Summary
Matching the treatment to an individual patient’s tumor state can increase therapeutic efficacy and reduce tumor recurrence. Circulating tumor cells (CTCs) derived from solid tumors are promising subjects for theragnostic analysis. To analyze how CTCs represent tumor states, we established cell lines from CTCs, primary and metastatic tumors from a mouse model and provided phenotypic and multiomic analyses of these cells. CTCs and metastatic cells, but not primary tumor cells, shared stochastic mutations and similar hypomethylation levels at transcription start sites. CTCs and metastatic tumor cells shared a hybrid epithelial/mesenchymal transcriptome state with reduced adhesive and enhanced mobilization characteristics. We tested anti-cancer drugs on tumor cells from a metastatic breast cancer patient. CTC responses mirrored the impact of drugs on metastatic rather than primary tumors. Our multiomic and clinical anti-cancer drug response results reveal that CTCs resemble metastatic tumors and establish CTCs as an ex vivo tool for personalized medicine.
Subject areas: Precision medicine, Cancer, Omics, Transcriptomics
Graphical abstract

Highlights
-
•
Primary, CTC and metastatic cell lines from mouse models were directly compared
-
•
Multiomic and phenotypic data indicate circulating cells resemble metastatic cells
-
•
CTCs and metastasis tumors from a patient similarly respond to anti-cancer drugs
-
•
CTCs are thus potentially useful for screening individual patient drug responses
Precision medicine; Cancer; Omics; Transcriptomics
Introduction
Metastasis is a complex cellular process that is associated with poor patient survival and is a major cause of cancer-related death (Siegel et al., 2020). Circulating tumor cells (CTCs) are cells disseminated from solid tumor sites into the bloodstream and represent a transition state that facilitates metastasis progression (Aceto et al., 2015). Thus, comprehensive characterization of circulating, primary, and metastatic tumor cells is essential for the elucidation of mechanisms underlying cancer metastasis and may provide important new clinical avenues. However, a systematic analysis and comparison of tumor cells at these stages using combined genomic, epigenetic, and transcriptomic approaches has not yet been reported.
To metastasize, tumor cells undergo a transition from the epithelial toward the mesenchymal state (Pastushenko et al., 2018; Tsai et al., 2012a). Metastasis is facilitated by the epithelial-mesenchymal transition (EMT) and colonization of a new tissue involves the reverse process, i.e., mesenchymal-epithelial transition (MET) (Lee et al., 2018; Tsai et al., 2012b). However, recent studies have shown that metastasizing cancer cells exist in an epithelial/mesenchymal (E/M) hybrid state (Cheung et al., 2013; Mekhdjian et al., 2017; Pastushenko et al., 2018; Puram, 2017). By doing so, cancer cells retain epithelial markers and take advantage of both phenotypes. This proposition is in line with the findings that metastatic cells often retain E-cadherin expression (Aceto et al., 2014; Cheung et al., 2013) and undergo collective migration during cancer invasion (Gaggioli et al., 2007; Hidalgo-Carcedo et al., 2011; Manning et al., 2015). A plausible way to resolve whether tumor cells use a complete or a partial/transient two-state transition to metastasize is to analyze the intermediate states en route from the primary to the metastatic site. CTCs represent an “intermediate state” for metastasis. A previous study has shown that the majority of CTCs in Lgr5Cre/ERKrasG12D/p53KO mice were triple-negative (VCAM1-/CD51-/CD61-), indicating that the cells were in an E/M hybrid state (Pastushenko et al., 2018). In line with this finding, we have recently demonstrated that the adhesion protein claudin-11 is expressed in CTC clusters of squamous cell carcinoma, supporting the existence of an E/M hybrid phenotype in CTCs (Li et al., 2019).
In addition to being an intermediate state for metastasis, CTCs possess important clinical features. CTCs can potentially provide an early-stage theragnostic opportunity to monitor tumor progression/evolution, and to predict the genotype and phenotype of metastatic tumors. Accordingly, several patient-derived CTC cell lines have been established (Cayrefourcq et al., 2015; Hodgkinson et al., 2014; Khoo et al., 2015; Klameth et al., 2017; Yu et al., 2014; Zhang et al., 2013). Studies on CTC-derived cell lines have provided insight into the origins of the tumorigenicity (Hodgkinson et al., 2014). For instance, the gene mutation landscape of cultured CTCs was reported to be concordant with the original tumor (Hwang et al., 2017) and to recapitulate the evolving features of the original tumor (Drapkin et al., 2018). In addition, patient CTC-derived xenograft mouse models have been established as a translational research platform to improve understanding of cancer biology and test novel therapies (Hidalgo et al., 2014). For patients with non-metastatic lung cancer, CTC-derived xenograft mouse models have been recently generated using CTCs isolated from primary tumor patient-derived xenograft (ptPDX) (Suvilesh et al., 2022). This approach enables tumor cells to expand in two generations of mice. The resulting CTC-derived xenograft mice were found to be more aggressive in tumor growth compared to their parental ptPDX and had a subset of tumor cells that matched patients’ non-xenografted metastases in transcriptome profiling (Suvilesh et al., 2022). Therefore, CTCs-derived xenograft mice can serve as models for aggressive tumors. To date, the study of such tumor models has been limited to the tumors growing at the site of inoculation, and the investigation of circulating and metastatic tumors originating from CTC-derived xenografts remains limited.
It has been reported that tumor cells disseminate into the bloodstream at early stages and throughout metastasis, rather than only at late stages of cancer progression (Podsypanina et al., 2008). CTC-based liquid biopsy has shown the potential usefulness of these cells for early cancer detection, including colon cancer (Tsai et al., 2016) and breast cancers (Trapp et al., 2018). The number of CTCs in the bloodstream is highly correlated with clinical outcomes in individual cancer patients. Quantitative analysis of CTCs indicates that these cells are a reliable and real-time biomarker that can monitor treatment efficacy as well as treatment guidance, for example, in first-line chemotherapy and endocrine therapy in hormone receptor-positive, HER2-negative breast cancer patients (Bidard et al., 2021). Ex vivo cultivation of CTC-derived cell lines further indicates that CTCs provide a minimally invasive method for ex vivo anti-cancer drug testing (Yu et al., 2014). Applications of CTC isolation and quantification, however, are limited to date, despite their clinical potential. The exact relationship between CTCs and metastatic tumors is still unclear, as is whether CTCs can act as a proxy for metastatic tumors for theragnostic analyses. The scarcity of CTCs and the high background of leukocytes have significantly hampered detailed analysis of these questions, as has difficulty with in vitro CTC cultivation (Yu et al., 2014).
To overcome these issues, we previously reported an efficient CTC isolation platform, CMx (“Cells captured in Maximum”). CMx comprises a noncovalent antibody-conjugated, lipid-coated surface combined with a micro-patterned chaotic-mixing microfluidics configuration (Chen, 2016; Lai et al., 2014; Wu et al., 2013; Yeh et al., 2018). Specifically, the biomimetic lipid membranes promote multivalent antibody and antigen interactions for CTC capture. Captured CTCs can be gently released from the non-covalently bound lipid surface using air foam to preserve cell viability. This platform was further improved (Yeh, 2018) and used to isolate CTCs in this work. The improved platform, LIPO, contains a liposome-lipid bilayer coating to further enhance the multivalency and maximize preservation of cell viability (Yeh, 2018).
Here, we describe a process that enabled us to establish CTC-derived xenograft mice to model aggressive metastatic tumors. From these metastatic mice we established primary, circulating, and metastatic tumor-derived cell lines and characterized these lines at the phenotypic, genomic, epigenomic, and transcriptomic levels. Finally, as a proof-of-concept, we used primary tumor and CTC samples from a metastatic breast cancer patient for ex vivo drug testing. The results demonstrated that the drug sensitivities of CTCs reflect those of metastatic tumors, whereas the response of cells sampled from the primary tumor resembled that of the primary tumor. These and other data indicate that CTCs and metastatic tumors are more similar to one another than to primary tumors in mouse cell lines at both the phenotypic and multiomic levels. In sum, these observations suggest that CTCs constitute an accessible cell type for efficient cancer drug screening to support personalized cancer therapy.
Results
Development of CTC-Derived metastatic xenograft mice and Cell lines
Because of technical challenges in generating primary, circulating, and metastatic tumor cell lines directly from advanced cancer patients, we set to establish CTC-derived xenograft mice to model aggressive metastatic tumors. The experimental design was illustrated in Figure 1A. In brief, the breast-cancer cell line 4T1-hHER2-Luc expressing human HER2 and the luciferase marker was inoculated into NOD/SCID-gamma (NSG) mice to generate cell-line-derived xenograft CDx mice. CTCs from the resulting mice were isolated and inoculated into the mammary fat pad of naive NSG mice to establish CTC-derived PDx models. The primary, circulating, and metastatic tumor cells of the CTC-derived PDx mice were harvested to establish the corresponding cell lines for analyses. To gain insight into tumor development and metastasis, the cell lines were characterized using phenotypic and multiomic assays. Technically, the first batch of mice served as a murine breast-cancer cell line-derived xenograft (CDx) orthotopic model system. They were considered as “patient” models whereas the second batch of mice served as a “patient-derived xenograft” (PDx) model system for further investigation.
Figure 1.
Establishment of primary, circulating and metastatic tumor cell lines
(A) Workflow for establishment and characterization of primary, circulating and metastatic tumor cell lines from a metastatic mouse model. The process included three steps: (1) The 4T1-hHER2-Luc cancer cell line was inoculated into NSG mice to generate the first batch of orthotopic spontaneous metastatic xenografts. These mice were called “patient” models. (2) CTCs derived from the “patient” mice were inoculated and in vivo expanded in a second batch of NSG mice to generate CTC-derived “patient” xenografts, CTC-PDx. (3) Primary, circulating, and metastatic tumor cells from CTC-PDx mice were used to establish respective cell lines. Subsequent phenotypic assays and multiomic analyses including exome, DNA methylome, and transcriptome sequencing of the resulting cell lines was performed.
(B) Isolation efficiency (mean ± SD) of 4T1-hHER2-Luc cancer cells by the LIPO platform without (Chip only) and with anti-mouse EpCAM antibody coating (Chip + Ab).
(C) Images of 4T1-hHER2-Luc cells released from the LIPO platform tested using the LIVE/DEAD assay. The 4T1-hHER2-Luc cells captured and subsequently released from the LIPO platform were assessed for viability using the LIVE/DEAD assay. BF: bright field; Calcein AM: green (live marker); EthD-1: red (dead marker).
(D) Representative immunostaining images of cells isolated from the blood of a CDx mouse using the LIPO platform were shown. Distinct classes of single CTCs, clustered CTCs, and white blood cells (WBC) were found. Green: anti-hHER2, Red: anti-mCD45, blue: DAPI. Scale bar: 10 μm.
(E and F) The numbers of single CTCs (E) and clustered CTCs (F) isolated from 75 μL blood from CDx mice in weeks 1, 3, and 4 after inoculation. Numbers in parentheses indicate the number of mice tested. ∗p< 0.05, ∗∗p< 0.01, ∗∗∗p< 0.001 (Student’s ttest, mean ± SD).
Establishing a 4T1-hHER2-Luc cell-capturing microfluidic platform
We confirmed expression of exogenous hHER2 and endogenous mouse pan-cytokeratin (pan-CK) in the parental 4T1-hHER2-Luc cells using RT-PCR and immunocytochemistry (Figure S1). We then examined the efficiency of the LIPO platform coated with the anti-mouse EpCAM antibody for 4T1-hHER2-Luc cell isolation (Yeh, 2018; Method details). Cell isolation efficiency (calculated as capture efficiency x release efficiency, Method details) of the platform coated with and without the antibody was 71.20 ± 0.06% and 0.27 ± 0.002%, respectively (Figure 1B). Notably, cells released from the LIPO platform after being captured had high viability, as evidenced by calcein AM staining and negative EthD-1 staining using a LIVE/DEAD assay (Figure 1C). The platform was used to isolate CTCs in subsequent studies.
