Abstract
Dipeptidyl peptidase-IV (DPP-IV) inhibitory peptides from fermented foods exhibit great potential to alleviate type 2 diabetes mellitus (T2DM). In this study, the DPP-IV inhibition activity of peptide extract from Chouguiyu was obviously enhanced after 4–8 d fermentation. A total of 125 DPP-IV inhibitory peptides in Chouguiyu were identified by peptidomics and were obtained from 46 precursor proteins, mainly including nebulin, titin, muscle-type creatine kinase, hemoglobin, and actin. After molecular docking with DPP-IV, four novel DPP-IV inhibitory peptides possessing the lowest docking energy were selected, including EPAEAVGDWR (D37), IPHESVDVIK (D22), PDLSKHNNHM (D35), and PFGNTHNNFK (D1). The DPP-IV inhibition activity of D37, D22, D35, and D1 were further verified after synthesis with the IC50 of 0.10 mM, 2.69 mM, 3.88 mM, and 8.51 mM, respectively, in accordance with their docking energies. Energy interaction showed that the structures of EP-, IPH-, -NHM, and PF- in these peptides were easy to connect with DPP-IV enzyme through hydrogen bond, salt bridge, and alkyl. The surface force including the H-bond interaction, hydrophobicity, aromatic interaction, and SAS, played a major role in the interaction between DPP-IV enzyme and peptides. The peptides that possess high hydrophobicity and can form strong hydrogen bond and salt bridge are potential DPP-IV inhibitory peptides using for T2DM remission.
Keywords: Chouguiyu, Dipeptidyl peptidase-IV, Peptide, Molecular docking, Hydrophobicity, Hydrogen bond
Abbreviations: (DPP-Ⅳ), Dipeptidyl peptidase Ⅳ; (T2DM), Type 2 diabetes mellitus; (AAs), Amino acids; (PLS-DA), Partial least squares-discriminant analysis; (IC), Interpolated charge; (SAS), Solvent accessible surface
Graphical abstract

Highlights
-
•
DPP-Ⅳ inhibition activity of peptide extract in Chouguiyu increased by fermentation.
-
•
The main precursor proteins of DPP-Ⅳ inhibitory peptides were nebulin and titin.
-
•
Inhibition mechanism was explored by energy interaction and surface force.
-
•
Docking energy was an effective index to select DPP-IV inhibitory peptides.
-
•
DPP-IV inhibitory peptides formed hydrogen bond and salt bridge with DPP-IV.
1. Introduction
Diabetes mellitus, especially Type 2 diabetes (T2DM), is a major health problem in the world affecting millions of people, and mainly occurs due to the different degrees of insulin deficiency and insulin resistance (Sagar et al., 2015). Glucagon-like peptide-1 (GLP-1) plays a crucial role in inhibiting glucagon release and in turn stimulates insulin secretion, thereby reducing the symptoms of diabetes (Li et al., 2016). As a serine protease in blood, dipeptidyl peptidase IV (DPP-IV) can rapidly degrade GLP-1, resulting in the inhibition of insulin secretion and increase of blood glucose (Song et al., 2017). Various DPP-IV inhibitors that are approved by Food and Drug Administration (FDA) are used as therapeutic drugs for type 2 diabetes mellitus (George and Joseph, 2014), such as sitagliptin, saxagliptin, and linagliptin. However, there are plenty of side effects in these drugs, such as joint pain, heart failure, headache, and nausea. Bioactive peptides are specific fragments of food proteins that possess various bioactivities, such as hypotensive, antitumor, and hpyerglycemic activities (Ishak et al., 2021; Wang et al., 2020). As a kind of bioactive peptides with hpyerglycemic activity, DPP-IV inhibitory peptides play a crucial role in the inhibition of DPP-IV, leading to the decrease of post-prandial glucose levels (Wang et al., 2020). These food protein-derived DPP-IV inhibitory peptides have been considered as a substitute for decreasing insulin deficiency and insulin resistance with no side effects (Wang et al., 2020). Most studies have concentrated on enzymatic hydrolysis food proteins by commercially available proteases to generate DPP-IV inhibitory peptides (Cian et al., 2022; Song et al., 2017). However, the restriction of their cutting sites results in the low diversity of DPP-IV inhibitory peptides, which limits the discovery of novel peptides with high DPP-IV inhibitory bioactivity.
Complex microbial communities in fermented foods can produce various proteases, contributing to the production of abundant peptides (Majumdar et al., 2016; Mouritsen et al., 2017). That makes it possible to discover novel DPP-IV inhibitory peptides with high bioactivity in fermented foods. However, much attention has been paid to umami peptides in fermented foods, and there is lack of study on the bioactive peptides, especially DPP-IV inhibitory peptides. Many studies have shown that the drugs with “N” centers in their structure, serving as proline mimics, possess good ability to inhibit the activity of DPP-IV enzyme (Sagar et al., 2015). Proline-rich protein substrate were selected for preparing DPP-IV inhibitory peptides (Nongonierma et al., 2017). Similarly, the peptides rich in proline at their N-terminal exhibit strong DPP-IV inhibitory activity (Song et al., 2017). Chouguiyu is a Chinese famous fermented fish product with abundant proline (Yang et al., 2020). However, the DPP-IV inhibitory peptides especially with specific location of proline have not been clarified in Chouguiyu. Nowadays, molecular docking is an important tool in drug discovery to predict the most favorable complex formed between a given protein receptor and a small molecule ligand (Li et al., 2020; Sousa et al., 2013). This technology has been widely used to screen non-peptide mimetic drugs with DPP-IV inhibitory activity, such as xanthine (Li et al., 2016), pyrazolo-pyrimidinones (Sagar et al., 2015), and triazole (Li et al., 2016). However, the relationship between DPP-IV enzyme and its inhibitory peptides has rarely been illustrated by molecular docking.
In this study, the DPP-IV inhibitory activities of peptide extract from Chouguiyu were evaluated. The sequence of peptides was identified by peptidomics to discover the DPP-IV inhibitory peptides based on the specific location of proline. Moreover, the precursor proteins were analyzed to determine the cleavage location of these peptides. The interactions between the DPP-IV enzyme and DPP-IV inhibitory peptides were studied using molecular docking, and the surface forces in their optimal binding conformation were also exhibited. The in vitro analysis was used to verify the results of DPP-IV inhibitory peptide virtual screening. The in vitro analysis was used to verify the results of DPP-IV inhibitory peptide virtual screening. This work is hopeful for providing comprehensive understanding into the inhibition mechanisms of DPP-IV inhibitory peptides in Chouguiyu. The core peptides can be developed into potential food supplements using for T2DM remission.
