Summary
More than half the world’s population is infected with human cytomegalovirus (HCMV), causing congenital birth defects and impacting the immuno-compromised. Many of the >170 HCMV genes remain uncharacterized, and this gap in knowledge limits the development of novel antivirals. In this study, we investigated the essential viral protein UL49 and found it displayed leaky late expression kinetics, and localized to nuclear replication compartments. Cells infected with mutant UL49 virus were unable to produce infectious virions and phenocopied other beta-gamma viral pre-initiation complex (vPIC) subunit (UL79, UL87, UL91, UL92, and UL95) mutant infections. RNA-seq analysis of vPIC mutant infections revealed a consistent diminution of genes encoding capsid subunits, including TRX2/UL85 and MCP/UL86, envelope glycoproteins gM, gL and gO, and egress-associated tegument proteins UL99 and UL103. Therefore, as a member of the vPIC, UL49 serves as a fundamental HCMV effector that governs viral gene transcription required to complete the replication cycle.
Subject areas: Molecular biology, virology, transcriptomics
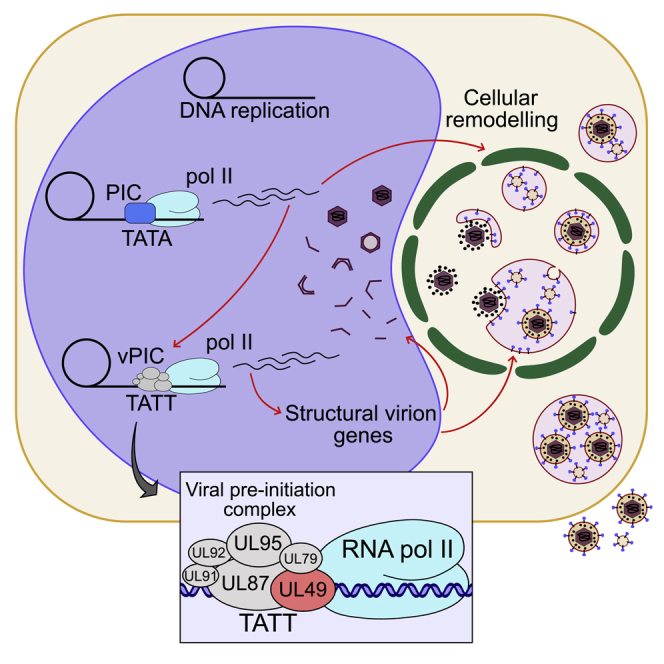
Graphical abstract
Highlights
-
•
Beta- and gamma-herpes viruses encode a viral pre-initiation complex (vPIC)
-
•
UL49, together with UL79, UL87, UL91, UL92, and UL95 Comprise the HCMV vPIC
-
•
UL49 is essential for HCMV replication and orchestrates late viral gene expression
-
•
Mutation of vPIC subunits reduces the transcription of structural virion components
Molecular biology; Virology; Transcriptomics
Introduction
Human cytomegalovirus (HCMV) is a pervasive human pathogen infecting much of the globe, with seropositivity greater than 90% in some populations (Cannon et al., 2010). HCMV establishes a latent lifelong, infection with periodic shedding of the virus providing a persistent source of transmission (Goodrum et al., 2012). Primary infection in healthy individuals is usually asymptomatic or mild, but presents serious risks for immunocompromised individuals such as organ transplant recipients (Ramanan and Razonable, 2013). During pregnancy, primary infection or reactivation of HCMV can lead to intrauterine transmission to the developing fetus and is the leading cause of infectious congenital birth defects (Bonalumi et al., 2011; Manicklal et al., 2013). Complications include sensorineural hearing and vision impairments, learning disabilities, and still birth in the most severe cases. There is currently no protective vaccine on the market, and only a handful of antiviral drugs available, with resistant strains constantly emerging. Therefore, understanding the fundamental biology of HCMV replication and pathogenesis will underpin novel therapeutic strategies.
HCMV is a member of the beta-herpes virinae subfamily of viruses, has a 236 kb linear double-stranded DNA (dsDNA) genome containing >170 open reading frames, many that remain uncharacterized (Dunn et al., 2003; Murphy and Shenk, 2008; Yu et al., 2003). The HCMV genome is packaged within an icosahedral nucleocapsid, surrounded by a thick layer of tegument proteins with a host cell-derived lipid envelope containing viral glycoproteins essential for cellular entry and membrane fusion (Nguyen and Kamil, 2018). Virion structure underpins the protracted replication cycle of HCMV which has nuclear and cytoplasmic stages. Nuclear stages predominantly take place in the replication compartment (RC), which is a virally induced inclusion (Caragliano et al., 2022). Here the viral genome is replicated by a rolling circle mechanism to produce concatemers, viral gene transcription takes place, capsids assemble, and genomes are cleaved and packaged (Muller et al., 2021). Maturing capsids exit the nucleus (Sanchez and Britt, 2021) and traffic into the cytoplasmic viral assembly compartment (vAC) for final regimentation, envelopment, and egress (Alwine, 2012).
HCMV has an elaborate transcription cascade, traditionally described by three major kinetic classes of gene expression: immediate-early (IE), delayed early (DE), and late (L), all of which are transcribed by host RNA polymerase II (pol II). L genes are additionally divided into leaky late (LL) and true late (TL) (Anders et al., 2007). LL products are expressed independently of DNA replication, but their levels remain low when DNA replication is inhibited, which is likely owing to low template stoichiometry. In contrast, TL products are defined as being entirely dependent on DNA replication, with DNA sequestration in RCs and chromatin modification likely contributing to TL dynamics (Gruffat et al., 2016). Recently, profiling of gene expression using proteomics (Weekes et al., 2014) and RNA-seq (Rozman et al., 2022) has defined 5 and 7 kinetic classes, respectively. These profiles are based on expression kinetics, but fail to describe the full complexity of transcriptional regulation. Transcription of all alpha-herpesvirus genes is exclusively initiated by the host pre-initiation complex (PIC), consisting of TATA-binding protein (TBP) and 5 additional general transcription factors (Gruffat et al., 2016), which recruit pol II to perform transcription (Gupta et al., 2016). For the beta- and gamma-subfamilies, a 6-member viral PIC (vPIC) is required to initiate transcription from a sub-set of late genes with promoters enriched in the non-canonical TATT box motif which subsequently recruits RNA pol II (Aubry et al., 2014; Davis et al., 2015; Gruffat et al., 2012, 2016). For HCMV, only 5 vPIC subunits have been characterized using infectious virus including UL79, UL87, UL91, UL92 and UL95 (Isomura et al., 2011; Omoto and Mocarski, 2013, 2014).
Previously, we concatenated HCMV gene annotations to identify late viral gene products including UL34 (Turner et al., 2022). Interestingly, UL49 also came up during the analysis; however, the previous investigation of its function has produced conflicting results. It has been reported that UL49 mRNA can be differentially spliced (Yang et al., 2015), and UL49 expression is required for viral replication (Zhang et al., 2010), with knockdown affecting viral DNA levels (Wang et al., 2013). Localization of UL49 has been reported in the cytoplasm of infected cells (Zhu et al., 2016), potentially indicating an envelopment or egress function. However, contrary to these observations, the gamma-herpes analogs of UL49 in Epstein-Barr virus (EBV) (Aubry et al., 2014) and Kaposi Sarcoma Associated Herpesvirus (KSHV) (Didychuk et al., 2020; Watanabe et al., 2020) are known to encode subunits of the 6-member vPIC. In addition, transient assays suggest that UL49 functions in the HCMV vPIC (Aubry et al., 2014). Therefore, to reconcile these conflicting reports, we took UL49 forward for comprehensive functional characterization.
Results
UL49 is essential for viral growth but dispensable for viral genome replication
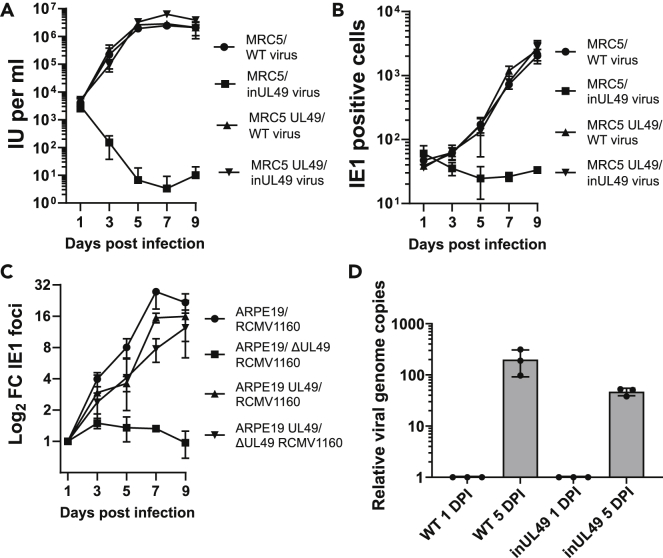
UL49 mutant virus (inUL49) was reconstituted by electroporating the inUL49 BAC (TS-353) (Table S1) (Yu et al., 2003), containing UL49 gene disruption by transposon insertion mutagenesis, into fibroblasts stably expressing UL49. To validate the inUL49 genome did not contain other mutations in essential genes, high and low MOI growth curves were assayed. This confirmed complete rescue, indicating that UL49 was essential for viral spread (Figures 1A and 1B). Similarly, UL49 was also found to be essential for the spread of Merlin HCMV (RCMV1160) (Stanton et al., 2010) in epithelial cells (Figure 1C). Next, to assess whether UL49 mutation impacted DNA replication, viral genome copies were assayed by qPCR at 1 and 5 days post-infection (DPI). Infections with inUL49 had approximately 50-fold more viral DNA copies at 5 DPI compared to 1 DPI (Figure 1D), confirming robust DNA replication in the absence of UL49; however, it did not reach WT amounts.