Monitoring CTC dynamics in a 4T1-hHER2-Luc cell line-derived xenograft (CDx)
As described in Figure 1A, to create a metastatic mouse model, parental 4T1-hHER2-Luc cells were inoculated into the inguinal mammary fat pads of NSG mice to follow spontaneous metastasis in 3–4 weeks (n = 12 for Group B, sacrificed 3 weeks after inoculation; n = 12 for Group C, sacrificed 4 weeks after inoculation; n = 4, negative control). Tumor progression in the resulting CDx mice was examined by monitoring tumor size and metastasis using the IVIS Spectrum in vivo imaging system before and after tumor cell inoculation at different time points (Figure S2A). Simultaneously, CTCs were isolated using the LIPO platform from 75 μL of blood drawn from each mouse. To discriminate CTCs from leukocytes, cells were co-stained with the anti-hHER2 antibody and the leukocyte-specific anti-mouse CD45 antibody. Cells with positive hHER2 and negative mCD45 staining were classified as CTCs, whereas cells negative for hHER2 and positive for mCD45 staining were leukocytes (Figure 1D). Both single and clustered CTCs were isolated, demonstrating the ability of the LIPO platform to isolate CTCs from low volumes of blood. The numbers of single CTCs (Figure 1E) and CTC clusters (Figure 1F) increased significantly in the third and fourth weeks after tumor cell inoculation, respectively, whereas almost no single or clustered CTCs were detected in the first week. The tumor size and metastasis condition of the CDx mice were also examined. In all 24 CDx mice, tumor mass started to increase continuously one week after inoculation (Figures S2A and S2B), whereas pulmonary metastasis was detected in the third-week post-inoculation (Figure S2A). These mice showed no significant change in body weight before serious metastasis in the fourth week (Figure S2E). In contrast, no tumorigenesis was detected in any negative control mouse (Figure S2A). Positive hHER2 expression in the primary tumors and metastatic lung tumors of all CDx mice was validated by RT-PCR and hHER2 immunohistochemistry assays (Figure S2), confirming that the tumors originated from inoculated parental 4T1-hHER2-Luc cells. Our data indicate that CTCs can be captured from an ultralow-volume of blood (75 μL) to monitor tumor progression in mice.
Successful development of xenografts from mouse-derived CTC-PDx
Next, we isolated CTCs derived from the “patient” CDx mice during early and late metastatic phases in the third and fourth weeks, respectively. We used the CTCs to generate CTC-patient-derived xenograft (CTC-PDx) mice (Figure 1A). To this end, approximately 6x102 and 1.1x103 CTCs were obtained from 750 μL of blood per mouse in Group B (3-week, early metastasis) and Group C (4-week, late metastasis), respectively. To implement tumor divergence, CTCs isolated from three randomly picked mice within the same Group were mixed and inoculated into the second batch of NSG mice to create No.1-4 and No. 5–8 CTC-PDx mice, respectively, as illustrated in Figure 1A. All resulting CTC-PDx mice showed tumor growth and spontaneous metastasis within three weeks of CTC inoculation—a 100% success rate (Figure S4). Our platform can, therefore, successfully capture and release viable CTCs from “patient” CDx mice that can then be used to generate in vivo CTC-PDx. This system potentially allows CTCs derived from human patients to be used for personalized clinical applications.
Development of primary/CTC/metastatic tumor cell lines from CTC-PDx
To gain insight into tumor evolution and metastasis, we generated long-term cell cultures from primary, circulating, and metastatic tumor cells from each CTC-PDx mouse. A total of nine cell lines, consisting of triple sets of primary tumor- (5-PRI, 7-PRI, and 8-PRI), CTC- (1-CTC, 3-CTC, and 6-CTC), and metastatic tumor- (3-META, 5-META, and 7-META) derived cell lines, were successfully established (Figure 2A and S5). These cell lines represented specific tumor stages, allowing us to examine and compare the phenotypic and molecular alterations between stages at systematic levels. Specifically, three matched pairs, 3-CTC and 3-Meta, 5-PRI and 5-META, and 7 PRI and 7-META, derived from No. 3, 5, and 7 mice, respectively, were used for tracking cancer cell progression in the same lineage. The cell lines at passage 2 were used for the following analyses, unless noticed otherwise.
Figure 2.
Phenotypic characterization of the primary tumor, CTC, and metastatic tumor-derived sublines and the parental 4T1-hHER2-Luc cell line
(A) Representative images of the parental 4T1-hHER2-Luc cell line (a), primary tumor-derived cell line 7-PRI (b), CTC-derived cell line 3-CTC (c), and metastasis-derived cell line 5-META (d).
(B) Cell cycle distribution of the parental cell line, and the indicated sublines 12 h after the 50 ng/mL nocodazole treatment is shown on the left; the average cell cycle distribution from the primary (PRI), circulating (CTC), and metastatic (META) tumor sublines after nocodazole synchronization is shown on the right (mean ± SD). No significant difference was observed among sublines using Chi-square analysis.
Phenotypic and multiomic analyses of primary, circulating, and metastatic tumor cell lines
Circulating and metastatic tumor cells spread less and are more adherent than primary tumor cells
We examined the morphology of these cell lines on tissue culture plates. A spreading and elongated cell shape with contiguous cell-cell adhesion was observed in the primary tumor-derived sublines, similar to that of the parental cell line (Figures 2Aa and b, and S5A). In contrast, cell spreading and cell-cell adhesion in CTC- and metastatic tumor-derived sublines was limited (Figures 2Ac and d, S5B, and S5C). Instead, more than 90% of CTCs and about 60-80% of metastatic tumor cells showed a spherical morphology. Despite the morphological difference, no distinct variation in the cell cycle distribution with nocodazole synchronization was found among the sublines (Figure 2B).
CTC-derived cell lines shares more common variants with metastatic tumor-derived cell lines than with primary tumor-derived cell lines
Because metastasis is a complex process involving microevolutionary shifts, including genome instability, epigenetic change, and transcriptomic alterations, we next examined genomic, epigenetic, and transcriptomic differences in all sublines and the parental line. Specifically, we analyzed the genomic profiles using whole exome sequencing (WES). The preexisting mutation in Trp53 was identified in all sublines and the parental line (Table S1), consistent with the previous finding that the 4T1 tumor cell had a frameshift insertion in Trp53 (Schrors et al., 2020). Tp53, the human counterpart of Trp53, is a tumor suppressor gene important for genome stability and is frequently mutated in breast cancer patients (Cancer Genome Atlas, 2012). Hundreds of INDEL and SNP mutations, which were absent in the parental line, were found in different sublines (Figure 3A and Table S1). The chromosomal distribution of the mutations is shown in a Circos plot (Figure 3B). Very few common mutations were identified within cell lines derived from the same tumor stage of different mice, even though they all originated from the parental 4T1-hHER2-Luc cell line (Figure 3A). This indicates that stochastic mutations occurred and accumulated at various stages during tumor cell proliferation and progression. By comparing sublines derived from the same mouse, the concurrent mutation analysis revealed that the CTC- and metastatic tumor cell-derived sublines shared more common variants (89 between 3-CTC and 3-META) than the primary and metastatic tumor cell-derived sublines (42 for between 5-PRI and 5-META; 14 between 7-PRI and 7-META) (Figure 3C), consistent with the idea that the genetic status of metastatic tumor cells is closer to CTCs than to the primary tumor.
Figure 3.
Whole exome NGS sequencing results of the primary, circulating and tumor cell lines derived from CTC-PDx mice
(A) Numbers of newly acquired genomic variants in each subline, and numbers of shared variants (intersect) among sublines.
(B) A Circos plot summarizing genomic variant density and distribution of the indicated tumor cell lines. The order of cell lines from outer to inner is 3-META, 5-META, 7-META, 1-CTC, 3-CTC, 6-CTC, 5-PRI, 7-PRI and 8-PRI. The black/red dot represents sparse/dense genomic variants in the chromosome coordinates shown on the outermost area of the plot.
(C) Concurrent mutation analyses between cell lines of the same lineage. The number in the intersection of two circles represents the number of common variants between the indicated cell lines. The percentage of overlap in No. 3, 4 and 7 lineages is 18.31%, 7.87%, and 1.74%, respectively. See extended data in Table S1.
The DNA methylome of CTC-derived sublines is more similar to metastatic than to primary tumor-derived sublines
Following genomic profiling, we used bisulfite sequencing to examine the DNA methylomes of primary, circulating, and metastatic tumor-derived sublines (Method details, Figure S6A). To facilitate the analysis, DNA methylation data from sublines at the same tumor stage were averaged (e.g., pooling all three primary tumor-derived sublines into one). A similar level of methylation density in CpG islands was observed among these sublines (Figure S6B). DNA methylation patterns near transcription start sites are associated with gene expression (Jones, 2012). We, therefore, analyzed methylomic changes (hyper- versus hypomethylation) around the transcription start site in sublines versus the parental line (Figure 4A). Both hypomethylation and hypermethylation distribution curves in all sublines showed maximal peaks near transcription start sites, with higher hypomethylation than hypermethylation levels. Notably, the hypomethylation curves of CTC- and metastatic tumor-derived sublines were much closer to each other than to primary tumor-derived cell lines. A similar trend was observed for the hypermethylation curve, albeit to a lesser degree.
Figure 4.
Methylome sequencing analysis of cancer cell lines derived from CTC-PDx mice
(A) The level and distribution of significant methylation (hypermethylation) and demethylation (hypomethylation) changes, relative to the parental cell line, for primary tumor (PRI)-, CTC- and Metastasis (META)-derived cell lines near the transcription start sites. Solid and dashed lines indicate hypomethylation and hypermethylation, respectively.
(B) Numbers of significant methylation changes between parental cell line (control) and primary tumor (PRI), CTC, and metastasis (META)-derived cell lines are shown.
(C) The relative methylation level of 76 specific CpG islands in primary tumor (PRI)-, CTC- and metastasis (META)-derived cell lines is shown. The 76 CpG island sites, which were only significantly different when comparing primary tumor and CTC lines, but not primary and tumor-metastasis lines, are arranged along the X axis. Average methylation changes relative to the parental cell for the 76 CpG islands are shown on the Y axis. A negative methylation value indicates lower methylation than the parental line. See extended data in Table S2.
Next, we compared differential methylation sites among the sublines. The methylome of CTC-derived sublines was significantly closer to metastatic tumor-derived sublines in terms of methylation sites, as there were only 67 diverse methylation sites in between, whereas there were 18,417 sites between CTC-derived sublines and primary tumor-derived sublines, and 2,203 sites between the metastatic tumor-derived sublines and primary tumor-derived sublines (Figure 4B and Table S2). The reduction of methylation diversity between primary and metastatic tumor-derived sublines suggested that epigenetic divergence occurred during tumor cell dissemination into the bloodstream and that such divergence was partially reduced during tumor colonization. To gain insight into the nature of such changes, we selected methylation sites that are differentially methylated (q <0.05, >25%) when primary tumor- and CTC-derived sublines are compared, but are similarly methylated in primary tumor- and metastatic tumor-derived lines. A total of 76 CpG islands fit the criteria (Figure 4C). The methylation level of these CpG sites in primary tumor, CTC, and metastatic tumor-derived sublines was measured relative to the parental cell line. These islands displayed a similar trend with decreasing methylation levels in primary tumor-derived sublines, further reduction in CTC-derived sublines, and slightly recovered methylation levels in metastatic tumor-derived sublines. As a result, CTC- and metastatic tumor-derived sublines exhibited a more closely related hypo-methylated state whereas primary tumor sublines had relatively distinct methylation levels. These data, together with the methylation change distribution pattern and level around the transcription start sites (Figure 4A), suggests that significant hypomethylation occurs during the transition from primary to CTCs in metastasis.