2. Material and methods
2.1. Preparation of peptide extract from Chouguiyu
Chouguiyu was produced using frozen mandarin fish (Siniperca chuatsi) with an average weight of 500 g in Anhui, China (Yang et al., 2022a, Yang et al., 2022b, Yang et al., 2022c). Clean fish were neatly arranged in fermentation tanks. The brine (6% salt and 0.02% spices) was added into the tanks until it soaked the fish. During the natural fermentation for 8 d, the Chouguiyu was obtained at 0 d (d0 group), 4 d (d4 group), and 8 d (d8 group) for peptide extract as described previously (Yang et al., 2022). Briefly, the Chouguiyu without skin (50 g) was homogenized for 0.5 h with 0.01 mol/L hydrochloric acid (100 mL). After centrifugation for 20 min at 4 °C and 7000×g, the supernatant was added with triploid ethanol, placed at 4 °C for 20 min, and centrifuged for 10 min at 4 °C and 16000×g to remove protein. The supernatant was lyophilized and stored at −80 °C after desalination.
2.2. Analysis of DPP-Ⅳ inhibitory activity
The DPP-IV inhibition assay was tested by DPP- IV inhibitor screening assay kit (KA1311, Abnova, China) as described previously (Guo et al., 2020). Briefly, each peptide extract (1 g/L) was mixed with assay buffer and DPP-IV in a 96-well plate. Then, the Gly-Pro-Aminomethylcoumarin (AMC) solution was added in each well and reacted at 37 °C for 30 min. The free AMC group was detected by the enzyme-labeled instrument (Sunrise-basic Tecan, Switzerland) at 355 nm (excitation) and 455 nm (emission). Sitagliptin at the same concentration (1 g/L) was used as positive control. The DPP- Ⅳ inhibition rate (I) of peptide extract was calculated using the following equation:
where, I (%) is the DPP- Ⅳ inhibition rate of peptide extract, Fi is the fluorescence of samples without addition of peptide extract or positive control, Ft is the fluorescence of samples with addition of peptide extract or positive control.
2.3. Peptide identification by peptidomics
Peptidomics was performed using ultra-high performance liquid chromatography tandem mass as studied previously (Murray et al., 2018) with slight modifications. The peptide samples were separated using the Easy-nLC™ 1200 UPLC (Thermo, USA), coupled with an ACQUITY UPLC peptide BEH C-18 column (1.7 μm, 2.1 mm × 150 mm, Waters, USA). Elution program was performed using solvent A (water solution with 2% acetonitrile and 0.1% formic acid) and solvent B (water solution with 80% acetonitrile and 0.1% formic acid) under 0–53 min, 5%B; 53–65 min, 23%B; 65–73 min, 29%B; 73–74 min, 38%B; 74–75 min, 48%B; 75–90 min, 100% B with a flow rate of 0.30 μL/min. Q Exactive™ HF-X tandem mass spectrometer (Thermo, USA) was used to analyze sequence and abundance of peptides. The instrument was operated in a single charge mode ([M + H]+) at the mass/charge (m/z) range of 350–1500. The peptide sequences were identified by Proteome Discoverer™ (Thermo, USA) according to the protein sequences of Siniperca chuatsi.
2.4. Physicochemical characteristic analysis of DPP-Ⅳ inhibitory peptides
ToxinPred was performed to analyze the toxicity of peptides using the website https://webs.iiitd.edu.in/raghava/toxinpred/multi_submit.php(Çağlar et al., 2021). ExPASy was used to check the isoelectric point and molecular weight of peptides on the link https://web.expasy.org/compute_pi/(Tu et al., 2018). The charge and hydrophobicity were searched from PepDraw (http://www.tulane.edu/~biochem/WW/PepDraw/)(Tu et al., 2021). The instability values were searched by ProtParam (https://web.expasy.org/protparam/)(Games et al., 2016). The novelty of peptides was predicted using database Pepbank (http://pepbank.mgh.harvard.edu/search/basic) (Yang et al., 2022a, Yang et al., 2022b, Yang et al., 2022c).
2.5. Molecular docking between DPP-Ⅳ enzyme and DPP-Ⅳ inhibitory peptides
The DPP-IV enzyme (PDB id:4PNZ) were downloaded from RCSB PDB (https://www.rcsb.org/) and optimized using Discovery Studio 2019 Client (San Diego, USA), including deleting water and ligands, supplement forcefield, and defining active binding sites. The structures of DPP-IV inhibitory peptides were constructed using Chem Draw (Cambridge soft, USA) and were then optimized using CFF partial charge method in Discovery studio 2019 Client. The molecular docking between DPP-IV enzyme and peptides was conducted by flexible ligand-flexible target module. The hypoglycemic drug sitagliptin was used as the positive control to dock with the DPP-IV enzyme.
2.6. Solid-phase synthesis and DPP-IV inhibitory activity of core DPP-IV inhibitory peptides
The core DPP-IV inhibitory peptides with a purity >95% were synthesized through the conventional Fmoc solid-phase synthesis method with APEX 396 automated peptide synthesizer (AAPPTEC, USA). DPP-IV inhibitory activity was measured in 96-well plates following the method described by previously (Vilcacundo et al., 2017) with some modifications. Briefly, the sample concentrations were set to 10 g/L, 1 g/L, 0.1 g/L, 0.01 g/L, and 0.001 g/L. A total of 25 μL sample and 25 μL H-Gly-Pro-p-nitroaniline (2 mmol/L) were mixed and incubated at 37 °C for 10 min, then were added with 50 μL DPP-IV enzyme solution (0.02 U/mL). After incubation at 37 °C for 60 min, the reaction was terminated by 100 μL sodium acetate buffer solution (1 mmol/L, pH 4.0). The absorbance was detected by the enzyme-labeled instrument at 405 nm to calculate the DPP-IV inhibitory activity of peptides.
2.7. Statistical analysis
All experiments were performed in triplicate and the data were expressed as mean ± standard deviation. The differences among treatments were analyzed by one-way analysis of variance (ANOVA) with multiple comparison Tukey test using the IBM SPSS Statistics (v20.0) (Zhao et al., 2021). The similarity of 125 DPP-IV inhibitory peptides in different groups was determined by partial least squares-discriminant analysis (PLS-DA) using Metabo Analyst v5.0 (Li et al., 2022a, Li et al., 2022b). The heatmap was drawn using MetaboAnalyst v5.0 (Li et al., 2022). The Sankey diagram was drawn by Origin (OriginLab Corp, USA). The peptide profile was drawn using Peptigram (Fu et al., 2018). Half-maximal inhibitory concentration (IC50) for DPP-IV inhibition of peptides was calculated by logarithmic regression analysis.