Figure 1.
UL49 is an essential viral protein dispensable for viral DNA replication
(A) Growth kinetics of inUL4AD 9169-GFP virus was measured by IE1 fluorescent focus assay of cell culture supernatants from WT and pBMN UL49-complementing MRC5 cells at various timepoints (MOI = 3, n = 3, bars = SD).
(B) Quantitation of low multiplicity spread of WT and inUL4AD 9169-GFP HCMV. At indicated timepoints, cells were fixed directly and stained for IE1 in WT and pBMN UL49-complementing MRC5 cells (MOI = 0.01, n = 3, bars = SD).
(C) Spread of ΔUL49 Merlin virus (RCMV1160) in WT and pBMN UL49-complementing ARPE-19 cells was quantified by fixing, staining, and counting IE1 positive cells at indicated timepoints (MOI = 0.01, n = 3, bars = SD).
(D) Intracellular HCMV genome copies at 5 DPI, relative to genome copies at 1 DPI for WT and inUL4AD 9169-GFP virus (MOI = 3, n = 3, bars = SD).
UL49 is expressed with leaky late kinetics and accumulates in the nucleus during infection
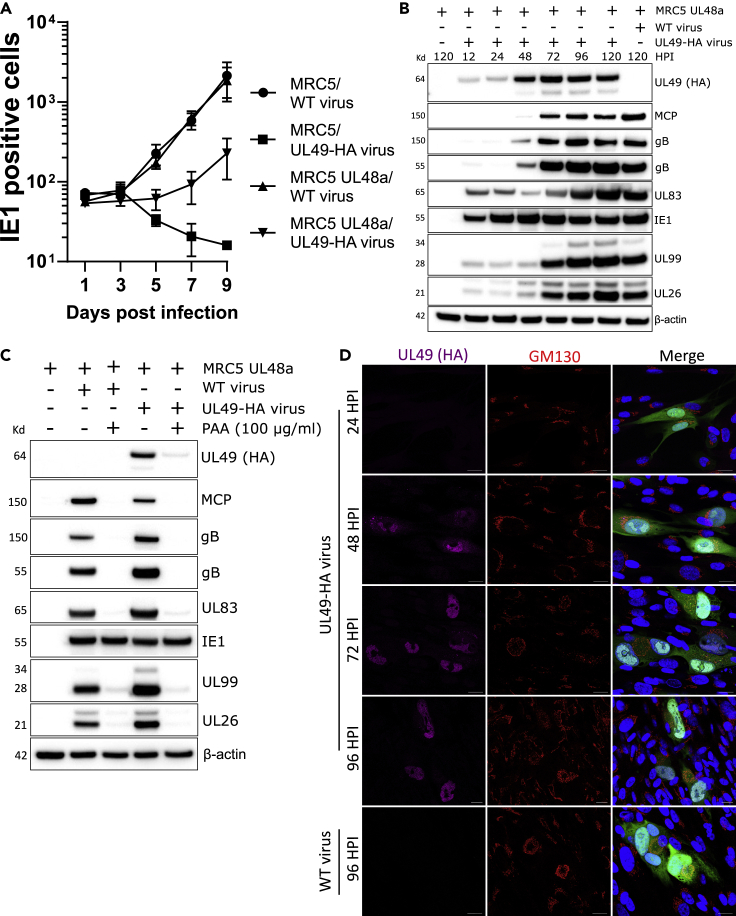
As commercial antibodies to UL49 proved unreliable, we constructed a BAC containing an HA-tagged UL49 to enable the characterization of the endogenous protein. The UL49 ORF has an n-terminal overlap with UL50 and c-terminal overlap with UL48a/SCP (Sijmons et al., 2014) which complicates the tagging strategy as the introduction of a tag to either termini would affect neighboring genes. We opted for a c-terminal tag and attempted to circumvent UL48a disruption by reintroducing the affected UL48a sequence after the UL49 STOP codon, along with additional linker sequence, to ensure the complete UL48a coding sequence is intact in the engineered BAC. Despite our best efforts, UL49-HA virus could not be reconstituted in WT cells, possibly owing to the UL48a promoter being disrupted by the HA tag and linker. To remedy this, a viable UL49-HA virus was successfully generated by reconstitution in UL48a complementing cells, with all subsequent UL49-HA experiments performed in the UL48a complementing background (Figure 2A).
Figure 2.
Characterization of UL49 as a nuclear localizing viral protein with leaky late expression kinetics
(A) Quantitation of low multiplicity spread of WT and UL49-HA (endogenous c-terminal HA tag) AD169-GFP HCMV. At indicated timepoints, cells were fixed directly and stained for IE1 in WT and pBMN UL48a/SCP-complementing MRC5 cells (MOI = 0.01, n = 3, bars = SD).
(B) Western blot analysis of UL49 expression kinetics. MRC5 pBMN UL48a cells were infected with HCMV AD169 UL49-HA virus, and lysed at indicated times post-infection. Mock and AD169-GFP WT control conditions were lysed at 5 DPI (MOI = 3). Membranes were probed with primary antibodies against HCMV viral proteins, HA, and beta-actin loading control.
(C) Western blot analysis of UL49 expression in MRC5 pBMN UL48a cells treated with phosphonoacetic acid (PAA), and subsequently infected with either AD169-GFP WT or AD169 UL49-HA HCMV (5 DPI, MOI = 3). Membranes were probed with primary antibodies against HCMV viral proteins, HA, and beta-actin loading control.
(D) Immunofluorescence time course analysis of MRC5 pBMN UL48a cells infected with HCMV AD169-GFP UL49-HA at indicated timepoints and WT control at 96 HPI. Samples were stained with HA (UL49) and GM130 antibodies (MOI = 0.1, scale bars = 20 μm).
Next, UL49 expression kinetics were investigated over 5 DPI. A weak UL49-HA signal was first detected 12-24 HPI, with robust protein expression observed between 48 and 120 HPI (Figure 2B). These kinetics showed that UL49-HA was robustly expressed after DNA replication between 24 and 48 HPI, but before maximum true late protein accumulation at 72 HPI, as demonstrated by MCP as well as the furin-cleaved and un-cleaved pools of gB (Figure 2B). Furthermore, to validate that UL49 expression was downstream of DNA replication, cells were treated with the DNA replication inhibitor phosphonoacetic acid (PAA), infected with UL49-HA virus, and expression assayed by Western blotting. Consistent with leaky late kinetics, minimal UL49 expression occurred in the absence of viral DNA replication (Figure 2C).
Finally, a time course analysis of UL49-HA intracellular localization using immunofluorescence confocal microscopy revealed cytoplasmic and nuclear localization by 48 HPI, with exclusive nuclear accumulation between 72 and 96 HPI (Figure 2D). However, the nuclear accumulation was not uniform, and instead UL49-HA localized to intranuclear structures consistent with RCs (Caragliano et al., 2022) (Figure 2D). Collectively, these results demonstrated that UL49 is a nuclear protein expressed with leaky late kinetics.