Transcriptome profiles in CTC-derived sublines are more similar to metastatic than to primary tumor-derived sublines
RNA sequencing of all sublines was performed to explore transcriptomic evolution during cancer progression. We compared the results with RNA-seq data from the breast cancer cell line 4T1 and other tumor cell types in the database using multi-dimensional scaling (MDS) analysis. All sublines were clustered with 4T1 and closely related to colon cancer cell lines, but segregated from pancreatic and lung cell lines (Figure 5Aa). To characterize tumor stage-specific properties of the sublines, RNA-seq data from the same stage were compared to measurements from different stages. A total of 245 differentially expressed genes (DE-Gs, with Padj< 0.05) were found (Table S3). When these genes were examined for clustering distance among nine sublines using correlation and average-linkage methods, the sublines of the same tumor stage displayed a closer relationship to one another than those of different tumor stages (Figure 5B). Notably, when sublines of different tumor stages were compared, the CTC and META groups were more similar to one another than to the PRI group (Figure 5B). The transcriptomes of circulating and metastatic tumor-derived sublines were, therefore, more closely related to one another than to primary tumor-derived cell lines. Likewise, when sublines of the same lineage were compared using the MDS analysis, the distance between META and CTC of mouse no. 3 was closer than META to PRI of No. 5 and No. 7 mice (Figure 5Ab).
Figure 5.
Results of RNA sequencing analysis and downstream validation of the 4T1-derived cancer cell lines
(A) (a) Multi-dimensional scaling (MDS) plot of nine sublines derived from the CTC-PDx mice compared to the 4T1 cell line, and pancreatic, lung, and colon tumor cell lines is shown on the left. A zoomed-in view of colon and breast tumor cell line clusters was shown on the right. Data for the 4T1 cell line (Breast Cancer) was obtained from the link: https://www.ebi.ac.uk/ena/browser/view/ERR454052; CT26 (Colon Cancer): https://www.ebi.ac.uk/ena/data/view/SRX5833199; MC38 (Colon Cancer): https://www.ebi.ac.uk/ena/data/view/SRX5833178;A-8 (Pancreatic Cancer) https://www.ebi.ac.uk/ena/data/view/SRX7114429; K14-L (Lung tumor): https://www.ebi.ac.uk/ena/data/view/SRX4606402; K14-H (Lung tumor): https://www.ebi.ac.uk/ena/data/view/SRX4606404. (b-c) MDS plots of META and PRI or CTC sublines from same lineage based on TPM values in RNA-seq results (b), or TPM of EMT-related genes (c).
(B) Hierarchical clustering analysis of primary, circulating and metastatic tumor-derived sublines. Differentially expressed genes (DE-Gs) based on the comparison between PRI, CTC and META cell lines using Padj<0.05 threshold were used to generate heatmap. The heatmap was generated using ClustVis (https://biit.cs.ut.ee/clustvis/). The clustering distance is calculated with correlation and average methods between cell lines (column) and genes (rows).
(C) The EMT score analysis of all sublines. The average EMT scores of PRI, CTC, and META-derived sublines and individual sublines were shown. An EMT score closer to 1.0 represent more mesenchymal-like (Mes) characteristic, whereas an EMT score closer to −1.0 represent more epithelial-like (Epi) characteristic.
(D and E) Tables showing the TPM values of selected epithelial markers (D, green color) and mesenchymal markers (E, blue color) in the indicated cell lines derived from CTC-PDx mice based on DESeq2 analysis of RNAseq data (mean ± SD; ∗: p< 0.05, ∗∗: p< 0.01, for PRI versus CTC or META; #: p< 0.05, ##: p< 0.01 for CTC versus META based on DESeq2 analysis and validated using qRT-PCR shown in Figure S8B). See extended data in Tables S3, S4, and S5.
We next analyzed DEG (Padj< 0.05) between sublines using Gene Ontology (GO) term enrichment analysis. The GO analysis revealed “cell adhesion” and “homophilic cell adhesion via plasma membrane adhesion molecules” as the category of biological process that were significantly altered (Figure S7). Of interest, they showed down-regulation and up-regulation, respectively, in circulating or metastatic tumor cell lines when compared with primary cell lines. Specifically, adhesion molecules afadin (Afdn), desmoglein3 (Dsg3), integrin beta 6 (Itgb6), periostin (Postn), Tnfaip6, EpCAM and cadherin 1 (Cdh1) in the category of “cell adhesion” were downregulated in CTC-derived sublines, whereas fibronectin leucine rich transmembrane protein 2 (Flrt2), meltrin alpha (Adam12), cadherin 17 (Cdh17) and integrin alph2 (Itga2) were downregulated specifically in metastatic tumor-derived sublines (Table S4). This result suggests progressive loss in the diversity of cell adhesion mechanisms from primary to circulating, and to metastatic tumor stages. In contrast, a distinct category of adhesion molecules involving homophilic cell adhesion was upregulated in circulating and metastatic tumor-derived sublines, compared with the primary tumor-derived sublines (Figure S7). These molecules included calsyntenin 3 (Clstn3), protocadherin gamma subfamily A (Pcdhga1), protocadherin alpha9 (Pcdha9), cadherin-like 24 (Cdh24), and cadherin11 (Cdh11) (Table S4). The switch of cadherin expression in epithelial cells from E-cadherin to N-cadherin or CDH11 has been associated with EMT and tumor progression (Hazan et al., 2004; Sarrio et al., 2008).
We then analyzed transcriptomic data with an emphasis on EMT-related genes. Specifically, we examined the EMT spectrum of each subline by calculating its EMT score, where a higher score indicates a more mesenchymal-like propensity of the cell (Tan et al., 2014). On average CTC- (0.1079) and metastatic tumor- (0.0436) derived sublines showed higher scores than primary tumor-derived lines (−0.1861). CTC-derived sublines showed the highest average score (Figure 5C). When compared between sublines of the same lineage, both 5-META and 7-META showed higher EMT scores than 5-PRI and 7-PRI sublines, respectively, whereas 3-CTC and 3-META had similar EMT scores. Moreover, MDS analysis of the transcript levels of mesenchymal and epithelial markers in these lineage-related sublines revealed that 3-CTC and 3-META are closer to each other than 5-PRI to 5-META and 7-PRI to 7-META (Figure 5Ac). Using migration and invasion transwell assays we observed enhanced migration and invasion in 3-CTC, with further enhancement in 5-META, as compared to 7-PRI (Figures S5D and S5E). These results support the notion that CTCs and metastatic tumors both lean more toward the mesenchymal state than primary tumor cell lines do.
Next, we examined and compared epithelial and mesenchymal markers between sublines. Most epithelial and mesenchymal markers were retained at a similar level in primary, circulating, and metastatic tumor cells (Figures 5D and 5E). The epithelial markers Krt19, EpCAM, Cdh1, Wnt7a, Muc1 were reduced in CTC- and/or metastatic tumor-derived sublines relative to primary tumor-derived lines (Figure 5D). Differential expression of these genes was confirmed by quantitative RT-PCR (qRT-PCR) (Figure S8B). In terms of mesenchymal markers, only Mmp2 and Mmp9 expression was reduced in CTCs, compared to primary tumor-derived cells (Figures 5E and S8B). Acta2, Cdh11, Mmp2, and Twist1 were increased in metastatic tumor-derived sublines relative to primary tumor- or CTC-derived sublines (Figures 5E and S8B). Together these data suggest the existence of an intermediate or hybrid E/M state (i.e., a partial transition), rather than complete conversion to a mesenchymal state in CTCs and metastatic tumors. To test this possibility, we examined the expression of “hybrid markers”, i.e., markers reported to be expressed in the intermediate or hybrid E/M state (Jolly et al., 2019; Liao and Yang, 2020). The hybrid markers, Vcam1 (Chen et al., 2011) and Emp3 (Evtimova et al., 2003), remained at a similar level throughout all stages, although Lamc2, Lamb3, Tnc and OvoI2 were reduced in CTC- and/or metastatic tumor-derived sublines and P4ha2 was decreased in CTC sublines but relatively increased in metastatic tumor-derived sublines (Figure S8A).
Use of CTCs for ex vivo analysis of anti-cancer drug efficacy
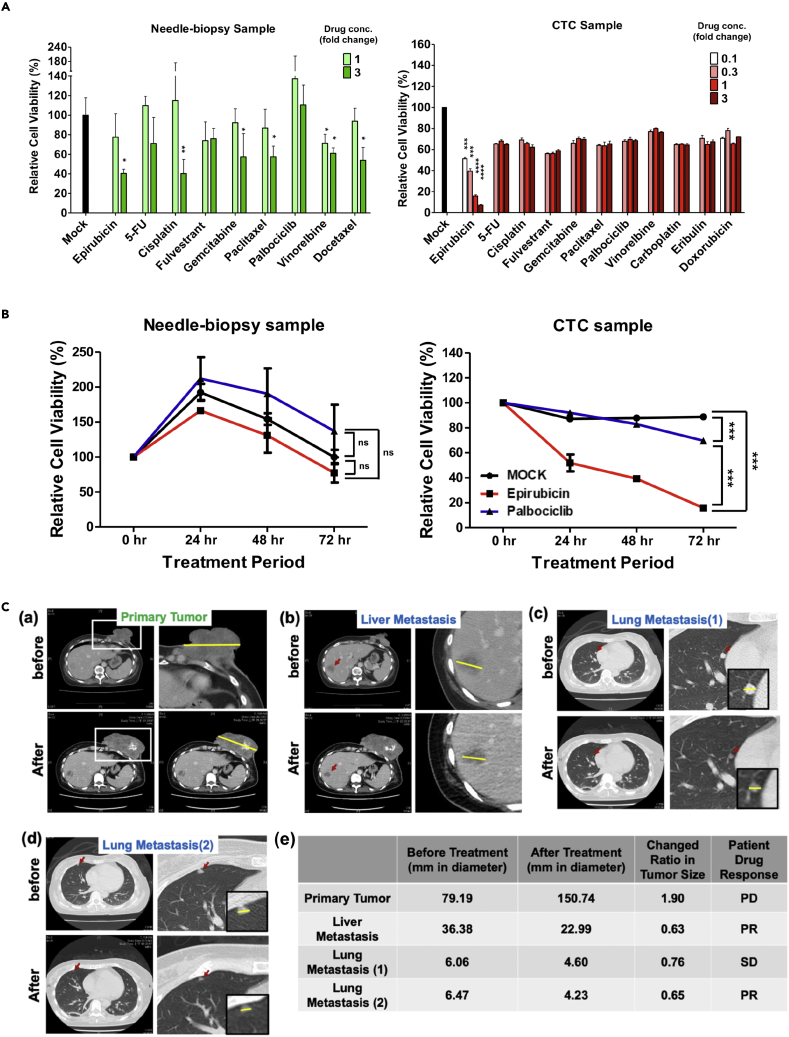
To test whether CTCs can be used in real-time clinical analysis, we isolated and cultured CTCs from patient blood and tested the effects of anti-cancer drugs on the cultivated cells. A breast cancer patient was recruited for proof-of-concept double-blind evaluation. A core-needle biopsy sample was taken from the primary tumor and blood-derived CTCs were collected and cultured to form three-dimensional (3D) spheroids from the patient before and after planned drug regimens. It has been well-documented that 3D cell culture better mimics the in vivo environment than two-dimensional cell monolayers and, as a result, more closely reflects the clinical therapeutic drug response (Imamura et al., 2015; Melissaridou et al., 2019). Representative spheroids cultured from primary tumor cells and CTCs are shown in Figure S9. The resulting spheroids were treated with various drugs at different dosages. Epirubicin (topoisomerase II inhibitor) treatment significantly reduced cell viability of CTC-, but not needle biopsy samples, at Cmax, the maximum serum concentration that the drug achieves in human blood (Figure 6A). Epirubicin treatment also showed clear dose-dependence (Figure 6A) and a time-dependent cell-killing effect in CTCs (Figure 6B). In contrast, no other chemotherapeutic drugs showed a significant differential response when tested on CTC-derived samples. Only vinorelbine showed a weak cell-killing effect on core needle biopsy compared with mock treatment at Cmax, although some other drugs had a significant effect at 3-fold Cmax (Figure 6A). However, vinorelbine’s killing effect was not significantly increased at 3-fold Cmax. The patient received a combinatorial drug treatment (epirubicin, cyclophosphamide, and 5-FU) for two months after our sample collection. After therapy, the patient showed a progressive response in the primary tumor, whereas the lung metastatic lesion (1) displayed a stable disease response, and liver metastasis and the lung metastatic lesion (2) had significant regression, with a size reduction >30% (Figure 6C). This result was consistent with the ex vivo drug response in needle–biopsy- and CTC-derived samples. In addition, the patient’s clinical history of failing to respond to palbociclib, a CDK4/6 inhibitor, matched well with our ex vivo drug tests which showed that palbociclib had no cytotoxic effect on either the needle biopsy or CTC samples at different concentrations tested (Figure 6A), although palbociclib showed a weak time-dependent cytotoxicity (Figure 6B). These results support the idea that the CTC 3D culture is similar to metastatic, but not primary, tumors in terms of drug response. The patient data are consistent with the above phenotypic and multiomics analysis which also indicates that CTC-derived sublines are more similar to metastatic tumors than to primary tumors. The results also demonstrated the potential utility of CTC-based ex vivo drug testing to predict clinical drug response in personalized medicine.