3. Results and discussion
3.1. DPP-IV inhibition rate of peptide extract in Chouguiyu
As a proline-rich fermented fish product, Chouguiyu can be used as a potential source for the extraction of DPP-IV inhibitory peptides. In this study, the DPP-IV inhibition rates of peptide extract from Chouguiyu at various fermentation groups are exhibited in Fig. 1. Interestingly, the peptide extract in Chouguiyu exhibited good DPP-IV inhibition activity as same as the positive drug sitagliptin. Higher DPP-IV inhibition rate was observed in the peptide extract after fermentation, especially in the d4 group (60.14%) than that in the d0 group. At the same concentration, the peptide extract in the d4 and d8 groups respectively achieved 67.07% and 28.03% of DPP-IV inhibition activity of sitagliptin.
Fig. 1.
DPP-IV inhibition rate of peptide extract in Chouguiyu during fermentation for 8 d. Bars labeled with different letters are statistically different (p < 0.05), as tested by one-way ANOVA and the Tukey test.
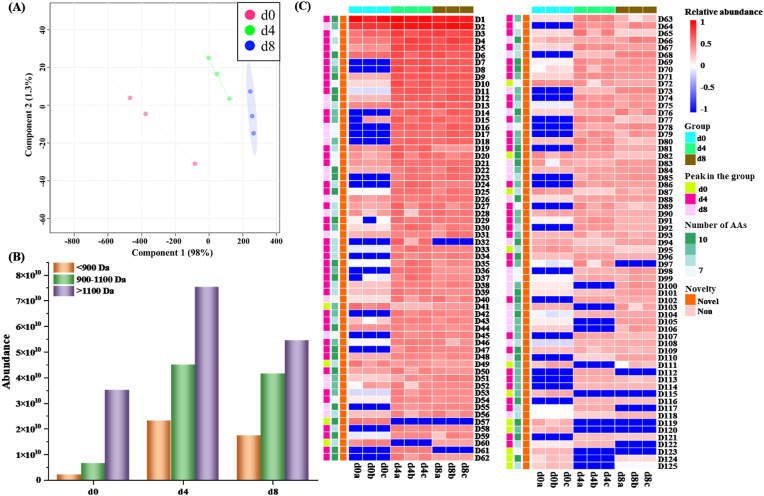
To confirm which peptides contributed to the DPP-IV inhibition activity, peptidomics was used to select the peptides with DPP-IV inhibitory bioactivity. Previous studies found that small-molecule peptides (generally ≤10 amino acids (AAs)) with various hydrophobic amino acids in the structure possessed good DPP-IV inhibitory activity (Hao et al., 2021; Lacroix and Li-Chan, 2013; Tu et al., 2021). Among hydrophobic amino acids, the presence of proline in the peptides contributed most to the inhibition of DPP-IV, especially when it exited at the first three amino acids at the N-terminal (Lacroix and Li-Chan, 2014; Lafarga, O Connor & Hayes, 2015; Song et al., 2017), such as PAL- (Ji et al., 2017), VPL (Rahfeld et al., 1991), IPQ- (Xu et al., 2019), and GGP- (Neves, Harnedy, O'Keeffe, Alashi, Aluko & FitzGerald, 2017). In this study, a total of 125 DPP-IV inhibitory peptides with total AAs ≤10 and proline at similar sites of N-terminal were discovered from Chouguiyu at different fermentation time. It was reported that DPP-IV inhibitory peptides in Sardine pilchardus below 1400 Da had the highest activity (Zhang et al., 2019). The largest molecular weight of DPP-IV inhibitory peptides in Chouguiyu was 1308.55 Da, which might be helpful in discovery of novel peptides with high DPP-IV inhibitory activity. The similarity of 125 DPP-IV inhibitory peptides in different groups was determined by PLS-DA. The Q2 of component 1 and component 2 was 0.70 and 0.41, respectively (Table S2). The PLS-DA plot showed that DPP-IV inhibitory peptides in the d0 group exhibited more difference with the d4 and d8 groups, while there was no marked difference between the d4 and d8 groups (Fig. 2A). The peptide abundance of three molecular weight distribution regions first increased and then decreased with the increasing fermentation time, and peaked in the d4 group, in accordance with the results of DPP-IV inhibition activity of peptide extract in different fermentation groups (Fig. 2B). The DPP-IV inhibitory peptides with the molecular weight distribution >1100 Da exhibited the highest abundance, while the peptides with the lowest abundance were found in <900 Da. The DPP-IV inhibitory peptides were named from D1 to D125 in order of their abundance (Fig. 2C and Table S1). The differential DPP-IV inhibitory peptides (|log2(fold change)| > 1) from Chouguiyu were compared in each group (Table S3). Comparative peptidomics analysis showed that between the d4 and d0 groups there were 118 differential DPP-IV inhibitory peptides, 99 of which were improved, while 19 of which were reduced in the d4 group. A total of 112 differential DPP-IV inhibitory peptides were observed between the d8 and d0 groups, among which 93 and 19 peptides were improved and reduced in the d8 group, respectively. Interestingly, most DPP-IV inhibitory peptides reached their peak in the d4 group. Among the 61 differential DPP-IV inhibitory peptides between the d8 and d4 groups, more peptides (37 peptides) possessed lower abundance in the d8 group than the d4 group, in accordance with the results of DPP-IV inhibitory activity from peptide extract in Chouguiyu.
Fig. 2.
Identification of DPP-Ⅳ inhibitory peptides from Chouguiyu at different fermentation time. (A) PLS-DA analysis of DPP-Ⅳ inhibitory peptides among the different fermentation groups. (B) Abundance of DPP-Ⅳ inhibitory peptides at different molecular weight distributions. (C) Heatmap analysis of DPP-Ⅳ inhibitory peptides during the fermentation process. The red, green, and blue squares respectively represent the peptides reaching their maximum on d0, d4, and d8. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Physicochemical characteristics of DPP-Ⅳ inhibitory peptides from Chouguiyu and their precursor protein analysis
The physicochemical properties of DPP-IV inhibitory peptides including isoelectric point, charge, hydrophobicity, solubility, toxicity and stability were further predicted in silico (Fig. 3 and Table 1). All the DPP-IV inhibitory peptides possessed hydrophobicity more than 8.17 kJ/mol. The strong hydrophobicity helped peptides easily interact with the hydrophobic active sites of DPP-IV enzyme, consequently resulting in the inactivation of DPP-IV enzyme (Lacroix and Li-Chan, 2013). The toxicity prediction showed that all the 125 DPP-IV inhibitory peptides were nontoxic. A total of 79 peptides displayed acidic isoelectric point, while only 46 peptides possessed basic isoelectric point. Most DPP-IV inhibitory peptides (95) possessed good water solubility. The stability of peptides in vivo played a key role in their biological activities (Singh and Vij, 2018). In this study, a total of 74 DPP-IV inhibitory peptides possessed stable structures, which were chosen for the analysis of molecular docking.