UL49 regulates late viral gene transcription akin to the beta-gamma viral pre-initiation complex
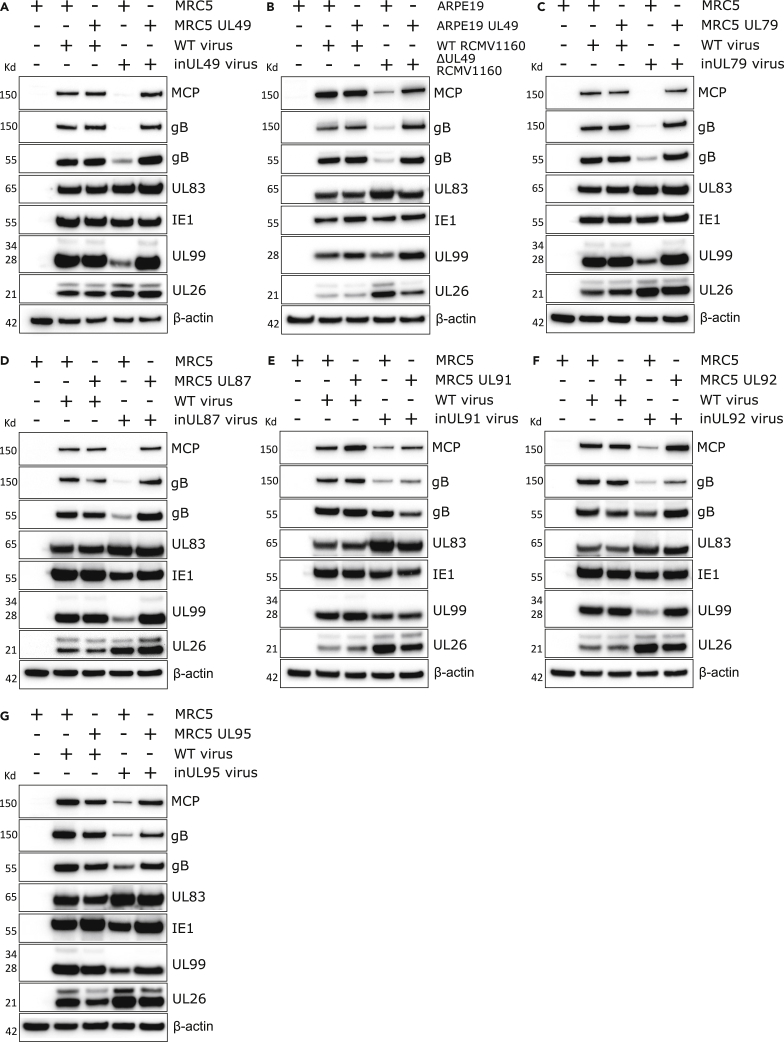
Given the nuclear localization and expression kinetics, next, we evaluated whether inUL49 impacted the expression of other viral proteins. Loss of UL49 did not alter expression levels of IE1 or tegument proteins UL26 and UL83, but did attenuate expression of the late viral protein UL99 (Figure 3A). Strikingly, MCP and gB had minimal expression in the absence of UL49 (Figure 3A). Similar defects were observed in ARPE-19 cells infected with ΔUL49 Merlin (Figure 3B).
Figure 3.
UL49 deletion phenocopies HCMV viral pre-initiation complex mutants
(A) Western blot analysis of MRC5 and pBMN UL49-complementing cell line lysates infected with HCMV AD169-GFP WT and inUL49 viruses (5 DPI, MOI = 3).
(B) Western blot analysis of ARPE-19 and pBMN UL49-complementing cell line lysates infected with HCMV WT Merlin (RCMV1160) and ΔUL49 Merlin viruses (5 DPI, MOI = 1).
(C-G) Western blot analysis of MRC5 and pBMN complementing cell line lysates infected with HCMV WT and inUL79, inUL87, inUL91, inUL92, and inUL95 mutant viruses, respectively (5 DPI, MOI = 3). Membranes were probed with primary antibodies against HCMV viral proteins and beta-actin loading control.
A comprehensive HCMV interactome was published during the completion of this project (Nobre et al., 2019). Examination of UL49 and its reciprocal interactions revealed associations in a network of viral proteins UL79, UL87, UL91, and UL95 (Isomura et al., 2011; Omoto and Mocarski, 2013, 2014), and at least 12 host RNA polymerase II subunits (Nobre et al., 2019; Perng et al., 2014). Together with the requirement for UL49 to induce the robust transcription of a luciferase reporter gene in HEK293 cells transfected with individual HCMV vPIC subunits (Aubry et al., 2014), we suspected that UL49 was the sixth member of the HCMV vPIC, as is the case in EBV and KSHV (Aubry et al., 2014; Didychuk et al., 2020; Watanabe et al., 2020).
To investigate furthermore, we first reconstituted stocks of inUL79 (TS-197), inUL87 (TS-430), inUL91 (TS-019), inUL92 (TS-356), and inUL95 (TS-307) mutant viruses using stable complementing cell lines (Table S1) (Yu et al., 2003). Infection of WT cells with inUL79 and inUL87 viruses revealed significantly attenuated expression of MCP and gB (Figures 3C and 3D), whilst the magnitude of reduction was less for inUL91, inUL92, and inUL95 infections (Figures 3E-3G). Taken together we concluded that UL49 is the sixth subunit of the HCMV vPIC.
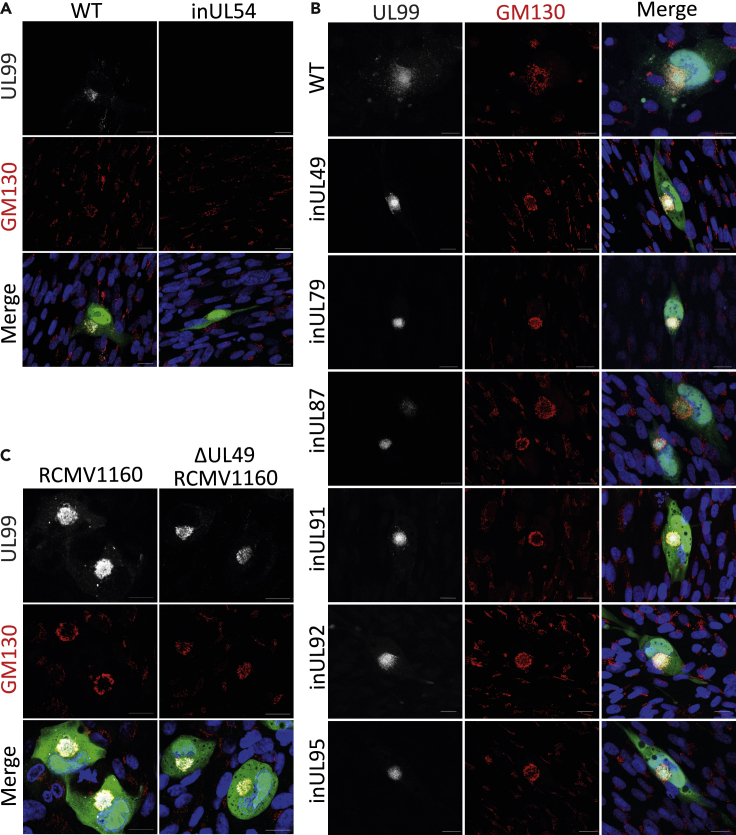
The viral pre-initiation complex is dispensable for viral assembly compartment formation
At late times during infection, coinciding with the onset of late gene expression, the cytoplasmic vAC forms (Alwine, 2012). Given the vPIC regulates a sub-set of late transcripts (Figures 3A-3G), and vAC formation requires viral DNA replication and subsequent late gene expression (Figures 2C and 4A) (Das et al., 2014), we investigated whether vPIC mutants could form a prototypic vAC. No defects were observed for any of the vPIC mutant viruses, as well as ΔUL49 Merlin, which all displayed prototypical vACs (Figures 4B and 4C). Interestingly, vPIC mutants did cluster UL99 more tightly in the center of the vAC and displayed a sharper Golgi ring morphology compared to WT (Figure 4B). This may be owing to the absence of maturing capsids in the cytoplasm which could redistribute vAC markers during envelopment and egress. Similarly, the concentrated UL99 localization likely reconciles the reduced UL99 expression observed by Western blot (Figures 3A-3G), with the comparably intense immunofluorescence signal in the mutant infections (Figure 4B). Taken together, we conclude that vAC formation requires the transcription of late genes not regulated by the vPIC.
Figure 4.
Viral assembly compartment formation in vPIC mutant infections
(A) Immunofluorescence analysis of the viral assembly compartment visualized by host GM130 and viral UL99 in WT MRC5 cells infected with HCMV AD169-GFP WT and inUL54 viruses (4 DPI, MOI = 0.1, scale bars = 20 μm).
(B) Immunofluorescence staining of GM130 and UL99 in WT MRC5 cells infected with HCMV AD169-GFP WT and vPIC mutant viruses (4 DPI, MOI = 0.1, scale bars = 20 μm).
(C) Immunofluorescence staining of GM130 and UL99 in WT ARPE-19 cells infected with either HCMV Merlin (RCMV1160) WT or ΔUL49 (4 DPI, MOI = 0.1. Scale bars = 20 μm).
