Abstract
Pseudomonas putida DOT-T1E is a solvent-tolerant strain able to grow in the presence of 1% (vol/vol) toluene in the culture medium. Random mutagenesis with mini-Tn5-′phoA-Km allowed us to isolate a mutant strain (DOT-T1E-42) that formed blue colonies on Luria-Bertani medium supplemented with 5-bromo-4-chloro-3-indolylphosphate and that, in contrast to the wild-type strain, was unable to tolerate toluene shocks (0.3%, vol/vol). The mutant strain exhibited patterns of tolerance or sensitivity to a number of antibiotics, detergents, and chelating agents similar to those of the wild-type strain. The mutation in this strain therefore seemed to specifically affect toluene tolerance. Cloning and sequencing of the mutation revealed that the mini-Tn5-′phoA-Km was inserted within the fliP gene, which is part of the fliLMNOPQRflhBA cluster, a set of genes that encode flagellar structure components. FliP is involved in the export of flagellar proteins, and in fact, the P. putida fliP mutant was nonmotile. The finding that, after replacing the mutant allele with the wild-type one, the strain recovered the wild-type pattern of toluene tolerance and motility unequivocally assigned FliP a function in solvent resistance. An flhB knockout mutant, another gene component of the flagellar export apparatus, was also nonmotile and hypersensitive to toluene. In contrast, a nonpolar mutation at the fliL gene, which encodes a cytoplasmic membrane protein associated with the flagellar basal body, yielded a nonmotile yet toluene-resistant strain. The results are discussed regarding a possible role of the flagellar export apparatus in the transport of one or more proteins necessary for toluene tolerance in P. putida DOT-T1E to the periplasm.
Organic solvents are extremely toxic for living organisms because they partition in the cell membranes and disorganize them by removing lipids and proteins, which eventually leads to cell death (6, 46). Following Inoue and Horikoshi's report (15) on the isolation of a Pseudomonas sp. strain able to grow on liquid medium with up to 50% (vol/vol) toluene, a number of Pseudomonas sp. strains have been isolated as able to grow in the presence of highly toxic organic solvents such as toluene (partition coefficient in an octanol-water mixture [logPow] = 2.5), styrene (logPow = 2.9), and xylenes (logPow = 3.4) (5–7, 42, 48). One of these isolates, named Pseudomonas putida DOT-T1E, has been shown to be not only toluene tolerant and able to grow in liquid culture medium in the presence of a second phase of these aromatic hydrocarbons, but also capable of using toluene, ethylbenzene, and other compounds as the sole C source.
The mechanisms underlying solvent tolerance are not fully understood yet, although a number of factors are claimed to be involved in this process. Several laboratories have recently shown that efflux pumps of the resistance-nodulation-cell division family are involved in the removal of toluene and other toxic compounds from the cell membranes (16, 21, 24, 34, 40, 41). In P. putida DOT-T1E, the ttgABC, ttgDEF, and ttgGHI operons encode efflux pumps that have been found to be involved in toluene tolerance: ttgB, ttgD, and ttgG mutants exhibited increased solvent sensitivity compared to the parental strain (34, 41, 42a). The ttgB and the ttgH mutants, but not the ttgD mutant, also showed increased sensitivity to antibiotics in comparison with the parental strain, suggesting that the TtgABC and TtgGHI pumps may exhibit a wider substrate specificity than the TtgDEF one. The TtgABC, TtgDEF, and TtgGHI pumps showed a high degree of similarity to the antibiotic efflux pumps MexAB/OprM, MexCD/OprJ, and MexEF/OprN of Pseudomonas aeruginosa PAO1 (25, 37, 38) and the AcrAB/TolC pump of Escherichia coli (1, 10, 28, 49). Although all these pumps were known to expel antibiotics, it has recently been shown that they are also able to remove organic solvents, although neither P. aeruginosa nor E. coli is able to withstand a second phase of toluene or ethylbenzene in liquid medium (1, 27, 49).
The finding that microbes such as P. aeruginosa with operational pumps are toluene sensitive suggests either that other as yet unidentified elements are involved in toluene tolerance or that the expression level and regulation of the pumps—a relatively unexplored research area—are also of importance for toluene tolerance in Pseudomonas spp. (22, 34). Other elements that have been proposed to be involved in solvent tolerance are the cis→trans isomerization of lipids and surface lipopolysaccharides (LPS). Our group and others have found a positive correlation between the degree of trans isomers of the C16:1,9 and C18:1,9 fatty acids and bacterial growth in the presence of toluene and other aromatic hydrocarbons (7, 13, 14, 18, 36, 40). Indeed, P. putida DOT-T1E cells growing in the absence of toluene exhibited a high proportion of cis isomers of fatty acids (cis/trans ≅ 7.5), whereas in the presence of toluene, the cis and trans isomers were equally abundant (cis/trans ≅ 1). The cti gene encoding the P. putida cis,trans-isomerase has been cloned (14, 18), and a cti knockout mutant of P. putida DOT-T1E was isolated and characterized (18). Growth of this mutant was delayed in the presence of organic solvents (18). It has also been suggested that LPS are a barrier to the entry of aromatic compounds through the cell membrane (36). We have generated LPS mutants of the solvent-tolerant P. putida DOT-T1E strain and found that LPS is not critical for solvent tolerance (19).
To further elucidate the process of solvent tolerance, we mutagenized P. putida DOT-T1E with a mini-Tn5-′phoA. Among the mutants, we looked for toluene-sensitive ones that conserved the wild-type pattern of lipids and resistance or sensitivity to antibiotics under different growth conditions. A mutant was found that exhibited a transposon insertion at the fliP gene, whose gene product is involved in flagellar assembly; as a consequence of this insertion, the cells were nonmotile. This unexpected finding led us to generate mutations in genes whose products are involved in flagellum biosynthesis. An flhB knockout mutant showed hypersensitivity to toluene and was also nonmotile. In contrast, the nonmotile mutant lacking the FliL protein, associated with the flagellar basal body, was toluene tolerant. These results indicated that the proteins of the flagellar export apparatus are neccesary for toluene tolerance.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture medium, and growth conditions.
