SUMMARY
Although central to evolution, the causes of hybrid inviability that drive reproductive isolation are poorly understood. Embryonic lethality occurs when eggs of the frog X. tropicalis are fertilized with either X. laevis or X. borealis sperm. We observed that distinct subsets of paternal chromosomes failed to assemble functional centromeres, causing their mis-segregation during embryonic cell divisions. Core centromere DNA sequence analysis revealed little conservation among the three species, indicating that epigenetic mechanisms that normally operate to maintain centromere integrity are disrupted on specific paternal chromosomes in hybrids. In vitro reactions combining X. tropicalis egg extract with either X. laevis or X. borealis sperm chromosomes revealed that paternally matched or over-expressed centromeric histone CENP-A and its chaperone HJURP could rescue centromere assembly on affected chromosomes in interphase nuclei. However, whereas the X. laevis chromosomes maintained centromeric CENP-A in metaphase, X. borealis chromosomes did not, and also displayed ultra-thin regions containing ribosomal DNA. Both centromere assembly and morphology of X. borealis mitotic chromosomes could be rescued by inhibiting RNA Polymerase I or by preventing collapse of stalled DNA replication forks. These results indicate that specific paternal centromeres are inactivated in hybrids due to disruption of associated chromatin regions that interfere with CENP-A incorporation, at least in some cases due to conflicts between replication and transcription machineries. Thus, our findings highlight the dynamic nature of centromere maintenance and its susceptibility to disruption in vertebrate interspecies hybrids.
Keywords: Centromere, hybrid incompatibility, speciation, Xenopus, chromosome segregation, CENP-A
Graphical Abstract
ETOC BLURB
Centromere incompatibilities in inviable Xenopus hybrids are sequence-independent and result from disruption of epigenetic pathways required for centromere maintenance.
INTRODUCTION
Hybridization between closely related species often leads to embryonic lethality accompanied by defects in genome stability and maintenance, but the cellular and molecular mechanisms underlying post-zygotic barriers that drive reproductive isolation and speciation are largely unknown1–4. Among animals, a number of studies of inviable hybrids resulting from crosses of related Drosophila species have revealed an important role for the centromere, the chromosomal site where the kinetochore assembles to mediate chromosome attachment to the mitotic spindle and segregation to daughter cells. Both centromere DNA sequence and protein components including the centromeric histone H3 variant, Centromere Protein A (CENP-A) are rapidly evolving5,6. Localization of exogenously expressed CENP-A to centromeres across Drosophila species was shown to require co-expression of its species-matched chaperone CAL1/HJURP, indicating that the CENP-A deposition machinery co-evolves7. In turn, kinetochore formation at centromeres depends on specific, epigenetic recognition and stabilization of CENP-A nucleosomes by other factors, including CENP-C, CENP-N, and M18BP18–16. Thus, co-evolution of centromere DNA and many associated proteins may generate barriers to hybrid viability by interfering with assembly of the chromosome segregation machinery.
Increasing evidence suggests that the chromatin environment also plays an important role in centromere assembly and that changes in the nuclear organization are related to hybridization outcomes. For example, disruption of the chromocenter, a domain containing the pericentromeric satellite DNA, is common among Drosophila hybrids and may underlie inviability17. Furthermore, known inviability factors such as hybrid male rescue (Hmr) and lethal hybrid rescue (Lhr) strongly impact chromosome segregation in Drosophila hybrids and have been reported to regulate transposable elements and heterochromatic repeats18,19, associate with chromatin chaperones adjacent to centromeres20, and to link pericentromeric and centromeric chromatin to maintain centromere integrity21. However, whether these factors play a direct role in centromere function is unclear22. Despite these advances, the relative contribution to hybrid inviability of diverging centromere sequences versus the activity and spatial organization of associated chromatin machineries that promote centromere assembly and maintenance is poorly understood.
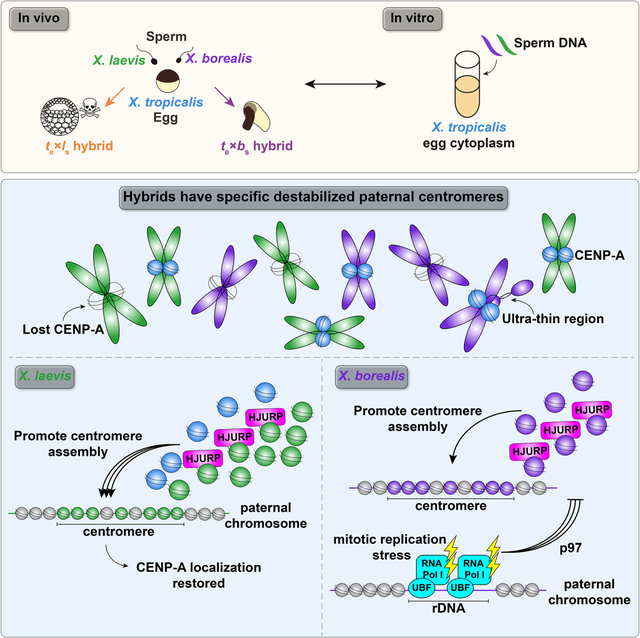
Among vertebrates, hybridization resulting in post-zygotic death has been more difficult to study. Xenopus frog species possess interesting evolutionary relationships that include past interspecies hybridization events. For example, hybridization and whole genome duplication of two X. tropicalis-like species produced the allotetraploids X. laevis and X. borealis, which each contain two distinct subgenomes termed L (long) and S (short) to indicate overall differences in chromosome length23. These closely related Xenopus species provide an ideal system to study the molecular basis of hybridization outcomes, since cross fertilization experiments are easily performed24,25, and mechanisms underlying hybrid incompatibility can be uniquely and powerfully investigated in vitro by combining the sperm chromosomes and egg extracts from different species. We showed previously that interspecies hybrids produced when X. laevis or X. borealis eggs are fertilized by X. tropicalis sperm are viable, while the reverse crosses die before gastrulation and zygotic gene activation by explosive cell lysis or exogastrulation, respectively25. The inviable hybrids displayed chromosome segregation defects during embryonic cleavages, characterized by lagging chromosomes, chromosome bridges, and formation of micronuclei. By whole genome sequencing, specific and distinct paternal chromosome regions were lost from both hybrids prior to embryo death. A fraction of X. laevis chromosomes failed to assemble centromeres/kinetochores, likely leading to spindle attachment defects and ultimately chromosome mis-segregation and embryo death25.
To better understand centromere-based Xenopus hybrid incompatibilities, here we combine genomic, in vitro, and in vivo analyses. We find that although core centromeric sequences are not conserved, X. tropicalis egg cytoplasm supports centromere assembly on X. laevis and X. borealis chromosomes. However, upon entry into metaphase, conflicts emerge that evict CENP-A from a subset of chromosomes. In the case of X. laevis, excess CENP-A and its chaperone HJURP can rescue this defect. In contrast, eviction of CENP-A from X. borealis chromosomes could be rescued by dissociating the rRNA polymerase Pol I or by preventing collapse of DNA replication forks. These results indicate that centromere incompatibility is driven primarily by centromere sequence-independent molecular conflicts that disrupt the epigenetic maintenance of CENP-A nucleosomes.
RESULTS
Core centromere sequence variation does not underlie Xenopus hybrid aneuploidy
We previously observed chromosome mis-segregation and loss of centromere and kinetochore proteins from a subset of chromosomes in hybrids generated by fertilizing X. tropicalis eggs with X. laevis sperm. Whole genome sequencing just prior to embryo death revealed consistent deletion of large genomic regions from two paternal chromosomes, 3L and 4L of the L subgenome25. We hypothesized that chromosome-specific aneuploidy resulted from divergent centromeric sequences on the affected chromosomes, rendering them incompatible with the maternal X. tropicalis centromeric histone CENP-A and its loading machinery. Recent characterization of X. laevis centromere sequences by chromatin immunoprecipitation with CENP-A antibodies and sequencing analysis (ChIP-seq) revealed a family of related sequences found in distinct combinations and abundances on different X. laevis chromosomes26. However, X. laevis centromeres 3L and 4L did not possess any distinguishing features in terms of size or composition. Thus, differences in core centromere DNA sequences do not appear to drive the specific chromosome mis-segregation events and genome loss observed in the inviable X. tropicalis/X. laevis hybrid.
To expand our analysis, we characterized a second inviable hybrid resulting from fertilization of X. tropicalis eggs with sperm from X. borealis, a frog species possessing an allotetraploid genome closely related to X. laevis23. These hybrids display specific and consistent genome loss from a different subset of paternal chromosomes including 1S, 5S, 4L, and 8L25. To determine the extent to which centromere sequences differed across the three Xenopus species, CENP-A ChIP-seq was similarly applied to X. tropicalis and X. borealis. We used an alignment-independent k-mer based analysis to identify sequence features of the highly repetitive centromeric arrays in each species without the need for a complete genome sequence (Fig. S1A). Comparing the enrichment value (normalized CENP-A k-mer counts/normalized input k-mer counts) revealed that the majority of individual k-mers are enriched in one species, but not the others (Fig. 1A). Furthermore, analysis of full-length sequencing reads that contained CENP-A enriched k-mers showed that CENP-A nucleosome-associated DNA sequences of the three species bear little relationship to one another (Fig. 1B). These findings reinforce the idea that incompatibilities leading to mis-segregation of specific chromosomes are not due to centromere sequence differences per se, and are consistent with a vast literature showing that centromere function is defined epigenetically in most eukaryotes, including vertebrates27–29.
Figure 1: Comparison of X. laevis, X. tropicalis, and X. borealis core centromere sequences.
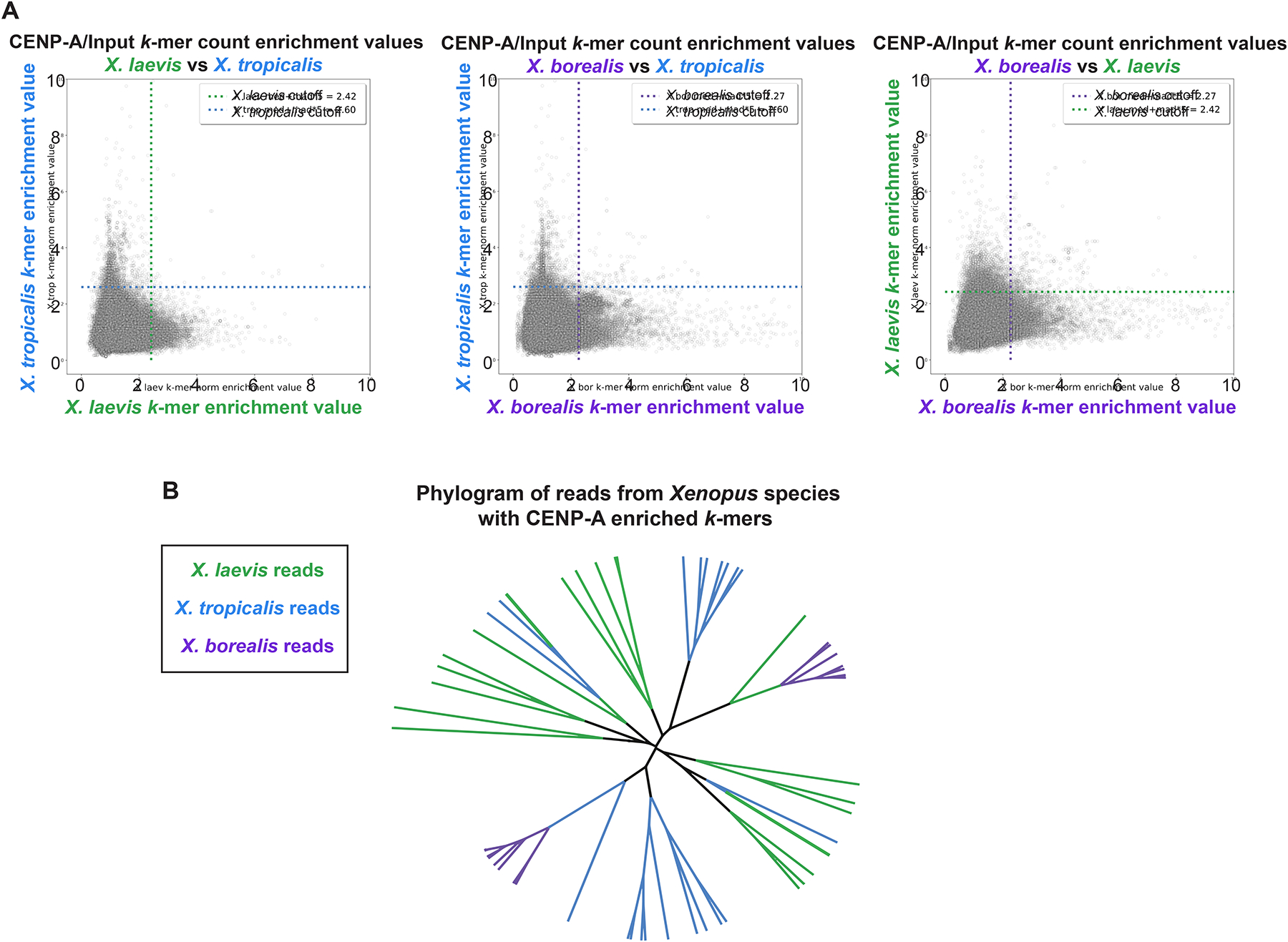
(A) Scatter plots of k-mer enrichment values (normalized CENP-A counts / normalized input counts) compared between species. Only k-mers found in both species are plotted. Dotted lines indicate enrichment value for each species that is five median absolute deviations above the median enrichment value to denote highly enriched k-mers, which are not well conserved across species.
(B) Phylogram of full-length sequencing reads from each Xenopus species. Branches are colored according to species of origin. Sequencing reads were selected first by the presence of at least 80 CENP-A enriched 25bp k-mers and then by hierarchical clustering. The phylogram illustrates a striking divergence of core centromere sequences.
