Abstract
Introduction
Pseudomonas aeruginosa (P. aeruginosa) and Acinetobacter baumannii (A. baumannii) can cause difficult-to-treat infections. We characterized molecular epidemiology of ceftazidime-resistant P. aeruginosa and carbapenem-resistant A. baumannii at a tertiary hospital in Ethiopia.
Materials and methods
Non-fermenting gram-negative bacilli (n = 80) isolated from admitted patients were subjected for species identification by MALDI-TOF. Pseudomonas species resistant to ceftazidime or meropenem, and Acinetobacter species resistant to meropenem, or imipenem were selected for whole genome sequencing. DNA extracted with EZ1 Advanced XL instrument (Qiagen, Hilden, Germany) was sequenced on Illumina (HiSeq2500) using libraries prepared by NEXTRA-kits (Illumina). Raw reads were assembled using SPAdes 3.13.0, and assembled genomes were used to query databases for resistome profile and sequence types.
Result
Among Pseudomonas species isolated, 31.7% (13/41), and 7.3% (3/41) were non-susceptible to ceftazidime, and meropenem, respectively. Carbapenem-resistance was 56.4% (22/39) among Acinetobacter species. Moreover, 92% (12/13) of Pseudomonas species non-susceptible to ceftazidime and/or meropenem, and 89.4% (17/19) of Acinetobacter species encoded multiple resistance genes for at least three classes of antimicrobials. The prevalent β - lactamase genes were blaOXA–486 (53.8%, 7/13), blaCTX–M–15 (23.0%, 3/13) among Pseudomonas, and blaGES–11 (57.8%, 11/19) among Acinetobacter. The blaOXA–51-like β - lactamase, blaOXA–69 (63.1%, 12/19) was the most prevalent carbapenemase gene among Acinetobacter isolates. Single isolates from both P. aeruginosa, and A. baumannii were detected with the blaNDM–1. Sequence type (ST)1 A. baumannii and ST274 P. aeruginosa were the prevalent sequence types. A cgMLST analysis of the ST1 A. baumannii isolates showed that they were closely related and belonged to the international clonal complex one (ICC1). Similarly, ST274 P. aeruginosa isolates were clonally related.
Conclusion
The prevalence of MDR isolates of Pseudomonas and Acinetobacter spp. was high. A. baumannii isolates were clonally spreading in the admission wards at the hospital. Emergence of blaNDM–1 in the intensive care, and surgical wards of the hospital is a severe threat that requires urgent intervention.
Keywords: ESBLs, carbapenemase, bla CTX–M–15 , bla GES–11 , bla NDM–1 , P. aeruginosa, A. baumannii, Ethiopia
Introduction
Pseudomonas aeruginosa (P. aeruginosa) and Acinetobacter baumannii (A. baumannii) are among the main causes of nosocomial infections (De Oliveira et al., 2020). They belong to the group of bacteria known as “ESKAPE” (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter species). Moreover, these group of bacteria are difficult to treat in most cases (Bergogne-Bérézin and Towner, 1996; De Oliveira et al., 2020; Ma et al., 2020).
The prevalence of antimicrobial resistant sub-populations of these strains has rapidly increased over the last few decades (Wong et al., 2017; Horcajada et al., 2019). Carbapenemase-producing A. baumannii (CRAB) and P. aeruginosa were listed top two of the three critical priority pathogens for which new antimicrobials are urgently needed (WHO, 2017). Several Acinetobacter and Pseudomonas species were previously reported from different clinical samples from both animal, and human infections (Chusri et al., 2014; Wong et al., 2017; Agnese et al., 2018). Many of them were resistant to multiple classes of antibiotics primarily by several intrinsic resistance mechanism they encode, and secondly by acquired resistance mechanisms (Bello-López et al., 2020; Meng et al., 2020). Studies have also shown that multidrug-resistant strains of P. aeruginosa and A. baumannii were the main drivers of hospital-acquired infections (Djahmi et al., 2014; Eichenberger and Thaden, 2019; Kazmierczak et al., 2020). A recent review of global epidemiology of carbapenemase-producing Gram-negative bacteria reported that carbapenemase-producing P. aeruginosa (CRPA) were associated with high mortality and morbidity among hospitalized patients with pneumonia and bloodstream infections in the United States (Brink, 2019). Though regional variations are common, colonization by carbapenem-resistant A. baumannii increased the risk of acquisition of bloodstream infection four-fold (Munoz-Price et al., 2016; Bassetti et al., 2017). In low-income countries like Ethiopia, comprehensive microbiological data is lacking.
The classical phenotyping methods commonly used in low-income countries cannot reliably define mechanism of resistance in both Pseudomonas and Acinetobacter species. Lack of sufficient standardized genotypic methods for detection and tracking of multidrug-resistant or extensively drug-resistant isolates was one of the challenges to understand epidemiology of antimicrobial resistance in sub-Saharan African countries (Eichenberger and Thaden, 2019). In most cases, global reports on antimicrobial resistance lack data from African countries. Despite discrepancies in availability of data, there is sufficient overall evidence that carbapenemase-producing Gram-negative bacilli have become a threat to global health. Rapid detection and tracking of any ongoing spread of resistant strains is necessary.
We aimed to analyze the phenotypic and molecular characteristics of Pseudomonas species and Acinetobacter species isolated from clinical samples at Jimma Medical Center (JMC), a tertiary hospital in Ethiopia.
Materials and methods
Isolation, identification, and selection of strains
As part of a large epidemiological study, a total of 1,087 clinical samples (urine, stools, wound secretions, and sputum) were collected from patients with suspected infections seeking medical care from June to October 2016 at JMC, Ethiopia. Pseudomonas species and Acinetobacter species were isolated on MacConkey agar and sheep blood agar. Species identification was performed by MALDI-TOF (Bruker Daltonik GmbH, Bremen, Germany) at Karolinska University Hospital (KUH), Clinical Microbiology laboratory, and a full panel of antimicrobial susceptibility testing was performed by using the EUCAST 2021 v11 guideline.1
Antimicrobial susceptibility testing
All Pseudomonas species and Acinetobacter species isolated were subjected to disk-diffusion susceptibility testing. Antibiotic discs of ceftazidime, meropenem, piperacillin-tazobactam, gentamicin, amikacin, ciprofloxacin was used for Pseudomonas species. Similarly, all Acinetobacter isolates were tested by using meropenem, imipenem, gentamicin, amikacin, ciprofloxacin, trimethoprim-sulfamethoxazole. Then, isolates with reduced susceptibility to ceftazidime and/or meropenem for Pseudomonas spp., and meropenem or imipenem for Acinetobacter spp. were selected for antimicrobial susceptibility testing using the newer antimicrobials (cefiderocol, ceftazidime-avibactam, ceftolozane-tazobactam, and imipenem-relebactam for Pseudomonas spp., and cefiderocol, meropenem, imipenem, and imipenem-relebactam for Acinetobacter spp. using microbroth dilution technique), and whole genome sequencing (WGS). Patients’ clinical data like admission, presence of underlying chronic illnesses, current use of antibiotics, and other factors was collected using a structured questionnaire.