Fig. 3.
Identification of precursor proteins for producing DPP-Ⅳ inhibitory peptides and characteristics of DPP-Ⅳ inhibitory peptides of Chouguiyu.
Table 1.
Molecular docking results of the 30 stable DPP-IV inhibitory peptides with the highest abundance.
| Name | Sequence | ΔEdocking (kcal/mol) | ΔEinteraction (kcal/mol) | ΔEbinding (kcal/mol) | ΔEvdw (kcal/mol) | ΔEele (kcal/mol) |
|---|---|---|---|---|---|---|
| D1 | PFGNTHNNFK | −106.778 | −76.137 | −393.385 | −33.933 | −416.936 |
| D2 | PFGNTHNNF | −90.879 | −64.772 | −382.306 | −27.852 | −416.651 |
| D3 | YPWTQRH | −84.245 | −64.880 | −243.322 | −28.733 | −259.438 |
| D5 | LPHDTPL | −48.494 | −47.987 | −157.232 | −26.909 | −183.112 |
| D7 | KPSAPKIP | −36.524 | −54.152 | −137.993 | −28.900 | −175.258 |
| D10 | DDPLLVH | −81.375 | −60.334 | −191.319 | −30.952 | −197.714 |
| D11 | EIPTKVPKAE | −96.214 | −68.462 | −240.629 | −40.383 | −270.890 |
| D12 | LDPEGTGTIK | −101.161 | −53.792 | −297.425 | −30.999 | −318.845 |
| D14 | KPSAPKIPD | −62.905 | −50.310 | −130.435 | −33.641 | −171.517 |
| D16 | DPIISDR | −79.292 | −72.330 | −187.298 | −25.291 | −200.557 |
| D18 | NPYKEIDVS | −104.337 | −60.506 | −280.125 | −32.964 | −304.800 |
| D20 | IPVVDDK | −68.758 | −62.933 | −108.907 | −27.844 | −128.317 |
| D21 | DTPEIVR | −80.034 | −41.771 | −202.268 | −22.282 | −222.421 |
| D22 | IPHESVDVIK | −111.038 | −65.437 | −294.789 | −39.491 | −301.470 |
| D23 | PEGTGTIKKQ | −103.547 | −53.732 | −303.534 | −30.870 | −328.638 |
| D24 | SLPHDTPL | −68.802 | −63.328 | −205.053 | −29.760 | −229.141 |
| D25 | IPPEKPIKIP | −73.950 | −90.147 | −186.272 | −41.299 | −231.023 |
| D26 | IPVNVDK | −73.015 | −58.851 | −178.553 | −29.799 | −189.573 |
| D31 | FPSIVGR | −55.094 | −34.694 | −155.809 | −19.807 | −169.726 |
| D33 | PDLSKHNN | −85.141 | −59.450 | −298.630 | −29.360 | −325.148 |
| D34 | NPYKEIDV | −92.101 | −70.969 | −234.603 | −33.366 | −248.249 |
| D35 | PDLSKHNNHM | −110.893 | −50.101 | −386.624 | −38.987 | −416.335 |
| D37 | EPAEAVGDWR | −115.415 | −82.482 | −310.680 | −25.919 | −331.430 |
| D38 | APKIPDGE | −81.200 | −63.232 | −119.896 | −26.928 | −147.389 |
| D39 | DNPGHPFI | −68.082 | −54.861 | −263.666 | −27.469 | −289.181 |
| D41 | PQTKTYFSH | −88.321 | −61.955 | −290.732 | −29.587 | −323.296 |
| D43 | LDPIISDR | −88.906 | −70.312 | −219.432 | −29.348 | −232.921 |
| D44 | DNPGHPFIMT | −92.205 | −75.595 | −340.436 | −34.494 | −368.833 |
| D45 | EVPEVYR | −90.566 | −75.773 | −182.124 | −27.160 | −192.760 |
| D47 | IPDGEKVDFD | −103.387 | −76.575 | −221.230 | −37.421 | −247.640 |
The main precursor proteins of 125 peptides were further analyzed using Sankey diagram (Fig. 3). A total of 46 kinds of precursor proteins (are represented by A in Fig. 3) were identified and most of them concentrated on nebulin, titin, muscle-type creatine kinase, hemoglobin, and actin, which were also considered as major proteins in mandarin fish. Interestingly, A17, A6, and A23 were the main precursor proteins to produce 16, 15, and 14 peptides, respectively. The peptides possessing high abundance were primarily from A23, A33, A44, A17, and A6.
3.3. Hydrolysis location of precursor proteins for producing DPP-IV inhibitory peptides
The DPP-IV inhibitory peptides can be obtained from the precursor proteins at multiple hydrolysis locations because of the catalytic action of various proteases from complex microorganisms in Chouguiyu. In this study, the DPP-IV inhibitory peptide profile hydrolyzed from the major precursor proteins was constructed based on corresponding hydrolysis sites (Fig. 4). The A17 and A6 respectively consisted of 2556 and 7376 amino acids, and were hydrolyzed into most kinds of DPP-IV inhibitory peptides at various locations. A23 had 382 amino acids, which were hydrolyzed into peptides at five distinct locations, including 2–11, 20–30, 62–71, 89–96, and 284–292. The hydrolysis locations of A24 for producing peptides were mainly concentrated on three locations, 99–106, 124–131, and 184–193. Similarly, A36 could be hydrolyzed at three locations, including 81–89, 151–157, and 733–742. There were 12 precursor proteins that can produce DPP-IV inhibitory peptides at only one distinct location, including A33 (35–45), A44 (44–57), A46 (37–46), A4 (17–29), A41 (25–34), A37 (236–245), A21 (997–1005), A18 (95–103), A22 (129–140), A3 (92–100), A32 (38–46), and A9 (152–161), while there were respectively two distinct hydrolysis locations in A19 (73–80 and 135–144), A26 (349–356 and 395–402), A31 (36–43 and 120–129), A7 (57–60 and 1020–1027), A15 (679–688 and 692–701), and A29 (251–260 and 292–299).
Fig. 4.
