Abstract
SARS-COV-2 (COVID-19) the virus that caused an epidemic of sever acute respiratory syndrome is what the world has been dealing with since Dec 2019. As the pandemic continues different variants that emerge during mutations have become the latest concern, with notable examples detected in South Africa, Brazil, and UK. Variants are complicated and each one is a collection of several mutations, all of which have the potential to change the virus in unexpected ways. Studying variants is imperative as they can lead the epidemic to the increase of population immunity. In the present study, we reviewed key mutations and concerning variants according to the WHO tracking Sars-Cov-2 program. Databases were searched through Feb to Mar 2022. Overall, 477 studies were extracted from databases, among them 165 studies included mutations, 239 included COVID-19 variants and 43 included both mutations and variants. At the final step of data screening 24 studies associated to mutations, 31 studies with the highlighted information on COVID-19 variants and 31 studies related to both mutations and variants were extracted for this review article. In conclusion, analyses of the genomic sequence of SARS-CoV-2 indicate that structural proteins are key molecules in the assembly of virus while NSPs can have different biochemical properties and possibly cellular functions.
Keywords: COVID 19, SARS-COV-2, Mutations, Concerning variants
Introduction
World has experienced the most demolishing pandemic of the 21st century from Dec 2019 in Wuhan, China where the first outbreak of the virus was detected (1). Up to now according to the WHO, 5.76 million deaths have occurred and 378 million cases have been detected globally, all associated to the Infection accounted by a novel coronavirus (SARS-COV-2) (https://www.who.int/emergencies/diseases/novel-coronavirus-2019).
The human project genome scientists detected foot prints of an ancient corona virus epidemic in the East Asia. David Enrad, a professor of Ecology and Evolutionary Biology, University of Arizona, USA has quoted that the human project genome discovering Corona virus footprints does not straightly manifest the ‘ancient virus’ but it exhibits natural selection signs and affect during ancient humans time. This project was able to track 42 variant encoding protein interacting viruses (VIPs) of about 900 generation. In which this study vividly illustrates 20,000 years back of East Asia human’s fitness process (2, 3). Paleo Virology also exhibits of confirmations that Coronavirus history goes back to more than 2000 years ago. It is under presumption that adapting potential unto new viruses infection can be described the Neanderthals and Humans inter-breeding. One of the last studies done by scientist has shown a specific region regarding COVID-19 which is on chromos number 3, and is also related to Neanderthals genes. This finding illustrates for us how imperative it is to reveal and discover information about pathogens elaboration, when analyzing and evaluating the disease pandemic (4, 5). Blood group type A has a higher chance to get infected and blood group type O exhibits lower chance to get infected by Sars-Cov-2 virus. Immune system reaction might be vary from men to women, since the X chromosome control number of immune system reactions. Therefore, women immune system may response more effective in compared to men (6).
Up to now, human outbreaks have encountered number of Coronaviruses related to the infectious diseases such as SARS in 2002–2003 and Middle East Respiratory Syndrome (MERS) in 2012. These infections are mainly responsible for respiratory and gastrointestinal tracts (7). In the beginning of this pandemic, there were few patient’s manifestation among children under 15 about 1.7%–2%, but after a while, some affected and death reports started to being announced from different countries (8).
Currently illumination of the molecular aspects of COVID-19 has become one of the first priorities of global health organizations since Dec 2019 and a lot of fruitful and positive discoveries and outcomes have been gained.