The bacterial strains used in this study are shown in Table 1. Plasmid pUT-′phoA-Km has the R6K origin of replication and encodes resistance to ampicillin and kanamycin. The latter marker together with ′phoA is part of the mini-Tn5 borne on this plasmid (41). Plasmid pRK600 was used as a helper; it encodes resistance to chloramphenicol and provides the tra functions for the mobilization of the pUT plasmid (41). For site-directed mutagenesis of the chromosomal fliL and flhB genes, plasmid pUNφ18, bearing a knockout fliL::Ω-Km gene, and plasmid pUC18, bearing a knockout flhB::Ω-Km, were used.
TABLE 1.
Strains used in this study
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| P. putida | ||
| DOT-T1E | Rifr Tolr | 41 |
| DOT-T1E-42 | Rifr Tols KmrfliP::Km | This study |
| DOT-T1E-PS23 | Rifr TolrfliL::Km | This study |
| DOT-T1E-PS50 | Rifr TolsfhlB::Km | This study |
| DOT-T1E-PS21 | Rifr Tolr | This study |
| E. coli | ||
| CC118λpir | Rifr, host strain to replicate plasmids bearing the R6K origin of replication | 40 |
| HB101 | Smr, host for plasmid pRK600 | 40 |
| JM109 | recA, used in cloning experiments | 2 |
Bacterial strains were routinely grown on liquid Luria-Bertani (LB) medium (2). Cultures were incubated at 30°C and shaken on an orbital platform operating at 200 strokes per min. Growth was usually determined as CFU on LB solid medium supplemented with appropriate antibiotics. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml for E. coli and 300 μg/ml for P. putida; carbenicillin (Cb), 300 μg/ml; chloramphenicol (Cm), 90 μg/ml; kanamycin (Km), 50 μg/ml; piperacillin (Pi), 100 μg/ml; rifampin (Rif), 20 μg/ml; and tetracycline, 15 μg/ml.
Isolation of toluene-sensitive Tn5-′phoA mutants of P. putida DOT-T1E.
About 5,000 mini-Tn5 transconjugants of P. putida DOT-T1E were obtained after triparental mating of the strain with E. coli CC118λpir(pUT-′phoA-Km) and E. coli HB101(pRK600). About 10% of the Km-resistant clones appeared as blue colonies in plates supplemented with 5-bromo-4-chloro-3-indolylphosphate (BCIP). Each individual Km-resistant blue transconjugant was tested for its ability to grow on LB medium supplemented with either 1% (vol/vol) toluene or 1% (vol/vol) octanol. One clone that failed to grow in the medium with 1% (vol/vol) toluene but that did grow in the presence of the same amount of octanol was called P. putida DOT-T1E-42 and was retained for further studies.
Cloning of mutation in P. putida DOT-T1E-42 and analysis of surrounding DNA sequence.
To clone the mutation in this strain, total DNA was digested with SphI and ligated to pUC18. Two Km-resistant colonies were obtained, both containing an identical plasmid carrying an SphI insert of about 3 kb. The resulting plasmid (pANA1) contained 1.7 kb of the mini-Tn5 plus 1.1 kb of chromosomal DNA. The DNA was sequenced by the dideoxy sequencing termination method and a primer located at the “O” end of the mini-Tn5. This made it possible to read outside the Km resistance gene and within the chromosomal insert. Based on the DNA sequence obtained, specific 20-mer primers were designed for further DNA walking. DNA was sequenced on both strands.
Rescue of wild-type P. putida DOT-T1E fli genes from a gene bank.
Wild-type genes were rescued from a DOT-T1E gene bank previously generated in our laboratory (41). The 1.1-kb SphI-NotI fragment of pANA1 was labeled with digoxigenin by standard procedures and used to screen the library. A single clone bearing a 10.5-kb P. putida DOT-T1E insert was found. The plasmid was called pANA9, its insert was sequenced on both strands, and the sequence was deposited in Genbank under accession number AF031418.
Generation of a P. putida DOT-T1E fliL::Ω-Km mutant.
Plasmid pANA9 was cut with SfiI, an enzyme that cuts within the fliL gene (position 4081 of the insert in sequence AF031418). The Klenow fragment and the four deoxynucleoside triphosphates (dNTPs) were used to fill in the ends and make them blunt (2). The 2-kb Ω-Km cassette of plasmid pHP45ΩKm (9) was obtained after digestion with EcoRI, and these ends were made blunt, as above, and ligated to the linearized pANA9 plasmid. The ligation was transformed into E. coli JM109, and cells were selected on LB plates supplemented with ampicillin and Km. After analysis by restriction enzymes, a clone carrying the correct plasmid, called pANA61, was selected. Electroporation was used to transfer pANA61 to P. putida DOT-T1E, whose colE origin of replication is not recognized in P. putida and which behaves like a suicide vector. However, because of identical sequences, pANA61 integration into the host chromosome via homologous recombination can be selected on LB solid medium with Rif, Km, and Pi. A merodiploid clone was grown overnight on LB to allow a second recombination event in which the wild-type gene was replaced by the mutant allele. For this selection, colonies were plated again on LB with Rif and Km. Among these colonies, those that did not grow in the presence of piperacillin were selected as putative resolved merodiploids. These mutants were checked again by Southern blot hybridization, cutting the chromosomal DNA with BamHI and using the 1,3-kb PstI fragment (positions 2901 to 4192) containing part of the fliK and fliL genes as a probe. One of the clones that exhibited the correct mutation was called P. putida strain DOT-T1E-PS23 and kept for further studies.