DPP-Ⅳ inhibitory peptide profile of representative precursor proteins during fermentation of Chouguiyu. Darker green indicates higher abundance of the peptides. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. Molecular docking between DPP-IV enzyme and DPP-IV inhibitory peptides
The 30 stable DPP-IV inhibitory peptides with the highest abundance were selected to evaluate the inhibition mechanisms for DPP-IV enzyme using molecular docking (Table 1). The interaction energies (ΔEinteraction) of most peptides were lower than −50 kcal/mol. The binding energies (ΔEbinding) between peptides and DPP-IV enzyme were all lower than −108.907 kcal/mol, indicating the stable binding structure (Wu et al., 2016). The electrostatic energy (ΔEele) was lower than the van der Waals energy (ΔEvdw), playing the major role in binding energy. The docking energies (ΔEdocking) of 30 DPP-IV inhibitory peptides were lower than −36.524 kcal/mol, especially D37, D22, D35, and D1 which possessed the ΔEdocking of −115.415, −111.038, −110.893, and −106.778 kcal/mol, respectively. Interestingly, these DPP-IV inhibitory peptides possessed lower docking energy with the receptor, compared with sitagliptin (-20.369 kcal/mol). Previous study found that lower docking energy indicated stronger affinity between the ligands and their receptor (Tu et al., 2018), probably resulting in more inhibition of DPP-IV enzyme activity. Interestingly, the abundance of D37, D35, and D1 in the d4 group was significantly higher than that in d0 and d8 groups (Table S3), contributing to the highest DPP-IV inhibition activity of peptide extract in the d4 group. Therefore, these four DPP-IV inhibitory peptides were chosen to explore the molecular interaction and surface force with DPP-IV enzyme. Interesting, these DPP-IV inhibitory peptides with sequence of EPAEAVGDWR (D37), IPHESVDVIK (D22), PDLSKHNNHM (D35), and PFGNTHNNFK (D1), exhibited good DPP-IV inhibition effect, possessing the IC50 of 0.10 mM, 2.69 mM, 3.88 mM, and 8.51 mM, respectively (Table 2). Under the same in vitro method, the IC50 on DPP-IV inhibition of D37 was significantly lower than the reported activity of silver carp muscle proteolytic peptide (AALEQTER, 0.6 mM) (Zhang et al., 2019). Besides, the DPP-IV inhibition activity of D37, D22, D35, and D1 was in accordance with their docking energy, suggesting the virtual selection of DPP-IV inhibition peptides according to their docking energy was accurate.
Table 2.
IC50 on DPP-IV enzyme of DPP-IV inhibitory peptides.
| Name | Peptide | IC50 (mM) |
|---|---|---|
| D37 | EPAEAVGDWR | 0.10 |
| D22 | IPHESVDVIK | 2.69 |
| D35 | PDLSKHNNHM | 3.88 |
| D1 | PFGNTHNNFK | 8.51 |
| Positive | Sitagliptin | 0.09 |
3.5. Molecular interaction between DPP-Ⅳ enzyme and DPP-Ⅳ inhibitory peptides
Molecular docking is widely applied to reveal the inhibition mechanism of DPP-IV inhibitory peptides (Gu et al., 2021). Generally, DPP-IV enzyme possesses four binding sites, including S1, S2, S1′, and S2’, among which S1 is a pocket with high hydrophobicity of residues Ser630, Asn710 and His740, while S2 plays important roles in the interactions at the residues of Glu205, Glu206, and Arg125 (Gu et al., 2021; Li et al., 2016). Besides, Tyr547 and Trp629 in the DPP-IV enzyme also play an important role in binding activity (Sagar et al., 2015; Wu et al., 2016). In this study, the energy interactions between the four DPP-IV inhibitory peptides and DPP-IV enzyme are exhibited in Fig. 5. Sixty-one residues in DPP-IV enzyme played a crucial role in the interaction with DPP-IV inhibitory peptides (Table S4). Interestingly, similar crucial residues such as Glu205, Glu206, Arg125, Tyr547, Trp629, Ser630, and His740 were found in this study to participate in the binding of four DPP-IV inhibitory peptides and sitagliptin. Conventional hydrogen bonds, carbon hydrogen bonds, and salt bridges were the major interactions between DPP-IV enzyme and DPP-IV inhibitory peptides, while for sitagliptin, conventional and carbon hydrogen bonds were the major interactions at the residues of Arg125, His126, Arg471 and Ser458 in DPP-IV enzyme. Interestingly, the DPP-IV inhibitory peptides formed more van der Waals with DPP-IV enzyme than sitagliptin, probably leading to more allosteric effects on the DPP-IV enzyme. The residues in DPP-IV enzyme for the formation of conventional hydrogen bonds were focused on Arg, Ser, His and Tyr, and the residues such as Arg and Tyr contributed to the formation of salt bridges, while Ser played a crucial role in the formation of carbon hydrogen bonds. A total of 3 carbon hydrogen bonds, 1 conventional hydrogen bond, 2 salt bridges, and 3 alkyl bonds were found between DPP-IV enzyme and D22. More salt bridges were found in D37 reaching 4, while more hydrogen bonds were observed in D1 and D35, respectively reaching 10 and 5. Interestingly, the hydrogen bonds, salt bridges, and alkyls formed between the DPP-IV enzyme and DPP-IV inhibitory peptides were concentrated on the specific structure (Fig. 5, Fig. S1 and Table S4). The structure of EP- in D37 connected with DPP-IV enzyme by salt bridge at Arg560 and carbon hydrogen bond at Ser473. The structure IPH- in D22 interacted with DPP-IV enzyme by alkyls (Trp62 and Lys463), salt bridge (Arg471), and carbon hydrogen bond (Glu408). The structure of -NHM in D35 formed three kinds of bonds, including Tyr456 for conventional hydrogen bond, Ser473 for carbon hydrogen bond, and Arg560 for salt bridge. For D1, the structure of PF- formed interaction with DPP-IV enzyme through hydrogen bonds, including two conventional hydrogen bonds (Glu408 and Phe461) and one carbon hydrogen bond (Leu410). Previous study found that hydrogen bond and pi-pi interaction formed between the DPP-IV inhibitory peptides and DPP-IV enzyme could well explain the inhibition of DPP-IV enzyme (Gu et al., 2021). Similar results were found in this study that the structures of EP-, IPH-, -NHM, and PF- in the DPP-IV inhibitory peptides played a crucial role in the inhibition of DPP-IV enzyme through hydrogen bond, salt bridge, and alkyl.
Fig. 5.
3D docking complexes and 2D diagrams between the four core DPP-Ⅳ inhibitory peptides and DPP-Ⅳ enzyme using molecular docking.
