Abstract
Background
: The Centers for Disease Control and Prevention (CDC) recommends 5–10 days of isolation for patients with COVID-19, depending on symptom duration and severity. However, in clinical practice, an individualized approach is required. We thus developed a clinical scoring system to predict viable viral shedding.
Methods
: We prospectively enrolled adult patients with SARS-CoV-2 infection admitted to a hospital or community isolation facility between February 2020 and January 2022. Daily dense respiratory samples were obtained, and genomic RNA viral load assessment and viral culture were performed. Clinical predictors of negative viral culture results were identified using survival analysis and multivariable analysis.
Results
: Among 612 samples from 121 patients including 11 immunocompromised patients (5 organ transplant recipients, 5 with hematologic malignancy, and 1 receiving immunosuppressive agents) with varying severity, 154 (25%) revealed positive viral culture results. Multivariable analysis identified symptom onset day, viral copy number, disease severity, organ transplant recipient, and vaccination status as independent predictors of culture-negative rate. We developed a 4-factor predictive model based on viral copy number (-3 to 3 points), disease severity (1 point for moderate to critical disease), organ transplant recipient (2 points), and vaccination status (-2 points for fully vaccinated). Predicted culture-negative rates were calculated through the symptom onset day and the score of the day the sample was collected.
Conclusions
: Our clinical scoring system can provide the objective probability of a culture-negative state in a patient with COVID-19 and is potentially useful for implementing personalized de-isolation policies beyond the simple symptom-based isolation strategy.
Keywords: COVID-19, Viral shedding, Predictive model, Isolation policy
1. Introduction
Since the global emergence of SARS-CoV-2 in 2019, the Centers for Disease Control and Prevention (CDC) has recommended an isolation and precaution period of 5 to 10 days for patients with COVID-19, depending on the severity and duration of the symptoms [1]. However, in clinical settings, large proportion of patients show detectable genomic viral copy numbers with occasionally shedding viable virus even after the recommended isolation period. These patients often have multiple underlying diseases, which may contribute to the individual-level heterogeneity of viable virus shedding [2]. Furthermore, the CDC guideline determines the duration of isolation and precaution regardless of vaccination status, despite several studies suggesting shorter viable viral shedding in vaccinated populations [3]. Therefore, an individualized approach is required to determine the isolation period for patients with COVID-19 [2].
The most valid method to evaluate viable virus shedding is culture-based virus isolation [4,5], which is, however, impractical because of its long running time and high cost [6]. Over the past two years, we conducted four prospective studies to explore the viable virus shedding kinetics of SARS-CoV-2 [3,[7], [8], [9]]. Based on the findings from these early studies, we developed a clinical prediction score system to estimate viable virus shedding, which can contribute to the determination of the isolation and precaution period in real clinical settings.
2. Methods
2.1. Study subjects
From February 2020 to January 2022, we carried out four independent prospective studies to evaluate the viable virus kinetics of SARS-CoV-2. The first cohort included patients admitted to a tertiary hospital in Seoul from February 2020 to December 2020 [9]. The second cohort included patients who had asymptomatic to mild disease, admitted to a community isolation facility in Seoul from January to February 2021 [7]. The third cohort included patients admitted to the community isolation facility from July to August 2021 [3, 8]. The final cohort was recruited from September 2021 to January 2022, when the Delta variant was prevalent in Korea and included patients with various degrees of severity and underlying conditions admitted to the hospital [10]. We performed genomic RNA PCR and culture-based virus isolation on dense respiratory samples to evaluate viable virus kinetics. All enrolled patients provided written informed consent, and studies were approved by institutional review board of Asan Medical Center (IRB 2021-0024).
Data about vaccination status and clinical characteristics, including disease severity, were prospectively collected. Disease severity was defined using the CDC severity criteria [1]. Vaccination status was classified into three groups: fully vaccinated, partially vaccinated, and unvaccinated. The fully vaccinated group included subjects for whom at least 2 weeks had elapsed since the last dose recommended by the CDC (2 doses of mRNA vaccines or ChAdOx1 nCoV-19 for the general population, 3 doses of mRNA vaccines or ChAdOx1 nCoV-19 for moderate to severely immunocompromised individuals)[11]. The unvaccinated group included patients who had never been vaccinated. Patients who did not belong to either group were classified into the partially vaccinated group.