Generation of a P. putida DOT-T1E flhB::Ω-Km mutant.
Plasmid pANA9 (10.3 kb) was cut with EcoRI, an enzyme that cuts in position 1 within the pUC18 polylinker and at position 5610 of the insert in sequence AF031418. Plasmid pANA50 (7.6 kb) is the result of the religation of the pANA9 fragment. Plasmid pANA50 contains a 4.7-kb insert encompassing the fliNOPQRflhB genes. pANA50 was cut with BpuAI, an enzyme that cuts at the 5′ end of the flhB gene (position 8229 in sequence AF031418). The Klenow fragment and the four dNTPs were used to fill in the ends and make them blunt (2). The 2-kb Ω-Km cassette of plasmid pHP45ΩKm (9) was obtained and treated as above. The Ω-Km fragment was ligated to the linearized pANA50 plasmid and transformed into E. coli JM109. The resulting Apr Kmr transconjugants were analyzed by digestion with restriction enzymes, and a clone carrying the correct plasmid, called pANA72, was selected. Plasmid pANA72 was introduced into P. putida DOT-T1E as above, and transformants incorporating the plasmid on the chromosome were selected on LB solid medium with Rif, Km, Cb, and Pi. One of the merodiploid clones was grown on LB for at least 20 generations to allow the second recombination event to occur. These clones were expected to be able to grow on LB plus Rif and Km but not in the presence of Cb and Pi. Seven clones were found as putative resolved merodiploids. These clones were checked by Southern blot hybridization, cutting the chromosomal DNA with BamHI and using a PCR probe labeled with digoxigenin, that contained 500 bp of the 3′ end of fliR and the first 500 bp of flhB. One of the clones that exhibited the correct mutation was called P. putida strain DOT-T1E-PS50 and kept for further studies.
RT-PCR.
P. putida DOT-T1E cells growing exponentially on LB medium were harvested by centrifugation (5,000 × g, 10 min). RNA from P. putida DOT-T1E was isolated with the RNeasy Total RNA kit (Qiagen). This RNA was treated with RNase-free Dnase I in the presence of an RNase inhibitor (Boehringer Mannheim) to avoid DNA contamination and RNA degradation. Reverse transcription (RT)-PCR was performed with the Titan OneTube RT-PCR system according to the manufacturer's instructions (Boehringer Mannheim). The annealing temperature used in the PCR experiments was 60°C, and the cycling conditions were as follows: 94°C for 30 s, 60°C for 30 s, and 68°C for 1 min. After 30 cycles, the sample was incubated at 68°C for 10 min. Positive and negative controls were included in all assays. The approximate locations of the primers used for RT-PCR are indicated in Fig. 2A. Primer sequences are available on request.
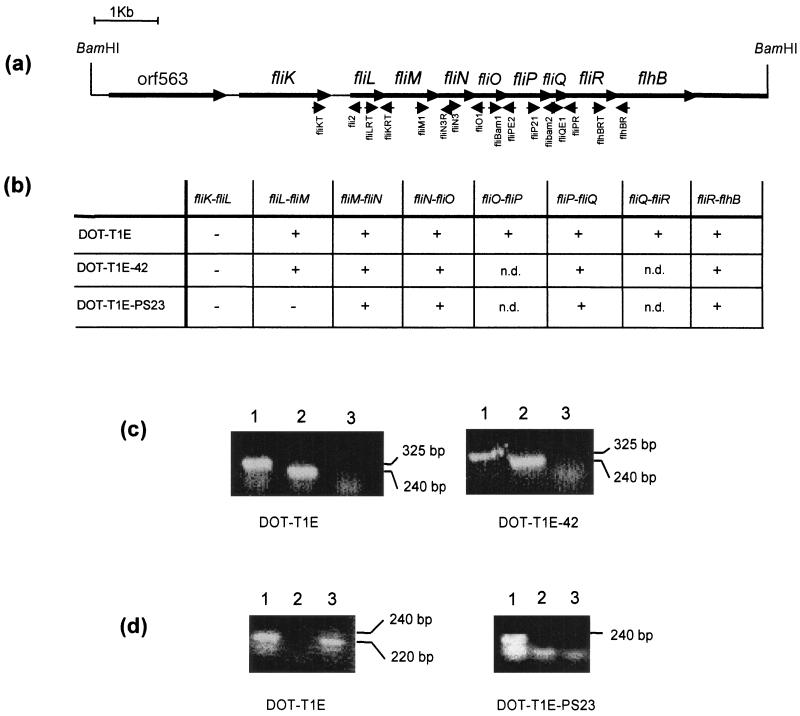
FIG. 2.
Physical map of fli-flh gene cluster in the solvent-tolerant P. putida DOT-T1E strain. (a) Gene order. The arrows indicate the direction of transcription. Small arrows indicate the positions of the primers used in RT-PCR. (b) Summary of RT-PCR results obtained with total RNA isolated from the indicated strains growing on LB. Symbols: +, RT-PCR yielded a fragment of the expected size when primers based on the indicated genes were used; −, RT-PCR failed to amplify RNA with the indicated primers; n.d., not determined. (c) Specific RT-PCR results with primers based on fliP and fliQ and on fliR and flhB. Total RNA was isolated from P. putida DOT-T1E and DOT-T1E-42, and RT-PCRs were done with primers based on fliP and fliQ that yielded a 325-nucleotide (nt) band (lane 1) and fliR and flhB that yielded a 240-nt band (lane 2). Lane 3, negative control using primers based on fliP and fliQ but in the absence of reverse transcriptase. (d) Specific RT-PCR with primers based on fliR and flhB and on fliL and fliM. RNA was isolated from P. putida DOT-T1E and DOT-T1-PS23 and RT-PCRs were done with primers based on fliR and flhB that yielded a 240-nt band (lane 1) and fliL and fliM that yielded a 220-nt band (lane 3). Lane 2, negative control using primers based on fliR and flhB but in the absence of reverse transcriptase.