DNA extraction, whole genome sequencing and analysis of genomic data
Genomic DNA was extracted using Qiagen kits on EZ1 automated DNA extractions system. The extracted DNA was quantified using Qubit™ 3.0 (Massachusetts, United States) and library preps were performed using NEXTRA-kit (Illumina) and sequenced using HiSeq2500 (Illumina). Raw reads were assembled using SPAdes ver. 3.13.0, and the assembled draft genomes were used for querying different databases, MLST-typing 2.0, hosted at center for genomic epidemiology, and detection resistome profile by using ResFinder 4.1.2 ,3 Epidemiologic analysis of relatedness between the isolates, and to other international isolates was performed by the minimum spanning tree using the isolate genomes deposited at the public domain for A. baumannii at4, and P. aeruginosa at.5 The Genome sequences were deposited at the NCBI, SRA database (Bioproject number: PRJNA593604, Biosample accession: SUB11593554).
Results
Clinical and demographic characteristics of the patient
From a total of 1,087 non-repeat clinical samples collected during the study period, non-duplicate, non-fermenting Gram-negative bacilli that belong to either Pseudomonas spp. or Acinetobacter spp. were isolated from 80 patents. Most of these patients, 73.7% (59/80) were male, and 26.3% (21/80) were female. Ninety percent (72/80) of these patients were admitted to different wards in the hospital [surgical ward (n = 52), intensive care unit (n = 6), pediatric ward (n = 3), and medical ward (n = 11)]. Clinical samples collected include urine for urinary tract infections, sputum for lower respiratory tract infections, wound swab for wound infections/abscess, and stools for diarrhea. Most of these patients were admitted to the hospital for other underlying diseases and developed infection after admission, and most of them were treated with locally available antimicrobial therapy. Among Pseudomonas isolates 68.2% (28/41) were from surgical ward, followed by 17.0% (7/41) from medical ward, and 9.7% (4/41) from the intensive care unit (Table 1A). Similarly, 69.2% (27/39) of the Acinetobacter species were isolated from the surgical ward and 20.5% (8/39) from the medical ward (Table 1B).
TABLE 1A.
Socio-demographic and clinical characteristics patients and isolation Pseudomonas species.
| Patient ID | Age | Sex | Inpatient/ outpatient |
Current diagnosis | *Current antibiotic | Specimen | Underlying disease | Pseudomonas species |
| I020 | 28 | M | Inpatient | Surgical site infection | CRO, MET | Wound swab | Surgical incision | P. aeruginosa |
| I032 | 28 | M | Inpatient | Urinary tract infection | CRO, MET | Urine | Trauma | P. aeruginosa |
| I038 | 30 | M | Inpatient | Urinary tract infection | CRO | Urine | Severe head injury | P. putida |
| I043 | 17 | M | Inpatient | Urinary tract infection | CRO | Urine | Aspiration pneumonia | P. aeruginosa |
| M019 | 70 | M | Inpatient | COPD | CRO, VAN | Sputum | Cor pulmonale | P. aeruginosa |
| M030 | 60 | M | Inpatient | Community-acquired pneumonia | CRO | Sputum | **COPD | P. aeruginosa |
| M074 | 50 | M | Inpatient | COPD | VAN, CIP | Sputum | Asthma | P. aeruginosa |
| M119 | 40 | M | Inpatient | Pneumonia | CRO | Sputum | Post TB fibrosis | P. aeruginosa |
| M304 | 40 | M | Inpatient | Community-acquired pneumonia | No | Sputum | Post TB fibrosis | P. aeruginosa |
| M334 | 35 | F | Inpatient | Severe community-acquired pneumonia | CRO | Sputum | No | P. aeruginosa |
| M521 | 60 | F | Outpatient | Community-acquired pneumonia | CRO | Sputum | **T2DM | P. aeruginosa |
| P014 | 14 | M | Inpatient | Wound infection | AUG | Wound swab | No | P. aeruginosa |
| P109 | 4 | F | Inpatient | Diarrhea | AMOX, GENT | Stool | SAM, pneumonia | P. aeruginosa |
| S007 | 32 | M | Inpatient | Necrotizing fasciitis | No | Wound swab | No | P. aeruginosa |
| S010 | 5 | M | Inpatient | Wound infection | CAF, CLO | Wound swab | Trauma | P. aeruginosa |
| S011 | 7 | M | Inpatient | Wound infection | AMP, CAF | Wound swab | Trauma | P. aeruginosa |
| S017 | 18 | M | Inpatient | Necrotizing fasciitis | CRO, MET | Wound swab | No | P. aeruginosa |
| S019 | 60 | M | Inpatient | Foot ulcer | CRO, MET | Wound swab | Diabetes mellitus | P. aeruginosa |
| S020 | 30 | M | Inpatient | Wound infection | CRO, MET | Wound swab | Trauma | P. aeruginosa |
| S036 | 17 | F | Inpatient | Wound infection | No | Wound swab | Unstable pelvis fracture | P. aeruginosa |
| S047 | 10 | M | Inpatient | Wound infection | AMP | Wound swab | Chronic osteomyelitis | P. aeruginosa |
| S048 | 15 | M | Inpatient | Wound infection | AMP, CAF | Wound swab | Fracture of femoral shaft | P. aeruginosa |
| S077 | 3 | M | Inpatient | Wound infection | AMP, GENT | Wound swab | Colostomy, imperforation | P. aeruginosa |
| S116 | 50 | F | Inpatient | Wound infection | AMOX, MET | Wound swab | Uterine cancer | P. aeruginosa |
| S129 | 77 | M | Inpatient | Wound infection | CRO, VAN | Wound swab | Amputation of leg | P. aeruginosa |
| S133 | 45 | M | Inpatient | Wound infection | CRO, MET | Wound swab | Neck injury trauma | P. aeruginosa |
| S114 | 25 | F | Inpatient | Acute kidney infection | CRO, MET | Urine | Rib fracture | P. aeruginosa |
| S155 | 21 | F | Inpatient | Wound infection | AMP, CAF | Wound swab | Infected skin graft | P. aeruginosa |
| S174 | 60 | M | Inpatient | Wound infection | CAF, AMP | Wound swab | Infected fracture site | P. aeruginosa |
| S192 | 75 | M | Inpatient | Urinary tract infection | CRO | Urine | **BOO | P. putida |
| S195 | 21 | F | Inpatient | Wound infection | CRO, MET | Wound swab | Skin graft infection | P. aeruginosa |
| S198 | 57 | M | Outpatient | Wound infection | CLO, CAF | Wound swab | Left femoral fracture | P. aeruginosa |