2.2. Measurement of viral load by real-time RT-PCR assay
Viral RNA was extracted from respiratory specimens using a QIAamp viral RNA Mini kit (Qiagen Inc., Hilden, Germany) followed by manufacturer's instruction. Multiplex RT-PCR assay mix (20 μL) included 0.1 μL of 200× enzyme mix, 4 μL of 5× master mix (LightCycler Multiplex RNA Virus Master, Roche, Basel, Switzerland), 500 nM of each S and N gene primer, 200 nM of S and 250 nM of N gene probes, 250 nM of internal control primers, 100 nM of internal control probes (Supplemental Table 1), and 5 μL of extracted RNA or in vitro-synthesized control RNA. We performed PCR amplification with a LightCycler 96 system (Roche) in the following conditions: reverse transcription at 50℃ for 10 min, initial denaturation at 95℃ for 5 min, 45 cycles of 2-step amplification, denaturation at 95℃ for 10 s, and final extension at 60℃ for 30 s. To generate calibration curves, serial dilutions from 107 to 5 copies/μL of synthetic control RNA were assayed in six independent sets of reactions. The detection limit of this assay was 5 copies/reaction (2.6 log copies/ml of specimen) and viral copy numbers were determined by plotting the Ct values against log copies/reaction.
2.3. Cell culture
Culture-based isolation of SARS-CoV-2 from the respiratory specimens that revealed positive genomic RNA results was performed by a plaque assay in a Biosafety Level 3 laboratory at Korea University College of Medicine, Seoul, South Korea. Prior to inoculation with respiratory specimens, Vero cells were cultured in 6-well plates at a density of 9 × 105 cells/well for 24 h. Specimens were serially diluted 10-fold using PBS, and 200 μl of the diluted samples were inoculated into Vero cells and incubated for 1 h (37℃, 5% CO2) with rocking every 15 min, and overlaid with 2 mL of Dulbecco's Modified Eagle Medium/Nutrient Mixture F12 (DMEM/F-12) medium containing 0.6% oxoid agar. Viral plaque formation was visualized by crystal violet staining after 72 h of incubation at 37℃ in a 5% CO2 incubator.
2.4. Outcome measures
The main outcome was the negative conversion of virus culture from daily dense respiratory samples. A prediction model for the culture-positive probability was developed. If there were more than one culture-positive sample on the same day, we selected the sample with the highest viral copy number.
2.5. Statistical analysis
Categorical variables were compared between groups using the χ2-test or the Fisher exact test. Continuous variables were compared between groups using ANOVA or the Kruskal–Wallis test if the distribution was not normal. The Kaplan-Meier method was used to construct survival curves for the probability of positive culture results from the symptom onset. All tests of significance were two-sided; P value <0.05 was considered to be significant. Statistical analyses were conducted using SAS® Version 9.4 (SAS Institute Inc., Cary, NC) and R software (version 4.1.1).
Since viral copy number is a time-dependent variable, we developed a Cox time-varying proportional hazard model using the counting process. To develop the prognostic model, variables significantly (p<0.2) related to the conversion of culture negativity were chosen as candidate predictive factors. The final model was then simplified with a backward elimination procedure using a removal criterion of 0.1. For each candidate predictor, we assessed the assumption of proportional hazards using the Schoenfeld residuals: No violations of the proportional hazard assumptions were found. Validation of training set was implemented through C-index, and bootstrap resampling method was implemented for internal validation. The performance of the final model was examined by assessing the c-index (0.841). Moreover, we performed an internal validation by using the bootstrap resampling method (500 resamples) to correct the c-index (0.826) for overoptimism and to calculate the shrinkage slope (0.902). For clinical purposes, the clinical scoring system was developed based on the final model using the method described by Sullivan et al. [12]. Viral copy numbers categories were defined as each category by regular intervals had one point score in the final prediction model, then the remaining variables’ scores were calculated based on this value.