See also Figure S1.
Interestingly, although protein sequence alignments of X. laevis, X. tropicalis, and X. borealis CENP-A showed that they are nearly 90% identical, divergence occurred in both the N-terminus and the CENP-A Targeting Domain (CATD) L1 loop region that provides specificity for recognition of the CENP-A/H4 complex by its dedicated chaperone HJURP7,30–32 (Fig. S1B). Together, these results suggest that as in Drosophila, CENP-A, and its chaperone may have co-evolved in Xenopus to strengthen the specificity of their interactions33. Importantly, however, although divergence and co-evolution of centromeres and associated proteins contributes to meiotic drive and can lead to hybrid inviability in flies34–36, such differences do not explain loss of centromere function on specific subsets of chromosomes in inviable Xenopus hybrids.
CENP-A eviction from a subset of chromosomes requires cell cycle progression
To better understand the process by which specific chromosomes lose centromere function in hybrids, we took advantage of the Xenopus egg extract system capable of recapitulating events of the embryonic cell division cycle in vitro. Interphase of these cell cycles lack gap (G) phases and consists entirely of S-phase when DNA replicates. During M-phase, chromosomes condense and assemble a kinetochore at each centromere37,38. To monitor centromere assembly, X. tropicalis, X. laevis, or X. borealis sperm nuclei were added to X. tropicalis egg extracts and probed for CENP-A at different stages of the cell cycle. Sperm chromosomes of all three species condensed and possessed single centromeric CENP-A foci when added directly to metaphase-arrested X. tropicalis extract (Fig. 2A, B), consistent with observations that sperm chromosomes contain CENP-A39,40. However, cycling the extract through interphase to allow sperm decondensation, nuclear envelope formation, and DNA replication in X. tropicalis egg cytoplasm resulted in no visible CENP-A on a subset of X. laevis and X. borealis mitotic chromosomes in the subsequent metaphase, whereas X. tropicalis centromeres were not affected (Fig. 2A, B).
Figure 2: Loss of centromeric CENP-A is cell cycle-dependent.
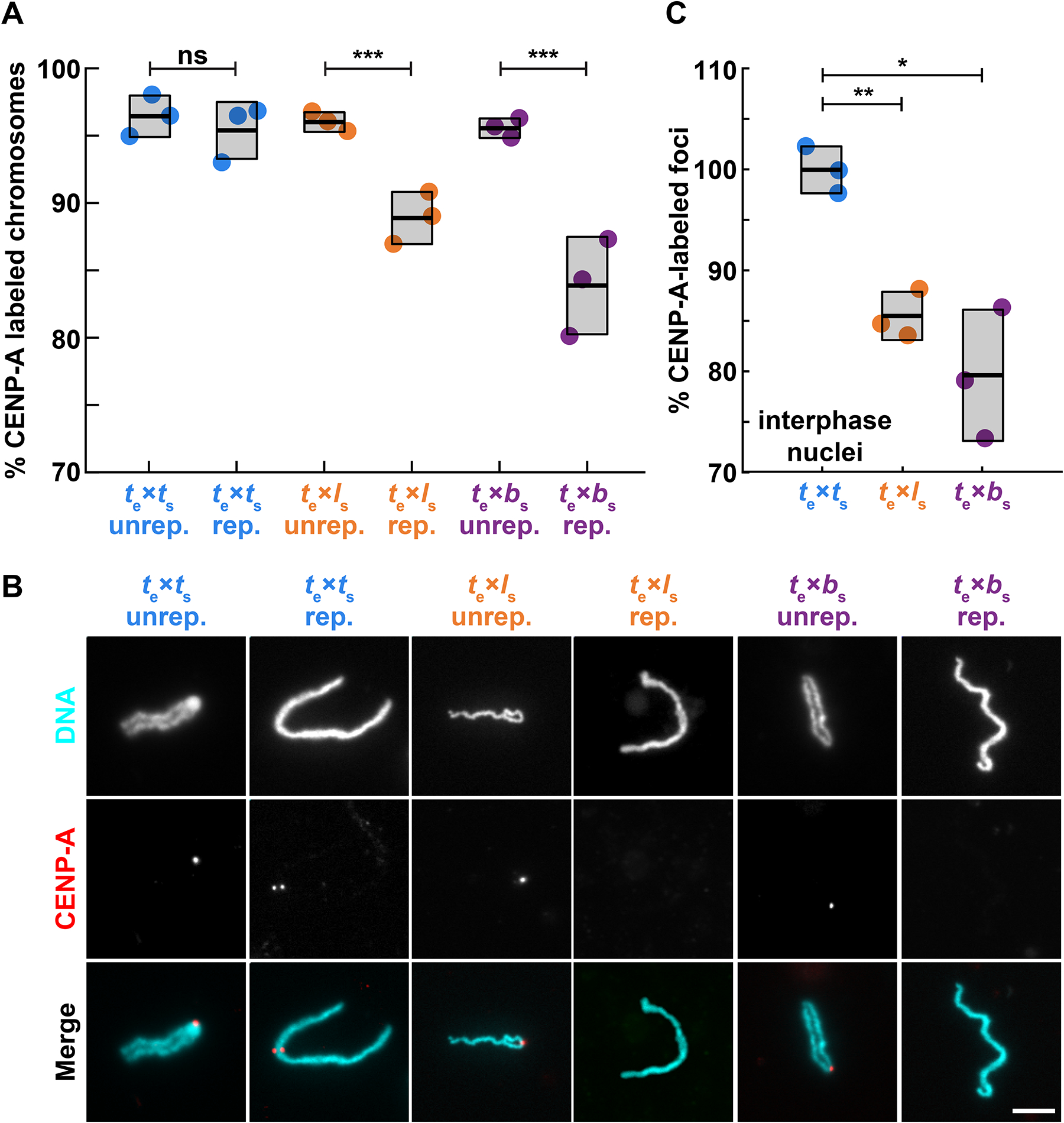
(A) Percentage of mitotic chromosomes with centromeric CENP-A staining in X. tropicalis egg extract. Over 95% of X. tropicalis, X. laevis, and X. borealis unreplicated sperm chromosomes added directly to metaphase-arrested X. tropicalis egg extracts possess centromeres, as indicated by immunofluorescence of the centromeric histone CENP-A. Following progression through the cell cycle, a fraction of replicated X. laevis and X. borealis mitotic chromosomes completely lose centromeric CENP-A foci. Unrep., unreplicated chromosomes; rep, replicated chromosomes. N = 3 extracts, N > 275 chromosomes per extract. p-values (left to right) by two-tailed two-sample unequal variance t-tests: 0.3356, 0.0008, 0.0004; ns, not significant.
(B) Representative images of mitotic unreplicated and replicated X. tropicalis, X. laevis, and X. borealis chromosomes formed in X. tropicalis egg extracts. The chromosomes shown here are not identified, but selected from a population of paternal chromosomes. DNA in cyan, CENP-A in red. Scale bar is 10 μm.
(C) Percentage of total expected CENP-A foci observed in nuclei formed in interphase X. tropicalis egg extract. X. laevis and X. borealis interphase nuclei both lose centromere foci during interphase, prior to entry into metaphase, whereas X. tropicalis nuclei do not. From N = 3 extracts, N > 64 nuclei per extract. p-values (top to bottom) by one-way ANOVA with Tukey post-hoc analysis: 0.0025, 0.0133.
Species nomenclature throughout figures denotes egg extract as subscript e and chromosomes as subscript s, for example te × ls indicates X. tropicalis egg extract combined with X. laevis sperm chromosomes. X. tropicalis is color-coded blue, while X. laevis and X. borealis hybrid combinations are orange and purple, respectively.
See also Figure S2.
To determine when in the cell cycle CENP-A was evicted from paternal chromosomes, we examined interphase nuclei in control and hybrid in vitro reactions. The expected number of centromere foci, 18 for X. laevis and X. borealis, decreased in X. tropicalis extract (Fig. 2C, Fig S2A, B). The loss of CENP-A localization from 2 or 4 paternal X. laevis and X. borealis chromosomes, respectively, corresponded very well to whole genome sequencing data of hybrid embryos in terms of the number of chromosomes affected25, and indicates that CENP-A is lost from this subset of paternal chromosomes during interphase.
The recent detailed characterization of X. laevis centromere sequences allowed us to test whether the centromere assembly defects observed in egg extract occurred on the same chromosomes disrupted in hybrid embryos25,26. Fluorescence in situ hybridization (FISH) probes in combination with CENP-A immunofluorescence identified X. laevis chromosomes 3L and 4L, the two chromosomes that lose large genomic regions in hybrid embryos, as those that also lose centromeric CENP-A staining when replicated in X. tropicalis egg extract (Fig. S2C–E). Thus, the in vitro system reproduces incompatibilities likely to underlie chromosome mis-segregation and ultimately genome loss observed in vivo. These results show that while all paternal sperm chromosomes initially possess CENP-A at their centromeres, a subset evict CENP-A during interphase, indicating that epigenetic mechanisms, likely involving the chaperone HJURP, operate to maintain CENP-A nucleosomes during DNA replication, as observed in cultured cells41. Such mechanisms enable hybrid centromere assembly despite evolutionary differences, but are disrupted on specific individual chromosomes.
CENP-A and its chaperone HJURP can rescue X. laevis centromere assembly
We next sought to determine whether enhancing centromere assembly by adding species-matched paternal factors could prevent CENP-A eviction and centromere loss from specific X. laevis and X. borealis chromosomes formed in X. tropicalis extracts. In vitro reactions were supplemented with paternally-matched proteins expressed in reticulocyte lysate, including CENP-A and its dedicated chaperone HJURP, at the onset of interphase (Fig. S3A–C). Whereas adding X. laevis CENP-A resulted in a partial rescue, CENP-A plus HJURP increased the percentage of replicated X. laevis mitotic chromosomes with CENP-A foci to control levels (Fig. 3A). In contrast, no combination of X. borealis centromere factors tested, including CENP-A, HJURP, and CENP-C42,43, restored CENP-A foci to replicated X. borealis mitotic chromosomes (Fig. 3B). Notably, however, examination of interphase nuclei in X. borealis sperm/X. tropicalis egg extract reactions prior to metaphase entry revealed that CENP-A localization was initially fully rescued, with the expected number of CENP-A-positive foci corresponding to the number of chromosomes (Fig. 3C). These results indicate that exogenous species-matched CENP-A and HJURP can restore proper centromere formation on all chromosomes during interphase of for both X. laevis and X. borealis, but that CENP-A is not maintained on a subset of X. borealis chromosomes upon entry into mitosis.
Figure 3: Driving CENP-A assembly rescues centromere localization in interphase, which persists on mitotic X. laevis, but not on X. borealis chromosomes.
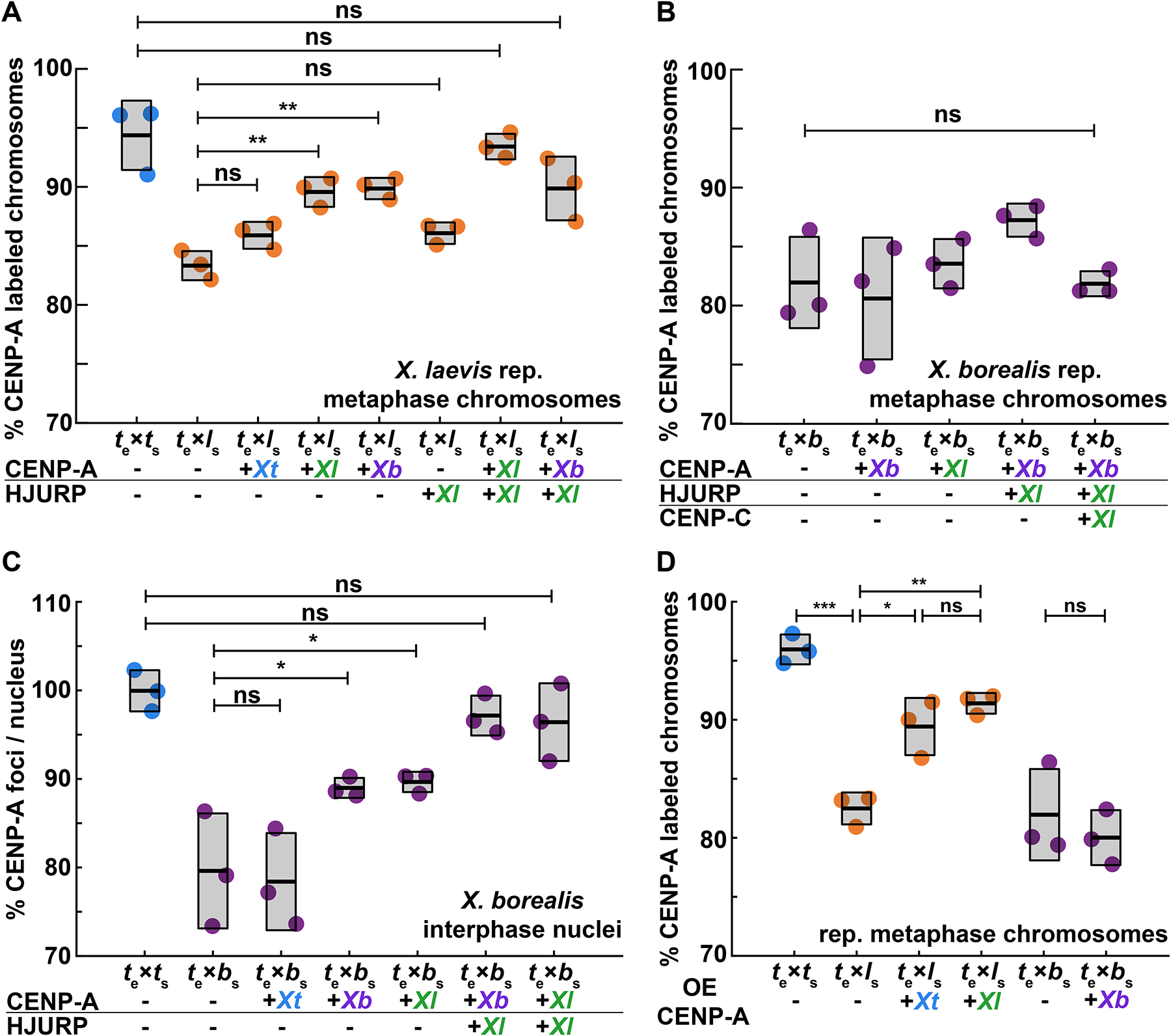
(A) Percentage of replicated X. laevis chromosomes with centromeric CENP-A staining in X. tropicalis extract supplemented with in vitro translated CENP-A and HJURP proteins from different Xenopus species. X. laevis chromosomes are fully rescued with species-matched centromere proteins. Quantification with N = 3 extracts, N > 315 chromosomes per extract. p-values (top to bottom) by one-way ANOVA with Tukey post-hoc analysis: 0.1734, 0.9999, 0.5522, 0.0057, 0.0086, 0.6281.