RNA-seq analysis identifies viral transcripts regulated by the vPIC
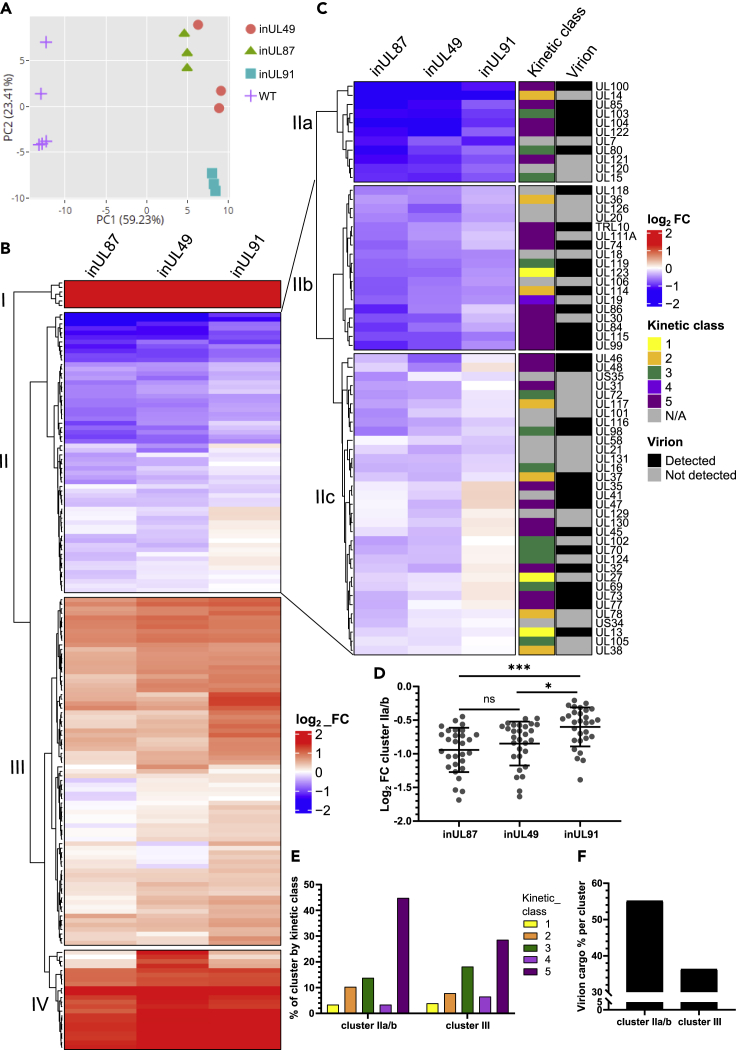
To reveal the full repertoire of viral transcripts regulated by the HCMV vPIC, we conducted RNA-seq analysis of cells infected with either inUL49, inUL87, or inUL91 viruses at 72 HPI. Principal component analysis clearly segregated the HCMV mutants from WT based on PC1, and inUL91 from inUL49 and inUL87 based on PC2 (Figures 5A, S1A and S1B). Differential expression analysis and hierarchical clustering revealed 3 clusters of viral genes that did not require the vPIC for expression (I, III, IV), and 1 gene cluster (II) that had attenuated expression in the vPIC mutant infections (Figure 5B, Data S1). Further sub-clustering of cluster II highlighted genes more sensitive to vPIC regulation (Figure 5C, top and middle panels), compared to genes with more modest vPIC regulation (Figure 5C, bottom panel). Despite genes in the latter group not achieving statistical significance for differential expression in this experiment (Figures S2A–S2C), many of these have been shown to be regulated by the vPIC including UL47, UL72, UL73/gN, and UL32/pp150 (Li et al., 2021; Omoto and Mocarski, 2013, 2014). Additionally, triplex capsid protein 1 (TRX1/UL46) is a notable transcript that falls into this sub-category.
Figure 5.
RNA-seq identification of viral genes regulated by the HCMV vPIC
(A) Principal component analysis of identified genes in cells infected with HCMV AD169-GFP WT or vPIC mutant viruses (3 DPI, MOI = 3, WT n = 5, mutants n = 3).
(B) Heatmap displaying the mean log2 fold-change of all viral transcripts compared to WT. Expression profiles were hierarchically clustered using complete linkage clustering and euclidean distance.
(C) Expanded heatmap of cluster II from B) further sub-clustered into IIa, IIb, and IIc with kinetic expression class (Weekes et al., 2014) and virion cargo (Turner et al., 2020) annotations. Annotations were not used for row clustering.
(D) The mean log2 expression change for each mutant compared to WT for transcripts in clusters IIa and IIb. Bars represent mean and SD, ∗∗∗p < 0.001, ∗p < 0.05, ns: not significant, one-way ANOVA with Tukey’s post-test.
(E) Distribution of genes within indicated clusters based on kinetic expression class.
(F) Proportion of genes within indicated clusters with virion cargo annotation. See also Figures S1 and S2.
Our initial characterization of vPIC mutant viruses revealed reduced attenuation of late protein expression for inUL91, inUL92, and inUL95 infections compared to inUL49, inUL79, and inUL87 (Figures 3A-3G). Similarly, RNA-seq analysis confirmed that inUL91 did not achieve the same magnitude of gene attenuation as inUL49 or inUL87 (Figures 5A-5C and S1B). These general observations were further quantified by plotting the mean log2 fold-change of cluster IIa/b transcripts for each mutant virus versus WT, which revealed a significant difference for the inUL91 condition (Figure 5D).
Next, we examined annotation based on kinetic expression class (Weekes et al., 2014), and assessed if the translated protein had been previously identified in the virion (Turner et al., 2020) (Figure 5C). Of the 29 genes found to be significantly regulated by the vPIC (Figure 5C, top and middle panels), 45% have late classification (kinetic class 5), and 14% have leaky late classification (kinetic class 3) (Figure 5E). Additionally, during typical infection and subsequent translation, 55% are present as infectious virion cargo (Figure 5F). For example, the capsid scaffolding protein UL80, TRX2/UL85, MCP/UL86, and the portal protein UL104 are regulated by the vPIC, as are a sub-set of envelope glycoproteins including gM/UL100, gL/UL115, and gO/UL74 (Figure 5C). Furthermore, egress-associated tegument proteins UL99 (Silva et al., 2003) and UL103 (Ahlqvist and Mocarski, 2011) were also significantly attenuated in vPIC mutants. A similar analysis of genes in cluster III revealed that 29% are classified as late (class 5) (Figures 5E and S2D), and only 36% of genes encode virion cargo (Figures 5F and S2D). Notable examples of late genes that are not regulated by the vPIC include UL25 (Battista et al., 1999), UL26 (Mathers et al., 2014; Munger et al., 2006), the multifunctional UL83 (Kalejta, 2008) and UL88 (Kumar et al., 2020) (Figure S2D).
Collectively, RNA-seq analysis 1) identified that the vPIC regulates expression of a large sub-set of late viral genes that encode virion components, and 2) within the 6 vPIC members, inUL49 and inUL87 impacted gene transcription more potently than inUL91.
Discussion
Previous analysis of HCMV gene annotations indicated a late molecular function for UL49 (Turner et al., 2022), and was of particular interest given its previous conflicting reports. Using transposon insertion mutants and endogenously tagged viruses, we showed that UL49 is a leaky late viral protein that localizes to RCs in the nucleus. Our data, together with interaction studies (Nobre et al., 2019; Perng et al., 2014) confirm that UL49 is the sixth member of the HCMV vPIC and regulates the transcription of structural virion and egress-associated transcripts.
The transcriptional program of HCMV is a complex and dynamic process requiring both host and viral regulation. To interrogate the vPIC regulated genes furthermore, we overlayed kinetic protein expression and virion composition data. This identified that structural virion constituents were over-represented in vPIC regulated transcripts and includes 6/7 capsid-associated gene products and 4/6 envelope glycoproteins (Figures 5B and 5C, S2A-S2C). Some tegument genes were also found to be vPIC-regulated, including the capsid-associated UL32 (Yu et al., 2017), the early acting virion cargo tegument UL48 (Kwon et al., 2017; Wang et al., 2006), and envelopment and egress-associated UL99 and UL103 (Figure 5C) (Ahlqvist and Mocarski, 2011; Sanchez et al., 2000; Seo and Britt, 2006; Silva et al., 2003). In contrast, we found that tegument genes UL25 (Battista et al., 1999), UL26 (Mathers et al., 2014; Munger et al., 2006), UL83 (Kalejta, 2008), and UL88 (Kumar et al., 2020) were not regulated by the vPIC (Figure S2D). Moreover, UL26, UL83, UL86/MCP, and UL99 exhibit similar late expression kinetics over the course of infection (Weekes et al., 2014), but only UL86 and UL99 are regulated by the vPIC (Figures 2B, 3A-3G, 5C and S2D). Conversely, UL83 expression is sensitive to regulation by IE2, whilst UL99 is not (Li et al., 2020). During the completion of this article, a comprehensive transcriptome analysis of HCMV gene expression using comparable methodology was published (Rozman et al., 2022). The authors defined 7 temporal expression classes during lytic infection which included sensitivity to PFA (viral DNA replication) and cycloheximide (translation inhibition). Their analysis highlighted that temporal expression and drug sensitivities do not fully align and reveal multiple modes of regulation. Our work and others (Li et al., 2021) investigating the vPIC regulation of gene expression adds an additional layer of specificity to the HCMV transcriptional cascade. Therefore, it is emerging that distinct networks of viral gene transcription are activated simultaneously and run concurrently, but with individual specificity.
For HCMV genes regulated by vPIC-dependent transcription, TATT-containing promoter motifs contribute some specificity. However, other non-canonical promoter motifs are also enriched at sites of vPIC occupancy (Li et al., 2021). This, together with the revelation that select viral promoters are driven by both host TBP PICs as well as vPICs, and may even compete for occupancy, illustrates a complex and nuanced mechanism of vPIC promoter occupancy (Spector et al., 2022). Future work is required to further refine our understanding of context-dependent vPIC specificity, and the viral transcriptional landscape more broadly over time. However, our study has demonstrated that the HCMV vPIC executes the transcription of primarily late-stage HCMV genes facilitating virion particle formation, envelopment, and egress, and contributes understanding to the emerging complexity of the HCMV transcriptional program. With the completion of our study, all HCMV vPIC subunits have now been characterized using reconstituted mutant viruses and found to localize to the nucleus to regulate late-stage viral transcription (Aubry et al., 2014; Isomura et al., 2011; Omoto and Mocarski, 2013, 2014). As it currently stands, a precise understanding of how all 6 members interact, and a complete complex structure remains elusive. Although UL79 was suggested to act as an elongation factor responsible for directing cellular RNA polymerase II for viral transcription (Perng et al., 2014), this has been refuted by Li et al. (2021). Interestingly, we observed that features of inUL49, inUL79, and inUL87 were more similar and resulted in stronger gene expression defects compared to inUL91, inUL92, and inUL95 (Figures 3A–3G, 5A–5D, S1B, and S2A–S2D). These results may shed light on specific functions of individual subunits in the complex.