Computer analysis.
DNA primary sequences were analyzed with several programs included in the DNA Strider 1.1. package. Homology searches were performed with the BLAST database search program.
RESULTS
Tolerance of P. putida DOT-T1E and DOT-T1E-42 to organic solvents, detergents, chelating agents, and antibiotics.
P. putida DOT-T1E-42 was isolated as a Km-resistant toluene-sensitive clone upon miniTn5-′phoA mutagenesis, although it resembled the wild-type strain in tolerance to heptane, propylbenzene, m-xylene, and octanol. The wild type and this mutant were able to form colonies on LB plates supplemented with chloramphenicol (90 μg/ml) or Cb (250 μg/ml). This finding contrasts with those reported previously for another toluene-sensitive derivative of DOT-T1E, called DOT-T1E-18, which did not form colonies in the presence of these high concentrations of chloramphenicol and Cb (41). This phenotypic difference clearly established that DOT-T1E-42 represented a distinct class of toluene-sensitive mutants.
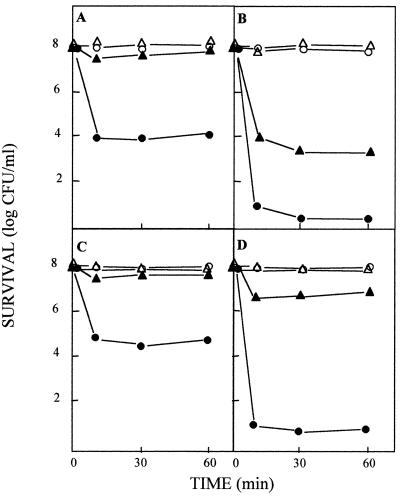
P. putida DOT-T1E was shown before to exhibit differential toluene tolerance depending on the growth conditions. In fact, whereas most cells tolerated a 0.3% (vol/vol) toluene shock upon growth on LB with toluene in the gas phase, only a low proportion (0.01%) of cells tolerated this shock when they were grown in the absence of toluene in the gas phase (41), (Fig. 1A). The mutant DOT-T1E-42 strain did not tolerate sudden exposure to 0.3% (vol/vol) toluene when grown on LB, and very small numbers of cells (0.01%) survived the shock even when grown with toluene in the gas phase (Fig. 1B).
FIG. 1.
Survival in response to toluene shocks of the wild-type P. putida DOT-T1E and a mutant derivative lacking the FliP, FliL, or FlhB protein. Cells were grown in 30 ml of LB (circles) or LB with toluene in the gas phase (triangles) until the culture reached an optical density at 660 nm of about 1. These cultures contained about 108 CFU/ml. The cultures were divided in two halves; to one we added 0.3% (vol/vol) toluene (solid symbols), and the other was kept as a control (open symbols). The number of viable cells was determined before toluene was added and 15, 30, and 60 min later. (A) P. putida DOT-T1E; (B) P. putida DOT-T1E-42; (C) P. putida DOT-T1E-PS23; (D) P. putida DOT-T1E-PS50.
It was previously shown that P. putida DOT-T1E grows on LB solid medium supplemented with 1 mM EDTA, 100 mM deoxycholate, 3% (wt/vol) Triton X-100, 1.5% (wt/vol) sodium dodecyl sulfate (SDS), or 15 g of p-hydroxybenzoate per liter. P. putida DOT-T1E-42 also grew on LB plates with one of these chemicals at the indicated concentrations. These results suggest that P. putida DOT-T1E-42 is sensitive only to toluene.
Functionality of the cis,trans-isomerase and efflux pumps in P. putida DOT-T1E-42.
Two elements are critical for solvent tolerance: cis→trans isomeration of unsaturated fatty acids, and the ability to extrude aromatic hydrocarbons (reviewed by Segura et al. [45]). The pattern of lipid composition of wild-type and DOT-T1E-42 cells grown on LB and LB plus 1% (vol/vol) heptane and toluene (supplied via the gas phase) was examined in cells growing exponentially. The results obtained were similar to those reported before for the wild-type strain (40) and suggest that DOT-T1E-42 did not exhibit any apparent damage with regard to fatty acid biosynthesis (not shown).
P. putida DOT-T1E was shown before to be able to extrude [14C]toluene from the cell membrane when cells were grown on either LB or LB plus toluene supplied via the gas phase, while the solvent-sensitive strain DOT-T1E-18 showed impaired aromatic hydrocarbon extrusion (41). We tested the extrusion of [14C]toluene in DOT-T1E-42 cells grown on LB that were exposed or not to toluene in the gas phase. Mutant cells extruded [14C]toluene less efficiently than the wild-type cells (Table 2). However, they still extrude part of the aromatic hydrocarbon, because in cells treated with 200 μM CCCP (carbonyl cyanide m-chlorophenylhydrazone) the amount of 14C accumulated in the cell membranes was three- to fivefold higher. These results indicated that in the mutant strain, functioning of the pump systems was compromised (21, 34, 40, 41, 45).
TABLE 2.
Accumulation of [14C]toluene in cells of P. putida DOT-T1E and its solvent-sensitive mutant DOT-T1E-42a
| Growth conditions | Amt of 14C in the cells (dpm/OD660)
|
|
|---|---|---|
| Wild type | DOT-T1E-42 | |
| LB | 5,300 ± 200 | 8,400 ± 400 |
| LB + toluene | 2,900 ± 500 | 5,500 ± 300 |
P. putida DOT-T1E and DOT-T1E-42 cells were grown on LB medium in the absence and in the presence of toluene supplied via the gas phase. Exponentially growing cells were harvested by centrifugation, washed with 10 volumes of LB, and suspended in 1.5 ml of LB to an optical density at 660 nm (OD660) of about 1. The cells were then incubated for 10 min at 30°C and exposed to 2 μCi of [14C]toluene (39). After 10 min, when the equilibrium level had been achieved, 250 μl of the cell suspension was filtered through a 0.45-μm Millipore filter and washed with 2 ml of LB medium. The filters were dried, and the amount of 14C associated with the cell pellet (disintegrations per minute) was determined in a Packard radiochemical detector. The data are the averages of three independent determinations.