(B) Percentage of replicated X. borealis chromosomes with centromeric CENP-A staining in X. tropicalis extract supplemented with in vitro translated centromere proteins from different Xenopus species. No combination or increased amounts of centromeric proteins CENP-A (CA), HJURP (HJ), and CENP-C (CC) restored CENP-A localization on X. borealis mitotic chromosomes. Quantification with N = 3 extracts, N > 216 chromosomes per extract. p-value by one-way ANOVA = 0.0786.
(C) Percentage of CENP-A-labeled centromeric foci in X. borealis nuclei assembled in X. tropicalis extract supplemented with in vitro translated centromere proteins from different Xenopus species. Driving centromere assembly with species-matched proteins fully restores formation of centromere foci in interphase, but CENP-A staining is subsequently lost in metaphase (panel B). Quantification with N = 3 extracts, N > 67 nuclei per extract. p-values (top to bottom) by one-way ANOVA: 0.9996, 0.0562, 0.0433, 0.9690, 0.9109.
(D) Percentage of replicated X. laevis or X. borealis chromosomes with centromeric CENP-A staining in X. tropicalis extract supplemented with excess (~80X endogenous levels) of in vitro translated X. laevis or X. tropicalis CENP-A. Whereas centromere staining is fully rescued on X. laevis mitotic chromosomes by CENP-A from either species, X. borealis centromere staining is not affected. Quantification with N = 3 extracts, N > 204 chromosomes per extract. p-values (top to bottom, then left to right) by one-way ANOVA with Tukey post-hoc analysis: 0.0042, 0.0001, 0.0249, 0.8845, 0.88946.
A-C: Centromere proteins were added at ~8X endogenous levels.
A-D: ns, not significant.
See also Figure S3.
The ability to mix and match egg extract, sperm chromosomes, and exogenous centromere assembly factors enabled evaluation of CENP-A/centromere compatibilities across species. For example, despite striking differences in core centromere sequences between X. laevis and X. borealis (Fig. 1), the CATDs of the two species’ CENP-A sequences are identical (Fig. S1B), and exogenous CENP-A from either species equivalently restored centromere assembly on X. laevis mitotic chromosomes replicated in X. tropicalis egg extract (Fig. 3A). Further, we observed that addition of excess exogenous X. tropicalis CENP-A could also increase the percentage of X. laevis mitotic chromosomes with centromere foci to control levels, although X. borealis chromosomes could not be rescued under any condition tested (Fig. 3B, D). Together, our results indicate that enhancing the pathway that drives CENP-A incorporation into centromeric chromatin can fully overcome whatever is destabilizing centromeres on specific X. laevis centromeres and raised the question of why the X. borealis chromosomes are refractory to this rescue.
X. borealis chromosome defects result from mitotic replication stress
A clue as to why X. borealis mitotic chromosomes behave differently than X. laevis chromosomes in the in vitro hybrid extract system emerged with observation of their morphology. Although a subset of replicated X. laevis mitotic chromosomes formed in X. tropicalis extract lacked centromeres, they otherwise appeared normal. In contrast, 7–10% of X. borealis mitotic chromosomes displayed ultra-thin regions of 2–3 μm in length following replication, although centromeres on these chromosomes appeared largely intact (Fig. 4A, B; Fig. S4A, B). We reasoned that incomplete DNA replication leading to fork stalling and subsequent collapse in mitosis, termed replication stress, caused the formation of fragile sites44,45. Consistent with this idea, adding low doses of the DNA polymerase inhibitor aphidicolin that leads to replication stress45–47 triggered formation of ultra-thin regions on X. tropicalis and X. laevis mitotic sperm chromosomes that had progressed through the cell cycle in X. tropicalis extract, and slightly exacerbated morphological defects of X. borealis chromosomes (Fig. S4C–E). Notably, however, the aphidicolin-induced replication stress did not affect CENP-A localization efficiency (Fig. S4F), indicating that replication stress per se does not interfere with CENP-A loading and maintenance. Thus, under all conditions tested, we found no correlation between replication stress indicated by ultra-thin regions and defects in centromere assembly on the same chromosome (Fig. S4B, E, and F).
Figure 4: Mitotic replication stress leads to X. borealis centromere and chromosome morphology defects.
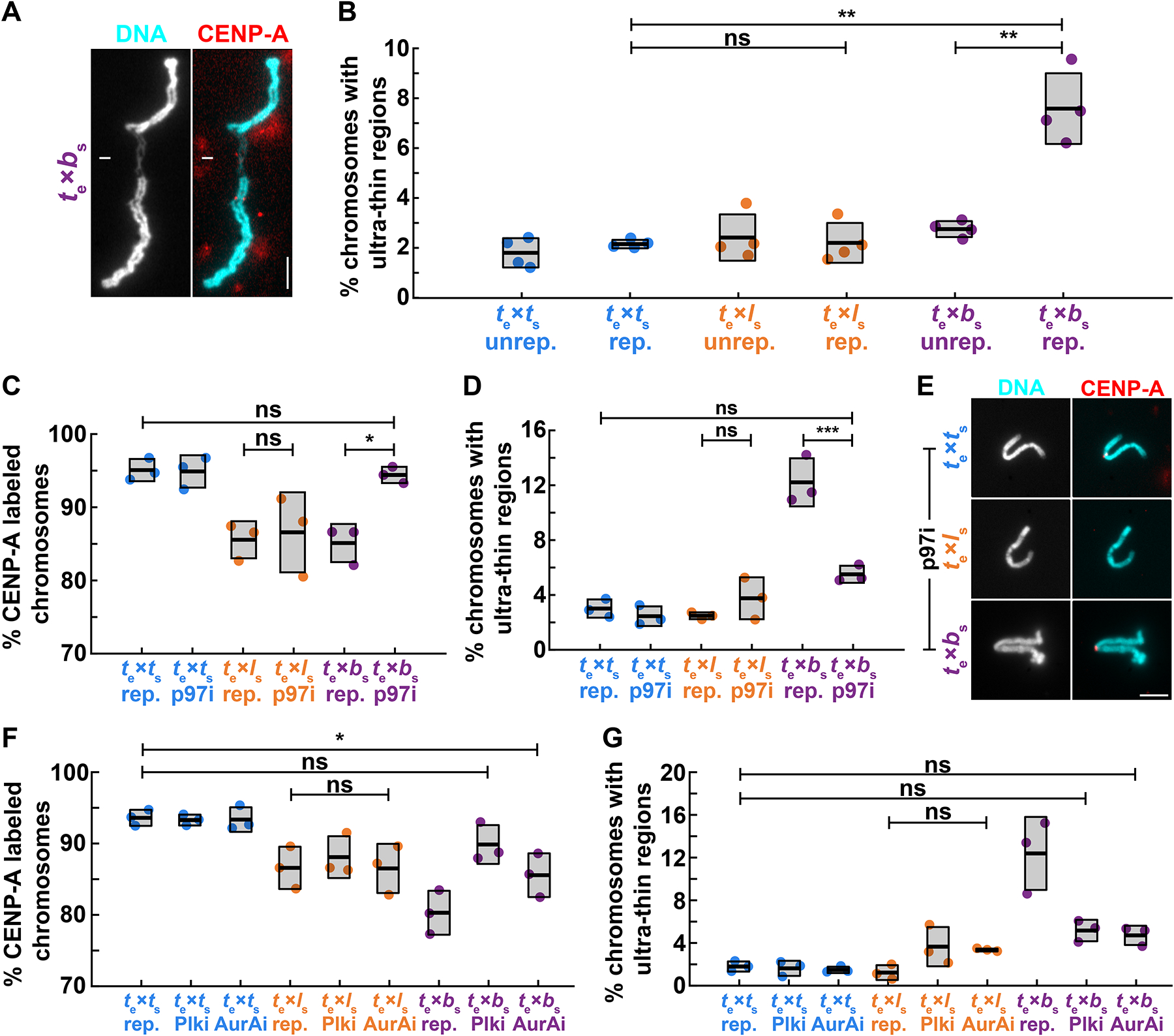
(A) Representative image showing an ultra-thin region of a mitotic X. borealis chromosome formed in X. tropicalis egg extract. Note that the chromosome has an intact centromere. DNA in cyan, CENP-A in red. Scale bar is 5 μm.
(B) Percentage of unreplicated and replicated mitotic chromosomes with ultrathin morphology defects in X. tropicalis extract. A low percentage of X. tropicalis, X. laevis or X. borealis unreplicated chromosomes display ultra-thin regions. After cycling through interphase, only X. borealis chromosomes exhibit a significant increase in this defect. Quantification with N = 3 extracts, N > 310 chromosomes per extract. p-values (top to bottom, then left to right) by one-way ANOVA with Tukey post-hoc analysis: 2.9352e-7, 0.9999, 1.6475e-6.
(C) Percentage of replicated chromosomes with centromeric CENP-A staining in X. tropicalis extracts treated with solvent control or 10 μM p97 ATPase inhibitor NMS-873 (p97i). Inhibition of p97 restores CENP-A staining on X. borealis mitotic chromosomes, but does not affect X. tropicalis or X. laevis chromosomes. p-values (top to bottom, then left to right) by one-way ANOVA with Tukey post-hoc analysis: 0.9997, 0.9978, 0.0204.
(D) Percentage of chromosomes with ultrathin regions in X. tropicalis extracts treated with solvent control or 10 μM p97 ATPase inhibitor NMS-873 (p97i). Inhibition of p97 rescues X. borealis chromosome morphology defects, but does not affect X. tropicalis or X. laevis chromosomes. p-values (top to bottom, then left to right) by one-way ANOVA with Tukey post-hoc analysis: 0.1114, 0.6903, 6.2572e-5.
(E) Representative images of mitotic replicated X. tropicalis, X. laevis, and X. borealis chromosomes following treatment with 10 μM p97 ATPase inhibitor NMS-873 (p97i). X. borealis chromosome morphology and centromere localization are rescued (bottom panels, compare to Fig. 4A, 2B images), similar to X. tropicalis, while X. laevis chromosomes have lost CENP-A staining (middle panels). DNA in cyan, CENP-A in red. Scale bar is 5 μm.
(F) Percentage of replicated chromosomes with centromeric CENP-A staining in X. tropicalis extracts treated with solvent control, 1 μM Polo-like kinase 1 inhibitor BI-2536 (Plk1i), or 1 μM Aurora A kinase inhibitor MLN-8237 (AurAi). CENP-A localization is fully or partially rescued on X. borealis mitotic chromosomes, whereas X. tropicalis or X. laevis chromosomes are not affected. p-values (top to bottom) by one-way ANOVA with Tukey post-hoc analysis: 0.0276, 0.7003, 0.9999.
(G) Percentage of chromosomes with ultrathin regions in X. tropicalis extracts treated with solvent control, 1 μM Polo-like kinase 1 inhibitor BI-2536 (Plk1i), or 1 μM Aurora A kinase inhibitor MLN-8237 (AurAi). Inhibition of Plk1 and AurA rescued X. borealis mitotic chromosome morphology defects, but did not affect X. tropicalis or X. laevis chromosomes. p-values (top to bottom) by one-way ANOVA with Tukey post-hoc analysis: 0.2882, 0.1525, 0.5887.
C, D: N = 3 extracts, N > 179 chromosomes per extract.
E, F: N = 3 extracts, N > 155 chromosomes per extract.
B-F: ns, not significant.
See also Figure S4.
To determine whether the X. borealis chromosome morphology and mitotic centromere defects were nevertheless linked, we treated the in vitro hybrid reactions with an inhibitor of the AAA ATPase p97 (Fig. S4C). p97 is a multifunctional chaperone that removes the DNA replication helicase and causes collapse of stalled replication forks in mitosis45,48. Remarkably, we observed a complete rescue of both CENP-A localization and chromosome morphology on X. borealis chromosomes upon treatment with the p97 inhibitor (Fig. 4C, D). Consistent with factors known to regulate the pathway of mitotic replication fork collapse and breakage45, Aurora A and Plk1 kinase inhibitors added to X. tropicalis extracts at low doses that avoided mitotic defects also rescued X. borealis chromosome morphology and CENP-A localization, but did not affect X. laevis or X. tropicalis chromosomes (Fig. 4E, F). Finally, X. laevis or X. tropicalis chromosomes treated with aphidicolin followed by p97 inhibition displayed very few chromosome defects (Fig. S4C, D). Combined, these data reveal that a subset of X. borealis chromosomes experience mitotic replication stress in X. tropicalis cytoplasm, and that this is coupled to CENP-A eviction. However, centromere loss occurs on a different subset of mitotic chromosomes than those with ultra-thin regions.