3.6. Surface force between DPP-Ⅳ enzyme and DPP-Ⅳ inhibitory peptides
The surface force between DPP-IV inhibitory peptides and DPP-IV enzyme is shown in Fig. 6. Aromatic interaction can be evaluated using face force and edge force (Fischer et al., 2008). In this study, the edge force was much stronger than the face force in domain of DPP-IV enzyme and DPP-IV inhibitory peptides. Obvious hydrogen bond (H-bond) interaction was observed in all the domains between DPP-IV enzyme and four DPP-IV inhibitory peptides. The density of interpolated charge (IC) is usually used to evaluate the binding strength of electronic structure (Yim and Klüner, 2008). In this study, the interpolated charge in the interaction region was much slighter, suggesting its weak effect on the interaction between the peptides and DPP-IV enzyme. Similarly, the ionizability was also slight, probably because of the lack of ionizable groups in the peptides and DPP-IV enzyme. Previous studies found that peptide hydrophobicity contributed most to the activity of DPP-IV inhibition (Guo et al., 2020; Song et al., 2017). In this study, all four DPP-IV inhibitory peptides possessed the obvious hydrophobicity (Table S1). Moreover, the interaction region between the DPP-IV enzyme and peptides exhibited clear hydrophobicity, especially near hydrophobicity amino acids, such as Pro, Phe, Tyr, Trp, Ile, Leu, Val in the peptides. Interestingly, the proline at second position of N-terminal showed stronger hydrophobicity than that at the first position, probably due to the less effect of hydrophilic group “-NH” in proline. Previous studies showed that peptides with proline at second position of N-terminal possessed stronger DPP-IV inhibition activity (Silveira et al., 2013; Song et al., 2017). It might result from the high hydrophobicity of proline at this position. Solvent accessible surface (SAS) is the area traced out by the probe sphere center which is related to the van der Waals (Connolly, 1983). In this work, obvious SAS was found in the interaction domain between the DPP-IV enzyme and peptides, mainly due to the abundant van der Waals. It was shown that the surface force including the hydrogen bond interaction, hydrophobicity, aromatic interaction, and SAS, played a major role in the inhibition of DPP-IV enzyme.
Fig. 6.
Surface force analysis between the four core DPP-Ⅳ inhibitory peptides and DPP-Ⅳ enzyme, including (A) aromatic interaction, (B) hydrogen bond, (C) interpolated charge, (D) hydrophobicity, (E) ionizability, and (F) solvent accessible surface.
Molecular interaction and surface force between the DPP-IV enzyme and DPP-IV inhibitory peptides could well explain the inhibition mechanisms of these peptides on DPP-IV enzyme. Lower docking energy indicated stronger affinity between DPP-IV enzyme and peptides, which provided a possibility for their interaction. The hydrophobicity of peptides was helpful in their docking to DPP-IV enzyme because the interaction location concentrated on the hydrophobic pocket in the enzyme. Among the interaction energies, hydrogen bond contributed most to the conformational changes of DPP-IV enzyme, followed by salt bridge and alkyl, eventually leading to the inactivation of DPP-IV enzyme. The peptides with high hydrophobicity and amino acid residues that can form strong hydrogen bond, salt bridge, and alkyl with DPP-IV enzyme may be potential DPP-IV inhibitory peptides using for T2DM remission.
4. Conclusion
Peptide extract of Chouguiyu exhibited a good inhibition effect on DPP-IV enzyme, especially in the fermented groups. A total of 125 DPP-IV inhibitory peptides were identified from Chouguiyu, and most DPP-IV inhibitory peptides in the fermented groups were more abundant than that before fermentation. The 125 peptides were hydrolyzed from 46 precursor proteins at various locations, which were focused on nebulin, titin, muscle-type creatine kinase, hemoglobin, and actin. All the DPP-IV inhibitory peptides possessed nontoxicity and good hydrophobicity, 95 peptides showed good water solubility, while only 74 peptides exhibited stable structures. Four novel DPP-IV inhibitory peptides with the lowest docking energy were selected, including EPAEAVGDWR, IPHESVDVIK, PDLSKHNNHM, and PFGNTHNNFK, with IC50 of 0.10 mM, 2.69 mM, 3.88 mM, and 8.51 mM, respectively. Arg, Ser, His and Tyr were the main residues in DPP-IV enzyme to form the hydrogen bond and salt bridge with the structures of EP-, IPH-, -NHM, and PF- in DPP-IV inhibitory peptides. The surface force including the H-bond interaction, hydrophobicity, aromatic interaction, and SAS, played a major role in the interaction between DPP-IV enzyme and peptides. The peptides with high hydrophobicity and amino acid residues of forming strong hydrogen bond, salt bridge, and alkyl may possess high inhibition activity of DPP-IV enzyme.
CRediT authorship contribution statement
Daqiao Yang: Methodology, Writing – original draft. Chunsheng Li: Writing – review & editing, Conceptualization. Laihao Li: Conceptualization. Yueqi Wang: Data curation. Shengjun Chen: Visualization. Yongqiang Zhao: Data curation. Xiao Hu: Visualization. Hui Rong: Data curation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was funded by the National Key R&D Program of China (2019YFD0901903), the Earmarked Fund for CARS (CARS-46), the Hainan Provincial Natural Science Foundation of China (322QN431), the Young S&T Talent Training Program of Guangdong Provincial Association for S&T, China (SKXRC202210), and the Central Public-interest Scientific Institution Basal Research Fund, CAFS (2020TD69).
Edited by, Dr. Quancai Sun
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crfs.2022.09.025.
Contributor Information
Chunsheng Li, Email: lichunsheng@scsfri.ac.cn.