Coronaviruses are enveloped RNA viruses with positive single stranded genome which belong to Coronaviridae family. There are four main subgroups of coronaviruses: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. SARS-COV-2 is classified in β coronavirus genus. RNA genome of single stranded CoVs is consisted of 29,891 nucleotides that encodes 9890 amino acids. The SARS-CoV-2 genome comprises several number of open reading frame (ORF) for the coding of structural (SP) and non-structural proteins (NSP) involved in viral life cycle and its pathogenic mechanisms (9). Coronaviruses have two types of protein on the basis of function: structural and nonstructural proteins. Structural proteins that help in making the structure of the virus includes: S (spike protein), E (envelope protein), M (membrane protein) and N (nucleocapsid protein) there are also sixteen non-structural proteins (nsp1-16) non-structural proteins that help in viral metabolism and interaction with host immune system (10). Coronaviruses like other viruses change during multiple replications and the change sometimes can cause a new pathogenic variant that may affect virus’s transmission, as well as disease intensity and vaccine efficiency (11). All SARS-COV-2 mutations is being tracked by the WHO, some of them as mentioned are identified as variants of concerns (VOC) due to the severity of the disease on public health and some of them are labeled as variants of interest (VOI). It is highly essential to monitor and trace the emergence of variants and firstly to do anything researchers need data and look forward to how these mutated viruses could affect global health effort.
Here, in this review we provide the latest SARSCOV-2 variants and related mutations up to Apr 2022.
Methods
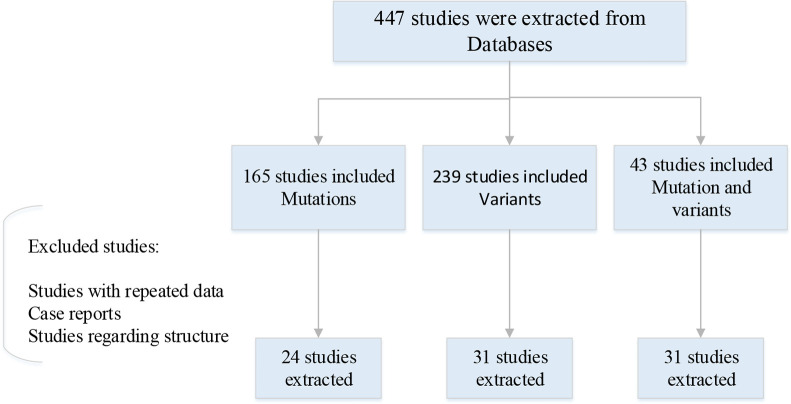
PubMed, Medline, Google Scholar and National databases were searched through Feb to Mar 2022. All the databases were searched using following keywords and terms: SARS-COV-2, COVID 19, and SARS-COV-2 ‘AND’ mutations. SARS-COV-2 ‘AND’ Concerning Variants. We also reviewed and rechecked the references sections for each of the studies to consolidate and identify further relevant studies. Then the applicable publications which were most relevant to notable mutations and variants were selected through our collected data (Fig. 1).
Fig. 1:
Information flow through the phase of review article. All the studies were published during 2019–2022
Results and Discussion
“SARS-COV-2 mutations”
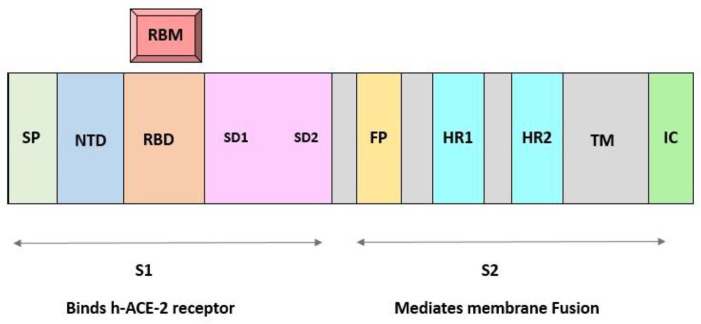
Spreading SARS-COV-2 various variants share several mutations that empower them in rising population while expanding their replication wellness. Most of these mutations occur in spike gene. In order to obtain a wide knowledge over SARS-COV-2 variants of concerns and variants of interests it is imperative to notice the mutation within viral properties (12). Spike proteins enhance the entry of the virus into the host cells by attaching to the particular host receptor. Therapeutics and vaccines are a part of clinical interventions for SARS-COV-2 infection (13). Monoclonal antibodies (mABs) have shown a clinical benefit in preventing infection by acting on the SARS-COV-2 spike protein (S) (14). The S protein consists of N-terminal subunit (S1) mediating receptor binding and C-terminal subunit S2 responsible for virus cell membrane fusion. During viral entry into cells, the receptor binding domain (RBD) of S1 engages the human ACE2 (15). Fig. 2 illustrates the molecular structure of COVID-19 spike protein in details.