Replication-transcription conflicts lead to centromere defects
The fragile sites observed on X. borealis chromosomes were reminiscent of secondary constrictions that occur at repetitive, late-replicating regions such as ribosomal DNA47,49 (rDNA). In Xenopus, the rDNA transcription machinery associates with mitotic chromosomes early in development and in egg extract50–53, even though rDNA transcription and nucleolus formation occur after zygotic genome activation54,55. We therefore tested whether ultra-thin regions of X. borealis chromosomes replicated in X. tropicalis extract contained rDNA by performing immunofluorescence using antibodies against RNA polymerase I (Pol I) and the rDNA transcription regulator upstream binding factor (UBF). Both proteins were consistently enriched on the ultra-thin regions of X. borealis mitotic chromosomes assembled in X. tropicalis egg extract (Fig. 5A, B).
Figure 5: Replication-transcription conflicts at rDNA on X. borealis chromosomes can be rescued by inhibiting RNA Pol I.
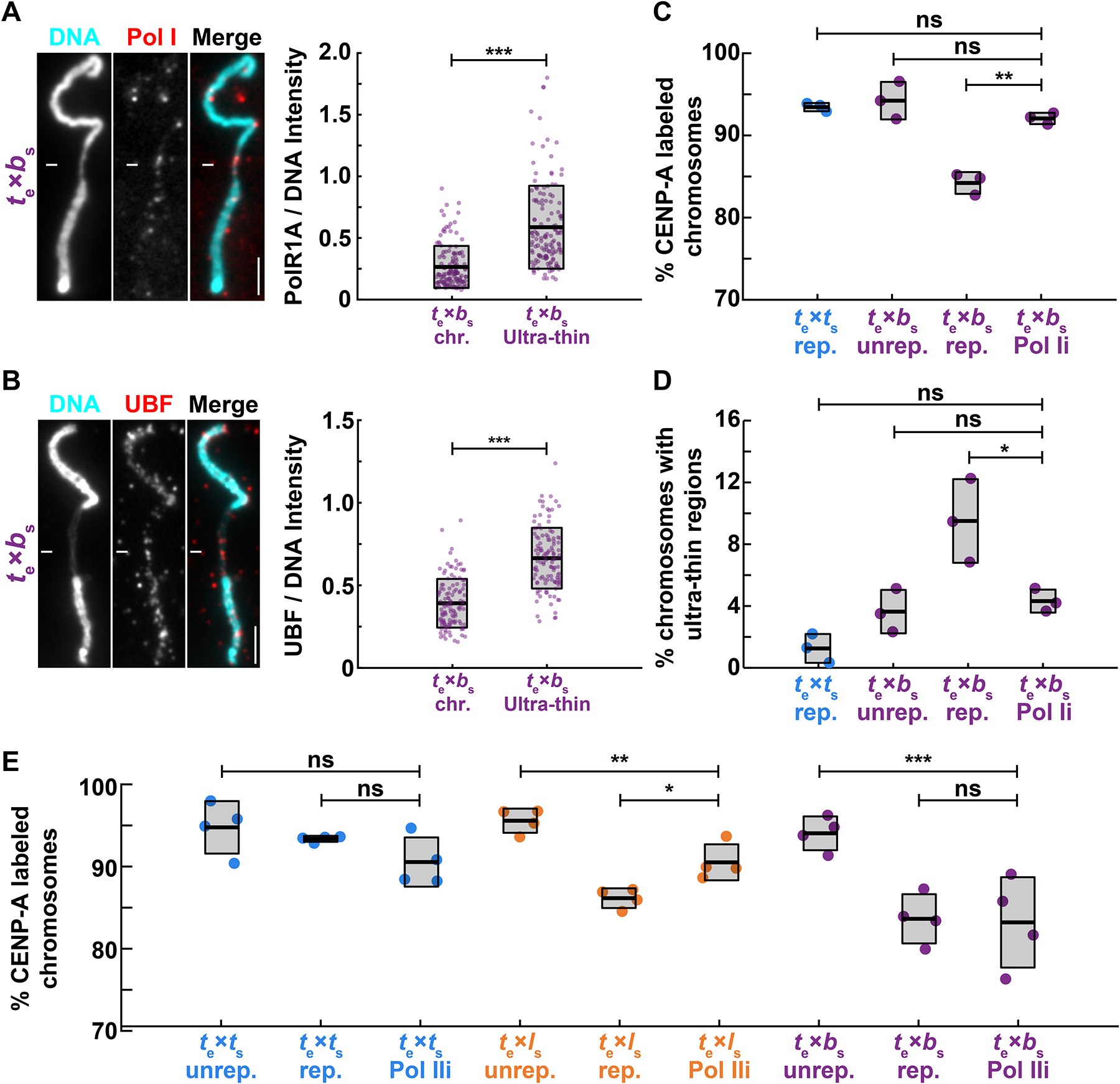
(A) Representative images and fluorescence intensity quantification of RNA Pol I staining relative to DNA on ultrathin and normal regions of X. borealis mitotic chromosomes, revealing enrichment of Pol I on ultra-thin regions. Quantification with N = 3 extracts, N = 140 chromosomes. p-value = 9.4793e-20 by two-tailed two-sample unequal variance t-tests.
(B) Representative images and fluorescence intensity quantification of UBF staining relative to DNA on ultrathin and normal regions of X. borealis mitotic chromosomes, revealing enrichment of UBF on ultra-thin regions. Quantification with N = 3 extracts, N = 62 chromosomes. p-value = 4.5004e-13 by two-tailed two-sample unequal variance t-tests.
(C) Percentage of mitotic chromosomes with centromeric CENP-A staining in X. tropicalis extracts treated with solvent control or 1 μM BMH-21 to inhibit RNA Pol I (Pol Ii), which fully rescues CENP-A localization on replicated X. borealis chromosomes. p-values (top to bottom) by one-way ANOVA with Tukey post-hoc analysis: 0.9794, 0.7979, 0.0005.
(D) Percentage of mitotic chromosomes with ultrathin regions in X. tropicalis extracts treated with solvent control or 1 μM BMH-21 (Pol Ii). Pol I inhibition also rescues X. borealis chromosome morphology defects. p-values (top to bottom) by one-way ANOVA with Tukey post-hoc analysis: 0.5078, 0.9999, 0.0469.
(E) Percentage of chromosomes with centromeric CENP-A staining in X. tropicalis extracts treated with solvent control or 25 μM triptolide to inhibit RNA Pol II (Pol IIi). X. laevis chromosomes are partially rescued, while X. tropicalis and X. borealis chromosomes are not affected. Quantification with N = 3 extracts, N > 322 chromosomes per extract. p-values (top to bottom, then left to right) by one-way ANOVA with Tukey post-hoc analysis: 0.4785, 0.8797, 0.0052, 0.0125, 0.0003, 0.9999.
A, B: DNA in cyan, Pol I in red. Scale bar is 5 μm.
C, D: N = 3 extracts, N > 172 chromosomes per extract.
C-E: ns, not significant.
See also Figure S5.
To test whether RNA Pol I occupancy at rDNA of X. borealis chromosomes contributed to the observed defects, X. tropicalis extract reactions were treated with the inhibitor BMH-21, which has been shown to dissociate the polymerase from chromatin56,57. Strikingly, X. borealis chromosome morphology defects as well as CENP-A localization were rescued (Fig. 5C, D). Interestingly, further analysis of CENP-A ChIP-seq reads revealed that, in contrast to other repetitive elements, rRNA and snRNA are specifically associated with X. borealis centromeres, showing a distinct enrichment not observed in X. laevis and X. tropicalis (Fig. S5A). Together, these data suggest that the replication stress experienced by X. borealis mitotic chromosomes occurs at rDNA loci, and that defects in rDNA chromatin dynamics act to destabilize a subset of X. borealis centromeres. In contrast, centromere formation on X. laevis chromosomes was not rescued by RNA Pol I inhibition (Fig. S5B, C), further indicating differences in the mechanisms underlying their incompatibility with X. tropicalis. However, we observed that inhibition of RNA polymerase II (Pol II) with triptolide partially rescued CENP-A localization to X. laevis chromosomes in X. tropicalis extract, whereas X. borealis chromosomes were not affected (Fig. 5E), and no species’ chromosomes were rescued by inhibition of RNA Pol III (Fig. S5D, E). Therefore, a common theme in hybrid incompatibility among Xenopus species may be replication-transcription conflicts that contribute to eviction of CENP-A from a subset of mitotic chromosomes. However, whereas this occurs at rDNA on X. borealis chromosomes and depends on RNA Pol I, X. laevis defects may be driven, at least in part, by RNA Pol II-induced defects. These observations lead to the model that epigenetic mechanisms promoting CENP-A incorporation at centromeres are disrupted by the presence or activity of RNA polymerases that cause under-replication at specific chromosome loci. Whereas X. laevis defects can be overcome by driving CENP-A incorporation at centromeres, X. borealis defects can only be rescued by blocking replication stress at rDNA, either by preventing fork collapse or by removing RNA Pol I.
Chromosome mis-segregation can be reduced in hybrid embryos, but inviability persists
To determine whether the incompatibility mechanisms identified through this work are responsible for hybrid inviability in vivo, we performed rescue experiments on cross-fertilized embryos. In vitro translated, paternally-matched CENP-A and HJURP proteins were microinjected into both blastomeres of the 2-cell hybrid embryo produced by fertilizing X. tropicalis eggs with X. laevis sperm, while X. tropicalis/X. borealis hybrid embryos were treated with RNA Pol I inhibitor BMH-21. Fewer micronuclei were observed in both cases, indicating a decrease in mitotic errors in hybrid embryos, although not to the low levels seen in wild-type X. tropicalis embryos (Fig. 6A, Fig. S6). Thus, the basis of chromosome defects identified using our in vitro egg extract assays also contribute to chromosome segregation defects in vivo. However, despite this partial rescue, treated hybrids died at the same time and in the same manner as untreated sibling controls (Fig. 6B, C, Video S1, S2). While it is possible that a complete rescue of chromosome segregation defects in the hybrid embryos is required for viability, we predict that other mechanisms that we have not yet identified also contribute, which can be uniquely addressed using a combination of in vitro and in vivo approaches in Xenopus.
Figure 6: Treatments that rescue CENP-A localization in egg extracts reduce micronuclei formation in hybrid embryos, but inviability persists.
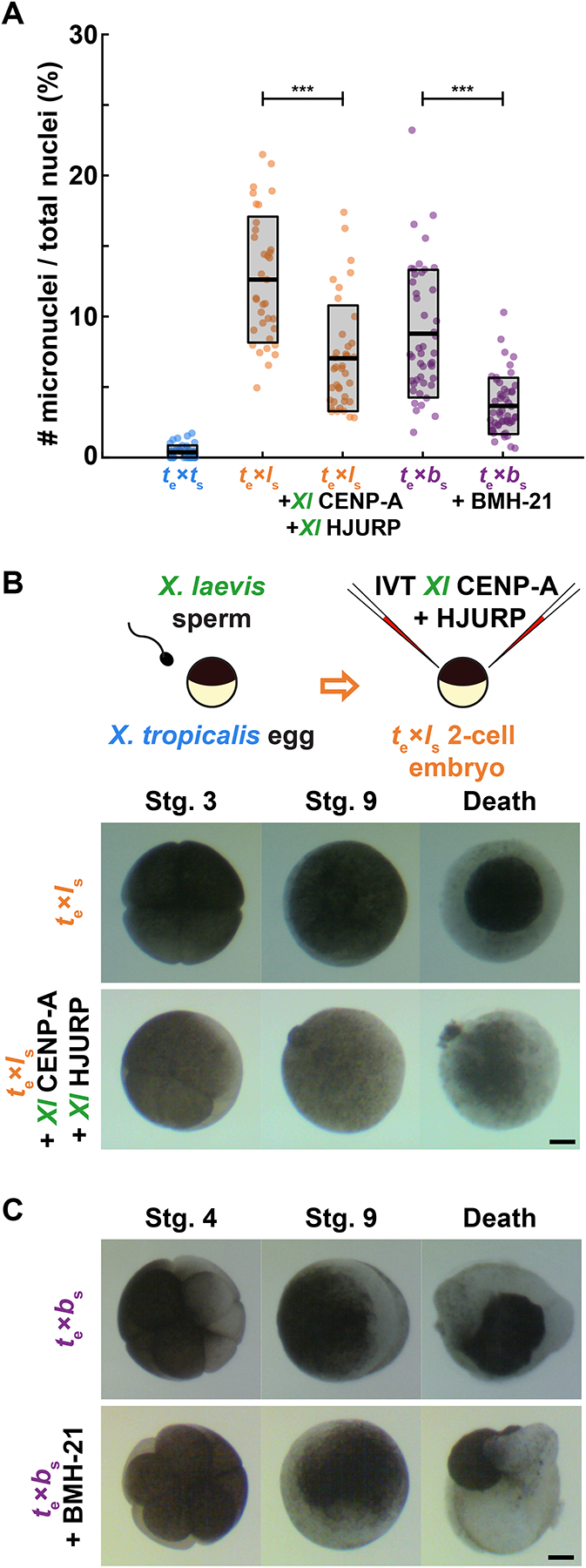
(A) Quantification of chromosome mis-segregation events as measured by the number of micronuclei compared to total nuclei in treated hybrid embryos. X. tropicalis eggs fertilized with X. laevis sperm were microinjected with X. laevis CENP-A/HJURP, while X. tropicalis eggs fertilized with X. borealis sperm were treated with Pol 1 inhibitor BMH-21. Embryos were fixed at stage 9 (7 hpf) just before gastrulation and hybrid death. The number of micronuclei was significantly reduced in both cases, but not to control levels measured in X. tropicalis eggs fertilized with X. tropicalis sperm. N = 3 clutches for each hybrid, N > 15 embryos and > 200 cells per embryo. p-values (left to right) by two-tailed two-sample unequal variance t-tests: 2.111e-7, 2.651e-9; ns, not significant.
