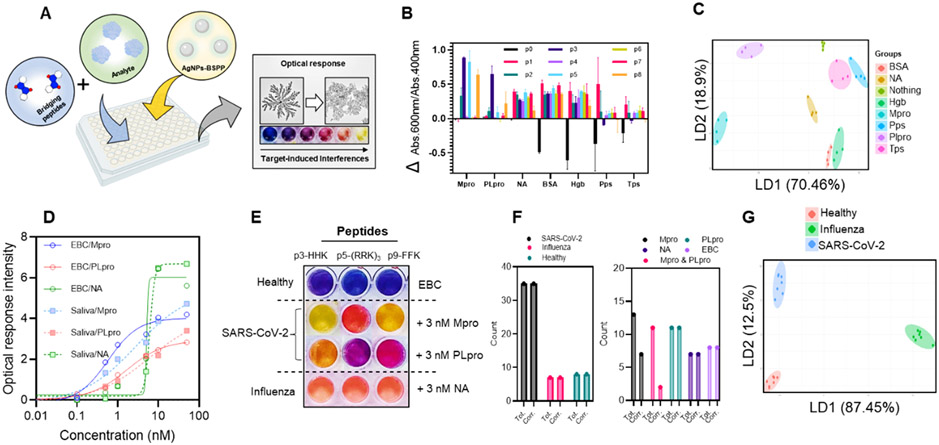
Abstract
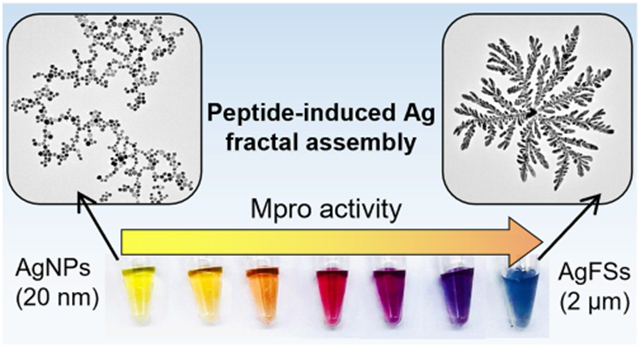
We report the peptide-programmed fractal assembly of silver nanoparticles (AgNPs) in a diffusion-limited aggregation (DLA) mode, and this change in morphology generates a significant color change. We show that peptides with specific repetitions of defined amino acids (i.e., arginine, histidine, or phenylalanine) can induce assembly and coalescence of the AgNPs (20 nm) into a hyperbranched structure (AgFSs) (~2 μm). The dynamic process of this assembly was systematically investigated, and the extinction of the nanostructures can be modulated from 400 to 600 nm by varying the peptide sequences and molar ratio. According to this rationale, two strategies of SARS-CoV-2 detection were investigated. The activity of the main protease (Mpro) involved in SARS-CoV-2 was validated with a peptide substrate that can bridge the AgNPs after the proteolytic cleavage. A subnanomolar limit of detection (0.5 nM) and the capacity to distinguish by the naked eye in a wide concentration range (1.25–30 nM) were achieved. Next, a multichannel sensor-array based on multiplex peptides that can visually distinguish SARS-CoV-2 proteases from influenza proteases in doped human samples was investigated.
Keywords: coalescence, colorimetric sensing, fractal structures, peptides, sensor-array silver nanomaterials
Graphical Abstract
Colorimetric biosensors have attracted increasing attention for the in vitro detection of various disease biomarkers. The use of simple spectrometry and/or naked eye detection1-5 makes them simpler and easier than traditional polymerase chain reaction (PCR) or enzyme-linked immunosorbent assay (ELISA) testing.6,7 Plasmonic metal nanostructures have been extensively investigated for this purpose8-10 because they possess efficient extinction coefficients and tunable localized surface plasmon resonance (LSPR) bands in the visible region depending on their size, morphology, or dielectric environment.11-14 Tuning the size and morphology of the nanoparticle via the analytes has been investigated by etching or growing a secondary material on the presynthesized nanoparticles to obtain a well-defined nanostructure and hence color change.15,16 However, these steps often use complicated redox chemistry during the sensing process that can interfere with the analyte.17
Tuning the interparticle distance of the individual nanoparticles in the ensemble in response to the analytes is another strategy.18 These systems often result in an irregular assembly (i.e., aggregation) with a relatively low sensitivity due to the broad LSPR and mono-colorimetric change.19,20 Ordered and programmable assembly can be obtained by modifying the nanoparticles with a recognition motif such as via antigen–antibody interactions21,22 and DNA pairing23-25 but requires specific surface ligands and elaborate assembling conditions.26,27 Biomarker-mediated assembly of nanoparticles into defined hierarchical microstructures with distinct optical properties remains relatively rare.
Here, we report the peptide-induced fractal assembly of silver nanoparticles (AgNPs) into hyperbranched structures via diffusion-limited aggregation (DLA). DLA is a theoretical model that describes the self-assembly of molecules driven by free energy minimization of the system mostly through diffusion.28-31 We identified peptides that could control the initiation and extent of DLA as a function of biomarker concentration. Surprisingly, the particles undergo coalescence during the assembly, which leads to the change of morphology of the material. We characterized this phenomenon with spectroscopy and microscopy and control peptides. Finally, the peptide-induced assembly of AgNPs into well-defined nanostructures—without complicated ligands or assembly conditions—could be used for colorimetric biosensors. We show two examples applied to the detection of SARS-CoV-2 virus: (i) the detection of the enzymatic activity of the main protease (Mpro) of SARS-CoV-2 and (ii) a multichannel sensor-array for the distinction between SARS-CoV-2- and influenza-infected external breath condensate (EBC) or saliva samples.
RESULTS AND DISCUSSION
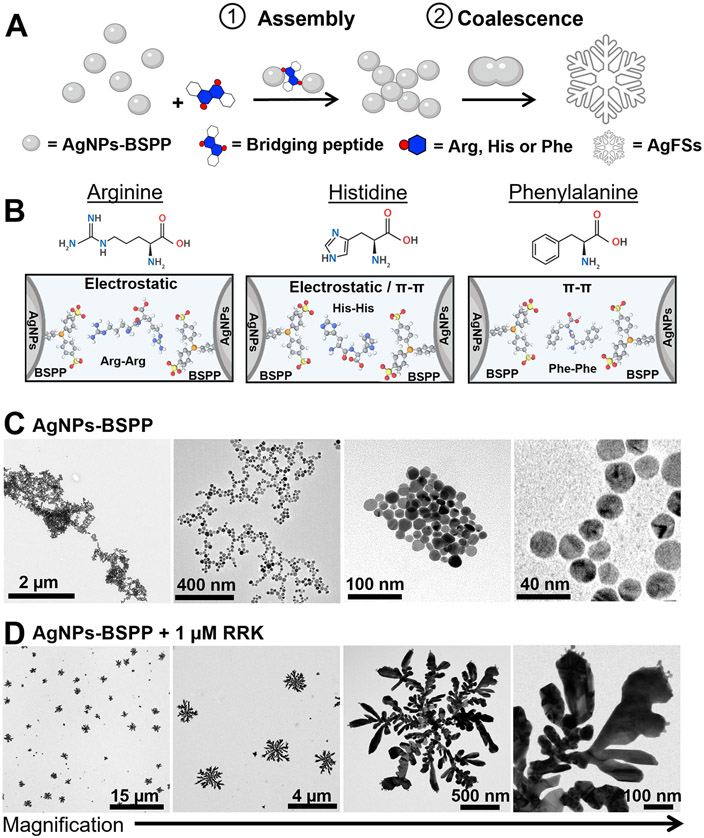
We first designed “bridging” peptides that contain at least two repetitions of amino acids (i.e., arginine, histidine, or phenylalanine) that can interact with the nanoparticles via electrostatic or hydrophobic interactions (Figure 1A and B). The peptides easily induce a highly structured assembly of AgNPs via DLA followed by a change in the particle morphology, thus producing hyperbranched fractal Ag nanostructures (AgFSs) (Figure 1C and D). Fractal structures are particularly interesting in optics or diagnostics because they express many nanometer-scale spatial regions of high local electric fields with a significantly enhanced absorption and scatter.32,33 They are usually obtained either by the chemical reduction34 or electrochemical deposition35 of a metal salt or by physical treatments of presynthesized nanoparticles such as solvent evaporation36 or microwave exposure.37
Figure 1.