3. Results
3.1. Patient characteristics
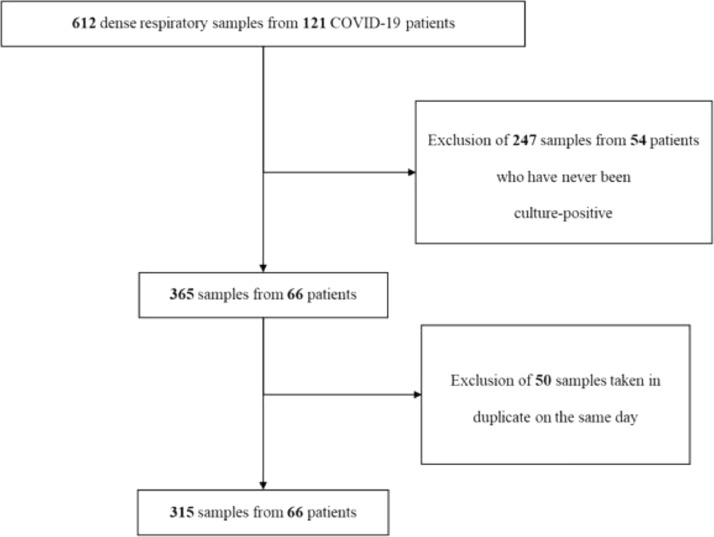
The study flow chart is shown in Fig. 1 . Among the four independent cohorts, 612 samples from 121 patients with varying degrees of severity and underlying diseases were collected. Of these, 247 samples from 55 patients that had never been culture-positive were excluded. Of the 365 samples obtained from the remaining 66 patients, 50 samples duplicated on the same date were excluded. Finally, 315 samples from 66 patients were analyzed.
Fig. 1.
Study flow chart.
The baseline clinical characteristics of the study subjects are shown in Table 1 . There was significant heterogeneity between the cohorts. The cohorts from the community isolation facility (cohorts 2 and 3) tended to consist of patients who were younger and less severely ill compared to the hospitalized cohorts (cohorts 1 and 4).
Table 1.
Baseline characteristics.
| Total (n=121) | Cohort 1 (n=19) | Cohort 2 (n=21) | Cohort 3 (n=39) | Cohort 4 (n=42) | p-value | |
|---|---|---|---|---|---|---|
| Study period | Feb 2020 – Jan 2022 | Feb 2020 – Dec 2020 | Jan 2021 – Feb 2021 | Jul 2021 – Aug 2021 | Sept 2021 – Jan 2022 | |
| Study location | Community isolation facility and hospital | Hospital | Community isolation facility | Community isolation facility | Hospital | |
| Variant dominancy | Pre-delta to delta | Pre-delta | Pre-delta | Delta | Delta | |
| Age (years ± SD) | 48.8 ± 17.6 | 57.6 ± 17.7 | 44.7 ± 13.5 | 39.8 ± 15.9 | 55.3 ± 16.7 | <0.001 |
| Sex | 0.11 | |||||
| Female | 69 (57) | 8 (42) | 12 (57) | 28 (72) | 21 (50) | |
| Male | 52 (43) | 11 (58) | 9 (43) | 11 (28) | 21 (50) | |
| Severity | <0.001 | |||||
| Asymptomatic to mild | 80 (66) | 3 (16) | 19 (90) | 34 (87) | 24 (57) | |
| Moderate | 18 (15) | 5 (27) | 2 (10) | 5 (13) | 6 (14) | |
| Severe to critical | 23 (19) | 11 (58) | 0 | 0 | 12 (29) | |
| Prior history of SARS-CoV-2 infection | 0 | 0 | 0 | 0 | 0 | >0.99 |
| Vaccination Status | <0.001 | |||||
| Not vaccinated | 84 (69) | 19 (100) | 21 (100) | 23 (59) | 21 (50) | |
| Partially vaccinated | 14 (12) | 0 | 0 | 11 (28) | 3 (7) | |
| Fully vaccinateda | 23 (19) | 0 | 0 | 5 (13) | 18 (43) | |
| Underlying Diseases or Conditions | ||||||
| Organ Transplant Recipient | 5 (4) | 1b (5) | 0 | 0 | 4c (10) | 0.13 |
| Hypertension | 31 (26) | 4 (21) | 4 (19) | 8 (21) | 15 (36) | 0.37 |
| Diabetes mellitus | 23 (19) | 6 (32) | 0 | 4 (10) | 13 (31) | 0.003 |
| Liver cirrhosis | 5 (4) | 1 (5) | 1 (5) | 0 | 3 (7) | 0.31 |
| Chronic renal insufficiency | 4 (3) | 0 | 0 | 0 | 4 (10) | 0.06 |
| Solid cancer | 9 (7) | 1 (5) | 0 | 0 | 8 (19) | 0.003 |
| Hematologic malignancy | 5 (4) | 3 (16) | 0 | 0 | 2 (5) | 0.02 |
| Pregnancy | 7 (6) | 0 | 0 | 0 | 7 (17) | 0.003 |
| SARS-CoV-2 variants | <0.001 | |||||
| Not defined | 40 (33) | 19 (100) | 21 (100) | |||
| Delta | 80 (66) | 39 (100) | 41 (98) | |||
| Omicron | 1 (0) | 1 (2) | ||||
| Sample characteristics | 612 | 155 | 144 | 151 | 162 | |