Figure 6.
Anti-cancer drug response of human patient-derived cultures
(A) Cell viability tests using needle biopsy and CTC cultures treated with different anti-cancer drugs. Needle-biopsy (left) and CTC (right) spheroid cultures were treated with the indicated anti-cancer drugs at a dose of 0.1, 0.3, 1 or 3 Cmax, and the relative cell viability of each treatment was measured 72 h after drug treatment. Relative cell viability was calculated by comparing the absolute luminescence intensity before (at time 0) and after (72 h) drug treatment and was used to determine the effectiveness of the chemotherapeutic drug. ∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.001 and ∗∗∗∗p<0.0001 denotes statistical significance comparing to Mock group (negative control, 0.5 % DMSO treatment) using Student’s ttest. Results are mean ± SEM (n = 3).
(B) A time-course of epirubicin and palbociclib effects on needle biopsy (left) and CTC (right) cultures. Relative cell viability of CTC-derived spheroids, compared to Mock and palbociclib treatment at a dose of 1 Cmax and at the indicated time points. Results are mean ± SEM (n = 3). ∗p<0.0001 (two-way ANOVA).ns: no significant difference.
(C) (a-d) CT images of the breast cancer patient before and after combinatorial treatment with epirubicin. (a) The CT images of the primary tumor before (upper) and after (lower) the treatment. The primary tumor is indicated by a white rectangle. The upper right figure is an enlargement of the upper left image. The tumor size (measured in length) is indicated by the yellow line. (b-d) CT images of liver metastasis (b), lung metastasis 1 (c), and lung metastasis 2 (d) before (upper) and after (lower) the treatment. An enlarged image of the metastasis is shown on the right, and the further enlarged image is shown in the black box. The metastatic tumor is indicated by the red arrow, and the size by the yellow line. (e) The disease tracking record and treatment response of the breast cancer patient. Tumor size before and after treatment was measured in mm as indicated by the yellow line shown in (a-d). Response rate was measured as size after treatment divided by size before treatment. Patient drug response is indicated as PD for progressive disease (>120%); SD, stable disease (<30%); PR, partial response (>30%).
Discussion
Here, we established a protocol using the LIPO platform to isolate CTCs and generate isogenic cell lines derived from primary tumors, CTCs, and metastatic tumors. We found characteristic molecular and behavioral changes as cells transition between states (Figure 7). Phenotypically, CTCs and metastatic tumors were more closely related to one another than to primary tumors, in that they shared similar morphology, increased proliferation, and enhanced migration and invasion ability. Genotypically and epigenetically, CTCs and metastatic tumors were alike in that they exhibited concurrent mutations and both exhibited reduced DNA methylation (hypomethylation). CTCs and metastatic tumors manifested higher EMT scores than primary tumors did. Although metastatic sublines and CTCs inclined toward a mesenchymal state, they were in an intermediate rather than a well-defined state. With the transformation from primary tumors, to CTC, and to metastatic tumors, cells lose their original properties and become more aggressive. The multiomic similarity between CTCs and metastatic tumors may facilitate the transition from CTCs to metastatic tumor cells and vice versa, and thereby promote tumor cell spreading. In addition, our results demonstrate that CTCs can serve as a potentially powerful tool for clinical theragnostics, and in particular that they can be used to monitor cancer progression in a mice model and as well as to assess drug efficacy.
Figure 7.
Summary of the qualitative characteristics of primary tumor, CTC, and metastatic tumor-derived sublines
As tumor metastasis progress, cells demonstrate reduced adhesion, increased migration/invasion, increased hypomethylation near transcription start sites (TSSs) and enhanced mesenchymal/reduced epithelial gene expression.-, + and ++ indicate very low, medium and high activity, respectively, and the activity increase accordingly to the number of symbol. The bigger the E and M size the stronger epithelial and mesenchymal characteristics, respectively, are. Bigger Hypo means more hypomethylation near TSSs.
The LIPO platform used in this work was able to capture and retrieve viable CTCs from blood samples with high isolation efficiency (Figure 1B). Notably, only 75 μL of whole blood was required to analyze CTCs and only 750 μL of whole blood was needed to establish mice carrying CTC-derived xenografts. On average, we estimated that only ∼400 CTCs are needed to generate CTC-derived xenografts, with metastasis detectable in 3–4 weeks, further accentuating the high bioactivity and viability of the retrieved CTCs. The success rate of generated CTC-derived xenografts was 100%, regardless of contamination during the primary culture process. In comparison to CTCs, over two orders of magnitude more of parental 4T1 cells (105 cells) were needed to obtain CDx xenografts with metastasis detectable within 3–4 weeks. This observation suggests that CTCs may be more tumorigenic than parental 4T1 cells. It is possible that fate/plasticity changes during development from primary tumor to CTCs in CDx mice enhanced the tumorigenicity of CTCs. Molecular analysis of these cells in CDx mice and parental cells may help understand the mechanisms underlying the plasticity changes with respect to increased tumorigenicity. In addition, characterization and comparison of tumor cells at the same stages across CDx and CTC-PDx mice may help understand the plasticity of tumor cells along their development and expansion from CDx to CTC-PDx mice. We note that because of technical issues cell lines were not successfully established from all of the CTC-PDx mice. Because of this, some sublines used in phenotypic and molecular characterization and comparison were necessarily derived from different mice. Therefore, the difference between mice might also contribute to the heterogeneity observed across sublines.
From a cancer biology perspective, one important feature of our device and approach is the ability to construct CTC-xenografts, CTC-derived cell lines, and cell lines of isogenic origin. In particular, the ability to construct a series of sublines at different stages in metastasis, including primary, circulating, and metastatic tumor-derived cell lines, opens up the possibility of studying cancer evolutionary pathway without the difficulties of analyzing human subjects. Our approach allowed us to conduct multiomic analyses on the evolution of primary, circulating, and metastatic tumors of the same lineages. Phenotypically, primary tumor-derived cell lines retained an adherent morphology, cell-cell contact, and epithelial gene expression. In contrast, CTC- and metastatic tumor-derived cell lines displayed a loss of cell-cell contact and enhanced 3D migration and invasion. These changes indicate that morphological alterations may expedite metastasis and antagonize anoikis (Zhao et al., 2012). Previous studies have indicated lack of universal genetic changes that drive metastasis. Indeed, the majority of established driver alterations occurred before dissemination (Birkbak and McGranahan, 2020). Consistently, we did not find any specific metastatic drivers in our isogenic primary, circulating, and metastatic tumor-derived sublines. The absence of universal drivers may indicate that genomic changes in metastasis are unstable random events. Nevertheless, multiomic analyses revealed that circulating and metastatic, but not primary, tumors, shared common genomic variants. This finding implies that the decisive event in metastasis might occur at the transition from the primary to the circulating tumor cell.
RNA-seq data revealed that most epithelial and mesenchymal markers were retained at a similar level in primary, circulating and metastatic tumor cells (Figures 5D and 5E). Consistently, biphenotypic or hybrid EMT has been shown in CTCs in breast cancer patients (Yu et al., 2013) and in pancreatic mouse models and patients through single cell RNA-seq and EMT in situ hybridization (Ting et al., 2014). However, certain epithelial markers such as Cdh1, EpCAM, Krt19, Muc1 and Wnt7a were decreased in CTC- and/or metastatic tumor-derived sublines, relative to primary tumor-derived ones; some mesenchymal markers, including Mmp2, and Mmp9 were decreased from primary tumor-to CTCs-derived sublines, and Acta2, Cdh11 and Mmp2, and Twist1 were enhanced from primary or circulatory to metastatic tumor-derived ones (Figures 5D, 5E, and S8B). These results suggest that there is not a complete two-state transition during tumor dissemination or colonization. On a separate note, we did not observe upregulation of all reported E/M hybrid state markers during metastasis. Instead, most of these markers were expressed at a similar level at different stages, and some were even down-regulated. Instead of the appearance of the so-called “hybrid-specific markers”, we found that the epithelial markers could be divided into two groups, one for canonical epithelial markers, and the other for noncanonical epithelial markers. The canonical markers, including Cdh1, EpCAM, and Krt19, are attenuated during metastasis. In contrast, the noncanonical markers, including Krt18 and syndecan family proteins, are retained throughout the entire metastatic process and hence can be considered potential hybrid markers. Whether this pattern is consistent across metastasis in different cancers and the role that these markers play in pathophysiology requires a large-scale, extensive study in the future.
Epigenetic regulation, including DNA methylation and histone modifications, has been shown to play important roles in tumor progression (Nebbioso et al., 2018). Our work indicates dynamic changes in DNA methylation during tumor progression, with significantly more changes between the transition from a primary tumor to CTCs than from CTCs to metastatic tumor. When differential methylated genes between primary tumor- and CTC-derived lines were analyzed using GO term enrichment analysis, the top biological processes identified included multicellular organism development (218 gene counts), cell differentiation (195), transmembrane transport (99) and cell adhesion (115). DNA methylation in the promotor region is associated with transcriptional down-regulation, whereas DNA methylation in the gene body is correlated with transcriptional up-regulation (Roadmap Epigenomics Consortium et al., 2015). Among the differentially expressed EMT genes between sublines listed in Figure 5D, EpCAM, Wnt7a, Krt19, Cdh1, Twist1, Mmp2, and Mmp9 showed differential methylation (Table S5). Some of the differential methylation patterns in EpCAM, Wnt7a and Krt19 were correlated with their downregulation from primary tumor-to circulating or to metastatic tumor-derived cell lines. For example, hypermethylation at −26905 and −26915 upstream of EpCAM and at −8846 upstream of Krt19 were correlated with their downregulation (Table S5). Hypomethylation at 6484 and 10,784 downstream of EpCAM and at multiple CpG sites downstream of Wnt7a were also correlated with their downregulation. Hypomethylation at −1594 and −1543 upstream of Twist1 were correlated with its upregulation from primary tumor-to circulating tumor-derived cell lines. Other methylation alterations appeared uncorrelated. Therefore, additional epigenetic changes, such as histone modifications or chromatin assembly, may be involved in transcriptomic differences between sublines. When differentially expressed genes (DE-Gs) (Table S3) between sublines were analyzed using the GO enrichment analysis, the top GO terms of biological processes identified, were those related to methylation on cytosine, histone assembly, and epigenetic regulation (Figure S7), and the affected genes were histone-coding genes (Table S4). These genes were downregulated in the metastatic tumor-derived sublines, compared with either primary tumor- or CTC-derived lines (Figure S7). Whether such downregulation is associated with epigenetic regulation in metastatic tumor-derived sublines requires further investigation.