Fig. 2:
Molecular structure of Spike protein, S1 and S2 region. S1 is for attachment and S2 is for fusion
SP: Signal peptide of S1/RBD: Receptor binding domain/RBM: Receptor binding motif (which is within RBD) / FP: Fusion peptides/HR: Hepta repeats
D614G
In late Feb 2020, SARS-COV-2 virus D614G point mutation within the spike genome of the virus spread rapidly through population. That nearly all the Covid19 viruses were having this mutation after a while. Initial research attempt showed this mutation has higher transmissibility and more pathogenic in comparison with original SARS-COV-2 (16, 17). The Spike protein mutation encoded by D614G, in the C terminal region of S1 domain directly related to S2. This mutation with altering glycine amino acid with aspartic acid, was dominating globally (18). This variant has the substitution of acid aspartic at the 614th position in the chain, to the amino acid glycine (19).
N501Y
The first emergence of this mutation was seen in United Kingdom and South Africa. N501Y mutation enhances ACE2 proximity and replication of the virus. This mutation is on the RBD and the affinity to the host cell receptor is notably increased by this mutation (20). The N501Y mutation consists of substitution of amino acid asparagine with tyrosine at position 501 in the spike protein (21). The N501Y has a 4-fold higher affinity to wild-type SARS-COV-2 to the ACE2 receptor (22). The N501Y also increase transmission of variants carrying the mutation. The transmission speed of variant form united Kingdome and South Africa was significantly higher than non-N501Y variant and swiftly became the dominant variant (23).
E484K
The E484K mutation ability to escape from neutralization could affect antibody-based countermeasures such as monoclonal antibodies and vaccines (23). E484K mutations has occurred in multiple variants. It was initially discovered in South Africa variant and later in the Brazilian variant (24). The E484K mutation occurs in spike protein and initiating the viral entry process. The mutation occurs at critical sites in the receptor binding motif of the RBD. As the central functional motif is relatively unchanged and directly impacts of binding to the ACE2 receptor. E484K consists of negatively charged amino acid (glutamic acid [E]) substituted with a positively- charged amino acid (Lysine [K]). The amino acid substitution occurs at position 484 in the receptor binding domain of the spike protein (25, 26).
Other RBD mutations
L452R, B.1.617.1, B.1.427/9, K417N/T, N439K, Y453F, S477N, T478K, F490S, S494P are all related to RBD mutations and enhancing RBD binding to the receptor (27).
NTD mutations
In different variants of concerns and variant of interests and individuals infected with extended SARS-COV-2 infections NTD deletions have been observed (28). Higher viral replication with deletion at 69–70 position have been associated in NTD mutations (27). One other deletion at position 141–146 and 242–244 participate in neutralizing of NTD antibodies. In L18F, D253Y and other NTD mutations reduction of susceptibility to NTD neutralizing antibodies occur (27, 29). Mutations close to the S1/S2 furin cleavage site and Non- spike mutations have been reported and seen in different variants of SARS-COV-2. While mutations such as Q675 H/R and P681H/R that seems to have a role in enhancement of S1/S2 cleavage in human cells to have an impact on SARS-COV-2. Hence non-spike mutations enhance transitivity of the virus to the host cells (30–32).
“SARS-COV-2 Concerning Variants”
Variants are labeled by their transmission rate and disease severity. Their lineage is an important factor for their classification as well as the mutation within the particular variants. Variants of concern with higher transmission, and increasing rate in hospitalization or morbidity are listed as below: (WHO Official Updates - Coronavirus Disease 2019)
Alpha Variant (B.1.1.7)
The SARS-COV-2 Alpha variant is one of the earliest mutated strains form the original SARSCOV-2. This variant was found in September 2020 in the United Kingdom (33). The key mutations acquired by the Alpha variant that make this variant, a variant of concern includes: several RBD mutations N501Y, P681H, at position 69–70 and 144 NTD deletions and several non-spike mutations (27, 34).