| Sample types | <0.001 | |||||
| Saliva | 494 (81) | 37 (24) | 144 (100) | 151 (100) | 162 (100) | |
| Nasopharyngeal swab | 63 (10) | 63 (41) | 0 | 0 | ||
| Sputum | 55 (9) | 55 (35) | 0 | 0 | ||
| Culture positivity | 154 (25) | 62 (40) | 29 (20) | 38 (25) | 24 (15) | <0.001 |
NOTE. Data are shown as number of patients (%) unless otherwise indicated.
a Including 3 patients who received the 3rd doses of vaccines. Two of them were immunocompromised (one with active chemotherapy against multiple myeloma, and the other received mycophenolate mofetil due to rheumatoid arthritis-related interstitial pneumonitis). The remaining one patient received the 1st booster dose of COVID-19 vaccine.
b One patient received allogeneic stem cell transplantation.
c Among 4 patients, one received deceased donor liver transplantation, two living donor kidney transplantation, and one allogeneic stem cell transplantation.
3.2. Predictive factor stratification, score model, and internal validation
The probability of the positive viral culture results depending on the duration of symptom onset is shown in Supplemental Figure 1 and Supplemental Table 2. Univariate and multivariable Cox models revealed that four variables were independently associated with the negative conversion of viral culture, namely viral copy number, moderate to critical severity, organ transplant recipient, and fully vaccinated status (Table 2 ). These variables were included in the model to predict viable virus shedding (Table 3 ).
Table 2.
Predictive factors of negative conversion rate of viral culture results.
| Characteristics | Univariate analysis | Multivariable analysis | ||||
|---|---|---|---|---|---|---|
| Culture-negative conversion rate | 95% CI | p-value | Culture-negative conversion rate | 95% CI | p-value | |
| Age | 1.00 | 0.99–1.01 | 0.66 | |||
| Male sex | 1.54 | 1.01-2035 | 0.03 | |||
| Hypertension | 0.98 | 0.58–1.67 | 0.95 | |||
| Diabetes mellitus | 0.89 | 0.48–1.65 | 0.70 | |||
| Liver cirrhosis | 0.89 | 0.54–1.46 | 0.63 | |||
| Chronic renal insufficiency | 0.79 | 0.31–2.08 | 0.63 | |||
| Solid cancer | 1.58 | 0.62–4.02 | 0.34 | |||
| Hematologic malignancy | 0.19 | 0.03–1.13 | 0.07 | |||
| Pregnancy | 1.49 | 0.65–3.43 | 0.35 | |||
| Viral copy number | 0.53 | 0.45–0.61 | <0.001 | 0.57 | 0.49–0.66 | <0.001 |
| Severity | ||||||
| Asymptomatic to mild | 1.00 | - | 0.02 | 1.00 | - | 0.053 |
| Moderate | 0.60 | 0.31–1.16 | 0.13 | 0.62 | 0.37–0.97 | 0.049 |
| Severe to critical | 0.45 | 0.24–0.83 | 0.01 | 0.52 | 0.25–1.10 | 0.07 |
| Immunocompromised | 0.31 | 0.12–0.88 | 0.03 | |||
| Organ transplant recipient | 0.27 | 0.08–0.91 | 0.03 | 0.62 | 0.10–0.90 | 0.03 |
| Vaccination status | ||||||
| Not vaccinated | 1.00 | - | <0.001 | 1.00 | - | <0.001 |
| Partially vaccinated | 0.98 | 0.49–1.99 | 0.96 | 0.98 | 0.46–2.14 | 0.95 |
| Fully vaccinated | 3.32 | 2.07–5.33 | <0.001 | 2.81 | 1.92–4.14 | <0.001 |
Table 3.
