Bradford protein quantification assay may be performed (see below for anticipated protein concentration and mitoDAMP yield).
Figure 11.
Isolated mitoDAMPs (supernatant; right), and non-mitoDAMP fraction (pelleted material; left, following 100,000 × g centrifugation
Expected outcomes
The volume of mitoDAMP preparation yielded by this protocol depends on the starting quantity of mouse livers that are processed for mitochondrial isolation. We generally resuspend the harvest from three livers in 1 mL of HBSS/protease inhibitor. At this ratio, yields of 10,000–20,000 μg/mL are achieved (see troubleshooting Section, below).
The mitoDAMP preparation is generally clear and pale yellow to pink in color (Figure 11). We store ∼100 μL aliquots of mitoDAMP preparations at this concentration, at −80°C until use in experiments. In our experiments, we treat both human and mouse lymphocytes with a range of doses, from 5 – 400 μg/mL; within this range, mitoDAMPs potently affect lymphocytes function without affecting cell viability (Westhaver et al., 2022). Further, we have found that intravenous injection of 800 μg of mitoDAMPs into mice was well tolerated, with no overt indication of pathology or illness. If performing in vivo assays, caution should be taken to ensure that mitoDAMPs are free from endotoxin, via routine limulus amebocyte lysate assay (Zhang et al., 2010; Westhaver et al., 2022).
To standardize the treatment dose of mitoDAMPs within and between experiments, and across multiple mitoDAMP preparations, we perform a standard Bradford protein quantification assay (Westhaver et al., 2022). We have found this to be a reliable method for normalizing between mitoDAMP preparations. Secondarily, you may wish to directly compare different mitoDAMP preparations within the same experiment. Thus, it is good practice to retain a few aliquots of older preparations so that they can be compared with future preparations. We find total protein concentrations of mitoDAMP to be consistently 10,000–20,000 μg/mL across preparations.
Limitations
Overall, we have found this protocol to be reliable and robust. Here, we describe the isolation of mitoDAMPs from mouse liver tissue, but this protocol can also be modified for the isolation of mitoDAMPs from cultured cells (described above, in materials and equipment section). The use of cultured cells for mitoDAMP isolation is considerably more labor intensive and costly; we have found that 1.2e8 Hep-G2 cells yield the same quantity of mitoDAMPs as one mouse liver. A further limitation of the use of cultured cells is the diversity in relative mitochondrial abundance across cell lines. We have used the human hepatoblastoma cell line Hep-G2 for isolation of human mitoDAMPs; a non-liver cell line may carry fewer mitochondria and thus require even more cultured cells.
This protocol requires the use of semi-specialized equipment, including a probe Sonicator and Ultracentrifuge capable of >100,000 × g. This equipment is common in laboratories specializing in molecular biology, virology or bacteriology.
We describe the preparation of a bulk heterogenous fraction of mitoDAMPs; further optimization may be required for the isolation of specific mitoDAMP fractions. For instance, mitochondrial DNA can be isolated from mitoDAMP preparations using a standard DNA isolation kit (Zhang et al., 2010). Please refer to our previous work for a complete list of the components of mitoDAMPs (Westhaver et al., 2022).
Finally, the mitochondrial isolation portion of this protocol is completed via a commercially available kit. This kit may be prohibitively expensive or unavailable in some areas and therefore inaccessible; alternative mitochondrial isolation kits are available but have not been used in our lab in conjunction with the protocol described here.
Troubleshooting
Problem 1
A layer of white fatty substance appears on surface of supernatant during mitochondrial isolation (Figure 12) (step 14).
Potential solution
Depending on age and body weight of mice (i.e., how much fat content the liver contains) a white fatty substance may appear on surface of supernatant. This can be gently removed by pipetting. You may choose to perform a second wash step (i.e., repeat steps 12–14). Alternatively, you may choose to use younger or leaner mice as a source for liver tissue. We have not found this lipid layer to affect downstream experiments with mitoDAMPs on lymphocytes.