Laihao Li, Email: laihaoli@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Çağlar A.F., Çakır B., Gülseren 0. LC-Q-TOF/MS based identification and in silico verification of ACE-inhibitory peptides in Giresun (Turkey) hazelnut cakes. Eur. Food Res. Technol. 2021;247(5):1189–1198. [Google Scholar]
- Cian R.E., Nardo A.E., Garzón A.G., Añon M.C., Drago S.R. Identification and in silico study of a novel dipeptidyl peptidase IV inhibitory peptide derived from green seaweed Ulva spp. hydrolysates. LWT--Food Sci. Technol. 2022;154 [Google Scholar]
- Connolly M.L. Solvent-accessible surfaces of proteins and. Nucl. Acids. Sci. New Series. 1983;221(4612):709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- Fischer F.R., Schweizer W.B., Diederich F. Substituent effects on the aromatic edge-to-face interaction. Chem. Commun. 2008;34(34):4031–4033. doi: 10.1039/b809058k. [DOI] [PubMed] [Google Scholar]
- Fu Y., Liu J., Hansen E.T., Bredie W.L.P., Lametsch R. Structural characteristics of low bitter and high umami protein hydrolysates prepared from bovine muscle and porcine plasma. Food Chem. 2018;257:163–171. doi: 10.1016/j.foodchem.2018.02.159. [DOI] [PubMed] [Google Scholar]
- Games P.D., DaSilva E.Q.G., Barbosa M.D.O., Almeida-Souza H.O., Fontes P.P., DeMagalhães-Jr M.J., Pereira P.R.G., Prates M.V., Franco G.R., Faria-Campos A., Campos S.V.A., Baracat-Pereira M.C. Computer aided identification of a Hevein-like antimicrobial peptide of bell pepper leaves for biotechnological use. BMC Genom. 2016;17(S12) doi: 10.1186/s12864-016-3332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R.E., Joseph S. A review of newer treatment approaches for type-2 diabetes: focusing safety and efficacy of incretin based therapy. Saudi Pharmaceut. J. 2014;22(5):403–410. doi: 10.1016/j.jsps.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Gao J., Shen Q., Gao D., Wang Q., Tangyu M., Mao X. Dipeptidyl peptidase-IV inhibitory activity of millet protein peptides and the related mechanisms revealed by molecular docking. LWT--Food Sci. Technol. 2021;138 [Google Scholar]
- Guo H., Richel A., Hao Y., Fan X., Everaert N., Yang X., Ren G. Novel dipeptidyl peptidase-IV and angiotensin-I-converting enzyme inhibitory peptides released from quinoa protein by in silico proteolysis. Food Sci. Nutr. 2020;8(3):1415–1422. doi: 10.1002/fsn3.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X., Yang W., Zhu Q., Zhang G., Zhang X., Liu L., Li X., Hussain M., Ni C., Jiang X. Proteolysis and ACE-inhibitory peptide profile of Cheddar cheese: effect of digestion treatment and different probiotics. LWT--Food Sci. Technol. 2021;145 [Google Scholar]
- Ishak N.H., Shaik M.I., Yellapu N.K., Howell N.K., Sarbon N.M. Purification, characterization and molecular docking study of angiotensin-I converting enzyme (ACE) inhibitory peptide from shortfin scad (Decapterus macrosoma) protein hydrolysate. J. Food Sci. Technol. 2021 doi: 10.1007/s13197-020-04944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W., Zhang C., Ji H. Two novel bioactive peptides from antarctic krill with dual angiotensin converting enzyme and dipeptidyl peptidase IV inhibitory activities. J. Food Sci. 2017;82(7):1742–1749. doi: 10.1111/1750-3841.13735. [DOI] [PubMed] [Google Scholar]
- Lacroix I.M.E., Li-Chan E.C.Y. Isolation and characterization of peptides with dipeptidyl peptidase-IV inhibitory activity from pepsin-treated bovine whey proteins. Peptides. 2014;54:39–48. doi: 10.1016/j.peptides.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Lacroix I., Li-Chan E. Inhibition of dipeptidyl peptidase (DPP)-IV and α-glucosidase activities by pepsin-treated whey proteins. J. Agric. Food Chem. 2013;61(31):7500–7506. doi: 10.1021/jf401000s. [DOI] [PubMed] [Google Scholar]
- Lafarga T., O Connor P., Hayes M. In silico methods to identify meat-derived prolyl endopeptidase inhibitors. Food Chem. 2015;175:337–343. doi: 10.1016/j.foodchem.2014.11.150. [DOI] [PubMed] [Google Scholar]
- Li C., Li W., Li L., Chen S., Wu Y., Qi B. Microbial community changes induced by a newly isolated salt-tolerant Tetragenococcus muriaticus improve the volatile flavor formation in low-salt fish sauce. Food Res. Int. 2022;156 doi: 10.1016/j.foodres.2022.111153. [DOI] [PubMed] [Google Scholar]
- Li C., Zhao Y., Wang Y., Li L., Huang J., Yang X., Zhao Y. Contribution of microbial community to flavor formation in tilapia sausage during fermentation with Pediococcus pentosaceus. LWT--Food Sci. Technol. 2022;154 [Google Scholar]
- Li G., Huan Y., Yuan B., Wang J., Jiang Q., Lin Z., Shen Z., Huang H. Discovery of novel xanthine compounds targeting DPP-IV and GPR119 as anti-diabetic agents. Eur. J. Med. Chem. 2016;124:103–116. doi: 10.1016/j.ejmech.2016.08.023. [DOI] [PubMed] [Google Scholar]
- Li Q., Han L., Zhang B., Zhou J., Zhang H. Synthesis and biological evaluation of triazole based uracil derivatives as novel DPP-4 inhibitors. Org. Biomol. Chem. 2016;14(40):9598–9611. doi: 10.1039/c6ob01818a. [DOI] [PubMed] [Google Scholar]
- Li X., Xie X., Wang J., Xu Y., Yi S., Zhu W., Mi H., Li T., Li J. Identification, taste characteristics and molecular docking study of novel umami peptides derived from the aqueous extract of the clam meretrix meretrix Linnaeus. Food Chem. 2020;312 doi: 10.1016/j.foodchem.2019.126053. [DOI] [PubMed] [Google Scholar]
- Majumdar R.K., Roy D., Bejjanki S., Bhaskar N. An overview of some ethnic fermented fish products of the Eastern Himalayan region of India. J. Ethnic Foods. 2016;3(4):276–283. [Google Scholar]
- Mouritsen O.G., Duelund L., Calleja G., Frøst M.B. Flavour of fermented fish, insect, game, and pea sauces: garum revisited. Int. J. Gastronomy Food Sci. 2017;9:16–28. [Google Scholar]
- Murray N.M., O'Riordan D., Jacquier J., O'Sullivan M., Holton T.A., Wynne K., Robinson R.C., Barile D., Nielsen S.D., Dallas D.C. Peptidomic screening of bitter and nonbitter casein hydrolysate fractions for insulinogenic peptides. J. Dairy Sci. 2018;101(4):2826–2837. doi: 10.3168/jds.2017-13853. [DOI] [PubMed] [Google Scholar]