Cloning of the mutation in P. putida DOT-T1E-42.
We cloned the mutation in DOT-T1E-42 and determined the nature of the mutated gene. The strategy is described in Materials and Methods. This approach yielded plasmid pANA1, which contained 1.7 kb of the mini-Tn5-′phoA-Km plus about 1.1 kb of the adjacent chromosomal DNA. Sequence analysis revealed that the ′phoA-Km cassette was inserted within a putative open reading frame homologous to fliP, a protein necessary for flagellar biosynthesis described in several species (4, 8, 26, 31, 32, 33, 35). Like fliP mutants of enterobacteria, the P. putida fliP mutant was nonmotile, in contrast to the wild-type strain. P. putida DOT-T1E showed a tuft of three to four polar flagella, whereas the DOT-T1E-42 mutant was nonflagellated (data not shown).
To unequivocally establish the nature of the fliP gene, the wild-type gene was rescued in pANA9. The sequence of the 10.5-kb BamHI insert of pANA9 was deposited in GenBank under accession number AF031418. Sequence analysis revealed the presence of 11 putative open reading frames, whose translated sequences were compared with those stored in several databanks. All of them showed high homology with different proteins related to flagella and chemotaxis in P. aeruginosa and enterobacteria. The gene order was found to be orf563 fliKLMNOPQRflhBA (Fig. 2a).
Transcriptional organization of fli genes: genes downstream of Tn5-′phoA insertion in P. putida DOT-T1E-42 are expressed.
Given the cluster structure of the fli genes and the lack of information on the transcriptional organization of these genes in P. putida, we could not rule out the possibility that the Tn5-′phoA insertion exerted a polar effect on downstream genes. As a way to establish the transcriptional organization of the fli genes in this strain, we isolated total RNA from P. putida DOT-T1E and DOT-T1E-42 and determined mRNA contiguity by RT-PCR. The primers used are given in Materials and Methods and are based on the 3′ and 5′ ends of two adjacent genes. The results obtained are shown in Fig. 2b, c, and d.
In the wild-type strain, the fli genes may be organized in at least two transcriptional units. RT-PCR yielded negative results when primers based on fliK and fliL were used for amplification. Internal primers within fliK and fliL gave positive results with the same sample; thus, the negative result cannot be attributed to the lack of mRNA. Thus, we concluded that fliK and fliL belong to different transcriptional units, with fliK transcribed presumably as a monicistronic mRNA. Because all other tested combinations of primers based on adjacent genes yielded positive results, we assumed that the other unit involved the fliLMNOPQRflhB genes. With mRNA isolated from DOT-T1E-42, we confirmed that the fliK gene constituted a transcriptional unit independent of the other fli genes. As expected, amplification with fliL and fliM, fliM and fliN and fliO primers (genes located upstream from fliP) with RT-PCR yielded positive results (Fig. 2b). Surprisingly, we also found RT-PCR products when we used fliP/fliQ and fliR/flhB primers (Fig. 2c), which suggests either that the mini-Tn5-′phoA insertion at fliP does not exert a drastic polar effect on genes located downstream or that multiple promoters are involved in expression of the fliQRflhBA genes. Although fine transcriptional analysis is needed to resolve the gene expression pattern, for the purpose of our study, the above results suggest that fliP is the gene responsible for the observed phenotypes of lack of motility plus sensitivity to solvents in P. putida DOT-T1E.
Replacement of mutant allele in P. putida DOT-T1E-42 with wild-type allele.
The suicide plasmid pANA9, bearing a 10.5-kb insert, was electroporated into P. putida DOT-T1E-42, and Pi-resistant clones were selected. These clones resulted from a single recombination event due to the integration of pANA9 within the host chromosome. After repetitive growth on LB medium, we spread cells on LB plates and searched for Km-sensitive clones. These clones were expected to result from the replacement of the mutant allele with the wild-type one. One such clone was found and called DOT-T1E-PS21. Southern blot was used to confirm the nature of the replacement (not shown). We found that DOT-T1E-PS21 cells had recovered motility and the ability to tolerate toluene shocks (not shown).
Are motility and solvent tolerance linked traits in P. putida DOT-T1E?
To answer this question, we generated mutations in fliL and flhB by inserting an Ω-Km interposon as described in Materials and Methods. We chose these genes because FliK, FliO, FliQ, FliR, FlhB, and FlhA, together with FliP, have been suggested to be part of the flagellar export apparatus, whereas FliL has been suggested to be a cytoplasmatic protein associated with the basal body (23, 44). The in vivo construction of the knockouts in fliL and flhB yielded DOT-T1E-PS23 and DOT-T1E-PS50 mutants, bearing an fliL::Ω-Km and flhB::Ω-Km insertion, respectively. Southern blots revealed the successful replacement of the wild-type fliL or flhB gene with the mutant fliL::Ω-Km or flhB::Ω-Km in DOT-T1E-PS23 and DOT-T1E-PS50, respectively (not shown). RT-PCR assays revealed that in DOT-T1E-PS23, in which the insertion was at the 5′ end of the fliL gene cluster, did not prevent the expression of the genes downstream of the Ω-Km insertion, as revealed by successful amplification with appropriate primers (Fig. 2b).