Fractal assembly of AgNPs and change of morphology. (A) Illustration of the peptide-induced assembly of the AgNPs-BSPP and their coalescence into AgFSs. (B) Example of amino acids that can be used to bridge the AgNPs-BSPP via electrostatic or hydrophobic interactions (pi–pi) with BSPP. TEM images of AgNPs-BSPP (C) before and (D) after the addition of 1 μM of the RRK peptide showing the change of morphology.
Characterization of the Fractal Assembly.
AgNPs with a core size of approximately 20 nm and coated with bis(p-sulfonatophenyl)phenylphosphine (BSPP) were synthesized via a seed-mediated growth procedure.38 BSPP was chosen for the phosphine–silver coordination and its negative charge and aromaticity. Detailed synthesis procedure and characterization of the AgNPs-BSPP can be found in the Supporting Information (Figures S1 to S7). We first demonstrated the DLA-mediated assembly by using a simple peptide sequence (i.e., RRK). Our assumption was that the two arginines could act like a bridge between the nanoparticles due to electrostatic interactions with the BSPP (Figure 1B). Transmission electronic microscopy (TEM) was used to confirm that the peptide RRK can be used to induce the fractal assembly of the AgNPs-BSPP. Figure 1C and D show the TEM images of AgNPs-BSPP (0.3 nM) before and after the addition of 1 μM RRK. This concentration was chosen because it corresponds to a density of 2.6 RRK/nm2, which is approximately twice the maximal density of peptide that could be theoretically carried (1.3/nm2). Fractal structures with a diameter of approximately 2 μm, primary branches of 1 μm, and secondary branches between 100 and 200 nm were observed (Figure 1D).
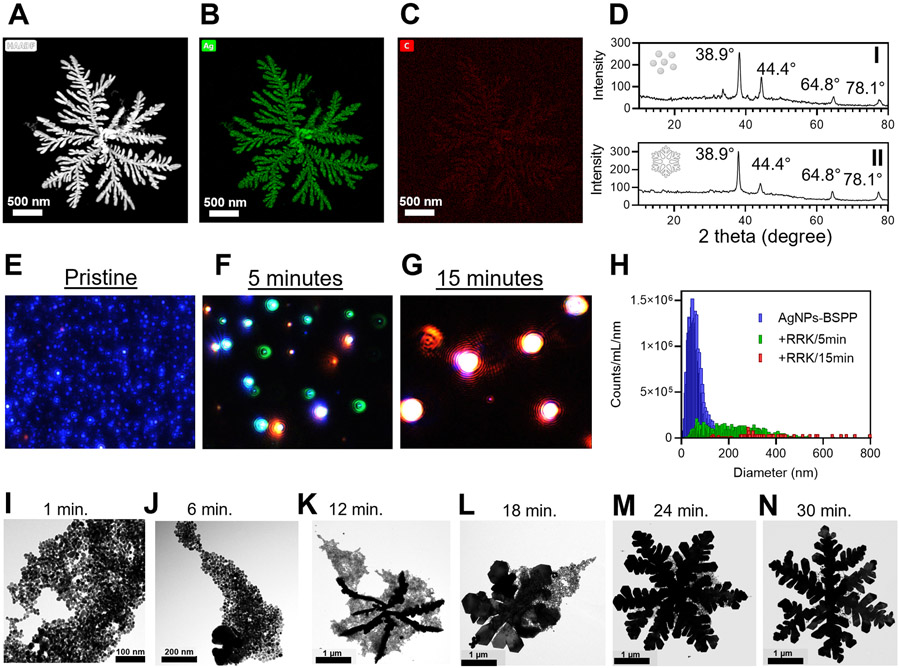
Energy-dispersive X-ray spectroscopy (EDX) revealed that the fractal structures were mostly silver (Figure 2A and B) with trace carbon from RRK (Figure 2C) and trace phosphorus and sulfur from BSPP (Figure S9e and f). X-ray diffraction (XRD) analysis revealed a face-centered cubic (FCC) crystal structure for the AgNPs-BSPP as peaks at 38.9°, 44.4°, 64.8°, and 78.1° corresponding to Ag(1,1,1), Ag(2,0,0), Ag(2,2,0), and Ag(3,1,1), respectively. The FCC crystal structure was maintained during the peptide-induced morphological change; that is, very similar XRD patterns were observed for the AgNPs-BSPP that reacted with 1 μM RRK (Figure 2D).
Figure 2.
Assembly characterization. (A, B, and C) EDX elemental mapping of AgNPs-BSPP after the reaction with 1 μM RRK showing the (B) silver and (C) carbon composition of the material. (D) Crystal structure of AgNPs-BSPP (I) before and (II) 30 min after the reaction with 1 μM RRK obtained by XRD. (E, F, G, and H) Multispectral advanced nanoparticle tracking analysis (MANTA) images of the AgNPs- BSPP (E) before or (F) 5 min and (G) 15 min after the addition of 1 μM RRK showing the size evolution and (H) the corresponding size distribution. (I, J, K, L, M, and N) TEM images showing the evolution of the assembly of AgNPs-BSPP upon the addition of 1 μM RRK at 4°c
The dynamic evolution of the assembly was evaluated with multispectral advanced nanoparticle tracking analysis (MANTA).39 MANTA measures scattering to size and count nanoparticles via three lasers with different wavelengths (e.g., blue, green, and red); the wavelength of scatter depends on nanoparticle size. For example, large nanoparticles (>300 nm) scatter more in green and red, while small nanoparticles (<200 nm) scatter in blue. Therefore, pristine 35 nm AgNPs-BSPP appeared blue (Figure 2E). After 5 min of interaction with 1 μM peptide RRK, green and red light scattering were observed because of the increased size of AgNPs-BSPP (Figure 2F). Finally, the formation of the large fractal structures caused red scattering (Figure 2G). Importantly, less than 10% of the initial AgNPs-BSPP remained after 5 min: the number of particles decreasing from ~1.5 × 1021 (pristine) to ~1.5 × 1020 (5 min). After 15 min, there were no remaining AgNPs-BSPP particles. Only particles with a mode size of several hundred nanometers could be detected (Figure 2H).
Similarly, the evolution of the RRK-induced assembly was monitored by TEM. Briefly, 1 μM RRK was added to the AgNPs-BSPP at 4 °C (to slow down the reaction), and TEM images were recorded over time. The temperature has a strong effect on the kinetics of the reaction per DLA (Figure S10).40 Only aggregated particles were observed at early stages (~1 min) (Figure 2I), but merging particles and larger structures were observed after a few minutes (~6 min) (Figure 2J). Fractal structures could be identified at later time points (~12 min) with the primary branches appearing (Figure 2K) and attracting the remaining spherical particles. Over time (from ~18 to ~30 min), the spherical particles merged with the fractal structures, thus increasing the fractal dimension with secondary branch formation (Figure 2L, M, and N). At longer time points, AgNPs-BSPP were no longer seen, in good agreement with MANTA and TEM (Figures S11 to S15).
Inductively coupled plasma (ICP) was used to highlight the role of redox chemistry in the formation of the AgFSs (Figure S16). The concentrations of silver ions for AgNPs-BSPP before and after the reaction with 1 μM RRK were compared. The total concentrations of silver ions were identical. The concentrations of silver ions in the supernatant were also very similar (less than 10% difference), suggesting that no or few free silver ions were used to build the AgFSs; that is, the AgFSs were built from AgNPs. AgNPs-BSPP purified from free silver ions were even more sensitive to AgFS formation upon addition of RRK (Figure S16).