- Neves A.C., Harnedy P.A., O'Keeffe M.B., Alashi M.A., Aluko R.E., FitzGerald R.J. Peptide identification in a salmon gelatin hydrolysate with antihypertensive, dipeptidyl peptidase IV inhibitory and antioxidant activities. Food Res. Int. 2017;100:112–120. doi: 10.1016/j.foodres.2017.06.065. [DOI] [PubMed] [Google Scholar]
- Nongonierma A.B., Hennemann M., Paolella S., FitzGerald R.J. Generation of wheat gluten hydrolysates with dipeptidyl peptidase IV (DPP-IV) inhibitory properties. Food Funct. 2017;8:2249–2257. doi: 10.1039/c7fo00165g. [DOI] [PubMed] [Google Scholar]
- Rahfeld J., Schierborn M., Hartrodt B., Neubert K., Heins J. Are diprotin A (Ile-Pro-Ile) and diprotin B (Val-Pro-Leu) inhibitors or substrates of dipeptidyl peptidase IV? Biochim. Biophys. Acta. 1991;1076(2):314–316. doi: 10.1016/0167-4838(91)90284-7. [DOI] [PubMed] [Google Scholar]
- Sagar S.R., Agarwal J.K., Pandya D.H., Dash R.P., Nivsarkar M., Vasu K.K. Design, synthesis and biological evaluation of novel pyrazolo-pyrimidinones as DPP-IV inhibitors in diabetes. Bioorg. Med. Chem. Lett. 2015;25(20):4428–4433. doi: 10.1016/j.bmcl.2015.09.015. [DOI] [PubMed] [Google Scholar]
- Silveira S.T., Martínez-Maqueda D., Recio I., Hernández-Ledesma B. Dipeptidyl peptidase-IV inhibitory peptides generated by tryptic hydrolysis of a whey protein concentrate rich in β-lactoglobulin. Food Chem. 2013;141(2):1072–1077. doi: 10.1016/j.foodchem.2013.03.056. [DOI] [PubMed] [Google Scholar]
- Singh B.P., Vij S. In vitro stability of bioactive peptides derived from fermented soy milk against heat treatment, pH and gastrointestinal enzymes. LWT--Food Sci. Technol. 2018;91:303–307. [Google Scholar]
- Song J.J., Wang Q., Du M., Ji X.M., Mao X.Y. Identification of dipeptidyl peptidase-IV inhibitory peptides from mare whey protein hydrolysates. J. Dairy Sci. 2017;100(9):6885–6894. doi: 10.3168/jds.2016-11828. [DOI] [PubMed] [Google Scholar]
- Sousa S.F., Ribeiro A.J.M., Coimbra J.T.S., Neves R.P.P., Martins S.A., Moorthy N.S.H.N., Fernandes P.A., Ramos M.J. Protein-ligand docking in the new millennium-A retrospective of 10 years in the field. Curr. Med. Chem. 2013;20:2296–2314. doi: 10.2174/0929867311320180002. [DOI] [PubMed] [Google Scholar]
- Tu M., Qiao X., Wang C., Liu H., Cheng S., Xu Z., Du M. In vitro and in silico analysis of dual-function peptides derived from casein hydrolysate. Food Sci. Hum. Wellness. 2021;10(1):32–37. [Google Scholar]
- Tu M., Wang C., Chen C., Zhang R., Liu H., Lu W., Jiang L., Du M. Identification of a novel ACE-inhibitory peptide from casein and evaluation of the inhibitory mechanisms. Food Chem. 2018;256:98–104. doi: 10.1016/j.foodchem.2018.02.107. [DOI] [PubMed] [Google Scholar]
- Vilcacundo R., Martínez-Villaluenga C., Hernández-Ledesma B. Release of dipeptidyl peptidase IV, α-amylase and α-glucosidase inhibitory peptides from quinoa (Chenopodium quinoa Willd.) during in vitro simulated gastrointestinal digestion. J. Funct.Foods. 2017;35:531–539. [Google Scholar]
- Wang K., Yang X., Lou W., Zhang X. Discovery of dipeptidyl peptidase 4 inhibitory peptides from Largemouth bass (Micropterus salmoides) by a comprehensive approach. Bioorg. Chem. 2020;105 doi: 10.1016/j.bioorg.2020.104432. [DOI] [PubMed] [Google Scholar]
- Wu Q., Du J., Jia J., Kuang C. Production of ACE inhibitory peptides from sweet sorghum grain protein using alcalase: hydrolysis kinetic, purification and molecular docking study. Food Chem. 2016;199:140–149. doi: 10.1016/j.foodchem.2015.12.012. [DOI] [PubMed] [Google Scholar]
- Wu W., Hao J., Domalski M., Burnett D.A., Pissarnitski D., Zhao Z., Stamford A., Scapin G., Gao Y., Soriano A., Kelly T.M., Yao Z., Powles M.A., Chen S., Mei H., Hwa J. Discovery of novel tricyclic heterocycles as potent and selective DPP-4 inhibitors for the treatment of type 2 diabetes. ACS Med. Chem. Lett. 2016;7(5):498–501. doi: 10.1021/acsmedchemlett.6b00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Yao Y., Xu X., Wang M., Pan M., Ji S., Wang L. Identification and quantification of DPP-IV-inhibitory peptides from hydrolyzed-rapeseed-protein-derived Napin with analysis of the interactions between key residues and protein domains. J. Agric. Food Chem. 2019;67(13):3679–3690. doi: 10.1021/acs.jafc.9b01069. [DOI] [PubMed] [Google Scholar]
- Yang D., Li C., Li L., Chen S., Hu X., Xiang H. Taste mechanism of umami peptides from Chinese traditional fermented fish (Chouguiyu) based on molecular docking using umami receptor T1R1/T1R3. Food Chem. 2022;389 doi: 10.1016/j.foodchem.2022.133019. [DOI] [PubMed] [Google Scholar]
- Yang D., Li C., Li L., Wang Y., Wu Y., Chen S., Zhao Y., Wei Y., Wang D. Novel insight into the formation mechanism of umami peptides based on microbial metabolism in Chouguiyu, a traditional Chinese fermented fish. Food Res. Int. 2022;157 doi: 10.1016/j.foodres.2022.111211. [DOI] [PubMed] [Google Scholar]
- Yang D., Li L., Li C., Chen S., Deng J., Yang S. Formation and inhibition mechanism of novel angiotensin I converting enzyme inhibitory peptides from Chouguiyu. Front. Nutr. 2022;9 doi: 10.3389/fnut.2022.920945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Liu S., Lv J., Sun Z., Xu W., Ji C., Liang H., Li S., Yu C., Lin X. Microbial succession and the changes of flavor and aroma in Chouguiyu, a traditional Chinese fermented fish. Food Biosci. 2020;37 [Google Scholar]
- Yim W., Klüner T. Atoms-in-molecules analysis for planewave DFT calculations-A numerical approach on a successively interpolated charge density grid. J. Comput. Chem. 2008;29(8):1306–1315. doi: 10.1002/jcc.20889. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Wang Y., Li C., Li L., Yang X., Wu Y., Zhao Y. Novel insight into physicochemical and flavor formation in naturally fermented tilapia sausage based on microbial metabolic network. Food Res. Int. 2021;141 doi: 10.1016/j.foodres.2021.110122. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu H., Hong H., Luo Y. Purification and identification of dipeptidyl peptidase IV and angiotensin-converting enzyme inhibitory peptides from silver carp (Hypophthalmichthys molitrix) muscle hydrolysate. Eur. Food Res. Technol. 2019;245(1):243–255. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.