| S209 | 18 | F | Inpatient | Wound infection | CRO | Wound swab | Burn wound | P. aeruginosa |
| S248 | 20 | F | Inpatient | Contaminated wound | Wound swab | No | P. aeruginosa | |
| S288 | 50 | M | Inpatient | Pneumonia | No | Sputum | Abdominal mass | P. aeruginosa |
| S319 | 40 | M | Inpatient | Urinary tract infection | No | Urine | BPH | P. aeruginosa |
| S325 | 37 | M | Inpatient | Wound infection | CAF, AMP | Wound swab | Compound distal fracture | P. aeruginosa |
| S328 | 60 | M | Inpatient | Pneumonia | CRO, MET | Sputum | 3rd degree burn | P. aeruginosa |
| S332 | 30 | M | Inpatient | Wound infection | CRO, MET | Wound swab | 2nd degree burn | P. fulva |
| S356 | 30 | M | Inpatient | Wound infection | CRO, MET | Wound swab | Infected palate of right knee | P. aeruginosa |
| S371 | 64 | M | Inpatient | Surgical site infection | CLO, CAF | Wound swab | Laparotomy | P. aeruginosa |
*Current antibiotics: CRO, ceftriaxone; Met, metronidazole; VAN, vancomycin; CLO, cloxacillin; CAF, chloramphenicol; CIP, ciprofloxacin; AMP, ampicillin; GENT, gentamicin.
**Underlying diseases: COPD, congestive obstructive pulmonary disease; T2DM, type-2 diabetes mellitus; BOO, Bladder outlet obstruction; BPH, Benign prostatic hyperplasia.
TABLE 1B.
Socio-demographic and clinical characteristics patients and isolation Acinetobacter species.
| Patient ID | Age | Sex | Inpatient/ outpatient |
Current diagnosis | *Current antibiotic | Specimen | Underlying disease | Bacterial species |
| I027 | 45 | F | Inpatient | Aspiration pneumonia | CRO, MET | Sputum | Stroke/hemiparalysis | A. baylyi |
| I030 | 28 | F | Inpatient | Skin infection | No | Wound swab | No | A. baumannii |
| M029 | 22 | M | Inpatient | Urinary tract infection | CRO | Urine | Retroviral infection | A. baumannii |
| M057 | 60 | F | Inpatient | COPD | No | Sputum | No | A. baumannii |
| M135 | 70 | F | Outpatient | Community-acquired pneumonia | CRO | Sputum | No | A. junii |
| M212 | 25 | M | Inpatient | Pneumonia | CRO, VAN | Sputum | Disseminated tuberculosis | A. calcoaceticus |
| M217 | 35 | M | Inpatient | Community-acquired pneumonia | No | Sputum | **COPD | A. schindleri |
| M328 | 50 | F | Outpatient | Severe community-acquired pneumonia | No | Sputum | **T2DM | A. baumannii |
| M344 | 17 | M | Outpatient | Pneumonia | No | Sputum | Electrical burn | A. junii |
| M431 | 50 | F | Inpatient | Pneumonia | CRO | Sputum | Rt&Lt femoral fracture | A. baumannii |
| P015 | 30 | M | Inpatient | Wound infection | CLO, CAF | Wound swab | Compound fracture of right tibia | A. baumannii |
| P038 | 35 | M | Outpatient | Wound infection | AMP, CAF | Wound swab | Infected fracture site | A. baumannii |
| S073 | 15 | M | Inpatient | Wound infection | CRO | Wound swab | Chronic osteomyelitis | A. baumannii |
| S082 | 45 | M | Inpatient | Wound infection | CIP | Wound swab | Femoral fracture | A. baumannii |
| S126 | 40 | M | Inpatient | Wound infection | CRO, MET | Wound swab | Surgical site infection | A. baumannii |
| S130 | 30 | M | Inpatient | Wound infection | CRO, MET | Wound swab | 3rd deg. burn | A. baumannii |
| S147 | 30 | F | Inpatient | Wound infection | CRO | Wound swab | Colostomy | A. baumannii |
| S161 | 50 | M | Inpatient | Wound infection | AMP, CAF | Wound swab | No | A. parvus |
| S165 | 8 | M | Inpatient | Wound infection | CAF, CLO | Wound swab | No | A. baumannii |
| S167 | 40 | M | Inpatient | Wound infection | CRO, MET | Wound swab | Scalp abscess | A. baumannii |
| S170 | 27 | M | Inpatient | Wound infection | CRO, MET | Wound swab | No | A. baumannii |
| S171 | 32 | M | Inpatient | Wound infection | CRO, MET | Wound swab | Retroviral infection | A. baumannii |
| S176 | 25 | F | Inpatient | Necrotic wound infection | CRO, MET | Wound swab | No | A. baumannii |
| S209 | 50 | M | Outpatient | Pneumonia | CRO, MET | Sputum | No | A. baumannii |
| S210 | 25 | M | Inpatient | Wound infection | CRO | Wound swab | No | A. baumannii |
| S212 | 40 | M | Inpatient | Wound infection | CRO, MET | Wound swab | Compound fracture left leg | A. baumannii |
| S217 | 48 | M | Inpatient | Pneumonia | CRO, MET | Sputum | colostomy/post-operation | A. baumannii |
| S219 | 39 | M | Inpatient | Wound infection | No | Wound swab | Tibia-fibular fracture | A. baumannii |
| S226 | 24 | M | Inpatient | Urinary tract infection | No | Urine | Urethral stricture | A. haemolyticus |
| S247 | 40 | M | Outpatient | Wound infection | AMP, CAF | Wound swab | Infected incision site | A. baumannii |
| S260 | 18 | F | Inpatient | Wound infection | CRO | Wound swab | Burn wound | A. baumannii |
| S267 | 30 | F | Inpatient | Wound infection | CRO, MET | Wound swab | Surgical site infection | A. baumannii |
| S270 | 26 | M | Inpatient | Wound infection | CRO, MET | Wound swab | Chronic osteomyelitis | A. baumannii |
| S275 | 48 | M | Inpatient | Wound infection | No | Wound swab | Compound fracture of femur | A. baumannii |
| S294 | 70 | M | Inpatient | Urinary tract infection | CRO | Urine | Bladder outlet obstruction | A. baumannii |
| S296 | 10 | M | Inpatient | Wound infection | AMP, CAF | Wound swab | Left femoral fracture | A. baumannii |
| S315 | 2 | M | Inpatient | Diarrhea | AMP, GENT, CLO | Stool | No | A. lwoffii |
| S327 | 29 | M | Inpatient | Severe community-acquired pneumonia | CRO, MET | Sputum | No | A. baumannii |
| S347 | 18 | F | Inpatient | Urinary tract infection | AMP, CAF | Urine | Cervical Spine fracture | A. baumannii |
*Current antibiotics: CRO, ceftriaxone; Met, metronidazole; VAN, vancomycin; CLO, cloxacillin; CAF, chloramphenicol; CIP, ciprofloxacin; AMP, ampicillin; GENT, gentamicin.