TEM analysis of the early stages of the assembly revealed many cases of particles merging together to form larger particles (Figure S17). The mechanism underlying the morphology change is believed to be the coalescence of the particles induced by the interfacial dissolution. To reduce the total surface area, the particles may coalesce to compensate for the loss of Ag atoms during the assembly; this is probably induced by complexation of the silver atoms with BSPP.41 BSPP likely induces release of silver ions into the solution during the assembly, thus leading to the fusion of the particles. This agrees well with Magdassi et al., who showed that AgNPs can behave as “soft” particles and undergo spontaneous coalescence in solution at room temperature after the modification of their surface properties with charge neutralization and desorption of surface-stabilizing ligands.42,43 It is particularly interesting as usually the shape transformation relies on light, heat, or excess reducing agent.44
The surface chemistry of the particles was investigated to further demonstrate the role of surface ligands in nanoparticle aggregation. AgNPs coated with thiolated poly(ethylene)glycol (HS-PEG40-COOH, Mw = 2000) ending by a carboxyl group were synthesized similar to the approach used for AgNPs-BSPP. AgNPs-S-PEG-COOH were insensitive to the addition of RRK even at high concentrations (>100 μM) (Figure S18B and C). The results show that the dense PEG layer, strongly anchored to the surface via a Ag─S bond, prevents any kind of assembly or coalescence of the AgNPs. AgNPs coated with other non-PEG thiolated ligands (i.e., mercaptobenzoic acid, MBA) were only weakly sensitive to RRK addition, thus confirming that a robust organic layer coating the particles protects them from coalescence (Figure S18).
Citrate-capped AgNPs (AgNPs-citrate) were also investigated as a control of “surface-accessible” AgNPs. AgNPs-citrate are probably the most commonly used AgNPs and are surrounded by citrate anions physiosorbed onto the surface. Addition of 100 μM RRK could induce assembly of AgNPs-citrate, and dense aggregates were observed (Figure S18). However, there was no change in the morphology, and fractal silver structures were not observed. Citrate anions likely remain adsorbed onto the surface during the assembly, and no silver atoms are removed from the surface unlike BSPP. This agrees well with Park et al., who showed that the strong adsorption of citrate anions on metal nanoparticles leads to a stabilized network that is extremely challenging to displace.45 Tan et al. have shown that BSPP can chelate silver ions, forming soluble BSPP-Ag+ complexes, resulting even in the dissolution of silver cores.46 Thus, we believe that the BSPP coating, unlike citrate, can desorb from the AgNPs during their peptide-mediated assembly, allowing the particles to undergo spontaneous coalescence after releasing Ag+ into the solution.
For comparison, gold nanoparticles coated with BSPP (AuNPs-BSPP) were also investigated. AuNPs-BSPP were produced from a ligand-exchange procedure starting from citrate-capped AuNPs (see Supporting Information Figure S19 for detailed synthesis). While the fractal assembly of AuNPs-BSPP was observed upon the addition of RRK, no change in morphology could be identified (Figure S19A). In the literature, the coalescence of bare gold nanocrystals in aqueous solution has been reported after formation of nanobridges that induce contact between different crystals and thus the rearrangement into a single crystal.47 This process is attributed to an accumulation of gold ions at the interface to promote the fusion of the gold nanocrystals. The BSPP coating is thus believed to protect the gold surface from ionic release: It cannot solubilize gold ions from the surface, and no coalescence is observed. In contrast, the BSPP coating on AgNPs helps to release the silver ions into solution. BSPP can efficiently solubilize the silver, thus facilitating particle fusion.
We note that AgNPs synthesized in the absence of BSPP (AgNPs-Bare), probably coated by only ascorbate, were also investigated: They were expected to be capable of coalescence as well. However, these particles were not sufficiently stable to be conveniently manipulated and were thus not suitable for use as a sensing reporter, as similar colorimetric signal was generated in the absence or presence of the target protein due to noncontrolled aggregation of the particles (Figure S18D and E). AgNPs-BSPP are thus believed to be a compromise between stability and tunability, thus leading to coalescence.
Control of the Assembly with Peptides.
The RRK-induced assembly of the AgNPs-BSPP was compared to other stimuli-induced aggregation such as in the presence of high ionic strength, pH variations, or halide addition. All of these conditions led to the aggregation/dissolution of the AgNPs-BSPP as their LSPR band first deformed and then decreased; no absorbance could be measured (Figure S20). We suggest that the RRK peptide can gently interact with the AgNPs-BSPP and act as a bridge between the particles, thus forcing them to assemble.
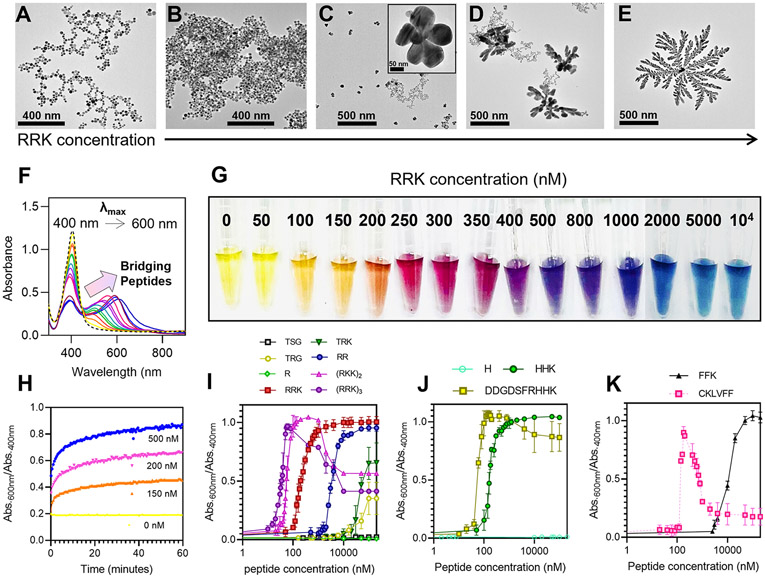
We further investigated the effect of the peptide concentration on the assembly: More peptide should induce more particle bridging. Different concentrations of the peptide RRK were added to the AgNPs-BSPP, and the assembly was characterized by TEM. The intensity of the assembly (i.e., the number of AgNPs that assemble as monitored by the change in absorbance) was proportional to the amount of peptide added. While a low amount of RRK (e.g., 100 nM) induced the aggregation of the particles (Figure 3B), a higher amount (e.g., 250 nM) induced the formation of ~200 nm flower-like structure (Figure 3C). At 400 nM, 500 nm dendritic structures were identified (Figure 3D), and large fractal structures over 1 μm appeared at 1 μM (Figure 3E). Importantly, the intensity of the assembly is not driven by the absolute concentration of peptides but the molar ratio of peptides per particles. Indeed, a linear relationship between the concentration of RRK needed for assembly and the concentration of the AgNPs-BSPP was obtained, thus confirming that the peptides bridge the particles (Figure S21). The intensity of the assembly has a direct impact on the optical properties of the AgNPs-BSPP.
Figure 3.
Peptide-mediated assembly. TEM images showing the different structures obtained for AgNPs-BSPP that reacted for 15 min with different concentrations of RRK: (A) 0 nM, (B) 150 nM, (C) 250 nM, (D) 400 nM, and (E) 1 μM. (F) Deformation of the UV–vis spectrum of AgNPs-BSPP 15 min after the addition of different concentrations of RRK from 0 nM to 10 μM (the arrow indicates the increase of peptides added) and (G) the corresponding pictures. (H) Evolution of the assembly intensity (Abs.600 nm/Abs.400 nm) over time for different concentrations of RRK added to AgNPs-BSPP. Evolution of the assembly intensity as a function of the peptide concentration for (I) arginine-based, (J) histidine-based, or (K) phenylalanine-based peptides.
The LSPR band of AuNPs-BSPP deforms strongly upon the addition of RRK with decreased 400 nm absorbance and an increase at 600 nm (Figure 3F). The deformation was proportional to the amount of peptide added and agrees well with TEM: The addition of more peptides led to a decrease in spherical AgNPs-BSPP (Abs. at 400 nm) and larger structures (Abs. at 600 nm). The morphology changes led to a wide range of colors (Figure 3G). The kinetics of the fractal assembly induced by the RRK sequence was studied by monitoring its intensity (Abs.600 nm/Abs.400 nm) over time by UV–vis spectroscopy (Figure 3H). After a few minutes (<3 min), the ratio Abs.600 nm/Abs.400 nm was already at 80% of its maximal value; the plateau was reached after approximately 20 min. The kinetics did not depend on the concentration of RRK added: the plateau was reached after 20 min for various concentrations (150, 200, and 500 nM). Only the value of the Abs.600 nm/Abs.400 nm ratio varied with RRK concentration. The kinetics of the assembly of AgNPs-BSPP exposed to more RRK concentrations can be found in the Supporting Information, Figure S22.