(B) Schematic of experiment and video frames of X. tropicalis eggs fertilized with X. laevis sperm microinjected at the two-cell stage with X. laevis CENP-A/HJURP, increasing centromeric protein concentration by ~44.5%. Microinjected hybrid embryos die at the same time and in the same manner as uninjected hybrid controls. N = 10 embryos across 4 clutches. Scale bar is 200 μm. See also Video S1.
(C) Video frames of X. tropicalis eggs fertilized with X. borealis sperm that were incubated from the two-cell stage with 1 μM RNA Pol I inhibitor, BMH-21. Treated hybrid embryos die at the same time and in the same manner as untreated hybrid controls. N = 12 embryos across 2 clutches. Scale bar is 200 μm. See also Video S2.
See also Figure S6.
DISCUSSION
Centromeric DNA sequences and centromere and kinetochore proteins have been shown to rapidly co-evolve, which is thought to counteract female meiotic drive and maintain faithful chromosome segregation5,36,58–60. Our study reveals very low conservation of core centromere DNA sequences across three Xenopus species, and differences in protein sequences of Xenopus CENP-A and its chaperone HJURP are also observed. However, robust epigenetic mechanisms must operate to maintain centromere compatibility in Xenopus hybrids, since many crosses are viable24,61–63, and only a subset of chromosomes display centromere/kinetochore defects in inviable hybrids25. Thus, neither differences in centromere sequences nor centromere/kinetochore proteins appear to contribute directly to Xenopus hybrid inviability, although more detailed analysis of X. borealis centromeres will be necessary to fully address this point.
The Xenopus egg extract and sperm chromosome reconstitution system uniquely allowed us to identify mechanisms by which centromere formation is disrupted on specific chromosomes in inviable interspecies hybrids. For X. tropicalis eggs fertilized with X. borealis sperm, in vitro experiments indicate that defects result from replication stress at rDNA, since both CENP-A localization and chromosome morphology can be rescued by either evicting RNA Pol I or preventing replication fork collapse by inhibiting the chaperone p9745,50,53. However, it is unclear why distinct subsets of paternal chromosomes appear to possess ultra-thin regions versus centromere defects. Given the observed enrichment of rRNA and snRNA repeats associated with X. borealis centromeres, we propose that clustering of repetitive elements including rDNA, pericentromeric, and centromeric repeats during interphase brings together different chromosomal loci and their associated machineries. Normally, such clustering is observed at chromocenters, which may function to stabilize centromeres and promote CENP-A deposition in early G1 of the cell cycle64,65. Although discrete chromocenters or other nuclear bodies such as nucleoli have not been observed to form in egg extracts, hybrid reactions may be revealing trans interactions that normally occur during interphase across rDNA loci, including the centromere-adjacent regions of four specific X. borealis chromosomes. Intriguingly, the p97 chaperone has been implicated in both CENP-A extraction from centromeres and activation of rDNA transcription in Arabidopsis66. While addition of excess CENP-A and its chaperone HJURP can rescue centromere assembly on these chromosomes during interphase, we propose that incomplete rDNA replication at the onset of mitosis due to Pol I occupancy and/or transcription locally recruits p97, which causes both fork collapse and CENP-A extraction from neighboring centromeres. Understanding how formation of fragile sites and centromere loss are related will require a complete X. borealis genome assembly that includes rDNA and other repetitive sequences.
Our findings highlight the dynamic interplay between machineries that promote and disrupt centromere assembly. For in vitro reactions reconstituting X. tropicalis eggs fertilized with X. laevis sperm, the disruption does not involve Pol I or replication stress. Centromere defects appear less severe in this hybrid reaction and can be fully rescued by addition of either species-matched or overexpressed CENP-A/HJURP and partially rescued by Pol II eviction, treatments that may reinforce epigenetic machineries that maintain centromeres. Thus, distinct mechanisms underlie centromere disruption in the two inviable hybrids, but defects in both cases are consistent with observations that aberrant polymerase occupancy or transcription adjacent to the centromere can compromise its assembly67–69.
An open question is how the incompatibilities we have characterized in vitro manifest in hybrid embryos in vivo. Whole genome sequencing of the X. tropicalis egg/X. laevis sperm hybrid just prior to embryo death combined with preliminary Hi-C analysis indicates that the long arms of chromosomes 3L and 4L have been largely eliminated, but the centromere persists on the short arm allowing it to be retained25 (unpublished data). One possible explanation is that under-replication of repetitive sequences adjacent to the centromere in this hybrid initially disrupts centromere assembly, but after chromosome breakage, the adjacent, troublesome sequences are removed and the centromere stabilizes on the short arm while the long arm lacking the centromere frequently ends up in micronuclei and is eventually degraded. Because micronuclei are observed throughout embryogenesis in both inviable hybrids25, multiple rounds of chromosome mis-segregation and instability likely occur that give rise to the terminal karyotype. In the X. tropicalis egg/X. borealis sperm inviable hybrid that experiences replication stress, a pathway involving p97-mediated extraction and degradation of the replicative helicase that leads to fork breakage and microhomology-mediated end joining events likely operates, which has been well characterized in Xenopus egg extracts45. Detailed genomic analysis of chromosome deletions and rearrangements in hybrid embryos will shed light on how replication-transcription conflicts give rise to specific chromosome defects, while additional in vitro experiments will reveal underlying molecular mechanisms.
Death of inviable Xenopus hybrids occurs at gastrulation when the zygotic genome undergoes widespread transcriptional activation, and the distinct death phenotypes observed upon fertilization of X. tropicalis eggs with either X. laevis or X. borealis sperm may be due to the different sets of genes affected by the loss of specific chromosomal loci. However, despite a reduction in micronuclei upon hybrid embryo treatments that rescued centromere formation in vitro, death was not delayed or the phenotypes altered in any way. Therefore, we hypothesize that other incompatibilities also contribute to hybrid inviability. In particular, mismatches between mitochondrial and nuclear-encoded genes have been shown to underlie inviability in some hybrids70,71.
In conclusion, our findings identify defects in epigenetic centromere maintenance that contribute to hybrid inviability. The combination of in vivo, in vitro, and genomic approaches possible in Xenopus promise to provide further mechanistic insights into the molecular basis of hybrid fates and speciation.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Rebecca Heald (bheald@berkeley.edu).
Materials Availability
All materials are available upon request. In general, plasmid constructs and antibodies are available for sharing.
Data and Code Availability
ChIP-seq data used in this study are publicly available at NCBI. Accession numbers are listed in the Key Resources Table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-xCENP-A | Straight Lab8,40 | N/A |
| Rabbit anti-POLR1A | Novus Biologicals | Cat#: NBP2-56122 |
| Mouse anti-UBTF, clone 6B6 | Abnova | Cat#: H00007343-M01; RRID: AB_607269 |
| Rabbit anti-Histone H3 | Abcam | Cat#: ab1791; RRID: AB_302613 |
| Mouse anti-Beta-tubulin E7 | Developmental Studies Hybridoma Bank | Cat#: E7; RRID: AB_2315513 |
| Mouse anti-c-myc, clone 9E10 | Sigma-Aldrich | Cat#: M4439; RRID: AB_439694 |
| Mouse anti-Ran | BD Biosciences | Cat#: 610340; RRID: AB_397730` |
| Alexa Fluor 488 | Invitrogen | Cat#: A-11008; RRID: AB_143165 |
| Alexa Fluor 568 | Invitrogen | Cat#: A-11011; RRID: AB_143157 |
| Goat anti-Rabbit IgG (H&L) Antibody DyLight 800 Conjugated | Rockland Immunochemicals | Cat#: 611-145-002-0.5; RRID: AB_11183542 |
| Donkey anti-Mouse IgG (H&L) Antibody DyLight 680 Conjugated | Rockland Immunochemicals | Cat#: 610-744-002; RRID: AB_1660920 |
| Bacterial and virus strains | ||
| XL1-Blue competent cells | Agilent | Cat#: 200249 |
| Chemicals, peptides, and recombinant proteins | ||
| Pregnant mare serum gonadotrophin | Calbiochem | Cat#: 367222 |
| Human chorionic gonadotrophin | Sigma-Aldrich | Cat#: CG10 |
| Hoechst 33342 | Invitrogen | Cat#: H3570 |
| Vectashield | Vector Labs | Cat#: H-1000 |
| Alexa Fluor 568-dUTP | Invitrogen | Cat#: C11399 |
| Random hexamers | Invitrogen | Cat#: 100026484 |
| Klenow (exo-) polymerase | New England Biolabs | Cat#: M0212S |
| Blocking reagent | Roche | Cat#: 11096176001 |
| Salmon sperm DNA | Invitrogen | Cat#: AM9680 |
| Aphidicolin | Sigma-Aldrich | Cat#: A0781 |
| BI-2536 | Selleck Chemicals | Cat#: S1109 |
| BMH-21 | Sigma-Aldrich | Cat#: SML1183 |
| ML-60218 | Sigma-Aldrich | Cat#: 557403 |
| MLN-8237 | Selleck Chemicals | Cat#: S1133 |
| NMS-873 | Sigma-Aldrich | Cat#: SML-1128 |
| Triptolide | Sigma-Aldridch | Cat#: T3652 |
| Critical commercial assays | ||
| TnT Sp6-coupled rabbit reticulocyte system | Promega | Cat#: L2080 |
| Protein A Dynabeads | Fisher | Cat#: 10-002-D |
| NEBNext Ultra II DNA library Prep Kit for Illumina | New England Biolabs | Cat#: E76452 |
| SuperScript III First Strand Synthesis system | Thermo Fisher Scientific | Cat#: 18080051 |
| RNeasy Mini kit | Qiagen | Cat#: 74104 |
| Deposited data | ||
| Xenopus laevis ChIP-seq | 26,73 | GSE153058 |
| Xenopus tropicalis ChIP-seq | 26,73 | GSE 199671 |
| Xenopus borealis ChIP-seq | This paper | PRJNA848409 |
| Experimental models: Organisms/strains | ||
| Xenopus laevis | Nasco | Cat#: LM00535 |
| Xenopus laevis | National Xenopus Resource | Cat#: NXR_0031 |
| Xenopus tropicalis | Nasco | Cat#: LM00822 |
| Xenopus tropicalis | National Xenopus Resource | Cat#: NXR_1018 |
| Xenopus borealis | Nasco | Cat#: LM00698 |
| Oligonucleotides | ||
| Primers (FCR monomer sequence amplification, pJET1.2 Sequencing Primers) FWD: CGACTCACTATAGGGAGAGCGGC REV: AAGAACATCGATTTTCCATGGCAG |
26 | N/A |
| Primers (X. laevis CENP-A amplification) FWD: CAAGCTTCGAATTCTGCAGTCGACTGCCACCATGAGACCGGGCTCCACTCC REV: GGGTTAATGAGGGACTGGGGTAAGAGCCTCTAGAACTATAGTGAGTCGTATTAC |
This paper | N/A |
| Primers (X. tropicalis CENP-A amplification) FWD: CAAGCTTCGAATTCTGCAGTCGACTGCCACCATGAGGCCTGGGTCTACTCC REV: (GAGTTACTGAGGGGTTGGGGTAAGAGCCTCTAGAACTATAGTGAGTCGTATTAC) |
This paper | N/A |
| Primers (X. borealis CENP-A amplification) FWD: TAAGCACTCGAGGCCATGAGATCGGGGTCCACTCC REV: AATCGTTCTAGAGGCTTACCCCAGTCCCTCATTAACCC |
This paper | N/A |
| Recombinant DNA | ||
| Plasmid: Full length X. laevis CENP-A in pCS2+ vector | This paper | N/A |
| Plasmid: Full length X. tropicalis CENP-A in pCS2+ vector | This paper | N/A |
| Plasmid: Full length X. borealis CENP-A in pCS2+ vector | This paper | N/A |
| Plasmid: Full length X. laevis GFP-xHJURP in pCS2+ vector | Straight Lab | ASP1640 |
| Plasmid: Full length X. laevis xCENP-C-myc in pCS2+ vector | Straight Lab | ASP867 |
| Plasmid: FCR monomer4 in pJET1.2 | Straight Lab26 | N/A |
| Plasmid: FCR monomer10 in pJET1.2 | Straight Lab26 | N/A |
| Plasmid: FCR monomer16 in pJET1.2 | Straight Lab26 | N/A |
| Plasmid: FCR monomer19 in pJET1.2 | Straight Lab26 | N/A |
| Software and algorithms | ||
| FIJI | 81 | https://imagej.net/software/fiji/ |
| Matlab | https://www.mathworks.com/products/matlab.html | |
| k-mer counting pipeline | 26 | https://github.com/straightlab/xenla-cen-dna-paper |
| cd-his-est | 74 | http://weizhong-lab.ucsd.edu/cd-hit/ |
| Geneious (7.1.4) | https://www.geneious.com/ | |
| RepeatMasker 4.0.9 | 75 | http://www.repeatmasker.org |
| Other | ||
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All frogs were used and maintained in accordance with standards established by the UC Berkeley Animal Care and Use Committee and approved in our Animal Use Protocol. Mature Xenopus laevis, X. tropicalis, and X. borealis frogs were obtained from Nasco (Fort Atkinson, WI) or the National Xenopus Resource (Woods Hole, MA). Xenopus frogs were housed in a recirculating tank system with regularly monitored temperature and water quality (pH, conductivity, and nitrate/nitrite levels). X. laevis and X. borealis were housed at 20–23°C, and X. tropicalis were housed at 23–26°C. All animals were fed Nasco frog brittle.
Chemicals
Unless otherwise states, all chemicals were purchased from Sigma-Aldrich, St. Louis, MO.
Frog care
X. laevis, X. tropicalis, and X. borealis females were ovulated with no harm to the animals with a 6-, 3-, and 4-month rest interval, respectively, as previously described72. To obtain testes, males were euthanized by over-anesthesia through immersion in ddH2O containing 0.15% MS222 (Tricaine; Sigma) neutralized with 5 mM sodium bicarbonate prior to dissection, and then frozen at −20°C.