**Underlying diseases: COPD, congestive obstructive pulmonary disease; T2DM, type-2 diabetes mellitus.
Species diversity and antimicrobial susceptibility
Species diversity
From a total of 41 isolates of Pseudomonas spp., 92.6% (38/41) were P. aeruginosa, and 7.3% (3/41) were another Pseudomonas spp. Among 39 isolates of Acinetobacter spp. (n = 39), 79.5% (31/39) were A. baumannii, and 20.5% (8/39) were other Acinetobacter spp. [A. junii which account for 5.1% (2/39), and A. baylyi, A. lwoffii, A. calcoaceticus, A. haemolyticus, A. parvus, and A. schindleri each account for 2.5% (1/39)].
Antimicrobial susceptibility pattern
Among isolates of Pseudomonas spp., 31.7%, (13/41) were non-susceptible to ceftazidime, and 7.3% (3/41) to meropenem. Among isolates of Acinetobacter spp., 56.4% (22/39) were carbapenem-resistant. Most of the ceftazidime-resistant Pseudomonas spp. were also resistant to piperacillin-tazobactam 84.6% (11/13), ciprofloxacin 30.7% (4/13), ceftazidime-avibactam 46.1% (6/13) and ceftolozane-tazobactam 53.8% (7/13). However, most of these isolates were susceptible to cefiderocol 84.7% (11/13), imipenem-relebactam 92.3% (12/13), and all Pseudomonas isolates were susceptible to amikacin (Table 2A). Most of the Acinetobacter isolates were resistant to meropenem, imipenem, and imipenem-relebactam 54.5%, (12/22). However, a lower rate of resistance was detected to ciprofloxacin 18.1% (4/22), gentamicin 27.2% (6/22), amikacin 18.1% (4/22), and cefiderocol 9.1% (2/22) (Table 2B).
TABLE 2A.
Antimicrobial susceptibility pattern of ceftazidime resistant Pseudomonas spp. isolated at JMC, Ethiopia.
| Bacterial species |
Type of antimicrobials used for antimicrobial susceptibility testing |
||||||||||
| Ceftazidime | Meropenem | Imipenem | Piperacillin-tazobactam | Ciprofloxacin | Amikacin | Cefiderocol | Ceftazidime-avibactam | Ceftolozane-tazobactam | Imipenem-relebactam | ||
| Pseudomonas aeruginosa (n = 10) | S (%) | 0 (0.0%) | 7 (70) | 8 (80) | 0 (0) | 7 (70) | 10 (100) | 9 (90) | 5 (50) | 6 (60) | 9 (90) |
| R (%) | 10 (100) | 3 (30) | 2 (20) | 10 (100) | 3 (30) | 0 (0) | 1 (10) | 5 (50) | 4 (40) | 1 (10) | |
| Other Pseudomonas species (n = 3) | S (%) | 1 (32.4) | 1 (32.4) | 3 (100) | 1 (32.4) | 2 (66.6) | 3 (100) | 2 (66.6) | 2 (66.6) | 0 (0) | 3 (100) |
| R (%) | 2 (66.6) | 2 (66.6) | 0 (0) | 2 (66.6) | 1 (32.4) | 0 (0) | 1 (32.4) | 1 (32.4) | 3 (100) | 0 (0) | |
S, susceptible; R, resistant, other Pseudomonas species [Pseudomonas putida (n = 2), Pseudomonas fulva (n = 1)].
TABLE 2B.
Antimicrobial susceptibility pattern of carbapenem resistant Acinetobacter species isolated at JMC, Ethiopia.
| Bacterial species |
Type of antimicrobials used for antimicrobial susceptibility testing |
||||||||
| Meropenem | Imipenem | Imipenem-relebactam | Cefiderocol | Ciprofloxacin | Trimethoprim/sulfamethoxazole | Gentamicin | Amikacin | ||
| Acinetobacter baumannii (n = 22) | S (%) | 5 (27.8) | 9 (50) | 9 (50) | 17 (94.4) | 14 (77.8) | 12 (66.7) | 12 (66.7) | 15 (83.3) |
| R (%) | 13 (72.2) | 9 (50) | 9 (50) | 1 (5.6) | 4 (22.2) | 6 (33.3) | 6 (33.3) | 3 (16.7) | |
| Other Acinetobacter spp. | S (%) | 1 (25) | 1 (25) | 1 (25) | 4 (100) | 1 (25) | 0 (0) | 4 (100) | 4 (100) |
| R (%) | 3 (75) | 3 (75) | 3 (75) | 0 (0) | 3 (75) | 4 (100) | 0 (0) | (100) | |
S, susceptible; R, resistant; Other Acinetobacter spp. (n = 4). (A. calcoaceticus, A. baylyi, A. junii, and A. lwoffii).
Resistome profile of Pseudomonas and Acinetobacter species
Resistome profiling showed that both Pseudomonas and Acinetobacter spp. encoded multiple β - lactamase genes. The blaOXA–486, blaOXA–50 and blaCTX–M–15 genes were most common among Pseudomonas isolates, and the blaOXA–51-like blaOXA–69, blaOXA–66, blaOXA–91, blaOXA–180 and blaGES–11 were the most common among Acinetobacter isolates (Table 3A).
TABLE 3A.