The effect of the peptide sequence was studied next; we assumed that peptides that can have more or stronger interactions with the particles would have more effect on the assembly. To study this, we next focused on the fractal assembly of AgNPs-BSPP induced by arginine-based peptides containing different amounts of arginine in the sequence (Figure 3I). Our results showed that repeating the RRK sequence leads to stronger assembly proportional to the number of repeat RRK units ((RRK)3 > (RRK)2 > RRK) probably because (RRK)3 offers more electrostatic interactions with AgNPs-BSPP than (RRK)2 or RRK. The decrease in the intensity of the assembly at high concentrations of (RRK)3 or (RRK)2 could also be explained by the repulsion forces between the initial aggregate that became too positively charged or by the saturation of the surface that occurs earlier with (RRK)2 and (RRK)3 than RRK. The C-terminal lysine could increase the affinity of the peptides for the particles; the RRK sequence has a stronger effect than only RR.
Arginine was also important to the assembly: If one arginine is replaced by threonine, the resulting TRK could induce only a minor assembly at high concentration (~100 μM). The necessity of repetition of two arginines was further confirmed by using peptides containing single or no arginine (TSG, R, TRG). The results suggest that only the side chain of arginine interacts significantly with the particles rather than the terminal amines. At least 2-fold repetition of arginine is then needed. For example, a bis-amine C12 alkane could not bridge the particles (see Figure S23). The confirmation of electrostatic interactions was shown by adding the RRK peptide to the AgNPs-BSPP at different pH values. The intensity of the assembly decreases as the pH becomes closer to the pKa of the arginine side chain (~12) (Figure S24).
BSPP is interesting because it can be involved in hydrophobic interactions in addition to electrostatic interactions. Other amino acids were investigated to trigger the assembly of the AgNPs-BSPP such as histidine- or phenylalanine-based peptides. Similarly, the effect of HHK was studied and was observed to be almost identical to RRK (Figure 3J). We thus hypothesized that a histidine-based peptide can interact with the particles via a combination of electrostatic and hydrophobic interactions. To test this hypothesis, we synthesized DDGDSFRHHK, which has no net positive charge (mostly zwitterionic) but still has a strong effect on the assembly. Histidine alone did not induce any assembly (Figure 3J). FFK induced strong assembly but at higher concentrations than RRK or HHK. These results indicate that the particles are less sensitive to hydrophobic interactions than electrostatic interactions or chelation (Figure 3K). However, increasing the hydrophobic nature of the peptide increases their effect on the particles. As an example, we show peptide CKLVFF, which has a higher affinity. The decrease in the intensity of the assembly at high concentrations is due to its hydrophobicity: This peptide can self-assemble in water and thus is unavailable to interact with the particles.48 The UV–vis spectra and pictures from the titration of all the peptides to the AgNPs-BSPP suspension can be found in the Supporting Information, Figures S25 to S39.
A change in particle morphology was observed regardless of the type of interaction (Figure S40). Also, minor differences in the reaction rate were observed for all of the interactions presented here. There was no more evolution of the LSPR band after 20 min (Figure S41). The addition of histidine- or phenylalanine-based peptides was performed on citrate-capped AgNPs to demonstrate the versatility of BSPP. Unlike AgNPs-BSPP, AgNPs-citrate are insensitive to the addition of HHK or FFK even at high concentrations (>100 μM) perhaps because the citrate anions can only be involved in electrostatic interactions, and AgNPs-citrate are most sensitive to the RRK sequence (Figure S42).
The peptide–particle interaction described here has interesting practical value in colorimetric biosensing. Indeed, a wide range of colorimetric signal can be obtained due to the fractal assembly of the particles. The intensity of the assembly can be tuned by modulating the peptide sequences or the peptide concentrations. The peptides are promising recognition elements that are easy and cheap to synthesize with perfect control over their sequences. We then show how this peptide–particle relationship can be employed for the detection of SARS-CoV-2 virus via two different strategies.49 Detecting SARS proteases could have value in rapid diagnosis50 or discriminating it from influenza.51
Detection of the Enzymatic Activity of the Main Protease of SARS-CoV-2 Virus.
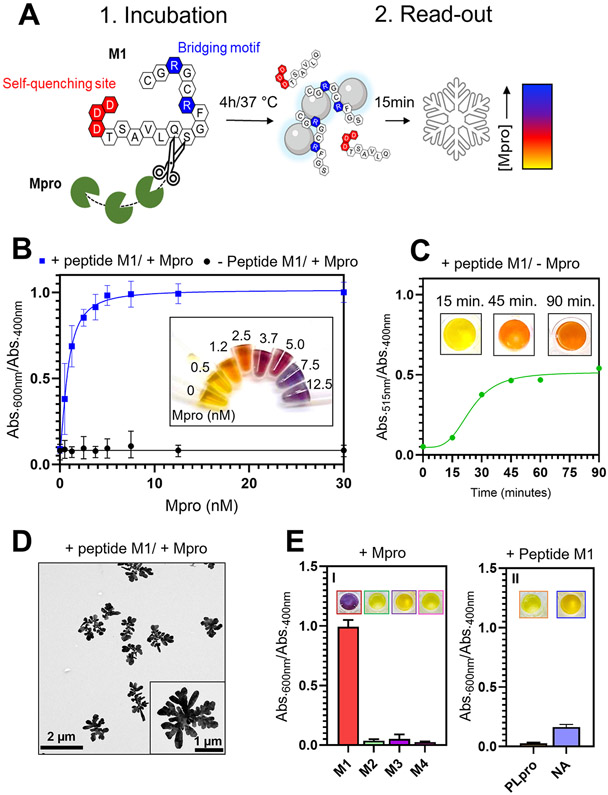
We first demonstrate the detection of the enzymatic activity of the Mpro produced by the SARS-CoV-2 virus. We designed peptide “M1” (Figure 4A) with a recognition sequence TSAVLQSGF where Mpro can cleave after the Q residue.52,53 This recognition sequence was conjugated with (i) a bridging motif (RCGRGC) and (ii) a negatively charged quenching site (DDD). The sequence had a net negative charge with negligible capability to induce the nanoparticle aggregation. In the presence of Mpro, M1 is cleaved and the fragment SGFRCGRGC is released, leading to the assembly of AgNPs-BSPP via the same bridging mechanism as described previously. The intensity of the assembly is proportional to the concentration of bridging peptides, and the optical change should be proportional to the initial concentration of Mpro (Figure 4A).
Figure 4.
Detection of the enzymatic activity of Mpro. (A) Illustration of the sensing mechanism with (1) the incubation of the peptide M1 and Mpro that after 4 h leads to the cleavage of M1 and (2) the read-out of Mpro activity with the AgNPs-BSPP that assemble due to the release of a “bridging fragment”. The optical responses are proportional to the concentration of Mpro. (B) Optical response (Abs.600 nm/Abs.400 nm) obtained 15 min after the addition of AgNPs-BSPP to Mpro samples that were incubated 4 h either with 5 μM M1 (blue) or in the absence of M1 (black). Inset shows the corresponding pictures. (C) Optical response (Abs.515 nm/Abs.400 nm) of the AgNPs-BSPP mixed with M1 in the absence of Mpro over time. Insets show pictures of the sample at 15, 45, and 90 min. (D) TEM images showing the reshaping of the AgNPs-BSPP when mixed with 12.5 nM Mpro incubated with M1. (E) Optical response at 15 min obtained for the detection of (I) Mpro incubated with either M1, M2, M3, or M4 or (II) PLpro and NA incubated with M1. Insets show the corresponding pictures.
To validate our proposed sensing mechanism, different concentrations of Mpro were incubated with M1 for 4 h followed by the addition of the AgNPs-BSPP as a read-out reporter. Figure 4B shows the optical responses (Abs.600 nm/Abs.400 nm) obtained 15 min after the addition of AgNPs-BSPP to all the samples. The signal intensity was proportional to the concentration of Mpro with a linear relationship between 0 nM and 2.5 nM and a limit of detection (LOD) of 500 pM (LOD by eye is ~1.2 nM). Mpro itself cannot induce assembly of the AgNPs-BSPP (Figure S43). The LOD we report in this Ag-based system is 1 order of magnitude lower than with Au nanoparticles.53 Another value of silver is the semiquantitative nature of the color change: 1.2–2.5 nM (orange), 3.7–5.0 nM (red), and 7.5 nM or greater (blue). The read-out time of 15 min was motivated by the fact that noncleaved M1 induces a slight assembly of the particles but only after 30 min. However, this assembly is much weaker than when M1 is cleaved as (i) the λmax of the secondary absorption peak is at 515 nm and (ii) the intensity of the ratio Abs.515 nm/Abs.400 nm is 50% lower than that of the ratio Abs.600 nm/Abs.400 nm. This induces a yellow-to-orange color change after 90 min (Figure 4C). This confirms the importance of the quenching site. A read-out time of 15 min ensures that the change of color observed is only due to peptide cleavage. Figure 4D shows the TEM image of the AgNPs-BSPP mixed with 12.5 nM Mpro incubated with M1, confirming the fractal assembly and the coalescence of the nanoparticles.