Prediction scores for negative viral culture results.
| Categories | Reference value | β coefficient | Score | ||
|---|---|---|---|---|---|
| -0.569 | |||||
|
Viral copy number (log copies/mL) [Ct value] |
>8 [<17.45] | 8.4 | 3 | ||
| 7–8 [20.78–17.45] | 7.5 | 2 | |||
| 6–7 [24.12–20.78] | 6.5 | 1 | |||
| 5–6 [27.45–24.12] | 5.5 (Reference) | 0 | |||
| 4–5 [30.79–27.45] | 4.5 | -1 | |||
| 3–4 [34.12–30.79] | 3.5 | -2 | |||
| <3 [>34.12] | 2.9 | -3 | |||
| Severity | Asymptomatic to mild | Reference | 0 | 0 | |
| Moderate | -0.473 | 1 | |||
| Severe to Critical | -0.657 | 1 | |||
| Organ transplant recipient | No | Reference | 0 | 0 | |
| Yes | -1.210 | 2 | |||
| Vaccination status | Unvaccinated | Reference | 0 | 0 | |
| Partially vaccinated | -0.025 | 0 | |||
| Fully vaccinated | 1.035 | -2 | |||
The predicted negative conversion rates at symptom onset days 3, 5, 7, 10, 14, and 20 are summarized in Table 4 . The predicted culture-negative rates at symptom onset day 10 in patients with scores -6, 0, and 5 were 100%, 86%, and 6%, respectively (Table 4).
Table 4.
Predicted culture-negative rates.
| DaysScore | 3 | 5 | 7 | 10 | 14 | 20 |
|---|---|---|---|---|---|---|
| -5 | 100 | 100 | 100 | 100 | 100 | 100 |
| -4 | 96 | 100 | 100 | 100 | 100 | 100 |
| -3 | 84 | 99 | 100 | 100 | 100 | 100 |
| -2 | 64 | 93 | 99 | 100 | 100 | 100 |
| -1 | 44 | 78 | 91 | 97 | 100 | 100 |
| 0 | 28 | 57 | 75 | 86 | 96 | 99 |
| 1 | 17 | 38 | 55 | 67 | 85 | 93 |
| 2 | 10 | 24 | 36 | 47 | 66 | 78 |
| 3 | 6 | 14 | 22 | 30 | 45 | 58 |
| 4 | 3 | 8 | 13 | 19 | 29 | 39 |
| 5 | 2 | 5 | 8 | 11 | 18 | 24 |
| 6 | 1 | 3 | 5 | 6 | 10 | 15 |
NOTE. Data are shown as % unless otherwise specified.
4. Discussion
Although culture-based isolation is considered the gold standard for ascertaining infectivity, a limited number of studies used it for SARS-CoV-2 [4]. Accordingly, the evidence to determine the period of isolation in patients who are immunocompromised or have severe COVID-19 is sparse, and establishing the isolation period for these patients is challenging [4]. Therefore, a predictive model allowing an individualized approach to assess the infectivity of patients with COVID-19 is increasingly needed. Our clinical scoring system to predict viable viral shedding may provide important information in real clinical settings and help determine the period of isolation and precaution. In particular, this model evaluates a quantified risk depending on the patient's sample collection time and risk factors. A quantified risk allows clinicians to adopt more objective and flexible isolation strategies for specific clinical environments.