The fliL::Ω-Km and flhB::Ω-Km mutants were nonmotile in soft agar plates (0.3%, wt/vol). Electron microscopy showed that the fliL mutant was nonflagellated, whereas the flhB mutant had lophotrichous flagella (data not shown). Their behavior with regard to toluene tolerance was different. In fact, while mutant DOT-T1E-PS23 was able to grow on 0.3% (vol/vol) toluene and behaved like the wild-type strain in response to sudden shocks with organic solvents, including toluene (Fig. 1C), the DOT-T1E-PS50 mutant was hypersensitive to sudden toluene shocks (Fig. 1D) and was unable to grow in LB in the presence of 0.3% (vol/vol) toluene. These results suggest that motility and toluene tolerance are not linked themselves but that the flagellar transport system is involved in toluene tolerance.
DISCUSSION
Our initial search for clones exhibiting sensitivity to toluene with a pattern of resistance to antibiotics similar to that of the wild type resulted in the isolation of one mutant out of 5,000 transconjugants analyzed. The analysis of this mutant led us to an unexpected finding: the mini-Tn5-′phoA insertion in P. putida DOT-T1E-42 occurred at the fliP gene, whose gene product seems to be a component of the flagellar export apparatus (32). Like fliP mutants of other microorganisms (4, 8, 20), the P. putida fliP mutant lost motility, suggesting that the P. putida fli genes are related to motility. However, the novel phenotype of solvent sensitivity indicates an unexpected connection between the FliP protein and toluene tolerance.
Our study shows that the fliP gene is located within a cluster of genes involved in flagellum biosynthesis and chemotaxis. At least two transcriptional units have been identified, one consisting of fliK and the other of fliLMNOPQRflhBA. RT-PCR assays revealed that in DOT-T1E-42 the genes downstream from fliP were expressed in spite of the insertion of a mini-Tn5-′phoA. This could be due to the existence of multiple promoters in the gene cluster, as described for E. coli (29, 31). The fact that in DOT-T1E-42 the genes upstream and downstream of fliP were expressed (Fig. 2b) suggests that the ′phoA-Km insertion does not exert important polar effects and that there is a specific role for the FliP protein in toluene tolerance in P. putida DOT-T1E. The role of FliP in solvent tolerance was further confirmed when replacement of the fliP::Tn5-′phoA allele with the wild-type gene resulted in recovery of motility and toluene tolerance.
The FliP protein has been found associated with the basal body MS ring, and its role seems to be facilitating the export of proteins from the cytoplasmic compartment to a compartment on the other side of the membrane, either the periplasm (as in the case of the flagellar rod proteins) or the cell exterior (as in the case of the hook and filament proteins). To determine whether the toluene sensitivity in the fliP mutant was specific for toluene tolerance or whether it resulted from loss of the flagellum, we generated fliL and flhB mutants. The FliL and FlhB proteins play different roles in flagellar structure. In fact, while FlhB is a 39-kDa cytoplasmic membrane protein involved in substrate specificity switching and part of the flagellar export apparatus system (33), the FliL protein is associated with the flagellar basal body (17, 39, 44). The P. putida DOT-T1E-PS23 (FliL mutant) and DOT-T1E-PS50 (FlhB mutant) mutants were nonmotile, but DOT-T1E-PS23 was as tolerant to sudden toluene shocks as the wild type, whereas DOT-T1E-PS50 was toluene sensitive. This indicates that solvent sensitivity in DOT-T1E-42 arises not from the loss of the flagellum itself (DOT-T1E-42 is not flagellated, whereas DOT-T1E-PS50 is), but from the absence of FliP. Therefore, it follows that FliP and FlhB play a direct or an indirect role in toluene tolerance, suggesting that an intact flagellar export system is required for toluene tolerance.
The higher toluene tolerance of DOT-T1E-42 and DOT-T1E-PS50 in induced cells versus uninduced cells could be because the expression of the ttgDEF and ttgGHI pump genes increased in response to solvents (34, 42a). Differences in solvent sensitivity between DOT-T1E-42 and DOT-T1E-PS50 may be due to altered stoichiometry of the flagellar transport components in the two mutants, although this remains to be tested.
Flagellar transport system proteins have homologues in type III transport systems, responsible for the export of virulence factors such as SpaP and SpaR proteins in Salmonella spp. (12), Spa24 and Spa29 in Shigella flexneri (43), Ysc proteins in Yersinia spp. (3), and Hrp in Pseudomonas solanacearum (47). Therefore, it is possible that the role of FliP and other flagellar transport proteins in toluene tolerance is to facilitate the transfer to the periplasmic space or to the outer membrane of a protein(s) involved in solvent exclusion. Because the P. putida FliP mutant was less efficient in toluene extrusion than the wild type, it is possible that FliP is involved in the transfer of the efflux pump components located in the periplasmic space (i.e., the TtgA, TtgD, and TtgG elements of the TtgABC, TtgDEF, and TtgGHI pumps [34, 41]) and/or in the outer membrane (i.e., TtgC, TtgF, and TtgI proteins). If this were the case, it would mean that the export of these proteins somehow utilizes the flagellum export system. This unusual case has, however, two precedents. One of them is the export to the outer medium of one of the virulence factors in Yersinia enterocolitica, a phospholipase (50). Mutants of this Yersinia sp. damaged in the flagellar export apparatus failed to export phospholipase and were less virulent than the wild type. In the second case, motility and pathogenicity have also been linked in Xenorhabdus, a bacterium symbiotically associated with nematodes of the steinernematide family. Cells with mutations in the flagellar master operon flhDC of Xenorhabdus nematophilus were nonmotile and exhibited reduced virulence due to decreased export of lipases and hemolysin (11). However, the authors did not investigate whether or not export in Xenorhabdus of these virulence proteins required the flagellar export apparatus.
Our results, together with the studies of virulence in Yersinia (50) and Xenorhabdbus (11), show that the flagellum system is coupled through unknown mechanisms to major networks involving bacterial physiology as well as motility.