Metastasis is a complex process. Our data suggest that CTCs and metastatic tumors are more similar to one another than to primary tumors, as manifested in phenotypic, methylomic, and transcriptomic profiles and anti-cancer drug response. Taken together, our study provides important insights regarding the characteristics of CTCs which will provide valuable guidance for the development of clinical theranostics for advanced-stage cancers.
Limitations of study
In this study, we generated and characterized primary, circulating and metastatic tumor cell lines from CTC-PDx models. We also generated cell lines from CDx mice but have not yet characterized them in detail. Further characterization and comparison of tumor cells at the same stages across CDx and CTC-PDx mice may help understand the plasticity of tumor cells along their development and expansion from CDx to CTC-PDx mice. We note that because of technical issues, cell lines were not successfully established from all of the CTC-PDx mice. Some sublines used in phenotypic and molecular characterization and comparison were necessarily derived from different mice. Therefore, the difference between mice might also contribute to the heterogeneity observed across sublines. In our clinical study, thirteen breast cancer patients were recruited. Only one patient showed distant metastasis and had paired primary and CTCs for the ex vivo anti-cancer drug test. Although our patient number is limited, the drug testing results showed that CTC responses mirrored the impact of drugs on metastatic rather than primary tumors. Nevertheless, more patient cases will help in a future study to address this limitation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-mouse EpCAM antibody | ThermoFisher Scicentific | Cat#13-5791-80; RRID:AB_1659715 |
| Rabbit anti-human-HER2 antibody | Agilent | Cat#A0485; RRID:AB_2335701 |
| Rat anti-mouse CD45-PE | eBioscience | Cat#12-0451-83; RRID:AB_465669 |
| Goat anti-rabbit Alexa Fluor 488 secondary antibody | Invitrogen | Cat#A-11029; RRID:AB_2534088 |
| Biological samples | ||
| Cancer patient whole blood and the core-needle biopsy samples | Chang-Gung Memorial Hospital and Academia Sinica | IRB serial No. 202000120A3 & IRB serial No. AS-IRB02-109398 |
| CTC-derived xenograft from CDx mice | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Accutase | Invitrogen | Cat# 00-4555-56 |
| vital dye Green 5-chloromethylfluorescein diacetate (CMFDA) | Thermo Fisher Scientific | Cat# C7025 |
| Pro-Long Gold Antifade Mountant with DAPI | Thermo Fisher Scientific | P36931 |
| Paraformaldehyde | Sigma-Aldrich | P6148 |
| endotoxin-free luciferin | Promega | E6551 |
| Collagenase | Sigma-Aldrich | C0130 |
| Nocodazole | Sigma-Aldrich | M1404 |
| propidium iodide | Sigma-Aldrich | P4170 |
| RNase A | Thermo Fisher Scientific | 12,091–021 |
| Critical commercial assays | ||
| High-Capacity cDNA Reverse Trancription Kit | Applied Biosystems | Cat#4368814 |
| Fast SYBR™ Green Master Mix | Applied Biosystems | Cat#4385612 |
| Illumine Truseq standard total RNA library preparation Kit | Illumina | 20,020,596 |
| LIVE/DEAD cell viability assay | Thermo Fisher Scientific | L3224 |
| Deposited data | ||
| RNA sequencing results | This paper | GEO ID: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159727 |
| whole exom sequencing data | This paper | Sequence Read Archive (SRA) ID: http://www.ncbi.nlm.nih.gov/bioproject/668668 |
| bisulfite sequencing data | This paper | BsSeq project on NCBI https://www.ncbi.nlm.nih.gov/bioproject/670066 |
| Mouse Reference genome mouse build 38, GRCm38 (mm10) | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.20/ |
| Experimental models: Cell lines | ||
| parental 4T1 cancer cells | ATCC | Cat# CRL-2539, RRID:CVCL_0125 |
| 4T1-hHER2-Luc cell line | This paper | N/A |
| Primary tumor cell lines from CTC-PDx mouse | This paper | N/A |
| CTC lines from CTC-PDx mouse | This paper | N/A |
| metastatic tumor cell lines from CTC-PDx mouse | This paper | N/A |
| Experimental models: Organisms/strains | ||
| NOD-SCID-gamma (NSG) mice | Genomics Research Center, Academia Sinica | N/A |
| 4T1-hER2-Luc-derived xenograft (CDx) mice | This paper | N/A |
| CTC-derived xenograft (PDx) mice | This paper | N/A |
| Oligonucleotides | ||
| Primers for Cdh1 qPCR: 5‘-GGGATTCGTTGCAGAAGGCG-3’ 5‘-TAGGCGATGGCAGCGTTGTA-3’ |
This paper | N/A |
| Primers for Epcam qPCR: 5‘-CCACGTGCTGGTGTGTCAAC-3’ 5‘-TTGAAGCGCAGTCTGCAAGC-3’ |
This paper | N/A |
| Primers for Krt19 qPCR: 5′-CCTCAATGATCGTCTCGCCTC-3′‘ 5’-CTTGGAGTTGTCAATGGTGGC-3′‘ |
This paper | N/A |
| Primers for Muc1 qPCR: 5’-CAGTACCAAGCGTAGCCCCT-3′‘ 5’-ACCAGCCCAGATCAGAGTGC-3′‘ |
This paper | N/A |
| Primers for Nid1 qPCR: 5’-TTCCCAGCAGACTTGTGCCA-3′‘ 5’-TCACCTTGCCATTGACCCGT-3′‘ |
This paper | N/A |
| Primers for Sdc2 qPCR: 5’-CGAGAACAGAGCTGACATCCGA-3′‘ 5’-GAAGCAGCACTAGTGAGTGGGA-3′‘ |
This paper | N/A |
| Primers for Wnt7a qPCR: 5’-AGTTCCGAAATGGCCGTTGG-3′‘ 5’-TCCTCCAGGATCTTGCTTCTCCT-3′‘ |
This paper | N/A |
| Primers for Acta2 qPCR: 5‘-CAGCGGGCATCCACGAAAC-3’ 5′-TTGCGTTCTGGAGGGGCAAT-3′‘ |
This paper | N/A |
| Primers for Cdh11 qPCR 5’-GCCGCCGACTTGTGAATGG-3′‘ 5’-CAAAGGGCCACAAAGCACAGT-3′‘ |
This paper | N/A |
| Primers for Mmp2 qPCR: 5’-CCACACCAACACTGGGACCT-3′‘ 5’-AAGGGACCTGTGGGCTTGTC -3′‘ |
This paper | N/A |
| Primers for Mmp9 qPCR: 5’-ACCTCCAACCTCACGGACAC-3′‘ 5’-CTCTCCCATCATCTGGGCGG-3′‘ |
This paper | N/A |
| Primers for Twist1 qPCR: 5’-TCCATGTCCGCGTCCCACTA-3′‘ 5’-CCCACGCCCTGATTCTTGTG-3′‘ |
This paper | N/A |
| Primers for Her2: 5’-TGGCATTTTTGCCGGAGAGC-3′ 5′-AATCCGTCCCCGAATGACCC-3′ |
This paper | N/A |
| Primers for Gapdh as the qPCR internal control: 5′-CGGGGTCCCAGCTTAGGTTC-3′ 5-‘GCCCAATACGGCCAAATCCG-3’ |
This paper | N/A |
| Software and algorithms | ||
| BWA 0.7.17-r1188 | Li (2013) | RRID:SCR_010910; https://sourceforge.net/projects/bio-bwa/ |
| GATK v4.1.0.0 Mutect2 | Data Sciences Platform at Broad institute | RRID:SCR_001876; https://gatk.broadinstitute.org/hc/en-us/articles/360051306691-Mutect2 |
| HISAT2 v2.1.0 | Kim et al., (2019) | RRID:SCR_015530; http://daehwankimlab.github.io/hisat2/ |
| StringTie | Pertea et al. (2016) | RRID:SCR_016323; v2.0.3; https://ccb.jhu.edu/software/stringtie/ |
| DESeq2 | Love et al. (2014) | RRID:SCR_015687; v1.26.0; https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| metaseqR2 | Fanidis and Moulos (2021) | https://doi.org/10.1093/bib/bbaa156 |
| EMT score | Tan et al. (2014) | http://www.csi.nus.edu.sg/bioinfo/index.php |
| GraphPad Prism software | GraphPad Software | RRID:SCR_002798; version 6.0c; https://www.graphpad.com/scientific-software/prism/ |
| TPM calculator | Vera Alvarez et al. (2019) | https://github.com/ncbi/TPMCalculator |
| Trimmomatic | Bolger et al. (2014) | RRID:SCR_011848; http://www.usadellab.org/cms/?page=trimmomatic |
| Bismark | Bioinformatics group at Babaham institute | RRID:SCR_005604; v0.14.4; https://www.bioinformatics.babraham.ac.uk/projects/bismark/ |
| FastQC | Bioinformatics group at Babaham institute | RRID:SCR_014583; https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| MultiQC | Ewels et al. (2016) | RRID:SCR_014982; https://multiqc.info/ |
| Methylkit | Akalin et al. (2012) | RRID:SCR_005177; https://www.bioconductor.org/packages/release/bioc/html/methylKit.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ying-Chih Chang (yingchih@gate.sinica.edu.tw).
Material availability
Cell lines generated in this study are available upon request. Primers used for qPCR are listed in the key resources table.
Experimental model and subject details
In vivo CTC-derived xenograft propagation and cell line establishment
Four weeks after 4T1-hHER2-Luc orthotopic inoculation to NSG mice, 750 μL of whole blood was collected through cardiac puncture after adequate anesthesia. The blood was processed using the LIPO platform for CTC purification. Total CTCs collected from three random mice were pooled together with Matrigel and directly injected into the mammary fat pad of alternative NSG mice for CTC-derived xenograft growth. The growth of CTC-derived xenograft and cancer metastasis was observed and monitored using the IVIS imaging system. At the end of the fourth week after CTC inoculation, the mice were sacrificed and both the primary tumor and lung metastatic cancer tissues were collected for further primary culture and corresponding cancer cell line establishment. In brief, the tissues were minced into 1× 1 mm fragments and digested with 200 U/mL collagenase (Sigma-Aldrich) overnight under 37°C with 5% CO2 humidified atmosphere. The digested tissues were filtrated using a 70 μm cell strainer (BD Biosciences) and washed using PBS after centrifugation. The pelleted cells were cultured on tissue culture plates using serum-free or complete RPMI-1640 medium. For CTC cell line establishment, around 750 μL of whole blood from each mouse was collected by cardiac puncture and processed using the CTC capture platform under a germ-free condition. The collected CTCs were cultured under the same culture conditions used for routine cell line cultivation. All primary cultured cells were passaged every other day with 80% confluency and photographed under a light microscope (CK40-F100; Olympus, Japan).