Problem 2
Low mitochondrial yield following mitochondrial isolation (step 15).
Potential solution
Mitochondrial concentration varies between tissues and cell types. It may be necessary to increase the starting amount of mouse liver tissue such that more mitochondria are isolated. If this is not possible, follow manufacturer’s protocol for troubleshooting. For instance, you may need to empirically determine the ideal number of strokes needed for Dounce homogenization, such that cell lysis is maximized and mitochondrial damage is minimized.
Problem 3
MitoDAMP preparation is dilute, as measured by Bradford protein quantification assay, and is therefore impractical for downstream applications (expected outcomes).
Potential solution
It is critical that mitochondrial isolation and sonication are performed on ice, and that all centrifugation cycles are maintained at 4°C. Delays in completion of the protocol or prolonged warm incubations may affect mitochondrial component integrity. Secondly, to concentrate final mitoDAMP preparation, decrease the volume of HBSS/protease inhibitor solution that mitochondria are resuspended in prior to sonication.
Finally, the vibration created by the sonicator generates heat; care must be taken to prevent denaturation of mitochondrial protein. If ice is held in a styrofoam container, care must be taken to prevent tube containing mitochondria from contacting the container during sonication. We have found that the resulting vibration of the tube against styrofoam will result in denaturing of the mitochondrial protein.
Problem 4
Mouse liver tissue for mitoDAMP isolation is not available to researcher, or mitoDAMPs derived from human mitochondria are required (before you begin).
Potential solution
Cultured cells can be used in place of mouse liver tissue as a source of isolated mitochondria. We have found that for the purposes of this protocol, 1.2e8 Hep-G2 cells is equivalent in mitochondrial yield to a single mouse liver, or approximately 1.2 g of wet tissue.
To use cultured HepG2 cells in place of mouse liver, follow manufacturers protocol for Mitochondria Isolation Kit for Cultured and then proceed with Sonication protocol once mitochondria have been isolated. Incubate pelleted cells from cell culture with Reagent A/BSA/Protease inhibitor solution for 2 min on ice (do not exceed 2 min of incubation), and then substitute pelleted cells for liver tissue into step 4 of the protocol. Proceed with remainder of protocol. We have found that the reagents included in the Mitochondria Isolation Kit for Tissue work equally well for cultured cells, using the Dounce homogenization method outlined in this protocol.
If human liver tissue is available, it may be substituted directly for mouse tissue in this protocol (see materials and equipment section for details), as the Mitochondrial Isolation Kit for Tissue (Thermo Fisher Scientific) used in this protocol is also suitable for human liver and skeletal muscle Zhang et al. (2010). However, further optimization may be required to determine whether relative mitochondrial yield is comparable to mouse liver tissue.
Problem 5
The Mitochondrial Isolation Kit for Tissue (Thermo Fisher Scientific) described in this protocol is not available to the researcher, or the researcher wishes to use an alternative method of mitochondrial isolation (before you begin).
Potential solution
In this protocol, we describe the use of the Mitochondrial Isolation Kit for Tissue (Thermo Fisher Scientific), which we have optimized for our purposes. We favor this kit for its relatively low cost and straightforward protocol, however, the researcher may choose to use alternative commercially-available kits or non-commercial isolation protocols.
The Mitochondrial Isolation Kit for Tissue (Thermo Fisher Scientific) relies on a hypotonic buffer and mechanical Dounce homogenization for cell disruption, followed by differential centrifugation to isolate mitochondria. Like other conventional mitochondria isolation methods, kit Reagent C contains mannitol and sucrose for functional stabilization of the mitochondria (Almeida and Medina, 1998), but does not rely on sucrose gradient centrifugation for purification. Alternatively, mitochondrial isolation can be achieved via conventional sucrose gradient-based techniques (Clayton and Shadel, 2014). However, we have not rigorously tested such alternatives and thus achieving suitable mitochondrial yield for subsequent mitoDAMP isolation may require additional optimization.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Jeanette Boudreau, at Jeanette.boudreau@dal.ca.