ACKNOWLEDGMENTS
We thank Patricia Godoy for assistance with determinations of fatty acids.
This work was supported by a grant from the European Commission (BT-CT97-2270) and a grant from the CICYT (BIO97-0641).
REFERENCES
- 1.Aono R, Tsukagoshi N, Yamamoto M. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J Bacteriol. 1998;180:938–944. doi: 10.1128/jb.180.4.938-944.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingstom R F, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1991. [Google Scholar]
- 3.Bergman T, Erickson K, Galyov E, Persson, C. C, Wolf-Watz H. The lcrB (yscN/U) gene cluster of Yersinia pseudotuberculosis is involved in Yop secretion and shows high homology to the spa gene clusters of Shigella flexneri and Salmonella typhimurium. J Bacteriol. 1994;176:2619–2626. doi: 10.1128/jb.176.9.2619-2626.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff D S, Weinreich M D, Ordal G W. Nucleotide sequences of Bacillus subtilis flagellar biosynthetic genes fliP and fliQ and identification of a novel flagellar gene, fliZ. J Bacteriol. 1992;174:4017–4025. doi: 10.1128/jb.174.12.4017-4025.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruden D L, Wolfram J H, Rogers R D, Gibson D T. Physiological properties of a Pseudomonas strain which grows with p-xylene in a two-phase (organic aqueous) medium. Appl Environ Microbiol. 1992;58:2723–2729. doi: 10.1128/aem.58.9.2723-2729.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Smet M J, Kingma J, Witholt B. The effect of toluene on the structure and permeability of the outer and cytoplasmic membranes of Escherichia coli. Biochim Biophys Acta. 1978;506:64–80. doi: 10.1016/0005-2736(78)90435-2. [DOI] [PubMed] [Google Scholar]
- 7.Diefenbach R, Keweloh H. Synthesis of trans unsaturated fatty acids in Pseudomonas putida P8 by direct isomerization of the double bond of lipids. Arch Microbiol. 1994;162:120–125. doi: 10.1007/s002030050112. [DOI] [PubMed] [Google Scholar]
- 8.Fan F, Ohnishi K, Francis N R, Macnab R M. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol Microbiol. 1997;26:1035–1046. doi: 10.1046/j.1365-2958.1997.6412010.x. [DOI] [PubMed] [Google Scholar]
- 9.Fellay R, Frey J, Krisch N. Interposon mutagenesis of soil and water bacteria, a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 10.Fralick J A. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Givaudan A, Lanois A. flhDC, the flagellar master operon of Xenohadbus nematophilus: requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J Bacteriol. 2000;182:107–115. doi: 10.1128/jb.182.1.107-115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groisman E A, Ochman D. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 1993;12:3779–3787. doi: 10.1002/j.1460-2075.1993.tb06056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heipieper H J, Diefenbach R, Keweloh H. Conversion of cis unsaturated fatty acids to trans, a possible mechanism for the protection of phenol-degrading Pseudomonas putida P8 from substrate toxicity. Appl Environ Microbiol. 1992;58:1847–1852. doi: 10.1128/aem.58.6.1847-1852.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holtwick R, Meinhardt F, Keweloh H. cis-trans isomerization of unsaturated fatty acids: cloning and sequencing of the cti gene from Pseudomonas putida P8. Appl Environ Microbiol. 1997;63:4292–4297. doi: 10.1128/aem.63.11.4292-4297.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue A, Horikoshi K. A Pseudomonas thrives in high concentrations of toluene. Nature. 1989;338:264–266. [Google Scholar]
- 16.Isken S, de Bont J A M. Active efflux of toluene in a solvent-resistant bacterium. J Bacteriol. 1996;178:6056–6058. doi: 10.1128/jb.178.20.6056-6058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenal V, White J, Shapiro L. Caulobacter flagellar function, but not assembly, requires FliL a non-polarly localized membrane protein present in all cell types. J Mol Biol. 1994;243:227–244. doi: 10.1006/jmbi.1994.1650. [DOI] [PubMed] [Google Scholar]
- 18.Junker F, Ramos J L. Involvement of the cis-trans isomerase Cti in solvent resistance in Pseudomonas putida DOT-T1. J Bacteriol. 1999;181:5693–5700. doi: 10.1128/jb.181.18.5693-5700.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Junker F, Rodríguez-Herva J J, Duque E, Ramos-González M I, Llamas M A, Ramos J L. A WbpL mutant of Pseudomonas putida DOT-T1E strain, which lacks the O-antigen side chain of lipopolysaccharides, is tolerant to organic solvent shocks. 2001. Extremophiles, in press. [DOI] [PubMed] [Google Scholar]
- 20.Katayama E, Shiraishi T, Oosawa K, Baba N, Aizawa S-I. Geometry of the flagellar motor in the cytoplasmic membrane of Salmonella typhimurium as determined by stereo-photogrammetry of quick-freeze deepetch replica images. J Mol Biol. 1996;255:458–475. doi: 10.1006/jmbi.1996.0038. [DOI] [PubMed] [Google Scholar]
- 21.Kieboom J, Dennis J J, de Bont J A M, Zylstra G J. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J Biol Chem. 1998;273:85–91. doi: 10.1074/jbc.273.1.85. [DOI] [PubMed] [Google Scholar]
- 22.Kieboom J, Dennis J J, Zylstra G J, de Bont J A M. Active efflux organic solvents by Pseudomonas putida S12 induced by solvents. J Bacteriol. 1998;180:6769–6772. doi: 10.1128/jb.180.24.6769-6772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kihara M, Homma M, Kutsukake K, Macnab R M. Flagellar switch of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989;171:3247–3257. doi: 10.1128/jb.171.6.3247-3257.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim K, Lee S, Lee K, Lim D. Isolation and characterization of toluene-sensitive mutants from toluene-resistant bacterium Pseudomonas putida GN73. J Bacteriol. 1998;180:3692–3696. doi: 10.1128/jb.180.14.3692-3696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Köhler T, Michea-Hamzehpour M, Henze V, Gotoh N, Curty L K, Pechère J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 26.Kuo S C, Koshland D E., Jr Sequence of the flaA (cheC) locus of Escherichia coli and discovery of a new gene. J Bacteriol. 1986;166:1007–1012. doi: 10.1128/jb.166.3.1007-1012.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Zhang L, Poole K. Organic solvent-tolerant mutants of Pseudomonas aeruginosa display multiple antibiotic resistance. J Bacteriol. 1998;180:2987–2991. [Google Scholar]