Studies completed in related gamma herpesviruses may also offer insights into structure and function. A genome-wide interaction study of KSHV identified ORF24 to be the essential DNA binding protein of the vPIC, which subsequently recruited the other 5 complex members (Davis et al., 2015). The EBV and HCMV orthologs of ORF24, BcRF1, and UL87, respectively, share the conserved TBP fold, so are likely to be the DNA binding proteins in their respective vPICs (Gruffat et al., 2012; Wyrwicz and Rychlewski, 2007). KSHV ORF24 has also been shown to recruit host RNA polymerase, based on conserved residues in the N-terminal domain binding to both the C-terminal domain of host pol II and promoter DNA (Castañeda et al., 2020). Recent characterization of KSHV ORF66, which is the ortholog of HCMV UL49, revealed it to be essential for virus production, gene expression from late promoters, and the C-terminus to be specifically required for interaction with ORF34 (HCMV UL95 ortholog) (Didychuk et al., 2020; Watanabe et al., 2020). Together, this work lays the foundation for detailed structural studies that pinpoint HCMV vPIC architecture, subunit interaction domains, and individual function and specificity.
In summary, we validated that UL49 is a member of the beta-gamma vPIC that regulates late-stage gene transcription. Together, the 6 member HCMV complex primarily regulates the expression of genes underpinning virion assembly, maturation, and egress. This transcriptional regulation is conserved across other beta and gamma herpesviruses, and the essential requirement for all subunits for virus propagation makes the vPIC an attractive antiviral target. Future work to resolve the complete structure of the complex and functional domains will provide valuable information for the future development of novel therapeutic interventions.
Limitations of the study
In this study, RNA-seq analysis was used to profile gene expression changes in vPIC mutant infections. A limitation of this approach is that it quantifies the total transcript pool and cannot distinguish alternative transcription initiation sites or splice variants. Alternative methods such as PRO-seq and PRO-cap could capture this additional information. Nonetheless, we were able to detect robust expression changes in vPIC mutant infected cells.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-UL99 | (Silva et al., 2003) | Clone 10B4 |
| Mouse anti-GM130 | BD Biosciences | Cat#610822 |
| Rabbit anti-GM130 | Abcam | Cat#ab52649 |
| Rabbit anti-HA epitope | Cell Signaling Technology | Cat#3724T |
| Mouse anti-IE1 | (Zhu et al., 1995) | Clone 1B12 |
| Mouse anti-HCMV gB | Abcam | Cat#ab6499 |
| Mouse anti-UL83 | (Nowak et al., 1984) | Clone 8F5 |
| Rabbit anti-MCP | Cusabio | Custom ordered, polyclonal |
| Mouse anti-UL26 | (Munger et al., 2006) | N/A |
| Mouse anti-HA epitope | Abcam | Cat#ab130275 |
| Mouse anti-β-actin | Sigma | Cat#A2228 |
| Goat anti-rabbit HRP conjugate | Bio-Rad | Cat#1706515 |
| Goat anti-mouse HRP conjugate | Bio-Rad | Cat#1706516 |
| Goat anti-mouse Alexafluor 568 conjugate | Invitrogen | Cat#A-11004 |
| Goat anti-rabbit Alexafluor 633 conjugate | Invitrogen | Cat#A-21070 |
| Bacterial and virus strains | ||
| HCMV AD169-GFP | (Yu et al., 2003; Yu et al., 2002) | N/A |
| HCMV Merlin (RCMV1160) | (Stanton et al., 2010) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Phosphonoacetic acid (PAA) | Sigma | Cat#284270 |
| Critical commercial assays | ||
| Gibson Hi-Fi DNA assembly cloning kit | NEB | Cat#E5520S |
| Deposited data | ||
| Raw and analysed RNA-seq data | This paper | GEO:GSE201208 |
| Experimental models: Cell lines | ||
| MRC-5 fetal lung fibroblasts | ATCC | CCL-171 |
| HEK293T embryonic kidney cells | ATCC | CRL-3216 |
| ARPE-19 retinal pigment epithelial cells | ATCC | CRL-2302 |
| Oligonucleotides | ||
| Primers for pBMN constructs, see Table S2. | This paper | N/A |
| Recombinant DNA | ||
| pBMN UL48a | This paper | N/A |
| pBMN UL49 | This paper | N/A |
| pBMN UL49 (Merlin) | This paper | N/A |
| pBMN UL54 | This paper | N/A |
| pBMN UL79 | This paper | N/A |
| pBMN UL87 | This paper | N/A |
| pBMN UL91 | This paper | N/A |
| pBMN UL92 | This paper | N/A |
| pBMN UL95 | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 9 | GraphPad Software Inc. | https://www.graphpad.com/scientific-software/prism/ |
| R Studio (v4.1.1) | R Studio | https://www.rstudio.com/ |
| RNAsik (version 1.5.5 = 1) | (Tsyganov et al., 2018) | https://github.com/MonashBioinformaticsPlatform/RNAsik-pipe |
| DE-Browser Shiny user interface (v1.21.1) | (Kucukural et al., 2019) | https://www.bioconductor.org/packages/devel/bioc/vignettes/debrowser/inst/doc/DEBrowser.html |
| ComplexHeatmap (v2.9.4) | ComplexHeatmap (v2.9.4) | https://github.com/jokergoo/ComplexHeatmap |
| EnhancedVolcano (v 1.11.5) | N/A | https://github.com/kevinblighe/EnhancedVolcano |
Resource availability
Lead contact
Further information and requests for resources should be directed to the lead contact, Rommel Mathias (rommel.mathias@monash.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cultured cells
Human primary fetal lung fibroblasts (MRC-5, ATCC CCL-171), HEK293T embryonic kidney cells (ATCC CRL-3216) and ARPE-19 retinal pigment epithelial cells (ATCC CRL-2302) were purchased directly from ATCC, cultured in Thermo Fisher Gibco DMEM (MRC5, 293T) or DMEM/F12 (ARPE-19) full media supplemented with 10% (v/v) Fetal Bovine Serum (Cell Sera), 10 U/ml penicillin and 10 U/ml streptomycin (Thermo Fisher Gibco). Cells were maintained at 37°C and 5% CO2 in a humidified incubator and passaged 1:3 to 1:5 every third day for MRC5 and ARPE-19.
Viruses
HCMV was reconstituted by electroporating bacterial artificial chromosomes (BACs) containing the AD169 genome or Merlin RCMV1160 genome (GFP, UL128 repaired) into MRC5 cells, together with pcDNA pp71 expression plasmid. All AD169 WT and mutant BACs were kindly provided by Prof. Thomas Shenk (Yu et al., 2002, 2003), and WT Merlin BACs by Dr Richard Stanton (Stanton et al., 2010). Transposon insertion positions for each AD169 mutant can be found in Table S1.
Method details
Virus isolation
Infected cellular supernatant was harvested, clarified by centrifugation at 1,500xg and underlaid with a sorbitol cushion (20% (m/v) D-sorbitol, 1X PBS, pH 7.4) prior to ultracentrifugation at 50,000xg for 1 h at 4°C (Britt, 2010). Virus pellets were resuspended in full media, titred by IE1 fluorescent focus assay using MRC5 (AD169) or ARPE-19 (Merlin) cells (described below), aliquoted and stored at −80°C. For HCMV infections, cell monolayers were grown overnight and infected in a low volume of media (adjusted depending on well format) with an MOI of 0.01, 0.1 or 3, depending on the experiment, for 2 h at 37°C with gentle agitation. After removal of the inoculum, fresh growth medium was added, and cells incubated for 12–120 h, depending on the experiment.
Immunofluorescence confocal microscopy
Cell monolayers were grown on 12 mm glass coverslips (no. 1.5, Menzel) in 24-well plates. At the indicated timepoints cells were fixed (4% PFA, PBS pH 7.4) and permeabilized (0.1% (v/v) Triton X-100 in PBS) for 15 min each. Cells were washed 3 times with 0.2% (v/v) Tween 20 in PBS and blocked at RT for 1 h (2% BSA, 0.2% Tween 20, 2.5% HSA in PBS). Primary mouse anti-UL99 (Clone 10B4 (Silva et al., 2003)) was diluted 1:100, and mouse anti-GM130 (610,822, BD Biosciences), rabbit anti-GM130 (ab52649, Abcam) and rabbit anti-HA epitope (3724T, Cell Signaling Technology) were diluted 1:500 in blocking buffer and incubated for 1 h at RT. Cells were washed 3 times in PBST and incubated with goat anti-mouse IgG antibody conjugated to Alexa Fluor 568 (A-11004, Invitrogen) or goat anti-rabbit IgG antibody conjugated to Alexa Fluor 633 (A-21070, Invitrogen) diluted 1:1000 with Hoechst (33342, Thermo Fisher) 1:2000 for 30 min at RT. Coverslips were washed again in PBST for 15 to 30 min, blotted dry on a Kimwipe and mounted on glass slides (Menzel) with Mowiol mounting media. Images were acquired on either a Leica inverted SP5 or SP8 using 63x oil immersion objective and sequential scan settings of individual channels (Fluor 633, Fluor 568, eGFP and hoechst) with a line average of 4 and frame average 1. Images were viewed and analyzed in LAS X core offline version (Leica).