Cell lines and cell culture
The 4T1-hHER2-Luc cell line was generated by integration of codon-optimized human HER2 and luciferase genes into the HER2-negative/luciferase-negative mouse breast cancer cell line 4T1-hHER2 (kindly provided by Dr. Wen-Hwa Lee and Dr. Chun-Mei Hu in the Genomics Research Center, Academia Sinica). The exogenous expression of hHER2 was verified through qRT-PCR and immunocytochemistry staining (Figure S1). Primary, circulating, and metastatic tumor cells from mouse-derived CTC-PDx mice were harvested simultaneously at the end of the fourth week after inoculation. Primary cultures were grown in the serum-free medium to select tumor cells for approximately 24–48 h and then cultured in the serum-containing medium for further growth. The 4T1-hHER2-Luc cell line and all cultured cells from mouse-derived CTC-PDx mice were cultured using RPMI-1640 medium (Gibco) supplied with 1% antibiotic-antimycotic solution (Gibco) and 10% fetal bovine serum (FBS, Gibco) at 37°C with 5% CO2 humidified atmosphere. The cells were detached using accutase (Invitrogen) and passaged on tissue culture plates (Corning, Corning, NY) every two days when reaching 80% confluency. CMFDA (Thermo Fisher Scientific) was used as a cell tracking dye for viable cell staining after cell release from the platform.
Patient sample
Thirteen breast cancer patients were recruited during the period of December 2020 to December 2021. The core-needle biopsy and whole blood samples were collected with signed informed consent forms. Only one patient (at age 37) showed distant metastasis and had paired primary and CTCs. Her ex vivo drug response and clinical treatment data were analyzed and compared in this study. Specifically, the EDTA vacutainer tube was used for a whole-blood sample collection from the metastatic cancer patient and ice-cold tissue preservation buffer was used for fresh core-needle biopsy sample delivery. All experiments using the patient’s samples were processed immediately after sample transfer and as a double-blind study. Patient recruitment, human sample collection, and ex vivo tests were executed by strictly following standard protocols reviewed by Institutional Review Board in both Chang-Gung Memorial Hospital (IRB serial No. 202000120A3) and Academia Sinica (IRB serial No. AS-IRB02-109398).
3D spheroid culture
Human patient-derived samples, core-needle biopsy samples or isolated CTCs using the LIPO platform, were seeded onto the 3D culture platform supplied with complete medium and cultured for 7–14 days. The 3D culture platform was prepared according to the previous results (Tsai et al., 2012a) and now is manufactured as R3CE (Acrocyte Therapeutics Inc., New Taipei City, Taiwan). Approximately, 135 CTC colonies were obtained from 1 mL of blood sample and 2000 tumor cell colonies were generated from 100 ul of the core-needle biopsy sample. Each well was supplemented with 200 μL Advanced DMEM/F-12 (Gibco) complete media with 10% FBS (Gibco), 1% antibiotic-antimycotic (Corning), 10 ng/mL FGF (Corning) and 100 μg/mL EGF (Gibco). The cell morphology was observed using a phase contrast microscope, before and after drug treatments.
Method details
Sample processing and CTC isolation on the LIPO microfluidic platform
The LIPO platform is a CTC isolation platform based on the CMx platform (Yeh, 2018). CMx contains a micro-pattern designed for chaotic mixing in the microchannel and a non-covalently bound anti-EpCAM conjugated lipid channel for multivalent antibody-antigen interaction and quick release (Chen et al., 2016). It has been used in clinical studies examining colorectal cancer (Gupta et al., 2019; Tsai et al., 2019), prostate cancer (Pang et al., 2018), and head and neck cancer (Li et al., 2019). The LIPO platform was further optimized to achieve maximal cell viability. Specifically, a liposomes-lipid bilayer was used as a surface coating instead of a lipid bilayer to facilitate detachment of surface-bound cells while preserving viability and bioactivity of eluted cells (Yeh, 2018). To capture CTCs derived from CDx or CTC-PDx or parental 4T1-hHER2-Luc cells, monoclonal anti-mouse EpCAM antibody was site-specifically biotinylated for target cell isolation as described previously (Chen et al., 2007; Lai et al., 2017) (ThermoFisher Scientific, Cat#13-5791-80, RRID:AB_1659715). A syringe pump (Harvard Apparatus PHD, 2000) with a syringe installed was connected using the tubing and adapter to the outlet of the microfluidic platform for blood sample processing and target cell isolation. Samples were loaded to the inlet of the platform and bound cells were released as previously reported (Lai et al., 2014). In brief, air-to-aqueous solution foam, produced by mixing BSA solution (BSA, UR) with air, was gently injected into the outlet of the platform to release bound cells. The released cells were collected for subsequent studies. To capture CTCs from the human patient blood, a similar procedure was followed except that anti-human EpCAM antibody was used.
Evaluation of cell isolation efficiency of the LIPO platform
Cultured cells were incubated with accutase (Invitrogen) at 37°C for 10 min to detach cells from the culture dish. The cell suspension was collected, re-suspended in cell culture medium and spiked into the microfluidic platform to evaluate isolation efficiency. To visualize cells, cancer cells were pre-stained with vital dye Green 5-chloromethylfluorescein diacetate (CMFDA, Thermo Fisher Scientific) at 37°C for 30 min and aliquoted into appropriate dilutions for the following experiments and enumeration. The eluted cells were washed using PBS, fixed with 4% formaldehyde, and mounted using DAPI-containing Pro-Long Antifade mounting medium (Thermo Fisher Scientific) for subsequent cell counting. The cell isolation efficiency was defined as the cell number released from the microfluidic platform divided by the cell number spiked into the microfluidic platform. The LIVE/DEAD cell viability assay was performed according to the manufacturer’s instruction with 2 μM of calcein AM for live cell staining and 10 μM of ethidium homodimer-1 for dead cells staining for 30 min before live/dead cell observation.
Immunofluorescence staining and image analysis
To perform immunofluorescence staining, the captured cells were first fixed with 4% paraformaldehyde (Sigma-Aldrich) followed with penetration by 0.1% Triton X-100 (EMD Millipore, Burlington, MA) with a PBS wash in between. After blocking with 10% normal goat serum (Jackson ImmunoResearch Laboratories, West Grove, PA), the captured CTC cells were subsequently stained with rabbit anti-human-HER2 antibody (Agilent, Santa Clara, CA, Cat#A0485, RRID:AB_2335701, 1:100 dilution) and rat anti-mouse CD45-PE (eBioscience, San Diego, CA, Cat#12-0451-83, RRID:AB_465669, 1:200 dilution), followed by the goat anti-rabbit Alexa Fluor 488 secondary antibody (Invitrogen, Cat#A-11029, RRID:AB_2534088,1:500 dilution). The immunofluorescence-stained samples were sealed with ProLong Gold Antifade Mountant with DAPI and photographed under a Nikon Ti-E fluorescence microscope. CTCs were verified and counted using Nikon NIS-Element AR software.
In vivo spontaneous metastasis animal experiment and ethics statement
The animal model experiment protocols were approved by the institutional animal care and use committee (IACUC) and performed according to the submitted IACUC protocol in Academia Sinica (IACUC protocol No. 17-11-1116). The 4–5-week-old female NOD-SCID-gamma (NSG) mice were bred and obtained from the animal facility of the Genomics Research Center, Academia Sinica. All experiments were maintained and operated under a specific pathogen-free environment with controlled temperature, humidity, and adequate daylight cycle. About 1x105 4T1-hHER2-Luc cells with Matrigel (Corning) in 1:1 dilution was orthotopically inoculated into the mammary fat pad of NSG mice. In vivo luciferase detection and monitoring were accomplished using endotoxin-free luciferin (concentration: 15 mg/mL, dosage: 10 μL/g, Promega, WI) and photographed using the IVIS spectrum in vivo imaging system (Caliper Life Sciences, Hopkinton, MA). The orthotopic tumor volume was measured and calculated as [(length x width2)/2] and body weight was recorded once every week. The tumor growth and cancer metastasis status were observed simultaneously at several time points using the IVIS imaging system. The time points included one day before, one week after, three weeks after, and four weeks after tumor inoculation. Orbital sinus sampling was used for blood sample collection and for subsequent experiments. 150 μL of whole blood was collected from each mouse and aliquoted equally for both CTC isolation and RT-PCR verification at each time point. Primary tumor and secondary metastatic lung tissue were collected for further paraffin embedding and followed by IHC staining confirmation after sacrifice.
Synchronization and cell cycle analysis
Cells were seeded on a tissue culture dish for 24 h and additional 50 ng/mL nocodazole (Sigma-Aldrich) was used for cell cycle synchronization. The cells were then cultured for 6 h in regular growth medium to release the cell cycle from G2/M phase. Synchronized cells were fixed with 75% alcohol at −20°C overnight. The fixed cells were stained with propidium iodide (PI, 20 μg/mL, Sigma-Aldrich) and RNase A (0.2 mg/mL, Thermo Fisher Scientific) for 20 min at room temperature. The cell cycle distribution after synchronization was analyzed by flow cytometry (BD FACSCalibur) afterward with at least 10,000 cells collected. The data between sublines were compared and analyzed using the chi-square analysis.
Transwell migration and invasion assay
About 1x105 cells were seeded on the cell culture insert (8 μm pore, Falcon, Corning) with serum-free medium, and the fitted culture dishes were supplied with 20% FBS containing culture medium as a chemoattractant for cell migration. An additional 20 μg of Matrigel (Corning) was precoated on the cell culture insert before cell seeding for the invasion assay. The cells were allowed to migrate or invade for 16 h and collected for further staining and quantitation. The non-migrating or non-invasive cells were wiped from the coating surface. The transwell migrated or invading cells were stained with 1% crystal violet and photographed under a light microscope. Five random views with three repeated assays were counted for further quantification and statistical analyses.
Whole exome sequencing (WES)
125 bp Illumina paired-end reads were sequenced on the HiSeq 2500. Mean depth of coverage for all genes was 200. 4T1-derived primary, CTC, and metastasis-derived cancer cell lines as well as parental 4T1 cancer cells (ATCC, Cat# CRL-2539, RRID:CVCL_0125) were utilized for both WES and bisulfite sequencing. Genomic DNAs from the parental 4T1-hHER2-Luc cell line, 4T1-derived primary tumors, CTCs, and metastasis tumors were analyzed by WES. Sequencing reads were cleaned and then aligned to the Mus musculus genome assembly GRCm38 (mm10) using the BWA algorithm (RRID:SCR_010910; bwa version 0.7.17-r1188). Then, the aligned data were sorted, validated and indexed. To observe genomic changes between different groups, 4T1-hHER2-Luc samples were nominated as normal and the others as tumor samples. On the basis of GATK best practice, somatic variants were identified through the Mutect2 algorithm (RRID:SCR_001876; GATK v4.1.0.0). Two variant filtering mechanisms were adopted. First, variants that did not PASS the FILTER value were eliminated. Second, variants with a non-zero AD value in parental 4T1-hHER2-Luc cells were removed to prevent false positive results. The remaining variants were deemed qualified for downstream analysis. The relative WES sequencing data were uploaded to Sequence Read Archive (SRA) server of NCBI under the following project ID: http://www.ncbi.nlm.nih.gov/bioproject/668668.
Bisulfite sequencing
For methylation profile analysis, sequence alignments were performed using the Bismark pipeline. Briefly, adapters at the 5′AND-3′ ends of reads were removed from chastity filtered fastq files using Trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic). The processed reads were aligned to the mouse genome (mm38), deduplicated, examined for coverage, and extracted to a CpG count matrix using Bismark v0.14.4 (RRID:SCR_005604; https://www.bioinformatics.babraham.ac.uk/projects/bismark/). Quality control and assurance (QC/QA) was performed using FastQC (RRID:SCR_014583; https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), Bismark, and MultiQC (RRID:SCR_014982; https://multiqc.info/). Bins with fewer than 10 counts or with coverage exceeding the 99th percentile were excluded. Only those with methylation changes >25% and q value <0.05 were selected for methylome analysis. Subsequently, differentially methylated region analysis was conducted using Methylkit (RRID:SCR_005177) in the R-environment. The parameters for calculate DiffMeth in methylKit were as follows: slim = TRUE, weighted.mean = TRUE, num.cores = 12. The relative bisulfite sequencing data were submitted to BsSeq project on NCBI (RRID:SCR_006472) server under the following ID: https://www.ncbi.nlm.nih.gov/bioproject/670066.