Various control experiments were performed to demonstrate that only the combination of Mpro and the peptide M1 could trigger the assembly of the particles (Table 1). Peptides M2, M3, or M4 are missing the arginines at the level of the bridging motif, and no assembly was observed when cleaved by Mpro (Figure 4E and Figure S44A). The specificity of this sensing platform was also investigated. Peptide M1 was incubated with either the papain-like protease (PLpro) of the SARS-CoV-2 virus or neuraminidase (NA) of the influenza virus.54 Figure 4E shows that none of these proteases induced a color change (Figure S44B). We suspect that this approach is modular for other proteases of interest.
Table 1.
Experimental Summary and Peptides Employed
| # | peptides | protease | LOD (nM) |
|---|---|---|---|
| M1 | DDDTSAVLQSGFRCGRGC | Mpro | 0.5 |
| M2 | DDDTSAVLQSGFACGAGC | Mpro | |
| M3 | DDDTSAVLQSGFAGGAGG | Mpro | |
| M4 | TSAVLQ | Mpro | |
| PLpro | DDDTSAVLQSGFRCGRGC | PLpro | |
| NA | DDDTSAVLQSGFRCGRGC | NA |
Sensor-Array for the Distinction between SARS-CoV-2 and Influenza-Infected Samples.
We next demonstrate a “chemical-nose” sensor-array for the distinction of SARS-CoV-2 versus influenza samples. Unlike specific sensing, the chemical nose works on the principle of selective binding between an analyte and an array of multiple receptors that will generate distinct responses for each analyte. The responses can thus be grouped in patterns that are specific to each analyte. Usually, array-based sensing strategies can maximize sensitivity because they can detect slight changes in complex environments. We used the peptides as receptors as described above.
The targets were Mpro, PLpro, and NA. These proteins are expressed by the viruses and are necessary for viral replication (Figure 5A). Samples containing the proteases have a different composition than healthy samples due to the presence of the viral proteins. This in turn affects peptide–particle interactions. Briefly, the sample is independently mixed with multiple bridging peptides (i.e., one peptide per well; Table 2), and the AgNPs-BSPP are then added. Multiple peptides—each with a specific affinity for the particles (defined as the concentration needed to induce an optical response of 0.5)—were used as receptors to obtain a specific pattern of optical responses for each sample. Low or high interferences lead to high or low assembly of the particles, respectively.
Figure 5.
Sensor-array mechanism and mode of response. (A) Illustration of the sensor-array with the optical response that is proportional to the interference caused by the analyte in the peptide–particles interaction. (B) Optical response pattern obtained for different proteins and eight peptides. Four replicates were performed for each protein. (C) Canonical score plot for the first two factors of the simplified optical response pattern of the sensor-array obtained by LDA against 10 nM of different proteins in water. Ellipses are 75% confidence. (D) Optical response intensity as a function of the concentration of proteins spiked in EBC or saliva. (E) Simplified picture of the sensor-array against healthy EBC spiked or not with 3 nM Mpro, PLpro, or NA. (F) Blind study results showing the total number of the different samples and the correctly identified samples. Tot. = total and Corr. = correct. (G) Canonical score plot for the first two factors of the simplified optical response pattern from the sensor-array obtained by the LDA against 3 nM Mpro and PLpro or NA spiked in healthy EBC. Ellipses are 99% of confidence.
Table 2.
Peptides Used in the Sensor-Array
| peptide | sequence | C 0.5 |
|---|---|---|
| P1 | RR | 3 μM |
| P2 | RRK | 200 nM |
| P3 | HHK | 180 nM |
| P4 | (RRK)2 | 55 nM |
| P5 | (RRK)3 | 16 nM |
| P6 | CKLVFF | 130 nM |
| P7 | DDGDSFRHHK | 60 nM |
| P8 | SGFRRGRR | 12 nM |
| P9 | FFK | 10 μM |
We first investigated the optical response produced for 10 nM Mpro, PLpro, or NA spiked in water, and we compared it to those of interfering proteins: hemoglobin (Hgb), bovine serum albumin (BSA), trypsin (Tps), and pepsin (Pps). Figure 5B shows the average optical response pattern for all of these proteins. A specific fingerprint was observed for each protein as well as a specific associated color pattern (Figure S45) followed by linear discriminant analysis (LDA).55,56 LDA is a tool that can differentiate different classes of objects via a linear combination of variables and maximizing the ratio of between-class variance to the within-class variance.57 Analysis of the 8 × 8 × 4 training matrix (eight peptides × eight conditions × four replicates; Table S1) generated canonical factors that are linear combinations of the optical response patterns (Table S1). Figure 5C shows the two-dimensional plot of the first two linear discriminants (70.46% and 18.9%). An ellipse with 75% confidence could be drawn without any overlaps. This finding shows that LDA can be used in a simplified representation of the optical response patterns.
The detection of Mpro, PLpro, and NA in complex media such as EBC or saliva was further evaluated. The optical responses were strongly inhibited in these media (Figures S46 and S47). After optimization, we found conditions in which the maximal optical response (i.e., blue sample) was obtained for each peptide in PCR-tested-negative EBC or saliva (Table S2). Negative EBC and saliva samples were spiked with different concentrations of Mpro, PLpro, or NA (from 0.1 to 50 nM) for the optimized sensor-array (Figures S48-S51). A LOD of 0.5 nM for Mpro and PLpro and 1.0 nM for NA was determined in EBC as well as in saliva (Figure 5D). Importantly, samples spiked with SARS proteases or influenza proteases could easily be distinguished by eye from 1 to 50 nM. Figure 5E shows a picture of the simplified sensor-array; that is, only the three most discriminating peptides (HHK, (RRK)3, and FFK) are represented for EBC spiked with 3 nM of protein. EBC spiked with NA and a mixture of Mpro and PLpro was easily differentiated from the other samples (Figure S52).
To demonstrate the robustness of this system, a blind study on 50 unknown samples was performed. Briefly, 50 samples of negative EBC were spiked with either 3 nM Mpro, PLpro, Mpro, and PLpro, NA or nothing. A reader blinded to the composition of each sample used images of the sensor-array to determine the protein composition of each sample and suggested the nature of the viral infection. The peptides used for the sensor-array can be seen in Table S3. An accuracy of 100% was obtained for samples spiked with SARS proteases, influenza proteases, or neither (Figure 5F). Despite an accuracy of 100% for the identification of PLpro-spiked samples, samples spiked with Mpro versus Mpro and PLpro could not be differentiated. This indicates that Mpro dominates the optical change. This could be improved by including other peptides in the sensor-array. Collectively, our sensor-array was validated for the efficient simple visual distinction of samples with SARS-CoV-2 or influenza biomarkers (Figures S54-S58 and Table S4).
From the blind study, 21 unknown samples were tested against our sensor-array (8 peptides) with the assistance of LDA (Table S5). Figure 5G shows the canonical score plot obtained by the LDA. A high degree of separation was obtained as ellipses of 99% of confidence were generated. The trained LDA was tested against nine supplementary unknown samples, and all could be correctly identified (Table S6).
Compared to other molecular- and nanotechnology-based analytical approaches for COVID-19 diagnosis,58 our method is more convenient and can be easily applied for point-of-care testing. Qiu et al. showed the detection of SARS-CoV-2 genes with two-dimensional gold nanoislands illuminated at their plasmonic resonance.59 Despite a high sensitivity (0.22 pM), this system is not suitable for field use. Moitra et al. showed the naked eye detection of RNA of SARS-CoV-2 with gold nanoparticles capped with oligonucleotides with a LOD of 0.18 ng/μL.60 However, extensive sample preparation is required for RNA extraction. Also, extracted RNA is easily degraded and could lead to misdiagnosis of COVID-19. The strength of our approach versus these competing systems is that it combines a colorimetric signal that can be detected even by the general public with the specificity and convenience offered by synthetic peptide probes.