Cloning and stable cell line generation
HCMV viral gene inserts were PCR amplified from isolated BAC DNA using Phusion high fidelity polymerase (ThermoFisher Scientific). The UL48a PCR product was digested with BamHI and SalI, then inserted into digested pBMN puro vector backbone (provided by Dr. Michael Lazarou) using traditional DNA ligation. All other pBMN plasmids were constructed using the Gibson Hi-Fi cloning kit (E5520S, NEB) according to the manufacturer’s instructions. Colonies were PCR screened with a forward sequencing primer and gene specific reverse Gibson primer for correct inserts, and further validated by restriction digest with BamHI and SalI. All inserts were Sanger sequence verified before cell line generation. A complete list of primer sequences used can be found in Table S2.
To generate stable cell lines, insert containing plasmids were isolated (PureLink plasmid miniprep kit, Invitrogen), and 4x106 293T cells (purchased from ATCC, and mycoplasma negative by PCR) transfected with 2 μg VSV-G, 4 μg gag/pol (retrovirus) packaging vector and 6 μg pBMN containing inserts using Lipofectamine 3000 (Invitrogen), according to manufacturer’s instructions. 16 h post transfection, medium was changed, and after 24 h supernatant containing retroviral particles was syringe-filtered through 0.45 μm Acrodisc filters (Pall) and added directly to cells (two harvests, 24 h apart). Cells were placed under selection (full media containing 3 μg/mL puromycin) for 5 days, changed at 48 h intervals, and then passaged as per ‘cells and viruses’.
IE1 fluorescent focus assay
Infected cell supernatant was collected, and cell debris removed by centrifugation at 500xg for 5 min. No low-speed centrifugation was used for virus stock titers as this was incorporated into the isolation protocol. Serial dilutions using full media (1:4) were performed, starting from neat samples to 4−5 for growth curves and 4−12 for virus stocks. 100 μL of each dilution was added to a 96-well reporter plate of confluent uninfected MRC5 or ARPE-19 cells. 24 HPI, cells were fixed and stained with mouse anti-IE1 primary (1:100, Clone 1B12 (Zhu et al., 1995)) and Hoechst as per “Immunofluorescence confocal microscopy”. Reporter plates were automatically imaged using a DMi8 (Leica) microscope with 10x objective. A focus map was constructed with a single point per well and autofocused using the Hoechst channel in LAS X navigator (Leica). A 3 × 3 tilescan was performed per well in hoechst, and IE1 channels with 0% image overlap and fill factor 75%. IE1 foci per well were manually counted in LAS X core offline version (Leica) at appropriate dilutions for IU/ml calculations.
BAC recombineering
BAC clone RCMV1160 (Merlin) was modified using bacterial homologous recombination. The UL49 coding sequence shares an overlap with UL50 on the 5′ end and UL48a on the 3’. Therefore, a partial deletion of UL49 was selected to not interfere with UL50 or UL48a. A kan resistance cassette was PCR amplified with primers containing homology arms specific to UL49 (Table S2). The linear PCR fragment was introduced by electroporation (Eppendorf) at 1660 V, 0.25 μF and 200 Ω in 0.1 mm gap cuvettes together with the pSC101-BAD-gbaA (tet resistance) plasmid containing the λ Red operon under an arabinose inducible promoter. pSC101-BAD-gbaA is heat sensitive and is lost at 37°C. Clones containing the correct insert were selected on LB plates containing kan and cml at 37°C to remove pSC101-BAD-gbaA. The kan cassette insertion deleted bases 96 to 593 of UL49 which was verified by sanger sequencing.
To create AD169 UL49-HA, the kan cassette was amplified as above with primers containing homology arms to the 3′ region of UL49 as well as the 5′ region of UL48a with HA-tag and linker sequence inserted upstream of the UL49 STOP codon (Table S2). Additional sequence in the linker was used to restore the ATG of UL48a and FRT sites flanking the Kan cassette for excision after recombination. The resulting PCR product was electroporated (as above) with pSC101-BAD-gbaA to create AD169 UL49-HA (Kan resistance). Correct clones were selected (as above) on cml/kan plates and screened by restriction enzyme digest. The kan cassette was deleted by electroporating the 706-Flp plasmid (GeneBridges) which expresses Flp recombinase at 30°C and is lost at 37°C. Correct clones were identified by triple streaking on cml, kan and tet plates. Deletion of the kan cassette and insertion of the HA tag was verified by sanger sequencing.
Densitometry-based protein quantification
Cell monolayers were washed once with PBS and lysed in LDS buffer (141 mM Tris, 2% (w/v) LDS, 10% (v/v) glycerol, 0.51 mM EDTA, 0.22 mM G250, 50 mM DTT, 50 mM TCEP, 50 mM chloroacetamide, pH 8.5) for 15 min on ice, tip sonicated and denatured at 95°C for 10 min 5 μL aliquots of samples and 5 μL BenchMark protein standard (Invitrogen) were separated by PAGE (NuPAGE 4–12% Bis-Tris gels), and the gels stained with SYPRO ruby (ThermoFisher Scientific) according to manufacturer’s instructions. Gels were imaged using a Typhoon Trio (GE Healthcare) at a resolution of 200 μm, with excitation (470 nm) and emission filters (610 nm) set. The protein concentration in each fraction was determined by relative densitometry of each lane compared to the BenchMark standard, as calculated by ImageQuant analysis toolbox software (GE Healthcare).
Western blotting
25 μg of each sample were subjected to PAGE as above, and proteins wet transferred in Tris-glycine buffer to PVDF membranes using a Mini-PROTEAN Tetra cell (BioRad) at 120 V for 60 min at 4°C. Membranes were blocked with 5% (w/v) skim milk in 1X TBST (150 mM NaCl, 50 mM Tris pH 7.4, 0.1% (v/v) Tween 20) for a minimum of 1 h at 4°C, membranes were incubated with primary antibodies: anti-HCMV gB (Abcam ab6499), anti-UL83 (Clone 8F5 (Nowak et al., 1984)), anti-UL99 (Clone 10B4 (Silva et al., 2003), anti-MCP (custom ordered rabbit polyclonal, Cusabio), anti-UL26 (Munger et al., 2006), anti-IE1 (Clone 1B12 (Zhu et al., 1995)), anti-HA (ab130275, Abcam) and anti-β-actin (Sigma, A2228) diluted 1:1000 in 5% skim milk and incubated overnight at 4°C. Membranes were washed 3 times with TBST, and appropriate anti-rabbit (#1706515, Bio-Rad) or anti-mouse (#1706516 or Bio-Rad) HRP-conjugated secondary antibodies diluted 1:5000 were incubated for 1 h at 4°C. Membranes were washed as above in TBST with a final wash in TBS, incubated in Clarity ECL substrate (Bio-Rad) for 1 min, and imaged using a Gel Doc imaging system (Bio-Rad). Images were viewed and analyzed using ImageJ.
Intracellular viral genome quantitation
Cells were infected at MOI = 3 as per ‘Cells and virus”. At indicated timepoints, cells were washed with PBS, lysed with TRIzol Reagent (Invitrogen), and total DNA isolated according to manufacturer’s instructions. Two step qPCR was performed using QuantiNova SYBR Green PCR master mix (Qiagen) in a Rotor-Gene-Q real-time PCR cycler (Qiagen). Initial heat denaturation was performed at 95°C for 5 min, then 40 cycles of 30 s denaturation (95°C) and 60 s anneal/extend (60°C). UL83 and ATG5 specific qPCR primer pairs were used for HCMV genome quantification and internal control, respectively (Table S2). The relative number of viral genomes was calculated using the ΔΔCt method, normalized to the 24-h control for both WT and ΔUL49 infections.
RNA-seq
Cells were infected at MOI = 3 as per “Cells and virus”, lysed in TRIzol at 72 HPI and total RNA extracted using the Direct-Zol MiniPrep kit (Zymo, R2050), according to the manufacturer’s instructions. Sequencing libraries were prepared using an MGIEasy stranded mRNA kit, and sequenced on an MGITech MGISEQ2000RS sequencer to generate 100b paired end reads.
Data were aligned to a custom reference genome using the RNAsik pipeline (version 1.5.5 = 1) (Tsyganov et al., 2018). The combined reference genome was generated by concatenating the fasta files from the X17,403.1 reference for HCMV (downloaded from NCBI) and the GRCh38 Ensembl reference (release 104). The annotation GFF3 for X17,403.1 was downloaded and modified to be compatible with the GRCh38 Ensembl GTF file before the two were concatenated to make a combined annotation GTF file. The RNAsik pipeline was run with the following parameters: ‘-align star -counts -paired -all’ and used the combined fasta file and gtf file as input for the ‘-fastaRef’ and ‘-gtfFile’ parameters. In brief, the RNAsik pipeline used STAR (Dobin et al., 2013) to align the raw fastq files to the reference genome and featureCounts (Liao et al., 2014) to count high quality aligned reads to annotated genes. The resulting count matrix was then analyzed for differential gene expression.