RNA sequencing
Total RNA was extracted from primary (5-PRI, 7-PRI, 8-PRI), circulating (1-CTC, 3-CTC, 6-CTC) and metastatic (3-Meta, 5-Meta and 7-Meta) tumor cells-derived sublines, and the parental 4T1-hHER2-Luc cell line. The library was prepared using illumine Truseq standard total RNA library preparation kit and followed by pair-end RNA sequencing using illumina HiSeq 2500 system. The RNA-Seq data analysis pipeline was run on the SeqsLab platform. First, sequencing data were analyzed using a pipeline comprised HISAT2 (v2.1.0; RRID:SCR_015530) for mapping reads to the mouse genome (GRCm38.96), StringTie (v2.0.3; RRID:SCR_016323) for transcript assembly and DESeq2 (v1.26.0; RRID:SCR_015687) for data normalization. The TPM (transcript per million, generated by TPM calculator) was calculated according to the TPMCalculator for RNA abundance quantification (Vera Alvarez et al., 2019). Then, MDS analysis was applied to illustrate inter-sample similarity. MDS plots in Figure 5A was based on TPM of each sample and generated using the metaseqR2(https://doi.org/10.1093/bib/bbaa156). The RNA sequencing results were archived in the NCBI Gene Expression Omnibus (GEO) dataset (RRID:SCR_005012) under the following ID: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159727.
Calculation of EMT score
To quantitatively score cell lines derived from CTC-PDx for their EMT status, the EMT score was calculated through the website (http://www.csi.nus.edu.sg/bioinfo/index.php) (Tan et al., 2014) using the RNA-seq data. The EMT score ranged from −1.0 (fully epithelial) to +1.0 (fully mesenchymal) and represents the EMT status of cells.
RNA extraction and qRT-PCR analysis
TRIzol reagent solution was used for isolating RNA, following the manufacturer’s protocol (Applied Biosystems, CA, USA). The PRI, CTC and META sublines at passages 21, 22, and 21, respectively, were used for the qRT-PCR analysis for epithelial and mesenchymal markers. The concentration of extracted RNA was measured using the Nano-drop ND2000 spectrophotometer (Thermo Fisher Scientific). Samples were then diluted in RNAse-free water to a final concentration of 0.25 μg/μL for qRT-PCR analysis. The reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit, according to the manufacturer’s protocol (Thermo Fisher Scientific). qPCR was then performed using Fast SYBR™ Green Master Mix, following the manufacturer’s protocol (Thermo Fisher Scientific). ΔCT and ΔΔCT were then calculated, and statistical analysis was performed using One-way ANOVA.
Anti-cancer drug test
Spheroids derived from the human patient’s core-needle biopsy sample or CTCs were cultured for drug tests. Anti-cancer drugs, including epirubicin, 5-fluorouracil (5-FU), fulvestrant, gemcitabine, paclitaxel, palbociclib, vinorelbine, carboplatin, eribulin, and doxorubicin, were purchased from MedChemExpress, and treated at a dose of 0.1, 0.3, 1 or 3 Cmax. All drugs were dissolved in DMSO, which were further diluted in media containing 10% FBS (final [DMSO] ≤ 0.25% (v/v)). Cell viability was measured using RealTime-Glo TM Cell Viability Assay, following the manufacturer’s protocol (Promega). Relative cell viability was calculated by comparing the absolute luminescence intensity before (at time 0) and after (24, 48, and 72 h) drug treatment and was used to determine the effectiveness of the chemotherapeutic drug.
Quantification and statistical analysis
All statistical analyses were accomplished using GraphPad Prism software (RRID:SCR_002798; version 6.0c, La Jolla, CA). The Student’s ttest was performed for statistical analysis and p ≤ 0.05 was considered as significant. All representative results are shown as mean ± SEM for at least three independent replicates of experiments.
Acknowledgments
We thank Prof. Wen-Hwa Lee of the University of California-Irvine and Academia Sinica for sharing the 4T1-hHER2-Luc murine breast cancer cell line. We thank Dr. Mei-Yeh Lu and the members of the High Throughput Genomics Core at Academia Sinica for conducting NGS library prep and Illumina sequencing. We thank the Genomics Center for the Clinical and Biotechnological Applications of Cancer Progression Research Center, National Yang-Ming University, Taipei, Taiwan, for technical services. The center is financially supported by the National Core Facility for Biopharmaceuticals (NCFB), Ministry of Science and Technology, Taiwan (MOST). We thank the National Core Facility for Biopharmaceuticals (NCFB, MOST 108-2319-B-492 -001) and National Center for High-performance Computing (NCHC) of National Applied Research Laboratories (NARLabs), Taiwan, for providing computational and storage resources.
Taiwan Ministry of Science and Technology (MOST) funded MOST-108-2314-B-010-020-MY3, MOST-108-2320-B-010-008 (MHY), MOST 110-2311-B-002-009-MY3 (YCW), MOST 104-2112-M-001-043-MY3, MOST 105-2627-M-001-006-, MOST 106-2627-M-001-005-, MOST 107-2627-M-001-001-, MOST 107-2119-M-001-039-, MOST 107-2112-M-001-040-MY3, MOST 108-2119-M-001-018- (CLG) and MOST 104-2113-M-001-015-MY3, MOST 106-2218-E-001-004, MOST 107-2218-E-001-003, MOST 107-2119-M-001-039, MOST 108-2119-M-001-018, and MOST 109-2823-8-001-003 (YCC). Academia Sinica, Taiwan, funded AS-105-TP-A04 (YCC) and AS-TP-109-M04 (CLG). National Institute of General Medical Sciences funded R35GM130332 (ARD). Stanford University funded 1216884-1-GWNDA (CWF, YCC). Taiwan Ministry of Education funded Pilot Study Abroad A+ Project (JYC, EL).
Author contributions
Each author’s contribution(s) to the article should be listed. Conceptualization: J.Y.C., Y.C.W., M.H.Y., and Y.C.C.; Methodology: J.Y.C., S.C.L., Y.J.H., C.Y.T., C.T.S., E.L., C.L.G., H.F.H., A.R.D., C.W.F., P.Y.Y., C.M.H., Y.C.W., M.H.Y., and YCC; Investigation & Execution: J.Y.C., S.C.L., Y.J.H., C.Y.T., C.T.S., E.L., Y.C.C., K.C.L., H.F.H., and H.H.C.; (Animal study: J.Y.C.; Cell phenotypic study: J.Y.C. and K.C.L.; Multiomic analysis: J.Y.C., K.C.L., C.Y.T., C.T.S., and H.F.H.; Clinical studies: H.H.C., S.C.L., and Y.J.H.); Supervision: Y.C.W., M.H.Y., and Y.C.C.; Writing—original draft: J.Y.C., C.Y.T., C.T.S., H.H.C., S.C.L., Y.J.H., J.W., H.F.H., Y.C.W., M.H.Y., and Y.C.C.; Writing—review & editing: J.Y.C., A.R.D., C.L.G., H.F.H., Y.C.W., M.H.Y., and Y.C.C.
Declaration of interests
Ying-Chih Chang is the founder and stockholder of Acrocyte Therapeutics Inc., New Taipei City, Taiwan. Other authors declare no conflicts of interest.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105081.
Contributor Information
Yi-Chun Wu, Email: yichun@ntu.edu.tw.
Muh-Hwa Yang, Email: mhyang2@vghtpe.gov.tw.
Ying-Chih Chang, Email: yingchih@gate.sinica.edu.tw.
Supplemental information
9 Tables S4 and S5
Data and code availability
WES sequencing data were deposited in the NCBI Sequence Read Archive (SRA) (ID: http://www.ncbi.nlm.nih.gov/bioproject/668668).
Bisulfite sequencingdata were deposited in the NCBI BsSeq project (ID: https://www.ncbi.nlm.nih.gov/bioproject/670066). RNA sequencing results were deposited in the NCBI Gene Expression Omnibus (GEO) library (ID: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159727). This paper does not report original code. Any additional information required to reanalyze the data reported in this article is available from the lead contacton request.