CONCLUSIONS
We report peptide-induced fractal assembly of AgNPs with applications in colorimetric biomarker detection. The peptides contain a “bridging motif’, i.e., repetitions of arginine, histidine, or phenylalanine residues, that induces the formation of hyperbranched silver nanostructures. While the morphology transition is usually induced by external factors such as light, heat, or pressure, the AgNPs-BSPP can undergo spontaneous coalescence here due to the BSPP coating of our particles that can desorb during the peptide-mediated assembly, releasing silver ions into the solution and making the silver surface accessible for coalescence. The fractal assembly was strongly dependent on the peptide sequences as well as the molar ratio of the peptide per particles. Meanwhile, the optical properties of the material can be simply tuned with the peptide design. This has important implications in colorimetric biosensing because peptides are promising affinity probes that are easy and cheap to produce with fine-tuning over their sequence. We demonstrate this approach with detection of SARS-CoV-2 proteases and a sensor-array to discriminate SARS-CoV-2 from influenza via spiked human samples. Despite the subnanomolar limits of detection shown here, future work is needed to further optimize the peptide sequences for better sensitivity. For example, a peptide containing twice the number of arginine compared to M1 might have a stronger effect on the assembly of AgNPs after the proteolytic cleavage by Mpro than M1.
EXPERIMENTAL SECTION
AgNPs-BSPP Synthesis.
BSPP-coated AgNPs (AgNPs-BSPP) were synthesized via a two-step procedure. First, silver seeds were produced by the reduction of silver nitrate (0.1 mM, 6 mL) with sodium borohydride (0.1 M, 60 μL) in a glass vial under magnetic stirring for 16 h. The seeds were then grown into nanoparticles by adding sodium ascorbate (15 mM, 400 μL), BSPP (5 mM, 200 μL), dropwise silver nitrate (1 mM, 4 mL), and then BSPP again (5 mM, 400 μL), and the resulting solution was stirred for 48 h at room temperature. Detailed synthesis procedure and characterization of the AgNPs-BSPP (UV–vis, TEM, XRD, EDX) can be found in the Supporting Information.
Peptide Synthesis.
Peptides TSG, TRG, TRK, RR, FFK, DDGDSFRHHK, SGFRRGRR, M1, M2, and M3 were made using standard Fmoc synthesis on an Aapptec Eclipse personal peptide synthesizer. Wang resin was used as the solid support for all peptides and was cleaved using a mixture of TFA/EODT/thioanisole/anisole (88:5:5:2, v/v) for 3 h under an argon blanket. The freshly cleaved peptide was precipitated in −80 °C diethyl ether and washed several times (3×) using centrifugation. After the final centrifugation cycle, diethyl ether was decanted, and the white peptide slurry was dissolved in an H2O/glacial acetic acid mixture (90:10, v/v) to ensure trifluoroacetic acid (TFA) salt replacement with acetate salt during lyophilization. The peptide was frozen, lyophilized overnight, and then stored at −20 °C under an argon blanket.
Peptide Purification.
Peptide purification was carried out using a Shimadzu LC-40 HPLC system equipped with an LC-40D solvent delivery module, photodiode array detector SPD-M40, and degassing unit DGU-403. The sample was dissolved in water and acetonitrile, applied on a Zorbax 300 BS, C18 column (5 mm, 9.4 × 250 mm) from Agilent, and eluted at 1.5 mL/min with a 40 min gradient from 10% to 95% acetonitrile in water (with 0.05% TFA). Preparative injections were monitored at 190, 220, and 254 nm. All products were purified by HPLC to reach a purity of >90%. Peptide synthesis was confirmed using electrospray ionization mass spectrometry (ESI-MS) via the Micromass Quattro Ultima mass spectrometer. ESI-MS and HPLC spectra can be found in the Supporting Information, Figure S8.
Peptide–Particle Interaction.
The modification of the optical properties of the particles was analyzed as follows: 250 μL of AgNPs-BSPP was mixed with different concentrations of peptides in a 96-well plate. After 15 min, the UV–vis spectra were recorded using a hybrid multimode microplate reader (Synergy H1 model, BioTek Instruments, Inc.).
Protease Detection.
Mpro was incubated with 5 μM of the peptide M1 in Tris·HCl buffer for 4 h at 37 °C. After incubation, 10 μL of the sample was added to 90 μL of AgNPs-BSPP (A = 0.8); pictures and UV–vis spectra were recorded 15 min later.
Sensor-Array.
In a 96-well plate, peptides were mixed with the target proteins in either water, EBC (10%), or saliva (0.1%). After 5 min, 250 μL of AgNPs-BSPP was added and UV–vis spectra were recorded. Negative EBC were collected from 7 people known to be COVID-19-negative by PCR, using a lab condensate tube setup. All the volunteers signed an informed consent form, and all work was approved by the UCSD IRB. We prepared 50 EBC samples containing either 30 nM Mpro and PLpro, 30 nM NA, or no dopant. An operator ignoring the composition of each samples performed the testing and identified the sample by eye.
Linear Discriminant Analysis.
LDA was computed with R statistical environment version 4.0.2 (R Core Team 2020, Vienna). LDA was performed using the following R package libraries: MASS,61 ggord, ggplot2,62 tydiverse,63 and caret.64 An 8 × 8 × 4 training matrix (eight peptides × 8 conditions × 4 replicates) was generated from the optical responses pattern of 256 samples of water spiked with different proteins (Mpro, PLpro, NA, BSA, Hgb, Pps, Tps) or nothing. The 8 × 5 × 7 (eight peptides × five conditions × seven replicates) training matrix for the influenza versus SARS-CoV-2 distinction was generated from the optical responses of the samples produced during the blind study. The training matrices were used to perform LDA. The 8 × 3 × 3 test matrix (eight peptides × three conditions × three replicates) was generated from 72 unknown samples of EBC spiked with either 30 nM Mpro/PLpro, NA, or nothing. The LDA was then used to correctly identify each sample using the training matrix for the influenza versus SARS-CoV-2 distinction.
Supplementary Material
ACKNOWLEDGMENTS
J. Jokerst acknowledges funding from NIH under grants R01DE031114, R21AG0657776-01S1, R21AI157957, and DP2HL137187-S1. We also acknowledge infrastructure support under NIH S10 OD023555. M.R. acknowledges the Wallonie-Bruxelles International (WBI) of the Fédération Wallonie-Bruxelles for financial support. Figures 1, 4, and 5 were made using BioRender.com.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.1c11643.
Materials, instrumentation, AgNP synthesis and characterization, peptide synthesis, assembly characterization, peptide-mediated assembly, UV–vis spectra of peptide titration, pictures of the sensor-array, training matrix, test matrix, optical response patterns, control protease detection (PDF)
The authors declare no competing financial interest.
Contributor Information
Maurice Retout, Department of NanoEngineering, University of California, San Diego, La Jolla, California 92093, United States.
Yash Mantri, Department of Bioengineering, University of California, San Diego, La Jolla, California 92093, United States.
Zhicheng Jin, Department of NanoEngineering, University of California, San Diego, La Jolla, California 92093, United States.
Jiajing Zhou, Department of NanoEngineering, University of California, San Diego, La Jolla, California 92093, United States.
Grégoire Noël, Functional and Evolutionary Entomology–Gembloux Agro-Bio Tech, University of Liège, 5030 Gembloux, Belgium.
Brian Donovan, Department of NanoEngineering, University of California, San Diego, La Jolla, California 92093, United States.
Wonjun Yim, Materials Science and Engineering Program, University of California, San Diego, La Jolla, California 92093, United States.
Jesse V. Jokerst, Department of NanoEngineering, Materials Science and Engineering Program, and Department of Radiology, University of California, San Diego, La Jolla, California 92093, United States.