Beta Variant (B.1.3.51)
The Beta variant was first found in May 2020 in South Africa as of July 2021(35). The key mutations acquired by the Beta variant that made this variant, a variant of concern includes: nine mutation in spike protein that N501Y, E484K and K417N are RBD mutations and NTD deletions at position 242–244. Sectional or whole viral evasion from mAB have been found in several studies (35) 34).
Gamma Variant (P.1)
Gamma variant was first found in Brazil in January 2021 (36). From the genetic sequencing of many virus samples from infected individuals discovered that Gamma variant has accumulated over 22 mutations with about 12 mutations on spike protein. RBD mutations include: L18F, N501Y, E484K and K 417T. NTD mutations are also discovered in gamma variant. This variant had higher rate of hospitalization and morbidity over 3 to 4 times in comparison with previous discovered variants (37).
Delta Variant (B.1.617.2)
Delta variant was first found in India in late 2020. Delta variant swiftly became the dominant strain in many countries. WHO at the early stages of Delta variant emergence announced that Delta variant is the most transmissible of the variants identified so far (WHO Official Updates - Coronavirus Disease 2019). Delta variant has 23 mutations, in which the key mutations include: Spike protein E484Q and L452R RBD mutation as well as P681R cleavage site mutation. Studies on this variant has shown several mutation on ORF3, ORF7 as well (38). This variant has had a higher transmission and infectious than other variants. Research conducted in Scotland showed that the prevalence of Delta variant among young ages. Symptoms that are common in patients with Delta virus include: Cough, Fever, and shortness of breath, vomiting, diarrhea, headache and sore throat, loss of taste, fatigue (39, 40).
Omicron (B.1.1.529)
This variant was first found in Nov 2021. Omicron is classified as a variant of concern since it has some concerning properties. This variant has large number of mutations, more than 50 that most of them are involved in immune escape or having higher transmissibility (41). This variant has so many mutations, and there is no link with any previous variants (42). What scientists have discovered is that it has more than 30 genetic changes in the spike protein (43). Regarding the emergence of this variant there are different thoughts, what is crystal clear is that the initial cases of this variant was first discovered in Southern Africa in Botswana, and one possibility on emergence of the virus is that this variant emerged in immunocompromised (Like HIV) patients who had Coronavirus for many months and that chronic Infection allowed the virus to slowly adapt inside the patient and become better suited to being inside human’s body. (Symptoms that are shown in patients with Omicron variant include: fever, cough and fatigue from mild to severe. Surprisingly loss of taste has been decreased in positive cases of this variant (44, 45).
SARS-CoV-2 is a positive- sense, single-stranded RNA virus, characterized as a member of the Betacoronaviruses (Beta-CoVs) of four genera (Alpha, Beta, Gamma and Delta) in the subfamily Orthocoronavirinae and the Coronaviridae family of the order Nidovirales (46). To date, two concerning variants Delta and Omicron are circulating around the world. Variants are characterized by their mutation constitute. Transmissibility, disease severity, and the ability to evade humoral immunity are all traits that distinguish different variants. Up to now various type of vaccines has gone through trials and until June 19th, 2021, approximately 21% of worldwide population has at least received one dose of vaccination (47).
Conclusion
Further studies should be conducted to better illustrate more detailed mechanisms of SARSCoV-2 participation in modulating viral replication and pathogenesis.
Journalism Ethics considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Footnotes
Conflict of interest
The authors declare that there is no conflict of interests.