- 28.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 29.Macnab R M. Flagella. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B Jr, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology Press; 1996. pp. 123–145. [Google Scholar]
- 30.Malakooti J, Ely B, Matsumura P. Molecular characterization, nucleotide sequence, and expression of the fliO, fliP, fliQ, and fliR genes of Escherichia coli. J Bacteriol. 1994;176:189–197. doi: 10.1128/jb.176.1.189-197.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malakooti J, Komeda Y, Matsumura P. DNA sequence analysis, gene product identification, and localization of flagellar motor components of Escherichia coli. J Bacteriol. 1989;171:2728–2734. doi: 10.1128/jb.171.5.2728-2734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minamino T, Macnab R M. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol. 1999;181:1388–1394. doi: 10.1128/jb.181.5.1388-1394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minamino T, Macnab R M. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J Bacteriol. 2000;182:4906–4914. doi: 10.1128/jb.182.17.4906-4914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosqueda G, Ramos J L. A set of genes encoding a second toluene efflux system in Pseudomonas putida DOT-T1E is linked to the tod genes for toluene metabolism. J Bacteriol. 2000;181:937–943. doi: 10.1128/jb.182.4.937-943.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohnishi K, Fan F, Schoenhals G J, Kihara M, Macnab R. The FliO, FliP, FliQ and FliR proteins of Salmonella typhimurium: putative components for flagellar assembly. J Bacteriol. 1997;179:6092–99. doi: 10.1128/jb.179.19.6092-6099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinkart H C, Wolfram J W, Rogers R, White D C. Cell envelope changes in solvent-tolerant and solvent-sensitive Pseudomonas putida strains following exposure to o-xylene. Appl Environ Microbiol. 1996;62:1129–1132. doi: 10.1128/aem.62.3.1129-1132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poole K, Heinrichs D E, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pryoverdine. Mol Microbiol. 1993;10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 38.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J-I, Li X Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB multidrug resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 39.Raha M, Sockett H, Macnab R M. Characterization of the FliL gene in the flagellar regulon of Escherichia coli and Salmonella typhimurium. J Bacteriol. 1994;176:2308–2311. doi: 10.1128/jb.176.8.2308-2311.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramos J L, Duque E, Rodríguez-Herva J J, Godoy P, Haïdour A, Reyes F, Fernández-Barrero A. Mechanims for solvent tolerance in bacteria. J Biol Chem. 1997;272:3887–3890. doi: 10.1074/jbc.272.7.3887. [DOI] [PubMed] [Google Scholar]
- 41.Ramos J L, Duque E, Godoy P, Segura A. Efflux pumps involved in toluene tolerance in Pseudomomonas putida DOT-T1E. J Bacteriol. 1998;180:3323–3329. doi: 10.1128/jb.180.13.3323-3329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramos J L, Duque E, Huertas M J, Haïdour A. Isolation and expansion of the catabolic potential of a Pseudomonas putida strain able to grow in the presence of high concentrations of aromatic hydrocarbons. J Bacteriol. 1995;177:3911–3916. doi: 10.1128/jb.177.14.3911-3916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42a.Rojas A, Duque E, Mosqueda G, Golden G, Hurtado A, Ramos J L, Segura A. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J Bacteriol. 2001;183:3967–3973. doi: 10.1128/JB.183.13.3967-3973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sasakawa C, Komatsu K, Tobe T, Suzuki T, Yoshikawa M. Eight genes in region 5 that form an operon are essential for invasion of epithelial cells by Shigella flexneri 2a. J Bacteriol. 1993;175:2334–2346. doi: 10.1128/jb.175.8.2334-2346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoenhals G J, Macnab R M. FliL is a membrane-associated component of the flagellar basal body of Salmonella. Microbiology. 1999;145:1769–1775. doi: 10.1099/13500872-145-7-1769. [DOI] [PubMed] [Google Scholar]
- 45.Segura A, Duque E, Mosqueda G, Ramos J L, Junker F. Multiple responses of gram-negative bacteria to organic solvents. Environ Microbiol. 1999;1:191–198. doi: 10.1046/j.1462-2920.1999.00033.x. [DOI] [PubMed] [Google Scholar]
- 46.Sikkema J, de Bont J A M, Poolman B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev. 1995;59:201–222. doi: 10.1128/mr.59.2.201-222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Gijsegem, Gough F C, Zischek C, Niqueux E, Arlat M, Genin S, Barberis P, German S, Castello P, Boucher C. The hrp gene locus of Pseudomonas solanacearum, which controls the production of a type III secretion system, encodes eight proteins related to components of the bacterial flagellar biogenesis complex. Mol Microbiol. 1995;15:1095–1114. doi: 10.1111/j.1365-2958.1995.tb02284.x. [DOI] [PubMed] [Google Scholar]
- 48.Weber F J. Adaptation of Pseudomonas putida S12 to high concentrations of styrene and other organic solvents. Appl Environ Microbiol. 1993;59:3502–3504. doi: 10.1128/aem.59.10.3502-3504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White D G, Goldman J D, Demple B, Levy S B. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young G M, Schmiel D H, Miller V L. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc Natl Acad Sci USA. 1999;96:6456–6461. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]