Sequence types and resistance genes observed among carbapenem-resistant Acinetobacter species isolated at Jimma Medical Center, Ethiopia (n = 19).
| Isolate ID | STs | Number and type of antimicrobial resistance genes |
||||||
| Carbapenemase | ESBL and other β -lactamases | Aminoglycosides | Trimethoprim | Sulfonamides | Tetracycline | Macrolides | ||
| I027AB* | ST2 | bla OXA–69 | bla GES–11 | aac(6′)-Ib3, aac(6′)-Ib-cr | dfrA7 | sul1 | ||
| I030AB | ST1 | bla OXA–69 | bla GES–11 | aac(6′)-Ib3, aac(6′)-Ib-cr | dfrA7 | sul1 | ||
| M217AB | ST1 | bla OXA–69 | bla GES–11 | aac(6′)-Ib3, aac(6′)-Ib-cr | dfrA1, | sul1 | ||
| M328AB | 164 | bla OXA–91 | blaCARB–5, blaCARB–49 | tet(39) | ||||
| P015AL | NA | blaCTX–M–15, blaOXA–1, | aac(6′)-Ib-cr | tet(A) | mdf(A), mph(A), mph(D) | |||
| S126AB | ST2090 | bla OXA–259/180 | ||||||
| S130AB | ST85 | blaNDM–1, blaOXA–94 | ant(2″)-Ia, aph(3′)-VI | sul2 | mph(E) msr(E) | |||
| S161AB | ST2 | bla OXA–66 | blaTEM–1D, | aac(3)-Ia, aph(6)-Id | sul1 | |||
| S167AB | ST1 | bla OXA–69 | bla GES–11 | aac(6′)-Ib-cr, aac(6′)-Ib3 | dfrA7 | sul1 | ||
| S170AB | ST164 | bla OXA–91 | blaCARB–49, blaCARB–16 | tet(39) | ||||
| S171AB | ST1 | bla OXA–69 | bla GES–11 | aac(6′)-Ib3, aac(6′)-Ib-cr | dfrA7 | sul1 | ||
| S176AB | ST1 | bla OXA–69 | bla GES–11 | aac(6′)-Ib3, aac(6′)-Ib-cr | dfrA7 | sul1 | ||
| S209AB | ST2 | bla OXA–66 | aac(3)-Ia, aadA1, aph(6)-Id | sul1sul2 | tet(B) | |||
| S212AB | ST1 | bla OXA–69 | bla GES–11 | aac(6′)-Ib3, aac(6′)-Ib-cr, | dfrA7 | sul1 | ||
| S270AB | ST1 | bla OXA–69 | bla GES–11 | aac(6′)-Ib3, aac(6′)-Ib-cr | dfrA7 | sul1 | ||
| S275AB | ST1 | bla OXA–69 | bla GES–11 | aac(6′)-Ib3, aac(6′)-Ib-cr | dfrA7 | sul1 | ||
| S296AB | ST1 | bla OXA–69 | bla GES–11 | aac(6′)-Ib3, aac(6′)-Ib-cr | dfrA7 | sul1 | ||
| S315AB | ST1 | bla OXA–69 | bla GES–11 | aac(6′)-Ib3, aac(6′)-Ib-cr | dfrA7 | sul1 | ||
| S327AB | ST1 | blaOXA–69 | ||||||
Moreover, two isolates, P. aeruginosa (n = 1) and A. baumannii (n = 1) carrying the blaNDM–1 carbapenemase gene were detected. Furthermore, the resistome profile shows that resistance genes to the antimicrobial classes of aminoglycosides, fluoroquinolones, and trimethoprim were prevalent among these two isolates (Tables 3A,B).
TABLE 3B.
Sequence types and resistance genes observed among ceftazidime-resistant Pseudomonas spp. isolated at Jimma Medical Center, Ethiopia (n = 13).
| Strain ID | STs | Number and type of antimicrobial resistance genes |
||||||
| Carbapenemase | β -lactamase genes | Aminoglycosides | Fluoroquinolones | Sulfonamides | Phenicols | Fosfomycin | ||
| I020PA* | 11 | blaOXA–486, blaPAO | aph (3′)-IIb | catB7 | fosA | |||
| I032PA | 2948 | bla NDM–1 | blaOXA–10, blaOXA–50 | aadA1, ant(2″)-Ia, aph(3′)-IIb | crpP | sul1 | catB7, cmlA1 | fosA |
| I038PP* | NA | blaCTX–M–15, blaOXA–1 | aac(6′)-Ib-cr, | |||||
| M019PA | 1228 | blavEB–1, blaOXA–486 blaOXA–10 | aadA1, aadA2 ant(2″)-Ia, aph(3′)-IIb | crpP qnrD1 | sul1 | catB7 cmlA1 | fosA | |
| M119PA | 274 | blaOXA–486, blaPAO, | catB7 | fosA | ||||
| M304PA | 274 | blaOXA–486, blaPAO | catB7 | fosA | ||||
| S116PA | 500 | blaOXA–486, blaPAO | crpP | catB7 | fosA | |||
| S114PP* | NA | blaCTX–M–15, blaOXA–10 blaOXA–1 | aac(3)-Iia, aac(3)-Ib aac(6′)-Ib, aac(6′)-Ib-cr, ant(3″)-Ia, aph(3″)-Ib | oqxB qnrVC1 | sul1 | |||
| S155PA | 274 | blaOXA–486, blaPAO | aph(3′)-IIb | catB7 | fosA | |||
| S248PA | 840 | blaOXA–486, blaPAO | aph(3′)-IIb | crpP | catB7 | fosA | ||
| S332PF** | NA | blaPER–1, | aph(6)-Id | qnrVC6 | sul1 | |||
| S356PA | 646 | blaCTX–M–15, blaOXA–494 blaOXA–50, blaTEM–1B | ant(2″)-Ia, aph(3′)-IIb aph(3′)-Ia, ph(6)-Id | sul1 | catB7, floR | fosA | ||
| S047PA | 244 | bla OXA–50 | aph(3′)-IIb | catB7 | fosA | |||
*A. baumannii; PA, Pseudomonas aeruginosa; PP**, Pseudomonas putida; PF**, Pseudomonas fulva.
Molecular epidemiology using cgMLST
Epidemiologic typing using the seven gene multi-locus sequence typing demonstrated that ST274 (23.0%, 3/13) among P. aeruginosa and ST1 (MLST-Pasteur) A. baumannii (63.1%, 12/19) were the most prevalent sequence types (Tables 3A,B). A cgMLST analysis showed that the ST1 A. baumannii isolates were highly similar with no allelic differences between them. Most of the Acinetobacter isolates belonged to the international clonal complexes, ICC1 (includes ST1) (n = 12) and ICC2 (n = 2) (Table 3A).