REFERENCES
- (1).Germain ME; Knapp MJ Optical Explosives Detection: From Color Changes to Fluorescence Turn-On. Chem. Soc. Rev 2009, 38, 2543–2555. [DOI] [PubMed] [Google Scholar]
- (2).Zhang Y; Jiao J; Wei Y; Wang D; Yang C; Xu Z Plasmonic Colorimetric Biosensor for Sensitive Exosome Detection via Enzyme-Induced Etching of Gold Nanobipyramid@MnO2Nanosheet Nanostructures. Anal. Chem 2020, 92, 15244–15252. [DOI] [PubMed] [Google Scholar]
- (3).Moon J; Kwon HJ; Yong D; Lee IC; Kim H; Kang H; Lim EK; Lee KS; Jung J; Park HG; Kang T Colorimetric Detection of SARS-CoV-2 and Drug-Resistant PH1N1 Using CRISPR/DCas9. ACS Sens. 2020, 5, 4017–4026. [DOI] [PubMed] [Google Scholar]
- (4).Saunders A; Zarzar L Structural Coloration from Total Internal Reflection at Microscale Concave Surfaces and Use for Sensing in Complex Droplets. Proc. SPIE 2020, 11292. [Google Scholar]
- (5).Medintz IL; Clapp AR; Mattoussi H; Goldman ER; Fisher B; Mauro JM Self-Assembled Nanoscale Biosensors Based on Quantum Dot FRET Donors. Nat. Mater 2003, 2, 630–638. [DOI] [PubMed] [Google Scholar]
- (6).Wilson R The Use of Gold Nanoparticles in Diagnostics and Detection. Chem. Soc. Rev 2008, 37, 2028–2045. [DOI] [PubMed] [Google Scholar]
- (7).Huang X; Liu Y; Yung B; Xiong Y; Chen X Nanotechnology-Enhanced No-Wash Biosensors for in Vitro Diagnostics of Cancer. ACS Nano 2017, 11, 5238–5292. [DOI] [PubMed] [Google Scholar]
- (8).Zhou W; Gao X; Liu D; Chen X Gold Nanoparticles for in Vitro Diagnostics. Chem. Rev 2015, 115, 10575–10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Cordeiro M; Carlos FF; Pedrosa P; Lopez A; Baptista PV Gold Nanoparticles for Diagnostics: Advances towards Points of Care. Diagnostics 2016, 6, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Xie X; Xu W; Liu X Improving Colorimetric Assays through Protein Enzyme-Assisted Gold Nanoparticle Amplification. Acc. Chem. Res 2012, 45 (9), 1511–1520. [DOI] [PubMed] [Google Scholar]
- (11).Jain PK; Eustis S; El-Sayed MA Plasmon Coupling in Nanorod Assemblies: Optical Absorption, Discrete Dipole Approximation Simulation, and Exciton-Coupling Model. J. Phys. Chem. B 2006, 110, 18243–18253. [DOI] [PubMed] [Google Scholar]
- (12).Kelly KL; Coronado E; Zhao LL; Schatz GC The Optical Properties of Metal Nanoparticles: The Influence of Size, Shape, and Dielectric Environment. J. Phys. Chem. B 2003, 107, 668–677. [Google Scholar]
- (13).Jain PK; Huang X; El-Sayed IH; El-Sayed MA Review of Some Interesting Surface Plasmon Resonance-Enhanced Properties of Noble Metal Nanoparticles and Their Applications to Biosystems. Plasmonics 2007, 2, 107–118. [Google Scholar]
- (14).Retout M; Jabin I; Bruylants G Synthesis of Ultra-Stable and Bioconjugable Ag, Au and Bimetallic Ag_Au Nanoparticles Coated with Calix[4]Arenes. ACS Omega 2021, 6, 19675–19684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Xu S; Jiang L; Liu Y; Liu P; Wang W; Luo X A Morphology-Based Ultrasensitive Multicolor Colorimetric Assay for Detection of Blood Glucose by Enzymatic Etching of Plasmonic Gold Nanobipyramids. Anal. Chim. Acta 2019, 1071, 53–58. [DOI] [PubMed] [Google Scholar]
- (16).Wang H; Rao H; Luo M; Xue X; Xue Z; Lu X Noble Metal Nanoparticles Growth-Based Colorimetric Strategies: From Monocolorimetric to Multicolorimetric Sensors. Coord. Chem. Rev 2019, 398, 113003. [Google Scholar]
- (17).Gao Z; Deng K; Wang XD; Miró M; Tang D High-Resolution Colorimetric Assay for Rapid Visual Readout of Phosphatase Activity Based on Gold/Silver Core/Shell Nanorod. ACS Appl Mater. Interfaces 2014, 6, 18243–18250. [DOI] [PubMed] [Google Scholar]
- (18).Wu C; Zhang R; Du W; Cheng L; Liang G Alkaline Phosphatase-Triggered Self-Assembly of Near-Infrared Nanoparticles for the Enhanced Photoacoustic Imaging of Tumors. Nano Lett. 2018, 18, 7749–7754. [DOI] [PubMed] [Google Scholar]
- (19).Alafeef M; Moitra P; Dighe K; Pan D RNA-Extraction-Free Nano-Amplified Colorimetric Test for Point-of-Care Clinical Diagnosis of COVID-19. Nat. Protoc 2021, 16, 3141–3162. [DOI] [PubMed] [Google Scholar]
- (20).Retout M; Valkenier H; Triffaux E; Doneux T; Bartik K; Bruylants G Rapid and Selective Detection of Proteins by Dual Trapping Using Gold Nanoparticles Functionalized with Peptide Aptamers. ACS Sens. 2016, 1, 929–933. [Google Scholar]
- (21).Hu T; Lu S; Chen C; Sun J; Yang X Colorimetric Sandwich Immunosensor for Aβ(1–42) Based on Dual Antibody-Modified Gold Nanoparticles. Sens. Actuators, B 2017, 243, 792–799. [Google Scholar]
- (22).Lesniewski A; Los M; Jonsson-NiedzioÌåka M; Krajewska A; Szot K; Los JM; Niedziolka-Jonsson J Antibody Modified Gold Nanoparticles for Fast and Selective, Colorimetric T7 Bacteriophage Detection. Bioconjugate Chem. 2014, 25, 644–648. [DOI] [PubMed] [Google Scholar]
- (23).Jazayeri MH; Amani H; Pourfatollah AA; Pazoki-Toroudi H; Sedighimoghaddam B Various Methods of Gold Nanoparticles (GNPs) Conjugation to Antibodies. Sensing and Bio-Sensing Research 2016, 9, 17–22. [Google Scholar]
- (24).Guo L; Xu Y; Ferhan AR; Chen G; Kim DH Oriented Gold Nanoparticle Aggregation for Colorimetric Sensors with Surprisingly High Analytical Figures of Merit. J. Am. Chem. Soc 2013, 135, 12338–12345. [DOI] [PubMed] [Google Scholar]
- (25).Sessler JL; Jayawickramarajah J Functionalized Base-Pairs: Versatile Scaffolds for Self-Assembly. Chem. Commun 2005, 15, 1939–1949. [DOI] [PubMed] [Google Scholar]
- (26).Sun X; Hagner M Novel Preparation of Snowflake-like Dendritic Nanostructures of Ag or Au at Room Temperature via a Wet-Chemical Route. Langmuir 2007, 23, 9147–9150. [DOI] [PubMed] [Google Scholar]
- (27).Gole MT; Yin Z; Wang MC; Lin W; Zhou Z; Leem J; Takekuma S; Murphy CJ; Nam SW Large Scale Self-Assembly of Plasmonic Nanoparticles on Deformed Graphene Templates. Sci. Rep 2021, 11, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Singh A; Khatun S; Gupta AN Anisotropy: Versus Fluctuations in the Fractal Self-Assembly of Gold Nanoparticles. Soft Matter 2020, 16, 7778–7788. [DOI] [PubMed] [Google Scholar]
- (29).Li X; Lenhart JJ; Walker HW Aggregation Kinetics and Dissolution of Coated Silver Nanoparticles. Langmuir 2012, 28, 1095–1104. [DOI] [PubMed] [Google Scholar]
- (30).He D; Bligh MW; Waite TD Effects of Aggregate Structure on the Dissolution Kinetics of Citrate-Stabilized Silver Nanoparticles. Environ. Sci. Technol 2013, 47, 9148–9156. [DOI] [PubMed] [Google Scholar]
- (31).Doyen M; Goole J; Bartik K; Bruylants G Amino Acid Induced Fractal Aggregation of Gold Nanoparticles: Why and How. J. Colloid Interface Sci 2016, 464, 60–166. [DOI] [PubMed] [Google Scholar]
- (32).Kim W; Safonov VP; Shalaev VM; Armstrong RL Fractals in Microcavities: Giant Coupled, Multiplicative Enhancement of Optical Responses. Phys. Rev. Lett 1999, 82, 4811–4814. [Google Scholar]