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This research was supported by the Natural Sciences and Engineering Research Council of Canada, Research Nova Scotia, the Nova Scotia Research Innovation Trust and the Canada Foundation for Innovation. L.P.W. is supported by a Dalhousie Faculty of Medicine Scholarship. L.K.M., S.N., A.N., and E.B.C. are trainees in the Cancer Research Training Program of the Beatrice Hunter Cancer Research Institute, with funds provided by the Dalhousie Medical Research Foundation (DMRF)’s Crease Endowment for Cancer Research to E.B.C. and funds provided by Craig’s Cause Pancreatic Cancer Society to A.N. L.K.M., S.N., and E.B.C. are recipients of Nova Scotia Graduate Scholarships. L.K.M. is additionally supported by a Genomics in Medicine Scholarship. S.N. and L.K.M. are additionally supported by a Killam Memorial Predoctoral Scholarship and the President’s award. S.N. is additionally funded by a CIHR Vanier award. A.N. is additionally funded by The Terry Fox Research Institute and Craig’s Cause Pancreatic Society. J.E.B. is the DMRF Cameron Cancer Scientist. The graphical abstract was created with biorender.com.
We gratefully acknowledge the support of the Dalhousie Carlton Animal Care Facility. We are grateful to Dr. I.P. Alwayn for his for his assistance in initiating this project. Dalhousie University is located in Mi’kma’ki, the ancestral and unceded territory of the Mi’kmaq.
Author contributions
Conceptualization, L.P.W., S.N., and J.E.B.; Methodology: L.P.W. and J.E.B.; Validation: L.P.W., S.N., A.N., L.K.M., E.B.C., and J.E.B.; Formal analysis: L.P.W., S.N., and J.E.B.; Investigation: L.P.W., S.N., A.N., L.K.M., and E.B.C.; Data curation: L.P.W., L.K.M., and S.N.; Writing-original draft: L.P.W. and J.E.B.; Writing Review & Editing: L.P.W., S.N., A.N., L.K.M., E.B.C., and J.E.B.; Visualization: L.P.W., S.N., and J.E.B. Supervision: J.E.B.; Project administration: L.P.W., S.N., and J.E.B.; Funding acquisition: J.E.B.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Lauren P. Westhaver, Email: lauren.westhaver@dal.ca.
Jeanette E. Boudreau, Email: jeanette.boudreau@dal.ca.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Almeida A., Heales S.J., Bolaños J.P., Medina J.M. A rapid method for the isolation of metabolically active mitochondria from rat neurons and astrocytes in primary culture. Brain Res. 1998;790:209–216. doi: 10.1016/s1385-299x(97)00044-5. [DOI] [PubMed] [Google Scholar]
- Clayton D.A., Shadel G.S. Purification of mitochondria by sucrose step density gradient centrifugation. Cold Spring Harb. Protoc. 2014;2014 doi: 10.1101/pdb.prot080028. pdb.prot080028. [DOI] [PubMed] [Google Scholar]
- Islinger M., Li K.W., Loos M., Liebler S., Angermüller S., Eckerskorn C., Weber G., Abdolzade A., Völkl A. Peroxisomes from the heavy mitochondrial fraction: isolation by zonal free flow electrophoresis and quantitative mass spectrometrical characterization. J. Proteome Res. 2010;9:113–124. doi: 10.1021/pr9004663. [DOI] [PubMed] [Google Scholar]
- Westhaver L.P., Nersesian S., Nelson A., MacLean L.K., Carter E.B., Rowter D., Wang J., Gala-Lopez B.L., Stadnyk A.W., Johnston B., Boudreau J.E. Mitochondrial damage-associated molecular patterns trigger arginase-dependent lymphocyte immunoregulation. Cell Rep. 2022;39:110847. doi: 10.1016/j.celrep.2022.110847. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Raoof M., Chen Y., Sumi Y., Sursal T., Junger W., Brohi K., Itagaki K., Hauser C.J. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.