METHODS DETAILS
CENP-A ChIP-seq and data analysis
CENP-A MNase ChIP-seq was performed as previously described26. Briefly, livers were extracted from adult X. borealis animals and flash frozen. Upon thawing, livers were diced on ice, rinsed in PBS, and buffer 1 (2.5 mM EDTA, 0.5 M EGTA, 15 mM Tris-HCl pH 7.4, 15 mM NaCl, 60 mM KCl, 15 mM sodium citrate 0.5 mM spermidine, 0.15 mM spermine, 340 mM sucrose, supplemented with 0.1 mM PMSF) was added and the tissue dounced using pestle A 12 times. A syringe with 18-gauge needle was backfilled with nuclei mixture and expelled into 2 mL tubes with additional buffer 1. Nuclei were spun at 6,000g for 5 min at 4°C, and washed 3 times with buffer 3 (2.5 mM EDTA, 0.5 M EGTA, 15 mM Tris-HCl pH 7.4, 15 mM NaCl, 60mM KCl, 15 mM sodium citrate 0.5 mM spermidine, 0.15 mM spermine, 340 mM sucrose, supplemented with 0.1 mM PMSF). Nuclei quality was checked and nuclei were counted by hemocytometer. ~5–10 million nuclei were used per IP reaction.
For MNase digestion, CaCl2 was added to each reaction tube to 5 mM together with 300 U of MNase. Digestion was performed at RT for 30 min and reaction was quenched with 10 mM EDTA and 5 mM EGTA. Nuclei were lysed with 0.05% IGEPAL CA-630 in ice for 10 min. Following an initial spin 1,500g 5 min 4°C, the pellet was resuspended in 500 μL buffer 3 + 200 mM NaCl and rotated overnight at 4°C to extract mononucleosomes. Samples were precleared, input fractions were taken and CENP-A mononucleosomes were isolated with 10 μg rabbit anti X. laevis CENP-A antibody prebound to protein A dynabeads in 200 μL TBST with rotation overnight at 4°C. Beads were washed and eluted, mononucleosomal DNA was isolated with Ampure beads, and sequencing libraries were prepared using NEBNext fit for Illumina sequencing which was performed on a NovaSeq instrument with paired end 150bp sequencing.
X. laevis and X. tropicalis CENP-A CHIP-seq datasets were used from previously described studies26,73. CENP-A ChIP and Input libraries from each species were processed to identify CENP-A enriched k-mers using the k-mer counting pipeline that normalizes k-mer counts by sequencing depth of each library (https://github.com/straightlab/xenla-cen-dna-paper). For this study 25bp k-mers were used and kmc was run with ci=10, indicating that k-mers must be found 10 times in the dataset to be considered. This was chosen so that more k-mers were identified from each species to make comparisons more likely.
A phylogram was generated using a method similar to that previously described26. From each species full length ChIP-seq reads were selected based on the presence of at least 80 CENP-A enriched k-mers. The reads from each species that met this criterion were then clustered by sequence similarity using cd-hit-est74 using sequential rounds of clustering by 98%, 95%, and 90% identical by sequence. The 20 top clusters from each species were then selected for phylogram generation using Geneious (7.1.4) Tree Builder with the following settings: Genetic Distance Model=Tamura-Nei, Tree building method=Neighbor-joining, Outgroup=No outgroup, Alignment Type=Global alignment, Cost Matrix=93% similarity. Colors for each species were added manually.
To identify repeat classes enriched in the CENP-A datasets from each species, RepeatMasker 4.0.975 was run using the giri Repbase library for Xenopus repeats on subsets of one million reads generated from CENP-A and Input sequencing libraries in triplicate. Counts for each repeat class were summarized and an enrichment score of CENP-A/Input was calculated for each pair of subsets. Enrichment scores for each repeat class were reported as a bar plot of the mean and standard deviation of the triplicates for each species.
Protein sequence alignments
Multiple sequence alignments were performed using Clustal Omega (default parameters). Sequence similarities were determined by pair-wise alignments using EMBOSS Needle (default parameters).
Xenopus egg extracts
X. laevis and X. tropicalis metaphase-arrested egg extracts and spindle reactions were prepared as previously described37,76,77. Briefly, freshly laid, metaphase II-arrested eggs were collected, dejellied, packed and crushed by centrifugation. The cytoplasmic layer was collected with a syringe and 18G needle, then supplemented with 10 μg/mL of leupeptin, pepstatin, and chymostatin (LPC), 20 μM of cytochalasin B, and energy mix (3.75 μM creatine phosphate, 0.5 μM ATP, 0.5 μM MgCl2, 0.05 μM EGTA). Typical reactions contained 20 μL CSF extract, sperm nuclei at a final concentration of 500 nuclei/μL, and rhodamine-labeled porcine brain tubulin at a final concentration of 50 μg/mL.
Chromosome immunofluorescence
Spindle reactions were prepared, spun-down, and processed for immunofluorescence as previously described37,76. Briefly, the extract reactions were fixed for 5–10 min with 2% formaldehyde and spun down at 5,500 rpm (5821.9 × g) for 20 min at 16°C. The coverslips were incubated for 30 s in cold methanol, washed in PBS + 0.1% NP40, and blocked overnight in PBS + 3% BSA at 4°C. We used rabbit anti-xCENPA, 1:5008,40, mouse anti-myc (9E10 clone, 1:500), rabbit anti-POLR1A (Novus Biologicals, 1:500), and mouse anti-UBTF (Abnova, 1:500) antibodies. Primary antibodies were added for 1 h in PBS + 3% BSA. After washing with PBS + 0.1% NP40, the coverslips were incubated with 1:1000 anti-rabbit or mouse secondary antibodies coupled to Alexa Fluor 488 or 568 (Invitrogen), respectively, for 30 min and then with 1:1000 Hoechst (Invitrogen) for 5 min. The coverslips were then washed and mounted for imaging with Vectashield (Vector Labs). Each presented dataset was obtained from three independent egg extracts.
Nuclear DNA FISH for FCR centromeric sequences
Nuclear DNA FISH using probes against various FCR monomers was performed as previously described26. Briefly, pJET1.2 plasmids containing 150 bp FCR monomer sequences were PCR-amplified and fluorescently labeled using random hexamer priming and Klenow (exo-) polymerase (New England Biolabs). Both Alexa Fluor 488 and 568 dUTP-conjugated fluorophores (Invitrogen) were used. Probes were desalted to remove unincorporated nucleotides, then precipitated and cleaned before resuspension in hybridization buffer (65% formamide, 5X SSC, 5X Denhardts with 1% blocking reagent (Roche), 0.5 mg/mL salmon sperm DNA added fresh). Each experiment used 4 uL of probe mixed with 4 uL of hybridization buffer.
Nuclei were assembled in egg extract, spun down onto coverslips, and probed with CENP-A antibody as previously described in78 and detailed above. Samples proceeded to FISH by fixation in 2.5% formaldehyde in PBS for 10 min, washed in PBS, and dehydrated with increasing concentrations of 70–100% ice-cold ethanol. Coverslips were blocked for 30 m in hybridization buffer. Probes were warmed and mixed with hybridization buffer before being added to samples, flipping coverslips onto glass slides for hybridization. These “sandwiches” were incubated at 80°C for 10 min, then incubated overnight at 37°C. Coverslips were removed from glass slides carefully with 4X SSC, washed thoroughly in SSC, stained with Hoechst and mounted with Vectashield (Vector Labs).
Protein expression in reticulocyte lysate
To generate plasmids for expression of species-specific X. laevis, X. tropicalis, and X. borealis CENP-A, total RNA was isolated from stage 9 embryos. Embryos were homogenized mechanically in TRIzol (Thermo Fisher Scientific) using up to a 30-gauge needle and processed according to manufacturer’s instructions. After resuspension in nuclease-free H2O, RNAs were cleaned using a RNeasy kit (Qiagen) according to manufacturer’s instructions, and cDNA was synthesized using the SuperScript III First Strand Synthesis system (Thermo Fisher Scientific) according to the manufacturer’s instructions. The X. laevis, X. tropicalis, and X. borealis CENP-A sequences were then PCR-amplified from the cDNA. The amplified sequence was then subcloned into a pCS2+ vector using Gibson assembly. The constructs were then amplified using XL1-Blue competent E. coli (Agilent).
The TnT Sp6-coupled rabbit reticulocyte system (Promega) was used for in vitro transcription/translation (IVT) of plasmid DNA according to the manufacturer’s protocol. 2–10% of the final egg extract reaction volume was added prior to addition of sperm nuclei; for CENP-A, this corresponds to 8–80 times endogenous protein levels.
Western blots
Increasing volumes of egg extracts and reticulocyte lysate were subject to SDS-PAGE and wet transferred to PVDF membranes. Blots were blocked with PBS + 0.1% Tween + 5% milk for 1 h, probed with primary antibodies diluted in PBS + 0.1% Tween + 5% milk for 1 h, rinsed 3x over a 10 m period with PBS + 0.1% Tween, then probed with secondary antibodies (Rockland Immunochemicals; goat anti-rabbit DyLight 800 and donkey anti-mouse DyLight 680, 1:10,000) diluted in PBS + 0.1% Tween for 30 m. Blots were scanned on an Odyssey Infrared Imaging System (Li-Cor Biosciences). Band intensities were quantified using FIJI.
Drug treatments
X. tropicalis extract was supplemented with the following drugs and concentrations: Aphidicolin (DNA replication inhibitor, 10 μg/mL, Sigma), BMH-21 (RNA Polymerase I inhibitor, 1 μM, Sigma), NMS-873 (p97 inhibitor, 10 μM, Sigma), MLN-8237 (Aurora A inhibitor, 1 μM, Selleck Chemicals), BI-2536 (Polo kinase 1 inhibitor, 1 μM, Selleck Chemicals), Triptolide (RNA Polymerase II inhibitor, 25 μM, Sigma).
Chromosome and nuclei imaging
Chromosomes were imaged using Micromanager 1.4 software79 and nuclei were imaged using Olympus cellSens Dimension 2 software on an upright Olympus BX51 microscope equipped with an ORCA-ER or ORCA-Spark camera (Hamamatsu Photonics) and Olympus UPlan 60X/NA 1.42 oil objective. All images across all datasets were taken using the same exposure settings.
In vitro fertilization and cross-fertilizations
In vitro fertilization and cross-fertilizations were performed as previously described25,72,80. X. laevis, X. borealis, and X. tropicalis males were injected with 500, 300, and 250 U, respectively, of human chorionic gonadotropin hormone (hCG, Sigma) 12–24 h before dissection. Testes were collected in Leibovitz L-15 Medium (Gibco, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS; Gibco) for immediate use. X. tropicalis females were primed with 10 U of hCG 12–18 h before use and boosted with 250 U of hCG on the day of the experiment. As soon as the first eggs were laid (~3 h after boosting), the male was euthanized and dissected. Two X. tropicalis testes or one X. laevis or X. borealis testis were added to 1 mL of L-15 + 10% FBS. X. tropicalis females were squeezed gently to deposit eggs onto glass Petri dishes (Corning) coated with 1.5% agarose in 1/10X MMR (1X MMR: 100 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgSO4, 0.1 mM EDTA, 5 mM HEPES-NaOH pH 7.6). Testes were homogenized using a pestle in L-15 + 10% FBS to create sperm solution. Any liquid in the Petri dishes was removed, and the eggs were fertilized with 500 uL of sperm solution per dish. Eggs were swirled in the solution to separate them and incubated for 5 min with the dish slanted. Dishes were flooded with ddH2O and incubated for 10 min. ddH2O was exchanged for 1/10X MMR and incubated for 10 min. The jelly coats were removed with a 3% cysteine solution (in ddH2O-NaOH, pH 7.8). After extensive washing with 1/10X MMR (at least four times), embryos were incubated at 23°C until the first cleavage at 1 hour post fertilization (hpf). Fertilized embryos were then sorted and placed in a mesh-bottomed dish for microinjection as described below.
Embryo microinjection
At stage 2 (2-cell embryo), embryos were transferred to a 1/9X MMR + 3% Ficoll. IVT reticulocyte lysate was backloaded into a needle pulled from a 1 mm glass capillary tube (TW 100F-4, World Precision Instruments) using a P-87 Micropipette Puller (Sutter Instrument). Embryos were placed in a mesh-bottomed dish and microinjected in both blastomeres with 2 nL of the IVT reticulocyte lysate using a Picospritzer III microinjection system (Parker) equipped with a MM-3 micromanipulator (Narishige). Injected embryos were transferred to a new dish and incubated at 23°C in 1/9X + 3% Ficoll for several hours, then buffer exchanged for 1/10X MMR overnight.
Embryo video imaging
Imaging dishes were prepared using an in-house PDMS mold designed to print a pattern of 0.9 mm large wells in agarose that allowed us to image six X. tropicalis embryos simultaneously within the 3 mm × 4 mm camera field of view for each condition. Embryos were imaged from stage 3 after microinjection. Treatment and control videos were taken simultaneously using two AmScope MD200 USB cameras (AmScope), each mounted on an AmScope stereoscope. Time-lapse movies were acquired at a frequency of one frame every 10 s for 20 h and saved as Motion JPEG using a Matlab (The MathWorks) script. Movie post-processing (cropping, concatenation, resizing, and addition of scale bar) was done using Matlab and FIJI81. All Matlab scripts written for this study are available upon request. Two of the scripts used here were obtained through the MATLAB Central File Exchange: ‘videoMultiCrop’ and ‘concatVideo2D’ by ‘Nikolay S’.