In multivariable analysis, 4 variables were found to be independently associated with a delayed negative conversion of culture-based viral isolation: viral copy number, severity, having received an organ transplant, and fully vaccinated status. These predictive factors are in line with previous studies evaluating viable viral kinetics [13], [14], [15], [16], [17]. The previous study demonstrated that infectious viral load is well correlated with genomic viral copy number [17]. However, genomic viral copy number does not reflect infectiousness of COVID-19, but rather correlates with them. Therefore, we developed the prediction model using genomic RNA copy number and days from the symptom onset with disease severity and vaccination status together. It is worth to note that it should be cautious to directly apply Ct value itself in our model as viral load without a standard curve using reference materials [18]. Interestingly, the univariate analysis revealed that male sex was statistically significantly associated with short duration of viable viral shedding. In contrast, the multivariable analysis exhibited this statistical significance was marginal (Supplemental Table 3). Considering the biologic plausibility and the inconsistent results from the previous studies [19, 20] about the sex effect on viable viral shedding, we excluded the sex variable in the final model. However, when male sex was included in the final model, the clinical prediction model revealed similar findings (Supplemental Tables 3, 4, 5).
Previous studies observed that low viral copy numbers especially beyond 7 days from the symptom onset were correlated with a lower chance of viable virus isolation [15, 16, 21, 22]. However, growing evidence suggests that substantial heterogeneity of viral kinetics exists between individuals with COVID-19, although the study could not assess the factors associated with this heterogeneity [2]. In this context, our study may provide a customized approach by using the easily accessible parameters depending on the symptom duration to de-isolation strategy and could mitigate the important health care burden related to isolation. That is, our clinical prediction model can be easily integrated in a medical calculator or infection control app. Especially in resource-limited setting for isolation such as many multi-patient rooms in the wards in South Korea, this objective scoring system may help the prioritizing of patients who need strict isolation. In addition, there is another potential benefit of decreasing unnecessary work restriction at the time of the shortage of medical staff.
Our study has several limitations. First, the cohorts used in the analysis were enrolled when the wild-type and Delta variants were prevalent in Korea. In the two early studies, we did not perform genomic typing of the isolated SARS-CoV-2, so information about SARS-CoV-2 variants was omitted [7, 9]. It is worth noting that strains other than wild-type started to be isolated in Korea in late 2020 [23]. Since variant strains were not included as factors in the multivariable analysis, the generalizability of this study in the era of the Omicron variant is limited. Furthermore, in our clinical model, vaccination was closely associated with shortening the viral shedding period, but there have been limited data on the effect of COVID-19 vaccination on the duration of viable viral shedding in the era of Omicron variant. The previous studies [24, 25] reported that fully vaccinated young individuals with mild Omicron variant infection had a relatively shortened viral shedding period of median 5 to 6 days, which suggests that viral shedding period may be shortened in fully vaccinated young individuals with Omicron variant infection. However, these studies did not compare vaccinated individuals with Omicron infection to unvaccinated individuals with Omicron infection, so it is difficult to draw a firm conclusion about the vaccination effect on the viable viral shedding of Omicron variant. So, the vaccination status might not be important or predicted culture-negative rates depending on the symptom onset might be higher during the Omicron-dominant era. Further studies are needed on this area. Second, most of our samples were from saliva (Table 1), whereas nasopharyngeal swabs may have been advantageous, being widely used in clinical practice and having more generalizability in hospital setting. However, obtaining daily nasopharyngeal swabs was logistically challenging in many aspects, while saliva could be a safe alternative for evaluating culture positivity by dense sampling [2, 26]. Third, our study did not include a sufficient number of patients who were immunocompromised. This limits the application of our clinical model for prediction of viral shedding period in immunocompromised patients with COVID-19, although these patients are the most challenging to determine their viable viral shedding period. In addition, patients who are moderately or severely immunocompromised have viral shedding for more than 20 days [4]. Therefore, our model's estimation of the probability of viable viral shedding up to 20 days after symptom onset has limited applicability to patients who are immunocompromised, and further studies including more patients who are immunocompromised, are needed to extend the estimate of the probability of viable viral shedding beyond 20 days after symptom onset to validate our clinical model in the immunocompromised patients. Fourth, several previous studies have demonstrated that effect of vaccination against COVID-19 wanes over time [27, 28]. However, we could not evaluate the waning effect of vaccination on the viable viral shedding. Further studies are needed on this issue. Fifth, we only included patients above 18 years old, which also limits the generalizability of this model into pediatric population. Finally, we did not perform external validation, so further studies are warranted to validate this prediction model.