References
- 1.Zhu N, Zhang D, Wang W, et al. (2020). A Novel Coronavirus from Patients with Pneumonia in China. N Engl J Med, 382(8): 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farhud DD, Bahadori M, Zarif-Yeganeh M. (2021). Evidence of the Ancestries of COVID-19 Virus in East Asia, More than 20,000 Years Ago. Iran J Public Health, 50(9): i–v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enard D, Petrov DA. (2020). Ancient RNA virus epidemics through the lens of recent adaptation in human genomes. Philos Trans R Soc Lond B Biol Sci, 375(1812): 20190575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farhud DD, Azari M, Mehrabi A. (2022). The History of Corona Virus, from Neanderthals to the Present Time: A Brief Review. Iran J Public Health, 51(3): 531–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Osail AM, Al-Wazzah MJ. (2017). The history and epidemiology of Middle East respiratory syndrome corona virus. Multidiscip Respir Med, 12:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farhud DD, Zokaei S. (2020). Fight against Viruses (COVID-19): Peace among Nations. Iran J Public Health, 49(Suppl 1):1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ullah H, Ullah A, Gul A, et al. (2021). Novel coronavirus 2019 (COVID-19) pandemic outbreak: A comprehensive review of the current literature. Vacunas, 22(2): 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsabouri S, Makis A, Kosmeri C, et al. (2021). Risk Factors for Severity in Children with Coronavirus Disease 2019: A Comprehensive Literature Review. Pediatr Clin North Am. 68(1): 321–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu A, Peng Y, Huang B, et al. (2020). Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe, 27(3): 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y, Yang C, Xu XF, et al. (2020). Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacologica Sinica, 41(9):1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malone B, Urakova N, Snijder EJ, et al. (2022). Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat Rev Mol Cell Biol, 23(1): 21–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antony P, Vijayan R. (2021). Role of SARS-CoV-2 and ACE2 variations in COVID-19. Biomed J, 44(3): 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belouzard S, Millet JK, Licitra BN, et al. (2012). Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses, 4(6): 1011–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brobst B, Borger J. (2022). Benefits And Risks Of Administering Monoclonal Antibody Therapy For Coronavirus (COVID-19). StatPearls. Treasure Island (FL), pp.: 6–8. [PubMed] [Google Scholar]
- 15.Cheng Y, He B, Yang J, et al. (2019). Crystal structure of the S1 subunit N-terminal domain from DcCoV UAE-HKU23 spike protein. Virology, 535: 74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becerra-Flores M, Cardozo T. (2020). SARSCoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int J Clin Pract, 74(8): e13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korber B, Fischer WM, Gnanakaran S, et al. (2020). Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell, 182(4):812–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson CB, Zhang L, Farzan M, Choe H. (2021). Functional importance of the D614G mutation in the SARS-CoV-2 spike protein. Biochem Biophys Res Commun, 538:108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Jackson CB, Mou H, et al. (2020). The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv, doi: 10.1101/2020.06.12.148726. [DOI] [Google Scholar]
- 20.Zhu X, Mannar D, Srivastava SS, et al. (2021). Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLoS Biol, 19(4):e3001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang JW, Toovey OTR, Harvey KN, et al. (2021). Introduction of the South African SARS-CoV-2 variant 501Y.V2 into the UK. J Infect, 82(4): e8–e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian F, Tong B, Sun L, et al. (2021). N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. Elife, 10:e69091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tegally H, Wilkinson E, Giovanetti M, et al. (2021). Detection of a SARS-CoV-2 variant of concern in South Africa. Nature, 592(7854):438–443. [DOI] [PubMed] [Google Scholar]