Discussion
In this study, nearly all isolates of both Pseudomonas and Acinetobacter spp. were from patients admitted to the hospital for more than 72 h. Nosocomial acquisition of MDR isolates of Pseudomonas spp. and Acinetobacter spp. is worrisome. The situation is complicated by limited availability of antimicrobial agents, lack of prescription guidelines, and insufficient standard routine microbiology laboratory services to support antibiotic selection. In such cases, the safety of patients admitted to the hospital can be severely compromised.
The prevalence of ESBL-producing strains among P. aeruginosa, and carbapenem non-susceptible isolates among A. baumannii strains was high in the present study. Previous phenotypic studies from Ethiopia also show that MDR strains of Pseudomonas and Acinetobacter were prevalent (Motbainor et al., 2020; Birhane Fiseha et al., 2021). However, most of these studies did not describe the genotypes and resistome profile of the isolates, and mechanism of resistance is difficult to compare between studies. So far, to our knowledge, there is no report of a genotypic study on the prevalence of ESBL-producing P. aeruginosa in Ethiopia. Generally, there are limited studies conducted on carbapenemase-producing P. aeruginosa in Africa. The few reports available are from northern Africa and mainly from Egypt (Osei Sekyere and Reta, 2020; Rizk et al., 2021). In northern Africa, the prevalence ranges from 0 to 96% (Gaballah et al., 2018). A finding from Uganda (7.4%) was comparable to the present study (Aruhomukama et al., 2019). But, one study from South Africa (51.0%) reported a higher prevalence of ceftazidime resistant P. aeruginosa compared to the present study (Hosu et al., 2021). The phenotypic studies from Ethiopia, and other African countries showed higher prevalence as compared to the present study.
In Ethiopia, a high prevalence of carbapenem-resistant Acinetobacter spp. was reported from one previous phenotypic study (Ayenew et al., 2021), which is comparable to the prevalence of carbapenem resistance herein. On the other hand, a systematic review and meta analysis on carbapenemase producing P. aeruginosa and A. baumannii in Africa showed that the lowest prevalence of carbapenemase-producing A. baumannii was 4.7% (n = 21), and the highest prevalence was 100% (n = 7) (Kindu et al., 2020). Studies conducted on P. aeruginosa and A. baumannii strains from Africa were limited to small sample sizes and were mainly phenotypic studies, which makes the comparison with genotypic studies difficult (Kindu et al., 2020; Osei Sekyere and Reta, 2020; Ayenew et al., 2021; Mekonnen et al., 2021). However, most available studies including a study for hospital environment (Solomon et al., 2017) reported higher prevalence of the MDR P. aeruginosa and A. baumannii in Ethiopia, calling for the application of genotypic methods to studies on mechanisms of resistance and spread.
The multiple genetic variants of antibiotic resistance observed among both Pseudomonas and Acinetobacter spp. pose a huge challenge on the limited therapeutic options available to low-income countries. Among Acinetobacter spp., the presence of the blaGES–11 (ESBL-genotype and weak carbapenemase) and the OXA-51-like (blaOXA–66 and blaOXA–69) carbapenemases encoding genes is a serious threat. The OXA-51-like intrinsic carbapenemase encoding ST1 A. baumannii Isolates were reported from India (Rose et al., 2021), but isolates from the present study encoded additionally the weakly carbapenem hydrolyzing blaGES–11 gene. The blaGES–11 ESBL-genotypes, and the blaOXA–51-like carbapenemases have not been previously reported from Ethiopia, and the present study is to our knowledge the first report of blaOXA–51 like and blaGES–11 from Ethiopia. Generally from Africa, only one study from Tunisia has documented clinical isolates of A. baumannii encoding the blaGES–11 (Chihi et al., 2016). The emergence of blaNDM–1 encoding isolates of A. baumannii at surgical ward, and P. aeruginosa at intensive care unit can severely compromise the safety of vulnerable patients admitted to the hospital. Moreover, detection of two isolates encoding the blaNDM–1 which showed resistance to the newer antimicrobials, A. baumannii for (cefiderocol and imipenem-relebactam), and P. aeruginosa for (cefiderocol, ceftazidime-avibactam, ceftolozane-tazobactam) compromises the already limited treatment alternatives for vulnerable groups of patients at this hospital.
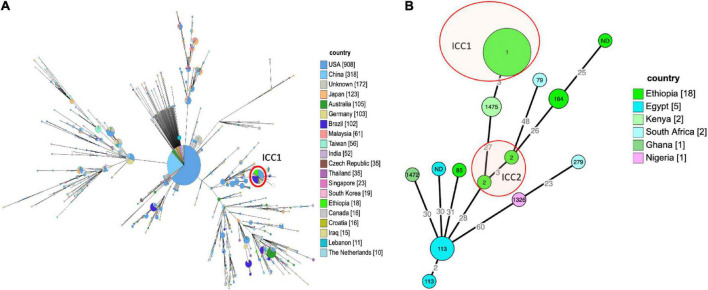
Sequence typing showed that both P. aeruginosa and A. baumannii strains were polyclonal. But, since a large proportion of A. baumannii isolates were ST1, the spread of A. baumannii isolates at this hospital might also be to some extent clonal. A previous study conducted at the same hospital had identified three strains of A. baumannii encoding blaNDM–1, and all of them belonged to the ST597. Furthermore, cgMLST analysis of A. baumannii with other international isolates in pubmlst showed that isolates in the current study were distinct from isolates from other African countries (Figure 1).6 However, these isolates were clustered with isolates from other countries like United States, and Brazil. Though isolates from this study were polyclonal, the most prevalent isolates were those that belong to the international cluster, CC1 and CC2.
FIGURE 1.
(A) Epidemiologic relatedness of A. baumannii isolates to the isolates collected from different geographical regions and deposited in the public database (pubmlst https://pubmlst.org/organisms/acinetobacter-baumannii~-- accessed on 26-09-2021), the isolates from this study (bright green) were clustered with the global ICC1 clone marked with red circle. The numbers in the circle show the sequence type of the isolates, the size of the circle is proportional to the number of isolates that belongs to that sequence type, and the color shows the country of origin for the isolates. The minimum spanning tree was constructed for isolates from different country of origin having ≥ 10 isolates recorded in the database. (B) Minimum spanning tree of 29 A. baumannii isolates from Africa, the tree was constructed based on core genome multi-locus sequence typing. There were no previous isolates from Ethiopia in PubMLST, all isolates labeled Ethiopia in the tree are from this study.