In conclusion, our clinical scoring system can provide the objective probability of negative culture status in a given patient with COVID-19 with genomic viral load and appears to be useful for implementing a de-isolation policy depending on individualized factors associated with viable viral shedding beyond the simple symptom-based isolation strategy recommended by the CDC.
Funding
This study was supported by a grants from the Korea National Institute of Health (Grant no. 2022-ER1609-00) and from the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT, Republic of Korea (NRF-2022M3A9I2017241 and NRF-2018M3A9H4056537).
Declaration of Competing Interest
The authors have declared that no conflict of interest exists.
Acknowledgments
The authors acknowledge the contribution of health care workers from Asan medical center.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2022.105319.
Appendix. Supplementary materials
References
- 1.Isolation and Precautions for People with COVID-19 | CDC. August 11, 2022 ed: Centers for disease control and prevention; 2022. https://www.cdc.gov/coronavirus/2019-ncov/your-health/isolation.html.
- 2.Ke R, Martinez PP, Smith RL, Gibson LL, Mirza A, Conte M, et al. Daily longitudinal sampling of SARS-CoV-2 infection reveals substantial heterogeneity in infectiousness. Nat. Microbiol. 2022;7:640–652. doi: 10.1038/s41564-022-01105-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung J, Kim JY, Park H, Park S, Lim JS, Lim SY, et al. Transmission and infectious SARS-CoV-2 shedding kinetics in vaccinated and unvaccinated individuals. JAMA Network Open. 2022;5 doi: 10.1001/jamanetworkopen.2022.13606. e2213606-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ending Isolation and Precautions for People with COVID-19: Interim Guidance. January 14, 2021 ed: centers for disease control and prevention; 2022. https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html.
- 5.Fontana LM, Villamagna AH, Sikka MK, McGregor JC. Understanding viral shedding of severe acute respiratory coronavirus virus 2 (SARS-CoV-2): Review of current literature. Infect. Control Hosp. Epidemiol. 2021;42:659–668. doi: 10.1017/ice.2020.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basile K, McPhie K, Carter I, Alderson S, Rahman H, Donovan L, et al. Cell-based culture informs infectivity and safe de-isolation assessments in patients with coronavirus disease 2019. Clin. Infect. Dis. 2021;73:ṅ. doi: 10.1093/cid/ciaa1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bae S, Kim JY, Lim SY, Park H, Cha HH, Kwon JS, et al. Dynamics of viral shedding and symptoms in patients with asymptomatic or mild COVID-19. Viruses. 2021;13 doi: 10.3390/v13112133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park S, Lim SY, Kim JY, Park H, Lim JS, Bae S, et al. Clinical and virological characteristics of SARS-CoV-2 B.1.617.2 (Delta) variant: a prospective cohort study. Clin. Infect. Dis. 2022 doi: 10.1093/cid/ciac239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JY, Bae JY, Bae S, Cha HH, Kwon JS, Suh MH, et al. Diagnostic usefulness of subgenomic RNA detection of viable SARS-CoV-2 in patients with COVID-19. Clin. Microbiol. Infect. 2022;28:101–106. doi: 10.1016/j.cmi.2021.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang SW, Kim JY, Park H, Lim SY, Kim J, Bae S, et al. Comparison of outward transmission potential between vaccinated and partially vaccinated or unvaccinated individuals with the SARS-CoV-2 delta variant infection. J. Infect. 2022;85:e69–e71. doi: 10.1016/j.jinf.2022.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical Guidance for COVID-19 Vaccination | CDC. April 21, 2022 ed: centers for disease control and prevention; 2022. https://www.cdc.gov/vaccines/covid-19/clinical-considerations/interim-considerations-us.html.