Embryo whole-mount immunofluorescence
Embryos were fixed at the desired stages for 1–3 h using MAD fixative (2 parts methanol, Thermo Fisher; 2 parts acetone, Thermo Fisher; 1 part DMSO, Sigma). After fixation, embryos were dehydrated in methanol and stored at −20°C. Embryos were then processed for immunofluorescence as previously described25. Briefly, embryos were gradually rehydrated in 0.5X SSC (1X SSC: 150 mM NaCl, 15 mM Na citrate, pH 7.0), then bleached with 2% H2O2 in 0.5X SSC with 5% formamide for 2 h under light. Embryos were washed with PBT (1X PBS, 0.1% Triton X-100, 2 mg/mL bovine serum albumin). Embryos were blocked in PBT supplemented with 10% goat serum and 5% DMSO for 1–3 h and incubated overnight at 4°C in PBT supplemented with 10% goat serum and primary antibodies. We used mouse anti-beta-tubulin (E7, Developmental Studies Hybridoma Bank, 1:300 dilution) and rabbit anti-histone H3 (Abcam, 1:500 dilution). Embryos were then washed 4 × 2 h in PBT and incubated overnight at 4°C in PBT supplemented with goat anti-mouse and goat anti-rabbit secondary antibodies coupled to Alexa Fluor 488 and 568 (Invitrogen). Embryos were then washed 4 × 2 h in PBT and gradually dehydrated in methanol. Finally, embryos were cleared in Murray’s clearing medium (2 parts benzyl benzoate, Sigma; 1 part benzyl alcohol, Sigma). Embryos were placed in a reusable chamber (Thermo Fisher) for confocal microscopy.
Confocal microscopy
Confocal microscopy was performed on an inverted Zeiss LSM 800 using the Zeiss Zen software, a Plan-Achromat 20X/0.8 air objective and laser power 0.5–2%, on multiple 1024×1024 pixel plans spaced of 0.68 μm in Z. Images are mean averages of two scans with a depth of 16 bits. Pinhole size always corresponded to 1 Airy unit.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of CENP-A localization on mitotic chromosomes was determined manually in a dataset of 100 images from one extract. Quantification of ultra-thin chromosomal regions was also determined manually in parallel from the same datasets. Only single chromosomes were counted. Each dataset had ~150–400 chromosomes. The average of each extract was calculated as a percentage of total chromosome number. Averages were plotted in Matlab, and statistical significance and p-values were determined with two-tailed, two-sample unequal variance t-tests or one-way ANOVA with Tukey post-hoc analysis in Microsoft Excel. The number of egg extracts used, individual chromosomes counted, and p-values are listed in the figure legends. For all box plots, the thick line inside the box indicates the average across biological replicates, and the upper and lower box boundaries indicate the standard deviation.
PolR1A and UBF fluorescent intensity on X. borealis ultra-thin chromosomes were quantified in FIJI by measuring the intensity of the stretched region specifically and comparing it to a random non-stretched region on the same chromosome. All intensity measurements were normalized to the samples’ Hoechst intensity.
Micronuclei in embryos were quantified at the relevant stages as the number of observed micronuclei divided by the number of nuclei, counted manually in FIJI. Statistical significance was determined by two-tailed, two-sample unequal variance t-tests.
Supplementary Material
X. tropicalis eggs were fertilized with X. laevis sperm and microinjected with in vitro translated paternally-matched CENP-A and HJURP (right) at stage 2. Embryos were imaged in separate dishes from stage 3 in 1/10X MMR. The movie plays 20 h in 15s (rate of 120 frames per second). Scale bar corresponds to 200 μm.
X. tropicalis eggs were fertilized with X. borealis sperm and incubated with 1 μM BMH-21 in 1/10X MMR (right) from stage 2. The movie plays 20 h in 15s (rate of 120 frames per second). Scale bar corresponds to 200 μm.
HIGHLIGHTS:
Divergent core centromeric sequences do not underlie Xenopus hybrid inviability
CENP-A eviction from specific paternal chromosomes requires cell cycle progression
Driving assembly can rescue and maintain X. laevis, but not X. borealis, centromeres
Mitotic replication stress and conflicts lead to X. borealis chromosome defects
ACKNOWLEDGEMENTS
We thank Daniel Rokshar, Austin Mudd, Sofia Medina-Ruiz, and Mariko Kondo for early access to X. borealis CENP-A, CENP-C, HJURP, and H3 sequences. We also thank students Elizabeth Turcotte, Costa Bartolutti, Justin Peng, and Christian Erikson for help with experiments to MK. We are grateful to the Welch, King, Dernberg, Karpen, Lewis, and Rokshar laboratories at UC Berkeley for sharing reagents, discussions, and expertise. We thank all past and present members of the Heald laboratory, Coral Y. Zhou, Gary Karpen, Dirk Hockemeyer, Rasmus Nielsen, and Mark J. Khoury for continuous support and fruitful discussions. M.K. was supported by a National Science Foundation (NSF) GRFP fellowship. O.K.S. was supported by a National Institutes of Health (NIH) T32 GM113854-02 and an NSF GRFP fellowship. A.F.S. was supported by NIH NIGMS R01 GM074728. R.H. was supported by NIH MIRA grant R35 GM118183 and the Flora Lamson Hewlett Chair.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Sanei M, Pickering R, Kumke K, Nasuda S, and Houben A (2011). Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybirds. PNAS 108, E498–E505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maheshwari S, and Barbash DA (2011). The Genetics of Hybrid Incompatibilities. Annu. Rev. Genet 45, 331–355. [DOI] [PubMed] [Google Scholar]
- 3.Fujiwara A, Abe S, Yamaha E, Yamazaki F, and Yoshida MC (1997). Uniparental chromosome elimination in the early embryogenesis of the inviable salmonid hybrids between masu salmon female and rainbow trout male. Chromosoma 106, 44–52. [DOI] [PubMed] [Google Scholar]
- 4.Gernand D, Rutten T, Varshney A, Rubtsova M, Prodanovic S, and Bru C (2005). Uniparental Chromosome Elimination at Mitosis and Interphase in Wheat and Pearl Millet Crosses Involves Micronucleus Formation, Progressive Heterochromatinization, and DNA Fragmentation. Plant Cell 17, 2431–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malik HS, and Henikoff S (2001). Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157, 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maheshwari S, Tan EH, West A, Franklin FCH, Comai L, and Chan SWL (2015). Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids. PLoS Genet. 11, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosin L, and Mellone BG (2016). Co-evolving CENP-A and CAL1 Domains Mediate Centromeric CENP-A Deposition across Drosophila Species. Dev. Cell 37, 136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moree B, Meyer CB, Fuller CJ, and Straight AF (2011). CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol 194, 855–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll CW, Silva MCC, Godek KM, Jansen LET, and Straight AF (2009). Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol 11, 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pentakota S, Zhou K, Smith C, Maffini S, Petrovic A, Morgan GP, Weir JR, Vetter IR, Musacchio A, and Luger K (2017). Decoding the centromeric nucleosome through CENP-N. Elife 6, e33442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chittori S, Hong J, Saunders H, Feng H, Ghirlando R, Kelly AE, Bai Y, and Subramaniam S (2018). Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science (80-.) 359, 339–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian T, Li X, Liu Y, Wang C, Liu X, Bi G, Zhang X, Yao X, Zhou ZH, and Zang J (2018). Molecular basis for CENP-N recognition of CENP-A nucleosome on the human kinetochore. Cell Res. 28, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LET, Van Duyne GD, Vinogradov SA, et al. (2015). CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science (80-.) 348, 699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.French BT, Westhorpe FG, Limouse C, and Straight AF (2017). Xenopus laevis M18BP1 Directly Binds Existing CENP-A Nucleosomes to Promote Centromeric Chromatin Assembly. Dev. Cell 42, 190–199.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shono N, Ohzeki JI, Otake K, Martins NMC, Nagase T, Kimura H, Larionov V, Earnshaw WC, and Masumoto H (2015). CENP-C and CENP-I are key connecting factors for kinetochore and CENP-A assembly. J. Cell Sci 128, 4572–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hori T, Shang WH, Hara M, Ariyoshi M, Arimura Y, Fujita R, Kurumizaka H, and Fukagawa T (2017). Association of M18BP1/KNL2 with CENP-A Nucleosome Is Essential for Centromere Formation in Non-mammalian Vertebrates. Dev. Cell 42, 181–189.e3. [DOI] [PubMed] [Google Scholar]
- 17.Jagannathan M, and Yamashita YM (2021). Defective Satellite DNA Clustering into Chromocenters Underlies Hybrid Incompatibility in Drosophila. Mol. Biol. Evol 38, 4977–4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomae AW, Schade GOM, Padeken J, Borath M, Vetter I, Kremmer E, Heun P, and Imhof A (2013). A Pair of Centromeric Proteins Mediates Reproductive Isolation in Drosophila Species. Dev. Cell 27, 412–424. [DOI] [PubMed] [Google Scholar]
- 19.Satyaki PRV, Cuykendall TN, Wei KHC, Brideau NJ, Kwak H, Aruna S, Ferree PM, Ji S, and Barbash DA (2014). The Hmr and Lhr Hybrid Incompatibility Genes Suppress a Broad Range of Heterochromatic Repeats. PLoS Genet. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anselm E, Thomae AW, Jeyaprakash AA, and Heun P (2018). Oligomerization of Drosophila Nucleoplasmin-Like Protein is required for its centromere localization. Nucleic Acids Res. 46, 11274–11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukacs A, Thomae AW, Krueger P, Schauer T, Venkatasubramani AV, Kochanova NY, Aftab W, Choudhury R, Forne I, and Imhof A (2021). The integrity of the HMR complex is necessary for centromeric binding and reproductive isolation in Drosophila. PLoS Genet. 17, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blum JA, Bonaccorsi S, Marzullo M, Palumbo V, Yamashita YM, Barbash DA, and Gatti M (2017). The Hybrid Incompatibility Genes Lhr and Hmr Are Required for Sister Chromatid Detachment During Anaphase but Not for Centromere Function. Genetics 207, 1457–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, et al. (2016). Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narbonne P, Simpson DE, and Gurdon JB (2011). Deficient induction response in a Xenopus nucleocytoplasmic hybrid. PLoS Biol. 9, e1001197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibeaux R, Acker R, Kitaoka M, Georgiou G, van Kruijsbergen I, Ford B, Marcotte EM, Nomura DK, Kwon T, Veenstra GJC, et al. (2018). Paternal chromosome loss and metabolic crisis contribute to hybrid inviability in Xenopus. Nature 553, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith OK, Limouse C, Fryer KA, Teran NA, Sundararajan K, Heald R, and Straight AF (2021). Identification and characterization of centromeric sequences in Xenopus laevis. Genome Res. 31, 958–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKinley KL, and Cheeseman IM (2016). The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol 17, 16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westhorpe FG, and Straight AF (2014). The Centromere: Epigenetic Control of Chromosome Segregation during Mitosis. Cold Spring Harb. Perspect. Biol 7, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panchenko T, and Black BE (2009). The epigenetic basis for centromere identity. [DOI] [PubMed]
- 30.Foltz DR, Jansen LET, Bailey AO, Yates JR, Bassett EA, Wood S, Black BE, and Cleveland DW (2009). Centromere-Specific Assembly of CENP-A Nucleosomes Is Mediated by HJURP. Cell 137, 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, and Almouzni-Pettinotti G (2009). HJURP Is a Cell-Cycle-Dependent Maintenance and Deposition Factor of CENP-A at Centromeres. Cell 137, 485–497. [DOI] [PubMed] [Google Scholar]
- 32.Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, et al. (2011). Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 25, 901–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosin LF, and Mellone BG (2017). Centromeres Drive a Hard Bargain. Trends Genet. 33, 101–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henikoff S, Ahmad K, and Malik HS (2001). The Centromere Paradox : Stable Inheritance with Rapidly Evolving DNA. Science (80-.) 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- 35.Malik HS, and Henikoff S (2009). Major Evolutionary Transitions in Centromere Complexity. Cell 138, 1067–1082. [DOI] [PubMed] [Google Scholar]
- 36.Kumon T, Ma J, Akins RB, Stefanik D, Nordgren CE, Kim J, Levine MT, and Lampson MA (2021). Parallel pathways for recruiting effector proteins determine centromere drive and suppression. Cell, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maresca TJ, and Heald R (2006). Methods for studying spindle assembly and chromosome condensation in Xenopus egg extracts. Methods Mol. Biol. (Clifton, NJ) 322, 459–474. [DOI] [PubMed] [Google Scholar]