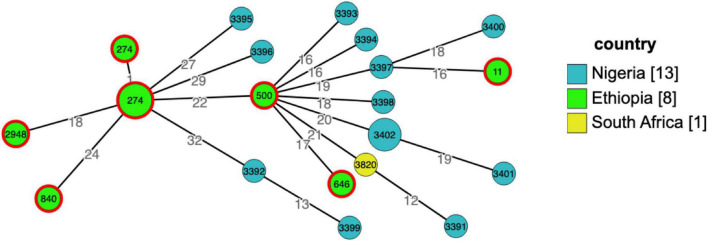
Although A. baumannii is a well-known major cause of nosocomial infections, knowledge of its genomic epidemiology and availability of reliable data regarding the genetic basis of antibiotic resistance is limited in low-income countries. Similarly, P. aeruginosa isolates were found to be polyclonal, and different from a collection of isolates other African counties found in pubmlst (see text footnote 5) (Figure 2).
FIGURE 2.
Grape tree generated using cgMLST from genomic collection of Pseudomonas aeruginosa isolates from Africa (n = 22), Ethiopia, Nigeria, and South Africa, pubmlst database for microorganisms (https://pubmlst.org/bigsdb?db=pubmlst_paeruginosa_isolates&page=query&genomes=1) accessed on 29-11-2021. The numbers in circles/nodes indicate sequence types of the isolates, size of the nodes is proportional to the number of isolates in a particular sequence type and the numbers on the branches are the number of allelic differences between the two neighboring nodes. From 10 isolates sequenced in this study, eight were included in the tree, two of the sequences failed quality required to upload to the pubmlst database.
The current study may serve as a baseline regarding local spread of international clones and alert clinicians and other health workers, researchers, and public health policy makers to the problem. Implementation of strict infection prevention and control strategies, and antimicrobial stewardship programs are highly desirable in the admission wards where the international clones are spreading. Furthermore, despite limitation of resources, the added value of next generation sequencing is in understanding the dynamics and mechanisms of spread of MDR bacterial clones.
Conclusion
The prevalence of MDR isolates is high among both in clinical isolates of Pseudomonas species and Acinetobacter species at Jimma medical center. Emergence of the blaNDM–1 in clinical isolates of P. aeruginosa and A. baumannii strains is worrisome. However, the susceptibility of P. aeruginosa strains to amikacin, cefiderocol, imipenem-relebactam and ceftolozane-tazobactam, and A. baumannii strains to amikacin and cefiderocol is important to consider as alternative options to carbapenems. The use of next generation sequencing is important to understand the mechanism of resistance and spread of resistant clones such as ICC1, and ICC2 A. baumannii strains detected at this hospital.
Data availability statement
The genome sequences were deposited at the NCBI, SRA database (PRJNA593604, Biosample accession: SUB11593554).
Ethics statement
The study obtained ethical approval from Addis Ababa University Institutional Review Board (AAU-IRB), Armauer Hansen Research Institute – ALERT Hospital Institutional Review Board (AHRI-ALERT-IRB), and Ethiopian National Ethics Review Committee (NERC). Patients were informed about the study and given written consent to participate in the study.
Author contributions
TS contributed to design, data acquisition, data analysis, and drafting of the manuscript. DA and YW contributed to data acquisition and write-up. AA contributed to the design, data acquisition, and revision of the manuscript. CG contributed to the overall design, data acquisition, supervision, drafting, and writing of the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
We acknowledge Isak Sylvin for technical support for genome assembly for both P. aeruginosa and A. baumannii isolates.
Footnotes
Funding
This work was supported by the Swedish International Development Agency (SIDA).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Agnese L., Marisa H. J., Ean-Yves M. (2018). Antimicrobial resistance in Acinetobacter spp. and Pseudomonas spp. Microbiol. Spectr. 6. 10.1128/microbiolspec.ARBA-0007-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruhomukama D., Najjuka C. F., Kajumbula H., Okee M., Mboowa G., Sserwadda I., et al. (2019). BlaVIM- and blaOXA-mediated carbapenem resistance among Acinetobacter baumannii and Pseudomonas aeruginosa isolates from the Mulago hospital intensive care unit in Kampala, Uganda. BMC Infect. Dis. 19:853. 10.1186/s12879-019-4510-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayenew Z., Tigabu E., Syoum E., Ebrahim S., Assefa D., Tsige E. (2021). Multidrug resistance pattern of Acinetobacter species isolated from clinical specimens referred to the Ethiopian Public Health Institute: 2014 to 2018 trend anaylsis. PLoS One 16:e0250896. 10.1371/journal.pone.0250896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassetti M., Carnelutti A., Peghin M. (2017). Patient specific risk stratification for antimicrobial resistance and possible treatment strategies in gram-negative bacterial infections. Expert Rev. Anti Infect. Ther. 15 55–65. 10.1080/14787210.2017.1251840 [DOI] [PubMed] [Google Scholar]
- Bello-López E., Rocha-Gracia R. D. C., Castro-Jaimes S., Cevallos M. Á, Vargas-Cruz M., Verdugo-Yocupicio R., et al. (2020). Antibiotic resistance mechanisms in Acinetobacter spp. Strains isolated from patients in a paediatric hospital in Mexico. J. Glob. Antimicrob. Resist. 23 120–129. 10.1016/j.jgar.2020.08.014 [DOI] [PubMed] [Google Scholar]
- Bergogne-Bérézin E., Towner K. J. (1996). Acinetobacter spp. As nosocomial pathogens: Microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9 148–165. 10.1128/cmr.9.2.148-165.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birhane Fiseha S., Mulatu Jara G., Azerefegn Woldetsadik E., Belayneh Bekele F., Mohammed Ali M. (2021). Colonization rate of potential neonatal disease-causing bacteria, associated factors, and antimicrobial susceptibility profile among pregnant women attending government hospitals in Hawassa, Ethiopia. Infect. Drug Resist. 14 3159–3168. 10.2147/idr.s326200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink A. J. (2019). Epidemiology of carbapenem-resistant Gram-negative infections globally. Curr. Opin. Infect. Dis. 32 609–616. [DOI] [PubMed] [Google Scholar]
- Chihi H., Bonnin R. A., Bourouis A., Mahrouki S., Besbes S., Moussa M. B., et al. (2016). GES-11-producing Acinetobacter baumannii clinical isolates from Tunisian hospitals: Long-term dissemination of GES-type carbapenemases in North Africa. J. Glob. Antimicrob. Resist. 5 47–50. 10.1016/j.jgar.2016.03.005 [DOI] [PubMed] [Google Scholar]