- 24.Yang WT, Huang WH, Liao TL, et al. (2022). SARS-CoV-2 E484K Mutation Narrative Review: Epidemiology, Immune Escape, Clinical Implications, and Future Considerations. Infect Drug Resist, 15:373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding C, He J, Zhang X, et al. (2021). Crucial Mutations of Spike Protein on SARS-CoV-2 Evolved to Variant Strains Escaping Neutralization of Convalescent Plasmas and RBD-Specific Monoclonal Antibodies. Front Immunol, 12:693775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jangra S, Ye C, Rathnasinghe R, et al. (2021). The E484K mutation in the SARS-CoV-2 spike protein reduces but does not abolish neutralizing activity of human convalescent and post-vaccination sera. medRxiv, doi: 10.1101/2021.01.26.21250543. [DOI] [Google Scholar]
- 27.Tao K, Tzou PL, Nouhin J, et al. (2021). The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet, 22(12): 757–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi B, Choudhary MC, Regan J, et al. (2020). Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N Engl J Med, 383(23):2291–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gangavarapu K, Latiff AA, Mullen JL, et al. (2022). Outbreak. info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. medRxiv, doi: 10.1101/2022.01.27.22269965. [DOI] [PMC free article] [PubMed]
- 30.Hodcroft EB, Domman DB, Snyder DJ, et al. (2021). Emergence in late 2020 of multiple lineages of SARS-CoV-2 Spike protein variants affecting amino acid position 677. medRxiv, doi: 10.1101/2021.02.12.21251658. [DOI] [Google Scholar]
- 31.Lubinski B, Fernandes MHV, Frazier L, et al. (2021). Functional evaluation of the P681H mutation on the proteolytic activation the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. bioRxiv, doi: 10.1101/2021.04.06.438731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Y, Gaudreault NN, Meekins DA, et al. (2021). Effects of Spike Mutations in SARSCoV-2 Variants of Concern on Human or Animal ACE2-Mediated Virus Entry and Neutralization. bioRxiv, doi: 10.1101/2021.08.25.457627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo CH, Morris CP, Sachithanandham J, Amadi A, et al. (2021). Infection with the SARS-CoV-2 Delta Variant is Associated with Higher Infectious Virus Loads Compared to the Alpha Variant in both Unvaccinated and Vaccinated Individuals. medRxiv, doi: 10.1101/2021.08.15.21262077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volz E, Mishra S, Chand M, et al. (2021). Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature, 593(7858): 266–269. [DOI] [PubMed] [Google Scholar]
- 35.Choi JY, Smith DM. (2021). SARS-CoV-2 Variants of Concern. Yonsei Med J, 62(11): 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cascella M, Rajnik M, Aleem A, et al. (2022). Features, Evaluation, and Treatment of Coronavirus (COVID-19). StatPearls. Treasure Island (FL).P.:11–12 [PubMed] [Google Scholar]
- 37.Ramesh S, Govindarajulu M, Parise RS, et al. (2021). Emerging SARS-CoV-2 variants: A review of its mutations, its implications and vaccine efficacy. Vaccines (Basel), 9(10):1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiehzadegan S, Alaghemand N, Fox M, et al. (2021). Analysis of the Delta Variant B.1.617.2 COVID-19. Clin Pract, 11(4):778–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menni C, Valdes AM, Polidori L, et al. (2022). Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: a prospective observational study from the ZOE COVID Study. Lancet, 399(10335): 1618–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohammadi M, Shayestehpour M, Mirzaei H. (2021). The impact of spike mutated variants of SARS-CoV2 [Alpha, Beta, Gamma, Delta, and Lambda] on the efficacy of subunit recombinant vaccines. Braz J Infect Dis, 25(4):101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao Y, Wang J, Jian F, et al. (2022). Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature, 602(7898):657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mallapaty S. (2022). Where did Omicron come from? Three key theories. Nature, 602(7895):26–8. [DOI] [PubMed] [Google Scholar]
- 43.Callaway E. (2021). Heavily mutated Omicron variant puts scientists on alert. Nature, 600(7887):21. [DOI] [PubMed] [Google Scholar]
- 44.Bhattacharyya RP, Hanage WP. (2022). Challenges in inferring intrinsic severity of the SARS-CoV-2 Omicron variant. N Engl J Med, 386(7):e14. [DOI] [PubMed] [Google Scholar]
- 45.Abbasi J. (2021). Researchers tie severe immunosuppression to chronic COVID-19 and virus variants. JAMA, 325(20):2033–2035. [DOI] [PubMed] [Google Scholar]
- 46.Pal M, Berhanu G, Desalegn C, et al. (2020). Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): An Update. Cureus, 12(3):e7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farhud DD, Zokaei SH. (2021). A Brief Overview of COVID-19 Vaccines. Iran J Public Health, 50(7):i–vi. [DOI] [PMC free article] [PubMed] [Google Scholar]