References
- Aceto N., Bardia A., Miyamoto D.T., Donaldson M.C., Wittner B.S., Spencer J.A., Yu M., Pely A., Engstrom A., Zhu H., et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;15:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aceto N., Toner M., Maheswaran S., Haber D.A. En route to metastasis: circulating tumor cell clusters and epithelial-to-mesenchymal transition. Trends Cancer. 2015;1:44–52. doi: 10.1016/j.trecan.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A., Mason C.E. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidard F.C., Jacot W., Kiavue N., Dureau S., Kadi A., Brain E., Bachelot T., Bourgeois H., Gonçalves A., Ladoire S., et al. Efficacy of circulating tumor cell count-driven vs clinician-driven first-line therapy choice in hormone receptor-positive, ERBB2-negative metastatic breast cancer: the STIC CTC randomized clinical trial. JAMA Oncol. 2021;7:34–41. doi: 10.1001/jamaoncol.2020.5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkbak N.J., McGranahan N. Cancer genome evolutionary trajectories in metastasis. Cancer Cell. 2020;37:8–19. doi: 10.1016/j.ccell.2019.12.004. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrefourcq L., Mazard T., Joosse S., Solassol J., Ramos J., Assenat E., Schumacher U., Costes V., Maudelonde T., Pantel K., Alix-Panabières C. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res. 2015;75:892–901. doi: 10.1158/0008-5472.CAN-14-2613. [DOI] [PubMed] [Google Scholar]
- Chen Q., Zhang X.H.F., Massagué J. Macrophage binding to receptor VCAM-1 transmits survival signals in breast cancer cells that invade the lungs. Cancer Cell. 2011;20:538–549. doi: 10.1016/j.ccr.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Sensitive and Specific Biomimetic Lipid Coated Microfluidics to Isolate Viable Circulating Tumor Cells and Microemboli for Cancer Detection. PLoS One. 2016;11 doi: 10.1371/journal.pone.0149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.C., Huang H.N., Lin C.T., Chen Y.F., King C.C., Wu H.C. Generation and characterization of monoclonal antibodies against dengue virus type 1 for epitope mapping and serological detection by epitope-based peptide antigens. Clin.Vaccine Immunol. 2007;14:404–411. doi: 10.1128/CVI.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.J., Gabrielson E., Werb Z., Ewald A.J. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 2013;155:1639–1651. doi: 10.1016/j.cell.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapkin B.J., George J., Christensen C.L., Mino-Kenudson M., Dries R., Sundaresan T., Phat S., Myers D.T., Zhong J., Igo P., et al. Genomic and functional fidelity of small cell lung cancer patient-derived xenografts. Cancer Discov. 2018;8:600–615. doi: 10.1158/2159-8290.CD-17-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evtimova V., Zeillinger R., Weidle U.H. Identification of genes associated with the invasive status of human mammary carcinoma cell lines by transcriptional profiling. Tumour Biol. 2003;24:189–198. doi: 10.1159/000074429. [DOI] [PubMed] [Google Scholar]
- Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanidis D., Moulos P. Integrative, normalization-insusceptible statistical analysis of RNA-Seq data, with improved differential expression and unbiased downstream functional analysis. Brief Bioinform. 2021;22:bbaa156. doi: 10.1093/bib/bbaa156. [DOI] [PubMed] [Google Scholar]
- Gaggioli C., Hooper S., Hidalgo-Carcedo C., Grosse R., Marshall J.F., Harrington K., Sahai E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- Gupta P., Gulzar Z., Hsieh B., Lim A., Watson D., Mei R. Analytical validation of the CellMax platform for early detection of cancer by enumeration of rare circulating tumor cells. J. Circ. Biomark. 2019;8 doi: 10.1177/1849454419899214. 1849454419899214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan R.B., Qiao R., Keren R., Badano I., Suyama K. Cadherin switch in tumor progression. Ann. N. Y. Acad. Sci. 2004;1014:155–163. doi: 10.1196/annals.1294.016. [DOI] [PubMed] [Google Scholar]
- Hidalgo M., Amant F., Biankin A.V., Budinská E., Byrne A.T., Caldas C., Clarke R.B., de Jong S., Jonkers J., Mælandsmo G.M., et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo-Carcedo C., Hooper S., Chaudhry S.I., Williamson P., Harrington K., Leitinger B., Sahai E. Collective cell migration requires suppression of actomyosin at cell-cell contacts mediated by DDR1 and the cell polarity regulators Par3 and Par6. Nat. Cell Biol. 2011;13:49–58. doi: 10.1038/ncb2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson C.L., Morrow C.J., Li Y., Metcalf R.L., Rothwell D.G., Trapani F., Polanski R., Burt D.J., Simpson K.L., Morris K., et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 2014;20:897–903. doi: 10.1038/nm.3600. [DOI] [PubMed] [Google Scholar]
- Hwang E., Uh J.H., Lee H.S., Lee C.H., Lee S.J., Ahn S.H., Son B.H., Lee J.W., Yu J.H., Kwon N.J., et al. Cancer gene panel analysis of cultured circulating tumor cells and primary tumor tissue from patients with breast cancer. Oncol. Lett. 2017;13:4627–4632. doi: 10.3892/ol.2017.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura Y., Mukohara T., Shimono Y., Funakoshi Y., Chayahara N., Toyoda M., Kiyota N., Takao S., Kono S., Nakatsura T., Minami H. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015;33:1837–1843. doi: 10.3892/or.2015.3767. [DOI] [PubMed] [Google Scholar]
- Jolly M.K., Somarelli J.A., Sheth M., Biddle A., Tripathi S.C., Armstrong A.J., Hanash S.M., Bapat S.A., Rangarajan A., Levine H. Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharmacol.Ther. 2019;194:161–184. doi: 10.1016/j.pharmthera.2018.09.007. [DOI] [PubMed] [Google Scholar]
- Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Khoo B.L., Lee S.C., Kumar P., Tan T.Z., Warkiani M.E., Ow S.G.W., Nandi S., Lim C.T., Thiery J.P. Short-term expansion of breast circulating cancer cells predicts response to anti-cancer therapy. Oncotarget. 2015;6:15578–15593. doi: 10.18632/oncotarget.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klameth L., Rath B., Hochmaier M., Moser D., Redl M., Mungenast F., Gelles K., Ulsperger E., Zeillinger R., Hamilton G. Small cell lung cancer: model of circulating tumor cell tumorospheres in chemoresistance. Sci. Rep. 2017;7:5337. doi: 10.1038/s41598-017-05562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-H., Choon Lim S., Wu L.-C., Wang C.-F., Tsai W.-S., Wu H.-C., Chang Y.-C. Site-specific antibody modification and immobilization on a microfluidic chip to promote the capture of circulating tumor cells and microemboli. Chem. Commun. 2017;53:4152–4155. doi: 10.1039/C7CC00247E. [DOI] [PubMed] [Google Scholar]
- Lai J.-M., Shao H.-J., Wu J.-C., Lu S.-H., Chang Y.-C. Efficient elusion of viable adhesive cells from a microfluidic system by air foam. Biomicrofluidics. 2014;8:052001. doi: 10.1063/1.4893348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.C., Lin J.C., Hwang W.L., Kuo Y.J., Chen H.K., Tai S.K., Lin C.C., Yang M.H. Macrophage-secreted interleukin-35 regulates cancer cell plasticity to facilitate metastatic colonization. Nat. Commun. 2018;9:3763. doi: 10.1038/s41467-018-06268-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.F., Chen J.Y., Ho Y.H., Hsu W.H., Wu L.C., Lan H.Y., Hsu D.S.S., Tai S.K., Chang Y.C., Yang M.H. Snail-induced claudin-11 prompts collective migration for tumour progression. Nat. Cell Biol. 2019;21:251–262. doi: 10.1038/s41556-018-0268-z. [DOI] [PubMed] [Google Scholar]
- Li H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv. 2013 https://arxiv.org/abs/1303.3997 Preprint at. [Google Scholar]
- Liao T.T., Yang M.H. Hybrid epithelial/mesenchymal state in cancer metastasis: clinical significance and regulatory mechanisms. Cells. 2020;9 doi: 10.3390/cells9030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning C.S., Hooper S., Sahai E.A. Intravital imaging of SRF and Notch signalling identifies a key role for EZH2 in invasive melanoma cells. Oncogene. 2015;34:4320–4332. doi: 10.1038/onc.2014.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhdjian A.H., Kai F., Rubashkin M.G., Prahl L.S., Przybyla L.M., McGregor A.L., Bell E.S., Barnes J.M., DuFort C.C., Ou G., et al. Integrin-mediated traction force enhances paxillin molecular associations and adhesion dynamics that increase the invasiveness of tumor cells into a three-dimensional extracellular matrix. Mol. Biol. Cell. 2017;28:1467–1488. doi: 10.1091/mbc.E16-09-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melissaridou S., Wiechec E., Magan M., Jain M.V., Chung M.K., Farnebo L., Roberg K. The effect of 2D and 3D cell cultures on treatment response, EMT profile and stem cell features in head and neck cancer. Cancer Cell Int. 2019;19:16. doi: 10.1186/s12935-019-0733-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebbioso A., Tambaro F.P., Dell'Aversana C., Altucci L. Cancer epigenetics: moving forward. PLoS Genet. 2018;14:e1007362. doi: 10.1371/journal.pgen.1007362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang S.-T., Chang Y.-H., Lin P.-H., Chang Y., Watson D., Segurado O., Lu S.-H., Wu J.-c., Lai J.-M., Shao H.-J., et al. Prospective clinical study of a prostate cancer (PCa) rule-out blood test for PSA gray zone patients using a sensitive circulating tumor cell assay. J. Clin. Oncol. 2018;36:143. doi: 10.1200/JCO.2018.36.6_suppl.143. [DOI] [Google Scholar]
- Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S., Van Keymeulen A., Brown D., Moers V., Lemaire S., et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 2017 doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., Lin D.T., Regev A., Bernstein B.E. Seeding and propagation of untransformed mouse mammary cells in the lung. Science. 2008;321:1841–1844. doi: 10.1126/science.1161621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrió D., Rodriguez-Pinilla S.M., Hardisson D., Cano A., Moreno-Bueno G., Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- Schrörs B., Boegel S., Albrecht C., Bukur T., Bukur V., Holtsträter C., Ritzel C., Manninen K., Tadmor A.D., Vormehr M., et al. Multi-omics characterization of the 4T1 murine mammary gland tumor model. Front. Oncol. 2020;10:1195. doi: 10.3389/fonc.2020.01195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- Suvilesh K.N., Nussbaum Y.I., Radhakrishnan V., Manjunath Y., Avella D.M., Staveley-O'Carroll K.F., Kimchi E.T., Chaudhuri A.A., Shyu C.R., Li G., et al. Tumorigenic circulating tumor cells from xenograft mouse models of non-metastatic NSCLC patients reveal distinct single cell heterogeneity and drug responses. Mol. Cancer. 2022;21:73. doi: 10.1186/s12943-022-01553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T.Z., Miow Q.H., Miki Y., Noda T., Mori S., Huang R.Y.J., Thiery J.P. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol. Med. 2014;6:1279–1293. doi: 10.15252/emmm.201404208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting D.T., Wittner B.S., Ligorio M., Vincent Jordan N., Shah A.M., Miyamoto D.T., Aceto N., Bersani F., Brannigan B.W., Xega K., et al. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 2014;8:1905–1918. doi: 10.1016/j.celrep.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp E., Janni W., Schindlbeck C., Jückstock J., Andergassen U., de Gregorio A., Alunni-Fabbroni M., Tzschaschel M., Polasik A., Koch J.G., et al. Presence of circulating tumor cells in high-risk early breast cancer during follow-up and prognosis. J. Natl. Cancer Inst. 2018;111:380–387. doi: 10.1093/jnci/djy152. [DOI] [PubMed] [Google Scholar]
- Tsai H.A., Shen C.N., Chang Y.C. Use of surface properties to control the growth and differentation of mouse fetal liver stem/prgenitor cell colonies. Biomacromolecules. 2012;13:3483–3493. doi: 10.1021/bm301074j. [DOI] [PubMed] [Google Scholar]
- Tsai J.H., Donaher J.L., Murphy D.A., Chau S., Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W.S., Chen J.S., Shao H.J., Wu J.C., Lai J.M., Lu S.H., Hung T.F., Chiu Y.C., You J.F., Hsieh P.S., et al. Circulating tumor cell count correlates with colorectal neoplasm progression and is a prognostic marker for distant metastasis in non-metastatic patients. Sci. Rep. 2016;6:24517. doi: 10.1038/srep24517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W.S., You J.F., Hung H.Y., Hsieh P.S., Hsieh B., Lenz H.J., Idos G., Friedland S., Yi-Jiun Pan J., Shao H.J., et al. Novel circulating tumor cell assay for detection of colorectal adenomas and cancer. Clin. Transl. Gastroenterol. 2019;10:e00088. doi: 10.14309/ctg.0000000000000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera Alvarez R., Pongor L.S., Mariño-Ramírez L., Landsman D. TPMCalculator: one-step software to quantify mRNA abundance of genomic features. Bioinformatics. 2019;35:1960–1962. doi: 10.1093/bioinformatics/bty896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Tseng, Tsai Antibody conjugated supported lipid bilayer for capturing and purification of viable tumor cells in blood for subsequent cell culture. Biomaterials. 2013 doi: 10.1016/j.biomaterials.2013.03.096. [DOI] [PubMed] [Google Scholar]
- Yeh Tethering Antibody on a Pegylated Liposome-Lipid Bilayer to Promote Flexible Chain Movement for Multivalent Antibody-Antigen Interactions and to Minimize Applied Force on Cells upon Release. Biophysical Journal. 2018;114 [Google Scholar]
- Yeh, Chen, Wang, Chang Promoting Multivalent Antibody Antigen Interactions by Tethering Antibody Molecules on a PEGylated Dendrimer- Supported Lipid Bilayer. Biomacromolecules. 2018 doi: 10.1021/acs.biomac.7b01515. [DOI] [PubMed] [Google Scholar]
- Yu M., Bardia A., Aceto N., Bersani F., Madden M.W., Donaldson M.C., Desai R., Zhu H., Comaills V., Zheng Z., et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Bardia A., Wittner B.S., Stott S.L., Smas M.E., Ting D.T., Isakoff S.J., Ciciliano J.C., Wells M.N., Shah A.M., et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ridgway L.D., Wetzel M.D., Ngo J., Yin W., Kumar D., Goodman J.C., Groves M.D., Marchetti D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl. Med. 2013;5:180ra48. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Wang L., Wang C.Y., Yu J., Guan K.L. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
9 Tables S4 and S5
Data Availability Statement
WES sequencing data were deposited in the NCBI Sequence Read Archive (SRA) (ID: http://www.ncbi.nlm.nih.gov/bioproject/668668).
Bisulfite sequencingdata were deposited in the NCBI BsSeq project (ID: https://www.ncbi.nlm.nih.gov/bioproject/670066). RNA sequencing results were deposited in the NCBI Gene Expression Omnibus (GEO) library (ID: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159727). This paper does not report original code. Any additional information required to reanalyze the data reported in this article is available from the lead contacton request.