- 12.Sullivan LM, Massaro JM, D'Agostino RB., Sr Presentation of multivariate data for clinical use: the framingham study risk score functions. Stat. Med. 2004;23:1631–1660. doi: 10.1002/sim.1742. [DOI] [PubMed] [Google Scholar]
- 13.van Kampen JJA, van de Vijver D, Fraaij PLA, Haagmans BL, Lamers MM, Okba N, et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19) Nat. Commun. 2021;12:267. doi: 10.1038/s41467-020-20568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folgueira MD, Luczkowiak J, Lasala F, Pérez-Rivilla A, Delgado R. Prolonged SARS-CoV-2 cell culture replication in respiratory samples from patients with severe COVID-19. Clin. Microbiol. Infect. 2021;27:886–891. doi: 10.1016/j.cmi.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 16.Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin. Infect. Dis. 2020;71:2663–2666. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakki S, Zhou J, Jonnerby J, Singanayagam A, Barnett JL, Madon KJ, et al. Onset and window of SARS-CoV-2 infectiousness and temporal correlation with symptom onset: a prospective, longitudinal, community cohort study. Lancet. Respir. Med. 2022 Aug 18;S2213-2600(22)00226-0. [DOI] [PMC free article] [PubMed]
- 18.Han MS, Byun JH, Cho Y, Rim JH. RT-PCR for SARS-CoV-2: quantitative versus qualitative. Lancet Infect. Dis. 2021;21:165. doi: 10.1016/S1473-3099(20)30424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu K, Chen Y, Yuan J, Yi P, Ding C, Wu W, et al. Factors associated with prolonged viral RNA shedding in patients with coronavirus disease 2019 (COVID-19) Clin. Infect. Dis. 2020;71:799–806. doi: 10.1093/cid/ciaa351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long H, Zhao J, Zeng HL, Lu QB, Fang LQ, Wang Q, et al. Prolonged viral shedding of SARS-CoV-2 and related factors in symptomatic COVID-19 patients: a prospective study. BMC Infect. Dis. 2021;21:1282. doi: 10.1186/s12879-021-07002-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 22.Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 2020;382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park AK, Kim IH, Kim J, Kim JM, Kim HM, Lee CY, et al. Genomic surveillance of SARS-CoV-2: distribution of clades in the Republic of Korea in 2020. Osong Public Health Res. Perspect. 2021;12:37–43. doi: 10.24171/j.phrp.2021.12.1.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boucau J, Marino C, Regan J, Uddin R, Choudhary MC, Flynn JP, et al. Duration of shedding of culturable virus in SARS-CoV-2 Omicron (BA.1) infection. N. Engl. J. Med. 2022 doi: 10.1056/NEJMc2202092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouton TC, Atarere J, Turcinovic J, Seitz S, Sher-Jan C, Gilbert M, et al. Viral dynamics of Omicron and Delta SARS-CoV-2 variants with implications for timing of release from isolation: a longitudinal cohort study. Clin. Infect. Dis. 2022 doi: 10.1093/cid/ciac510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.To KK, Tsang OT, Yip CC, Chan KH, Wu TC, Chan JM, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020;71:841. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin EG, Lustig Y, Cohen C, Fluss R, Indenbaum V, Amit S, et al. Waning immune humoral response to BNT162b2 Covid-19 vaccine over 6 months. N. Engl. J. Med. 2021;385:e84. doi: 10.1056/NEJMoa2114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chemaitelly H, Tang P, Hasan MR, AlMukdad S, Yassine HM, Benslimane FM, et al. Waning of BNT162b2 vaccine protection against SARS-CoV-2 infection in Qatar. N. Engl. J. Med. 2021;385:e83. doi: 10.1056/NEJMoa2114114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.