- Chusri S., Chongsuvivatwong V., Rivera J. I., Silpapojakul K., Singkhamanan K., McNeil E., et al. (2014). Clinical outcomes of hospital-acquired infection with Acinetobacter nosocomialis and Acinetobacter pittii. Antimicrob. Agents Chemother. 58 4172–4179. 10.1128/AAC.02992-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Oliveira D. M. P., Forde B. M., Kidd T. J., Harris P. N. A., Schembri M. A., Beatson S. A., et al. (2020). Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 33:e00181-19. 10.1128/CMR.00181-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djahmi N., Dunyach-Remy C., Pantel A., Dekhil M., Sotto A., Lavigne J. P. (2014). Epidemiology of Carbapenemase-producing Enterobacteriaceae and Acinetobacter baumannii in Mediterranean Countries. Biomed. Res. Int. 2014:305784. 10.1155/2014/305784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberger E. M., Thaden J. T. (2019). Epidemiology and Mechanisms of resistance of extensively drug resistant gram-negative bacteria. Antibiotics 8:37. 10.3390/antibiotics8020037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballah A., Elbaradei A., Elbaradei A. (2018). Emergence of blaVEB and blaGES among VIM-producing Pseudomonas aeruginosa clinical isolates in Alexandria, Egypt. Acta Microbiol. Immunol. Hung. 66 131–142. 10.1556/030.65.2018.044 [DOI] [PubMed] [Google Scholar]
- Horcajada J. P., Montero M., Oliver A., Sorlí L., Luque S., Gómez-Zorrilla S., et al. (2019). Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 32:e00031-19. 10.1128/CMR.00031-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosu M. C., Vasaikar S. D., Okuthe G. E., Apalata T. (2021). Detection of extended spectrum b-lactamase genes in Pseudomonas aeruginosa isolated from patients in rural Eastern Cape Province, South Africa. Sci. Rep. 11 1–8. 10.1038/s41598-021-86570-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak K. M., de Jonge B. L. M., Stone G. G., Sahm D. F. (2020). Longitudinal analysis of ESBL and carbapenemase carriage among Enterobacterales and Pseudomonas aeruginosa isolates collected in Europe as part of the International Network for Optimal Resistance Monitoring (INFORM) global surveillance programme, 2013–17. J. Antimicrob. Chemother. 75 1165–1173. 10.1093/jac/dkz571 [DOI] [PubMed] [Google Scholar]
- Kindu M., Derseh L., Gelaw B., Moges F. (2020). Carbapenemase-Producing non-glucose-fermenting gram-negative Bacilli in Africa, Pseudomonas aeruginosa and Acinetobacter baumannii: A systematic review and meta-analysis. Int. J. Microbiol. 2020:9461901. 10.1155/2020/9461901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. X., Wang C. Y., Li Y. Y., Li J., Wan Q. Q., Chen J. H., et al. (2020). Considerations and caveats in combating ESKAPE pathogens against nosocomial infections. Adv. Sci. 7:1901872. 10.1002/advs.201901872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekonnen H., Seid A., Molla Fenta G., Gebrecherkos T. (2021). Antimicrobial resistance profiles and associated factors of Acinetobacter and Pseudomonas aeruginosa nosocomial infection among patients admitted at Dessie comprehensive specialized Hospital, NorthEast Ethiopia: A cross-sectional study. PLoS One 16:e0257272. 10.1371/journal.pone.0257272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Liu H., Lan T., Dong L., Hu H., Zhao S., et al. (2020). Antibiotic resistance patterns of Pseudomonas spp. isolated from raw milk revealed by whole genome sequencing. Front. Microbiol. 11:1005. 10.3389/fmicb.2020.01005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motbainor H., Bereded F., Mulu W. (2020). Multi-drug resistance of blood stream, urinary tract and surgical site nosocomial infections of Acinetobacter baumannii and Pseudomonas aeruginosa among patients hospitalized at Felegehiwot referral hospital, Northwest Ethiopia: A cross-sectional study. BMC Infect. Dis. 20:92. 10.1186/s12879-020-4811-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Price L. S., Rosa R., Castro J. G., Laowansiri P., Latibeaudiere R., Namias N., et al. (2016). Evaluating the impact of antibiotic exposures as time-dependent variables on the acquisition of carbapenem-resistant Acinetobacter baumannii. Crit. Care Med. 44 e949–e956. 10.1097/CCM.0000000000001848 [DOI] [PubMed] [Google Scholar]
- Osei Sekyere J., Reta M. A. (2020). Genomic and resistance epidemiology of gram-negative bacteria in Africa: A systematic review and phylogenomic analyses from a one health perspective. mSystems 5:e00897-20. 10.1128/msystems.00897-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk S. S., Elwakil W. H., Attia A. S. (2021). Antibiotic-resistant Acinetobacter baumannii in low-income countries (2000–2020): Twenty-one years and still below the radar, is it not there or can they not afford to look for it? Antibiotics 10:764. 10.3390/antibiotics10070764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose S., Shamanna V., Underwood A., Nagaraj G., Prasanna A., Govindan V., et al. (2021). Molecular dissection of carbapenem-resistant Acinetobacter baumannii circulating in Indian hospitals using whole genome sequencing. bioRxiv [Preprint]. 451. 10.1101/2021.07.30.454432 [DOI] [Google Scholar]
- Solomon F. B., Wadilo F., Tufa E. G., Mitiku M. (2017). Extended spectrum and metalo b-lactamase producing airborne Pseudomonas aeruginosa and Acinetobacter baumanii in restricted settings of a referral hospital: A neglected condition’. Antimicrob. Resist. Infect. Control 6:106. 10.1186/s13756-017-0266-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2017). WHO publishes list of bacteria for which new antibiotics are urgently needed. Available online at: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed September 3, 2021). [Google Scholar]
- Wong D., Nielsen T. B., Bonomo R. A., Pantapalangkoor P., Luna B., Spellberg B., et al. (2017). Clinical and pathophysiological overview of Acinetobacter infections: A century of challenges. Clin. Microbiol. Rev. 30 409–447. 10.1128/CMR.00058-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequences were deposited at the NCBI, SRA database (PRJNA593604, Biosample accession: SUB11593554).