Data quality control, filtering and differential expression (DE) analysis was performed using DE-Browser Shiny user interface (v1.21.1) (Kucukural et al., 2019) in R Studio (v4.1.1). Low count filtering parameters were set to CPM <3 in all 14 samples and normalised using Trimmed Mean of M-values (TMM). Batch effect correction was set to “none”. Relative log expression was plotted using the complete dataset with default settings. Principal component analysis was performed on viral transcripts only with DE-Browser default settings. DE analysis was performed between the 5 WT replicates and 3 replicates of each vPIC mutant sample with DE method: “Limma”, Normalization: “TMM”, Fit Type: “Is” and Norm. Bet. Arrays: “none” set. The output from each comparison was exported to Excel (Data S1) and mean log2 fold changes of all viral genes were consolidated for heatmap analysis. Fold change heatmaps were constructed using ComplexHeatmap (v2.9.4) (Gu et al., 2016) with default parameters. “cluster columns” was set to FALSE and “row split” set to 3 or 4, depending on the heatmap. Kinetic class and virion cargo data were matched to gene IDs in Excel and used to construct the heatmap annotations. Volcano plots were constructed using EnhancedVolcano (v 1.11.5) with log2 FC and adjusted p values (Data S1). “FCcutoff” was set to 0.5 and “pCutoff” to 0.001.
Quantification and statistical analysis
Statistical analysis and data plots were generated using Graphpad Prism 9, DE-Browser Shiny user interface (v1.21.1) (Kucukural et al., 2019) and R Studio (v4.1.1). For virology experiments, data are presented as the mean with SD of three replicates, with all relevant experimental parameters outlined in the figure legends. For RNA-seq analysis, data filtering, transformation, PCA and differential expression analysis was performed using DE-Browser with inbuilt parameters as outlined in the experimental methods. Adjusted p values and fold changes presented in the volcano plots (Figures S2A–S2C) were taken directly from the DE-Browser session output and available in Data S1. Log2 FC > 0.5 and p < 0.001 was considered significant. The mean log2 expression change for each mutant compared to WT for transcripts in clusters IIa and IIb were analyzed using one-way ANOVA with Tukey’s post-test, p < 0.05 was considered significant.
Acknowledgments
We would also like to thank the Monash Micro Imaging Plat-form, the Monash Genome Modification Plat-form, the Monash Bioinformatics Plat-form, and Micromon Genomics. D.L.T. was supported by an Australian Government Research Training Program (RTP) Stipend, RTP Fee-Offset Scholarship, and Postgraduate Publications Award through Monash University. This work was supported by the National Health & Medical Research Council of Australia (# APP1100737 to R.A.M.) and the Australian Research Council (# DP170102020 to S.J.T.). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the article.
Author contributions
R.A.M. conceived the project; R.A.M. and D.L.T. oversaw and directed experiments; D.L.T., S.F., and S.S. designed and completed molecular biology and virology experiments; D.L.T., A.A.B., B.E.R., and S.J.T. designed and performed RNA-seq analysis; and D.L.T. and R.A.M. wrote the article with input from all authors.
Declaration of interests
The authors have declared that no competing interests exist.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105168.
Supplemental information
Data and code availability
-
•
The complete RNA-seq dataset is publicly available in the Gene Expression Omnibus (GEO) with identifier #GSE201208.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- Ahlqvist J., Mocarski E. Cytomegalovirus UL103 controls virion and dense body egress. J. Virol. 2011;85:5125–5135. doi: 10.1128/JVI.01682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwine J.C. The human cytomegalovirus assembly compartment: a masterpiece of viral manipulation of cellular processes that facilitates assembly and egress. PLoS Pathog. 2012;8:e1002878. doi: 10.1371/journal.ppat.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders D.G., Kerry J.A., Pari G.S. In: Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Arvin A., Campadelli-Fiume G., Mocarski E., Moore P.S., Roizman B., Whitley R., Yamanishi K., editors. Cambridge University Press; 2007. DNA synthesis and late viral gene expression. [PubMed] [Google Scholar]
- Aubry V., Mure F., Mariamé B., Deschamps T., Wyrwicz L.S., Manet E., Gruffat H. Epstein-Barr virus late gene transcription depends on the assembly of a virus-specific preinitiation complex. J. Virol. 2014;88:12825–12838. doi: 10.1128/JVI.02139-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battista M.C., Bergamini G., Boccuni M.C., Campanini F., Ripalti A., Landini M.P. Expression and characterization of a novel structural protein of human cytomegalovirus, pUL25. J. Virol. 1999;73:3800–3809. doi: 10.1128/JVI.73.5.3800-3809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonalumi S., Trapanese A., Santamaria A., D'Emidio L., Mobili L. Cytomegalovirus infection in pregnancy: review of the literature. J. Prenat. Med. 2011;5:1–8. [PMC free article] [PubMed] [Google Scholar]
- Britt W.J. Human cytomegalovirus: propagation, quantification, and storage. Curr. Protoc. Microbiol. 2010 doi: 10.1002/9780471729259.mc14e03s18. Chapter 14, Unit 14E.13. [DOI] [PubMed] [Google Scholar]
- Cannon M.J., Schmid D.S., Hyde T.B. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev. Med. Virol. 2010;20:202–213. doi: 10.1002/rmv.655. [DOI] [PubMed] [Google Scholar]
- Caragliano E., Bonazza S., Frascaroli G., Tang J., Soh T.K., Grünewald K., Bosse J.B., Brune W. Human cytomegalovirus forms phase-separated compartments at viral genomes to facilitate viral replication. Cell Rep. 2022;38:110469. doi: 10.1016/j.celrep.2022.110469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda A.F., Didychuk A.L., Louder R.K., McCollum C.O., Davis Z.H., Nogales E., Glaunsinger B.A. The gammaherpesviral TATA-box-binding protein directly interacts with the CTD of host RNA Pol II to direct late gene transcription. PLoS Pathog. 2020;16:e1008843. doi: 10.1371/journal.ppat.1008843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Ortiz D.A., Gurczynski S.J., Khan F., Pellett P.E. Identification of human cytomegalovirus genes important for biogenesis of the cytoplasmic virion assembly complex. J. Virol. 2014;88:9086–9099. doi: 10.1128/jvi.01141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis Z.H., Verschueren E., Jang G.M., Kleffman K., Johnson J.R., Park J., Von Dollen J., Maher M.C., Johnson T., Newton W., et al. Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol. Cell. 2015;57:349–360. doi: 10.1016/j.molcel.2014.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didychuk A.L., Castañeda A.F., Kushnir L.O., Huang C.J., Glaunsinger B.A. Conserved Cx(n)C motifs in Kaposi's sarcoma-associated herpesvirus ORF66 are required for viral late gene expression and are essential for its interaction with ORF34. J. Virol. 2020;94:012999–e1319. doi: 10.1128/jvi.01299-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W., Chou C., Li H., Hai R., Patterson D., Stolc V., Zhu H., Liu F. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA. 2003;100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrum F., Caviness K., Zagallo P. Human cytomegalovirus persistence. Cell Microbiol. 2012;14:644–655. doi: 10.1111/j.1462-5822.2012.01774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruffat H., Kadjouf F., Mariamé B., Manet E. The Epstein-Barr virus BcRF1 gene product is a TBP-like protein with an essential role in late gene expression. J. Virol. 2012;86:6023–6032. doi: 10.1128/JVI.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruffat H., Marchione R., Manet E. Herpesvirus late gene expression: a viral-specific pre-initiation complex is key. Front. Microbiol. 2016;7:869. doi: 10.3389/fmicb.2016.00869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Gupta K., Sari-Ak D., Haffke M., Trowitzsch S., Berger I. Zooming in on transcription preinitiation. J. Mol. Biol. 2016;428:2581–2591. doi: 10.1016/j.jmb.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isomura H., Stinski M.F., Murata T., Yamashita Y., Kanda T., Toyokuni S., Tsurumi T. The human cytomegalovirus gene products essential for late viral gene expression assemble into prereplication complexes before viral DNA replication. J. Virol. 2011;85:6629–6644. doi: 10.1128/JVI.00384-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalejta R.F. Tegument proteins of human cytomegalovirus. Microbiol. Mol. Biol. Rev. 2008;72:249–265. doi: 10.1128/MMBR.00040-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukural A., Yukselen O., Ozata D.M., Moore M.J., Garber M. DEBrowser: interactive differential expression analysis and visualization tool for count data. BMC Genom. 2019;20:6. doi: 10.1186/s12864-018-5362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Cruz L., Sandhu P.K., Buchkovich N.J. UL88 mediates the incorporation of a subset of proteins into the virion tegument. J. Virol. 2020;94:e00474-20–e00420. doi: 10.1128/JVI.00474-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K.M., Oh S.E., Kim Y.E., Han T.-H., Ahn J.-H. Cooperative inhibition of RIP1-mediated NF-κB signaling by cytomegalovirus-encoded deubiquitinase and inactive homolog of cellular ribonucleotide reductase large subunit. PLoS Pathog. 