- (33).Shalaev VM; Markel VA; Poliakov EY; Armstrong RL; Safonov VP; Sarychev AK Non Linear Optical Phenomena in Nanostructured Fractal Materials. J. Nonlinear Opt. Phys. Mater 1998, 7, 131–152. [Google Scholar]
- (34).Parab H; Jung C; Woo MA; Park HG An Anisotropic Snowflake-like Structural Assembly of Polymer-Capped Gold Nanoparticles. J. Nanopart. Res 2011, 13, 2173–2180. [Google Scholar]
- (35).Cheng Z; Qiu Y; Li Z; Yang D; Ding S; Cheng G; Hao Z; Wang Q Fabrication of Silver Dendrite Fractal Structures for Enhanced Second Harmonic Generation and Surface-Enhanced Raman Scattering. Opt. Mater. Express 2019, 9, 860–869. [Google Scholar]
- (36).Ansari JR; Singh N; Ahmad R; Chattopadhyay D; Datta A Controlling Self-Assembly of Ultra-Small Silver Nanoparticles: Surface Enhancement of Raman and Fluorescent Spectra. Opt. Mater 2019, 94, 138–147. [Google Scholar]
- (37).Noroozi M; Zakaria A; Moksin MM; Wahab ZA; Abedini A Green Formation of Spherical and Dendritic Silver Nanostructures under Microwave Irradiation without Reducing Agent. Int. J. Mol. Sci 2012, 13, 8086–8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Ziegler C; Eychmüller A Seeded Growth Synthesis of Uniform Gold Nanoparticles with Diameters of 15–300 Nm. J. Phys. Chem. C 2011, 115, 4502–4506. [Google Scholar]
- (39).Moore C; Wing R; Pham T; Jokerst JV Multispectral Nanoparticle Tracking Analysis for the Real-Time and Label-Free Characterization of Amyloid-β Self-Assembly in Vitro. Anal. Chem 2020, 92, 11590–11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Röder H; Bromann K; Brune H; Kern K Diffusion-Limited Aggregation with Active Edge Diffusion. Phys. Rev. Lett 1995, 74, 3217–3220. [DOI] [PubMed] [Google Scholar]
- (41).Tai JT; Lai CS; Ho HC; Yeh YS; Wang HF; Ho RM; Tsai DH Protein-Silver Nanoparticle Interactions to Colloidal Stability in Acidic Environments. Langmuir 2014, 30, 12755–12764. [DOI] [PubMed] [Google Scholar]
- (42).Magdassi S; Grouchko M; Berezin O; Kamyshny A Triggering the Sintering of Silver Nanoparticles at Room Temperature. ACS Nano 2010, 4, 1943–1948. [DOI] [PubMed] [Google Scholar]
- (43).Grouchko M; Popov I; Uvarov V; Magdassi S; Kamyshny A Coalescence of Silver Nanoparticles at Room Temperature: Unusual Crystal Structure Transformation and Dendrite Formation Induced by Self-Assembly. Langmuir 2009, 25, 2501–2503. [DOI] [PubMed] [Google Scholar]
- (44).Yu P; Huang J; Tang J Observation of Coalescence Process of Silver Nanospheres during Shape Transformation to Nanoprisms. Nanoscale Res. Lett 2012, 7, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Park JW; Shumaker-Parry JS Strong Resistance of Citrate Anions on Metal Nanoparticles to Desorption under Thiol Functionalization. ACS Nano 2015, 9, 1665–1682. [DOI] [PubMed] [Google Scholar]
- (46).Tan YN; Yang J; Lee JY; Wang DIC Mechanistic Study on the Bis(p-Sulfonatophenyl)Phenylphosphine Synthesis of Monometallic Pt Hollow Nanoboxes Using Ag*-Pt Core-Shell Nanocubes as Sacrificial Templates. J. Phys. Chem. C 2007, 111, 14084–14090. [Google Scholar]
- (47).Jin B; Sushko ML; Liu Z; Jin C; Tang R In Situ Liquid Cell TEM Reveals Bridge-Induced Contact and Fusion of Au Nanocrystals in Aqueous Solution. Nano Lett. 2018, 18, 6551–6556. [DOI] [PubMed] [Google Scholar]
- (48).Pizzi A; Dichiarante V; Terraneo G; Metrangolo P Crystallographic Insights into the Self-Assembly of KLVFF Amyloid-Beta Peptides. Biopolymers 2017, 110, 23088. [DOI] [PubMed] [Google Scholar]
- (49).Komarova NL; Schang LM; Wodarz D Patterns of the COVID-19 Pandemic Spread around the World: Exponential versus Power Laws. J. R. Soc., Interface 2020, 17, 20200518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Tang Y; Schmitz JE; Persing DH; Stratton CW Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J. Clin. Microbiol 2020, 58, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Ali I; Alharbi OML COVID-19: Disease, Management, Treatment, and Social Impact. Sci. Total Environ 2020, 728, 138861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Moore C; Borum RM; Mantri Y; Xu M; Fajtová P; O’Donoghue AJ; Jokerst JV Activatable Carbocyanine Dimers for Photoacoustic and Fluorescent Detection of Protease Activity. ACS Sens. 2021, 6, 2356–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Jin Z; Mantri Y; Retout M; Cheng Y; Zhou J; Jorns A; Fajtova P; Yim W; Moore C; Xu M; Creyer M; Borum R; Zhou J; Wu Z; He T; Penny W; O’Donoghue A; Jokerst J A Charge-Switchable Zwitterionic Peptide for Rapid Detection of SARS-CoV-2 Main Protease. Angew. Chem., Int. Ed 2022, 61, e202112995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).V’kovski P; Kratzel A; Steiner S; Stalder H; Thiel V Coronavirus Biology and Replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol 2021, 19, 155–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).De M; Rana S; Akpinar H; Miranda OR; Arvizo RR; Bunz UHF; Rotello VM Sensing of Proteins in Human Serum Using Conjugates of Nanoparticles and Green Fluorescent Protein. Nat. Chem 2009, 1, 461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).You CC; Miranda OR; Gider B; Ghosh PS; Kim IB; Erdogan B; Krovi SA; Bunz UHF; Rotello VM Detection and Identification of Proteins Using Nanoparticle-Fluorescent Polymer “chemical Nose” Sensors. Nat. Nanotechnol 2007, 2, 318–323. [DOI] [PubMed] [Google Scholar]
- (57).Fletcher R; Reeves CM The Use of Multiple Measurements in Taxonomic Problems. Ann. Hum. Genet 1936, 7, 179–188. [Google Scholar]
- (58).Kailasa SK; Mehta VN; Koduru JR; Basu H; Singhal RK; Murthy ZVP; Park TJ An Overview of Molecular Biology and Nanotechnology Based Analytical Methods for the Detection of SARS-CoV-2: Promising Biotools for the Rapid Diagnosis of COVID-19. Analyst 2021, 146, 1489–1513. [DOI] [PubMed] [Google Scholar]
- (59).Qiu G; Gai Z; Tao Y; Schmitt J; Kullak-Ublick GA; Wang J Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277. [DOI] [PubMed] [Google Scholar]
- (60).Moitra P; Alafeef M; Alafeef M; Alafeef M; Dighe K; Frieman MB; Pan D; Pan D; Pan D Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Venables WN, Ripley BD Random and Mixed Effects. In Modern Applied Statistics with S; Springer: New York, 2002; pp 271–300. [Google Scholar]
- (62).Wickham H Ggplot2. In Elegant Graphics for Data Analysis; Springer: New York, 2009; p 35. [Google Scholar]
- (63).Wickham H; Averick M; Bryan J; Chang W; McGowan L; Francois R; Grolemund G; Hayes A; Henry L; Hester J; Kuhn M; Pedersen T; Miller E; Bache S; Müller K; Ooms J; Robinson D; Seidel D; Spinu V; et al. Welcome to the Tidyverse. Journal of Open Source Software 2019, 4, 1686. [Google Scholar]
- (64).Kuhn L; Page K; Ward J; Worrall-Carter L The Process and Utility of Classification and Regression Tree Methodology in Nursing Research. Journal of Advanced Nursing 2014, 70, 1276–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.