- 38.French BT, and Straight AF (2017). The Power of Xenopus Egg Extract for Reconstitution of Centromeres and Kinetochore Function. Prog Mol Subcell Biol 56, 59–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernad R, Sánchez P, Rivera T, Rodríguez-Corsino M, Boyarchuk E, Vassias I, Ray-Gallet D, Arnaoutov A, Dasso M, Almouzni G, et al. (2011). Xenopus HJURP and condensin II are required for CENP-A assembly. J. Cell Biol 192, 569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milks KJ, Moree B, and Straight AF (2009). Dissection of CENP-C – directed Centromere and Kinetochore Assembly. Mol. Biol. Cell 20, 4246–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zasadzińska E, Huang J, Bailey AO, Guo LY, Lee NS, Srivastava S, Wong KA, French BT, Black BE, and Foltz DR (2018). Inheritance of CENP-A Nucleosomes during DNA Replication Requires HJURP. Dev. Cell, 348–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, and Straight AF (2008). Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol 183, 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roure V, Medina-Pritchard B, Lazou V, Rago L, Anselm E, Venegas D, Jeyaprakash AA, and Heun P (2019). Reconstituting Drosophila Centromere Identity in Human Cells. Cell Rep. 29, 464–479.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gómez-González B, and Aguilera A (2019). Transcription-mediated replication hindrance : a major driver of genome instability. Genes Dev. 33, 1008–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng L, Wu RA, Sonneville R, Kochenova OV, Labib K, Pellman D, and Walter JC (2019). Mitotic CDK Promotes Replisome Disassembly, Fork Breakage, and Complex DNA Rearrangements. Mol. Cell 73, 915–929.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kabeche L, Nguyen HD, Buisson R, and Zou L (2018). A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation SUPP. Science (80-.) 359, 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durkin SG, and Glover TW (2007). Chromosome Fragile Sites. Annu. Rev. Genet 41, 169–192. [DOI] [PubMed] [Google Scholar]
- 48.Maric M, Maculins T, De Piccoli G, and Labib K (2014). Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science (80-.) 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Durica DS, and Krider HM (1977). Studies on the ribosomal RNA cistrons in interspecific Drosophila hybrids. I. Nucleolar dominance. Dev. Biol 59, 62–74. [DOI] [PubMed] [Google Scholar]
- 50.Roussel P, André C, Comai L, and Hernandez-Verdun D (1996). The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J. Cell Biol 133, 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gébrane-Younès J, Fomproix N, and Hernandez-Verdun D (1997). When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. J. Cell Sci 110, 2429–2440. [DOI] [PubMed] [Google Scholar]
- 52.Bell P, and Scheer U (1997). Prenucleolar bodies contain coilin and are assembled in Xenopus egg extract depleted of specific nucleolar proteins and U3 RNA. J. Cell Sci 110, 43–54. [DOI] [PubMed] [Google Scholar]
- 53.Bell P, Mais C, McStay B, and Scheer U (1997). Association of the nucleolar transcription factor UBF with the transcriptionally inactive rRNA genes of pronuclei and early Xenopus embryos. J. Cell Sci 110, 2053–2063. [DOI] [PubMed] [Google Scholar]
- 54.Shiokawa K, Kurashima R, and Shinga J (1994). Temporal control of gene expression from endogenous and exogenously- introduced DNAs in early embryogenesis of Xenopus laevis. Int. J. Dev. Biol 38, 249–255. [PubMed] [Google Scholar]
- 55.Newport J, and Kirschner M (1982). A major developmental transition in early xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell 30, 675–686. [DOI] [PubMed] [Google Scholar]
- 56.Peltonen K, Colis L, Liu H, Trivedi R, Moubarek MS, Moore HM, Bai B, Rudek MA, Bieberich CJ, and Laiho M (2014). A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell 25, 77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Colis L, Peltonen K, Sirajuddin P, Liu H, Sanders S, Ernst G, Barrow JC, and Laiho M (2014). DNA intercalator BMH-21 inhibits RNA polymerase I independent of DNA damage response. Oncotarget 5, 4361–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malik HS, Vermaak D, and Henikoff S (2002). Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc. Natl. Acad. Sci. U. S. A 99, 1449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pontremoli C, Forni D, Pozzoli U, Clerici M, Cagliani R, and Sironi M (2021). Kinetochore proteins and microtubule-destabilizing factors are fast evolving in eutherian mammals. Mol. Ecol 30, 1505–1515. [DOI] [PubMed] [Google Scholar]
- 60.Hooff JJ, Tromer E, Wijk LM, Snel B, and Kops GJ (2017). Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Rep. 18, 1559–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Robertis EM, and Black P (1979). Hybrids of Xenopus laevis and Xenopus borealis express proteins from both parents. Dev. Biol 68, 334–339. [DOI] [PubMed] [Google Scholar]
- 62.Woodland HR, and Ballantine JEM (1980). Paternal gene expression in developing hybrid embryos of Xenopus laevis and Xenopus borealis. J. Embryol. Exp. Morphol 60, 359–372. [PubMed] [Google Scholar]
- 63.Bürki E (1985). The expression of creatine kinase isozymes in Xenopus tropicalis, Xenopus laevis laevis, and their viable hybrid. Biochem. Genet 23, 73–88. [DOI] [PubMed] [Google Scholar]
- 64.Brändle F, Frühbauer B, and Jagannathan M (2022). Principles and functions of pericentromeric satellite DNA clustering into chromocenters. Semin. Cell Dev. Biol [DOI] [PubMed] [Google Scholar]
- 65.Stellfox ME, Bailey AO, and Foltz DR (2013). Putting CENP-A in its place. Cell. Mol. Life Sci 70, 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mérai Z, Chumak N, García-Aguilar M, Hsieh TF, Nishimura T, Schoft VK, Bindics J, Iusarz L, Arnoux S, Opravil S, et al. (2014). The AAA-ATPase molecular chaperone Cdc48/p97 disassembles sumoylated centromeres, decondenses heterochromatin, and activates ribosomal RNA genes. Proc. Natl. Acad. Sci. U. S. A 111, 16166–16171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rošić S, Köhler F, and Erhardt S (2014). Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Biol 207, 335–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grenfell AW, Heald R, and Strzelecka M (2016). Mitotic noncoding RNA processing promotes kinetochore and spindle assembly in Xenopus. J. Cell Biol 214, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bobkov GOM, Gilbert N, and Heun P (2018). Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. J. Cell Biol 217, 1957–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee HY, Chou JY, Cheong L, Chang NH, Yang SY, and Leu JY (2008). Incompatibility of Nuclear and Mitochondrial Genomes Causes Hybrid Sterility between Two Yeast Species. Cell 135, 1065–1073. [DOI] [PubMed] [Google Scholar]
- 71.Ma H, Marti Gutierrez N, Morey R, Van Dyken C, Kang E, Hayama T, Lee Y, Li Y, Tippner-Hedges R, Wolf DP, et al. (2016). Incompatibility between Nuclear and Mitochondrial Genomes Contributes to an Interspecies Reproductive Barrier. Cell Metab. 24, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kitaoka M, Heald R, and Gibeaux R (2018). Spindle assembly in egg extracts of the Marsabit clawed frog, Xenopus borealis. Cytoskeleton 75, 244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bredeson JV, Mudd AB, Medina-Ruiz S, Mitros T, Smith OK, Miller KE, Lyons JB, Batra SS, Park J, Berkoff KC, et al. (2021). Conserved chromatin and repetitive patterns reveal slow genome evolution in frogs. bioRxiv, 2021.10.18.464293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fu L, Niu B, Zhu Z, Wu S, and Li W (2012). CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smit A, Hubley R, and Green P RepeatMasker Open-4.0 http://www.repeatmasker.org.
- 76.Hannak E, and Heald R (2006). Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat. Protoc 1, 2305–2314. [DOI] [PubMed] [Google Scholar]
- 77.Brown KS, Blower MD, Maresca TJ, Grammer TC, Harland RM, and Heald R (2007). Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. J. Cell Biol 176, 765–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Levy DL, and Heald R (2010). Nuclear Size Is Regulated by Importin A and Ntf2 in Xenopus. Cell 143, 288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edelstein AD, Tsuchida M. a, Amodaj N, Pinkard H, Vale RD, and Stuurman N (2014). Advanced methods of microscope control using μManager software. J. Biol. Methods 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gibeaux R, and Heald R (2019). Generation of Xenopus Haploid, Triploid, and Hybrid Embryos. Methods Mol. Biol 1920, 303–315. [DOI] [PubMed] [Google Scholar]
- 81.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
X. tropicalis eggs were fertilized with X. laevis sperm and microinjected with in vitro translated paternally-matched CENP-A and HJURP (right) at stage 2. Embryos were imaged in separate dishes from stage 3 in 1/10X MMR. The movie plays 20 h in 15s (rate of 120 frames per second). Scale bar corresponds to 200 μm.
X. tropicalis eggs were fertilized with X. borealis sperm and incubated with 1 μM BMH-21 in 1/10X MMR (right) from stage 2. The movie plays 20 h in 15s (rate of 120 frames per second). Scale bar corresponds to 200 μm.
Data Availability Statement
ChIP-seq data used in this study are publicly available at NCBI. Accession numbers are listed in the Key Resources Table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-xCENP-A | Straight Lab8,40 | N/A |
| Rabbit anti-POLR1A | Novus Biologicals | Cat#: NBP2-56122 |
| Mouse anti-UBTF, clone 6B6 | Abnova | Cat#: H00007343-M01; RRID: AB_607269 |
| Rabbit anti-Histone H3 | Abcam | Cat#: ab1791; RRID: AB_302613 |
| Mouse anti-Beta-tubulin E7 | Developmental Studies Hybridoma Bank | Cat#: E7; RRID: AB_2315513 |
| Mouse anti-c-myc, clone 9E10 | Sigma-Aldrich | Cat#: M4439; RRID: AB_439694 |
| Mouse anti-Ran | BD Biosciences | Cat#: 610340; RRID: AB_397730` |
| Alexa Fluor 488 | Invitrogen | Cat#: A-11008; RRID: AB_143165 |
| Alexa Fluor 568 | Invitrogen | Cat#: A-11011; RRID: AB_143157 |
| Goat anti-Rabbit IgG (H&L) Antibody DyLight 800 Conjugated | Rockland Immunochemicals | Cat#: 611-145-002-0.5; RRID: AB_11183542 |
| Donkey anti-Mouse IgG (H&L) Antibody DyLight 680 Conjugated | Rockland Immunochemicals | Cat#: 610-744-002; RRID: AB_1660920 |
| Bacterial and virus strains | ||
| XL1-Blue competent cells | Agilent | Cat#: 200249 |
| Chemicals, peptides, and recombinant proteins | ||
| Pregnant mare serum gonadotrophin | Calbiochem | Cat#: 367222 |
| Human chorionic gonadotrophin | Sigma-Aldrich | Cat#: CG10 |
| Hoechst 33342 | Invitrogen | Cat#: H3570 |
| Vectashield | Vector Labs | Cat#: H-1000 |
| Alexa Fluor 568-dUTP | Invitrogen | Cat#: C11399 |
| Random hexamers | Invitrogen | Cat#: 100026484 |
| Klenow (exo-) polymerase | New England Biolabs | Cat#: M0212S |
| Blocking reagent | Roche | Cat#: 11096176001 |
| Salmon sperm DNA | Invitrogen | Cat#: AM9680 |
| Aphidicolin | Sigma-Aldrich | Cat#: A0781 |
| BI-2536 | Selleck Chemicals | Cat#: S1109 |
| BMH-21 | Sigma-Aldrich | Cat#: SML1183 |
| ML-60218 | Sigma-Aldrich | Cat#: 557403 |
| MLN-8237 | Selleck Chemicals | Cat#: S1133 |
| NMS-873 | Sigma-Aldrich | Cat#: SML-1128 |
| Triptolide | Sigma-Aldridch | Cat#: T3652 |
| Critical commercial assays | ||
| TnT Sp6-coupled rabbit reticulocyte system | Promega | Cat#: L2080 |
| Protein A Dynabeads | Fisher | Cat#: 10-002-D |
| NEBNext Ultra II DNA library Prep Kit for Illumina | New England Biolabs | Cat#: E76452 |
| SuperScript III First Strand Synthesis system | Thermo Fisher Scientific | Cat#: 18080051 |
| RNeasy Mini kit | Qiagen | Cat#: 74104 |
| Deposited data | ||
| Xenopus laevis ChIP-seq | 26,73 | GSE153058 |
| Xenopus tropicalis ChIP-seq | 26,73 | GSE 199671 |
| Xenopus borealis ChIP-seq | This paper | PRJNA848409 |
| Experimental models: Organisms/strains | ||
| Xenopus laevis | Nasco | Cat#: LM00535 |
| Xenopus laevis | National Xenopus Resource | Cat#: NXR_0031 |
| Xenopus tropicalis | Nasco | Cat#: LM00822 |
| Xenopus tropicalis | National Xenopus Resource | Cat#: NXR_1018 |
| Xenopus borealis | Nasco | Cat#: LM00698 |
| Oligonucleotides | ||
| Primers (FCR monomer sequence amplification, pJET1.2 Sequencing Primers) FWD: CGACTCACTATAGGGAGAGCGGC REV: AAGAACATCGATTTTCCATGGCAG |
26 | N/A |
| Primers (X. laevis CENP-A amplification) FWD: CAAGCTTCGAATTCTGCAGTCGACTGCCACCATGAGACCGGGCTCCACTCC REV: GGGTTAATGAGGGACTGGGGTAAGAGCCTCTAGAACTATAGTGAGTCGTATTAC |
This paper | N/A |
| Primers (X. tropicalis CENP-A amplification) FWD: CAAGCTTCGAATTCTGCAGTCGACTGCCACCATGAGGCCTGGGTCTACTCC REV: (GAGTTACTGAGGGGTTGGGGTAAGAGCCTCTAGAACTATAGTGAGTCGTATTAC) |
This paper | N/A |
| Primers (X. borealis CENP-A amplification) FWD: TAAGCACTCGAGGCCATGAGATCGGGGTCCACTCC REV: AATCGTTCTAGAGGCTTACCCCAGTCCCTCATTAACCC |
This paper | N/A |
| Recombinant DNA | ||
| Plasmid: Full length X. laevis CENP-A in pCS2+ vector | This paper | N/A |
| Plasmid: Full length X. tropicalis CENP-A in pCS2+ vector | This paper | N/A |
| Plasmid: Full length X. borealis CENP-A in pCS2+ vector | This paper | N/A |
| Plasmid: Full length X. laevis GFP-xHJURP in pCS2+ vector | Straight Lab | ASP1640 |
| Plasmid: Full length X. laevis xCENP-C-myc in pCS2+ vector | Straight Lab | ASP867 |
| Plasmid: FCR monomer4 in pJET1.2 | Straight Lab26 | N/A |
| Plasmid: FCR monomer10 in pJET1.2 | Straight Lab26 | N/A |
| Plasmid: FCR monomer16 in pJET1.2 | Straight Lab26 | N/A |
| Plasmid: FCR monomer19 in pJET1.2 | Straight Lab26 | N/A |
| Software and algorithms | ||
| FIJI | 81 | https://imagej.net/software/fiji/ |
| Matlab | https://www.mathworks.com/products/matlab.html | |
| k-mer counting pipeline | 26 | https://github.com/straightlab/xenla-cen-dna-paper |
| cd-his-est | 74 | http://weizhong-lab.ucsd.edu/cd-hit/ |
| Geneious (7.1.4) | https://www.geneious.com/ | |
| RepeatMasker 4.0.9 | 75 | http://www.repeatmasker.org |
| Other | ||