2017;13:e1006423. doi: 10.1371/journal.ppat.1006423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Ball C.B., Collins G., Hu Q., Luse D.S., Price D.H., Meier J.L. Human cytomegalovirus IE2 drives transcription initiation from a select subset of late infection viral promoters by host RNA polymerase II. PLoS Pathog. 2020;16:e1008402. doi: 10.1371/journal.ppat.1008402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Hu Q., Collins G., Parida M., Ball C.B., Price D.H., Meier J.L. Cytomegalovirus late transcription factor target sequence diversity orchestrates viral early to late transcription. PLoS Pathog. 2021;17:e1009796. doi: 10.1371/journal.ppat.1009796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Manicklal S., Emery V.C., Lazzarotto T., Boppana S.B., Gupta R.K. The "silent" global burden of congenital cytomegalovirus. Clin. Microbiol. Rev. 2013;26:86–102. doi: 10.1128/cmr.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers C., Schafer X., Martínez-Sobrido L., Munger J. The human cytomegalovirus UL26 protein antagonizes NF-κB activation. J. Virol. 2014;88:14289–14300. doi: 10.1128/JVI.02552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C., Alain S., Baumert T.F., Ligat G., Hantz S. Structures and divergent mechanisms in capsid maturation and stabilization following genome packaging of human cytomegalovirus and herpesviruses. Life. 2021;11:150. doi: 10.3390/life11020150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J., Yu D., Shenk T. UL26-deficient human cytomegalovirus produces virions with hypophosphorylated pp28 tegument protein that is unstable within newly infected cells. J. Virol. 2006;80:3541–3548. doi: 10.1128/JVI.80.7.3541-3548.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Shenk T. Human cytomegalovirus genome. Curr. Top. Microbiol. Immunol. 2008;325:1–19. doi: 10.1007/978-3-540-77349-8_1. [DOI] [PubMed] [Google Scholar]
- Nguyen C.C., Kamil J.P. Pathogen at the Gates: human cytomegalovirus entry and cell tropism. Viruses. 2018;10 doi: 10.3390/v10120704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobre L.V., Nightingale K., Ravenhill B.J., Antrobus R., Soday L., Nichols J., Davies J.A., Seirafian S., Wang E.C., Davison A.J., et al. Human cytomegalovirus interactome analysis identifies degradation hubs, domain associations and viral protein functions. Elife. 2019;8:e49894. doi: 10.7554/eLife.49894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak B., Sullivan C., Sarnow P., Thomas R., Bricout F., Nicolas J.C., Fleckenstein B., Levine A.J. Characterization of monoclonal antibodies and polyclonal immune sera directed against human cytomegalovirus virion proteins. Virology. 1984;132:325–338. doi: 10.1016/0042-6822(84)90039-4. [DOI] [PubMed] [Google Scholar]
- Omoto S., Mocarski E.S. Cytomegalovirus UL91 is essential for transcription of viral true late (γ2) genes. J. Virol. 2013;87:8651–8664. doi: 10.1128/JVI.01052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto S., Mocarski E.S. Transcription of true late (γ2) cytomegalovirus genes requires UL92 function that is conserved among beta- and gammaherpesviruses. J. Virol. 2014;88:120–130. doi: 10.1128/JVI.02983-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng Y.C., Campbell J.A., Lenschow D.J., Yu D. Human cytomegalovirus pUL79 is an elongation factor of RNA polymerase II for viral gene transcription. PLoS Pathog. 2014;10:e1004350. doi: 10.1371/journal.ppat.1004350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan P., Razonable R.R. Cytomegalovirus infections in solid organ transplantation: a review. Infect. Chemother. 2013;45:260–271. doi: 10.3947/ic.2013.45.3.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozman B., Nachshon A., Levi Samia R., Lavi M., Schwartz M., Stern-Ginossar N. Temporal dynamics of HCMV gene expression in lytic and latent infections. Cell Rep. 2022;39:110653. doi: 10.1016/j.celrep.2022.110653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V., Britt W. Human cytomegalovirus egress: overcoming barriers and co-opting cellular functions. Viruses. 2021;14 doi: 10.3390/v14010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V., Sztul E., Britt W.J. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-golgi-intermediate compartment. J. Virol. 2000;74:3842–3851. doi: 10.1128/jvi.74.8.3842-3851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J.-Y., Britt W.J. Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J. Virol. 2006;80:5611–5626. doi: 10.1128/JVI.02630-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijmons S., Van Ranst M., Maes P. Genomic and functional characteristics of human cytomegalovirus revealed by next-generation sequencing. Viruses. 2014;6:1049–1072. doi: 10.3390/v6031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M.C., Yu Q.-C., Enquist L., Shenk T. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 2003;77:10594–10605. doi: 10.1128/jvi.77.19.10594-10605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector B.M., Parida M., Li M., Ball C.B., Meier J.L., Luse D.S., Price D.H. Differences in RNA polymerase II complexes and their interactions with surrounding chromatin on human and cytomegalovirus genomes. Nat. Commun. 2022;13 doi: 10.1038/s41467-022-29739-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton R.J., Baluchova K., Dargan D.J., Cunningham C., Sheehy O., Seirafian S., McSharry B.P., Neale M.L., Davies J.A., Tomasec P., et al. Reconstruction of the complete human cytomegalovirus genome in a BAC reveals RL13 to be a potent inhibitor of replication. J. Clin. Invest. 2010;120:3191–3208. doi: 10.1172/JCI42955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsyganov K., James Perry A., Kenneth Archer S., Powell D. RNAsik: a Pipeline for complete and reproducible RNA-seq analysis that runs anywhere with speed and ease. J. Open Source Softw. 2018;3:583. doi: 10.21105/joss.00583. [DOI] [Google Scholar]
- Turner D.L., Korneev D.V., Purdy J.G., de Marco A., Mathias R.A. The host exosome pathway underpins biogenesis of the human cytomegalovirus virion. Elife. 2020;9:e58288. doi: 10.7554/eLife.58288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D.L., Templin R.M., Barugahare A.A., Russ B.E., Turner S.J., Ramm G., Mathias R.A. UL34 deletion restricts human cytomegalovirus capsid formation and maturation. Int. J. Mol. Sci. 2022;23:5773. doi: 10.3390/ijms23105773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Loveland A.N., Kattenhorn L.M., Ploegh H.L., Gibson W. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 2006;80:6003–6012. doi: 10.1128/JVI.00401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li Y., Zhao G., Wu Y., Zhang X., Li H., Zhou T. Inhibition of human cytomegalovirus DNA replication by small interfering RNAs targeted to UL49. Acta Biochim. Biophys. Sin. 2013;45:401–407. doi: 10.1093/abbs/gmt025. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Nishimura M., Izumi T., Kuriyama K., Iwaisako Y., Hosokawa K., Takaori-Kondo A., Fujimuro M. Kaposi's sarcoma-associated herpesvirus ORF66 is essential for late gene expression and virus production via interaction with ORF34. J. Virol. 2020;94:e01300–e01319. doi: 10.1128/jvi.01300-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekes M.P., Tomasec P., Huttlin E.L., Fielding C.A., Nusinow D., Stanton R.J., Wang E.C.Y., Aicheler R., Murrell I., Wilkinson G.W.G., et al. Quantitative temporal viromics: an approach to investigate host-pathogen interaction. Cell. 2014;157:1460–1472. doi: 10.1016/j.cell.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrwicz L.S., Rychlewski L. Identification of herpes TATT-binding protein. Antiviral Res. 2007;75:167–172. doi: 10.1016/j.antiviral.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Yang G., Li W., Liao W., Zhang X., Zou Y., Dai J., Li Y., Jing C., Zhou T. Identification of the alternative splicing of the UL49 locus of human cytomegalovirus. BioMed Res. Int. 2015;2015:280276. doi: 10.1155/2015/280276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Silva M.C., Shenk T. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA. 2003;100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Smith G.A., Enquist L.W., Shenk T. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 2002;76:2316–2328. doi: 10.1128/jvi.76.5.2316-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Jih J., Jiang J., Zhou Z.H. Atomic structure of the human cytomegalovirus capsid with its securing tegument layer of pp150. Science. 2017;356:eaam6892. doi: 10.1126/science.aam6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Li H., Li Y., Zeng Z., Li S., Zhang X., Zou Y., Zhou T. Effective inhibition of HCMV UL49 gene expression and viral replication by oligonucleotide external guide sequences and RNase P. Virol. J. 2010;7:100. doi: 10.1186/1743-422X-7-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., Yuan J., Li H.J., Zeng Z.F., Luo Z.W., Li S.Q., He C.Q., Jia X.F., Zhang X., Zuo H., et al. Human cytomegalovirus UL49 encodes an early, virion-associated protein essential for virus growth in human foreskin fibroblasts. Arch. Virol. 2016;161:1273–1284. doi: 10.1007/s00705-016-2780-4. [DOI] [PubMed] [Google Scholar]
- Zhu H., Shen Y., Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 1995;69:7960–7970. doi: 10.1128/JVI.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The complete RNA-seq dataset is publicly available in the Gene Expression Omnibus (GEO) with identifier #GSE201208.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.