Summary
Background
The hospital environment serves as a reservoir of microorganisms which may be associated with healthcare-associated infections (HCAI). The study of environmental contamination with microorganisms is a method for the assessment of hospital environmental hygiene. We sought to evaluate the environmental colonisation of a national reference hospital unit, using the total aerobic colony count (ACC) and the isolated microorganisms, as assessment tools.
Methods
A cross-sectional study was conducted in the Paediatric Intensive Care Unit (PICU) of the Hospital Central de Maputo during a four-week period in 2018. Surfaces and air were sampled before and after room cleaning, using swabs and passive air method. Those samples were processed at the microbiology laboratory where total ACC levels were evaluated, and microorganisms were isolated, identified and assessed for antibiotic susceptibility.
Discussion
Comparison of the total median ACC of the indoor air (287 cfu/m3 before and 195 cfu/m3 after) and surfaces (0.38 cfu/cm2 before and 0.33 cfu/cm2 after) before and after room cleaning did not show significant differences (P>0.05). Microorganisms of epidemiological importance, including coagulase negative staphylococci (CoNS), Klebsiella pneumoniae and Serratia odorifera were isolated and all of these three were multi-drug resistant (MDR).
Conclusion
The results showed controlled contamination levels on high touch surfaces in the patient environment and a high level of contamination of the indoor air suggesting deficiencies in the PICU environmental decontamination process. There was evidence of the presence of fungi and MDR species of epidemiological importance in the context of HCAI.
Keywords: Contamination, Environment, Surfaces, Healthcare-associated infections
Introduction
Healthcare-associated infections (HCAI) are a public health problem which result in significant costs and increased morbidity and mortality [1] especially in developing countries [2,3]. The WHO reported a prevalence of HCAI of 7.5% in high income countries [4] and, due to the scarcity of data, the prevalence for developing countries was estimated to be between 5.1% to 19.1% [4]. The HCAI burden is worsened by the worldwide crises of antimicrobial resistance, which, represents a challenge for infection treatment especially in healthcare facilities in low-resource countries. This issue has been associated with empirical antimicrobial administration due to a lack of diagnostic capacity [5,6] and the widespread use of antibiotics without the need for a prescription [7,8]. Many studies have enhanced the awareness of the role of the hospital environment as a reservoir for pathogenic microorganisms and the importance and benefits of evaluation of hospital environment hygiene [[9], [10], [11], [12]]. An interventional study conducted in an intensive care unit (ICU) in Argentina, showed a reduction of acquisition and HCAI caused by multi-drug resistant organisms (MDR) from 4.3% before to 2% after an intervention [13]. The intervention consisted in the allocation of an environment hygiene nurse full time in the ICU, who was dedicated to the control and monitoring of staff hygiene and ward cleaning practices [13].
Ward surfaces may become contaminated by infected patients or indirectly by vehicles such as healthcare workers' hands [14]. Surfaces such as bed rails, supply cart, hand washing sink are classified as high touch surfaces because more than other surfaces these are exposed to a higher interaction with the patient and with healthcare workers [15,16].
The most frequently isolated microorganisms in the hospital environment include coagulase negative staphylococci (CoNS), Klebsiella pneumoniae, Acinetobacter baumannii and Enterobacter spp. and fungi including Aspergillus spp. Many of the bacteria isolated are resistant to β-lactam and cephalosporin antibiotics of second generation [2,14,17,18]. These microorganisms are considered of epidemiological importance for HCAI, because they have been isolated in previous HCAI outbreaks or reported in previous studies of HCAI, and are resistant to first line or multiple antibiotics, or represent an remerging pathogen [14].
Hospital cleaning assessment is a method for the evaluation of the surfaces total aerobic colony count (total ACC) which comprises the concentration of microorganisms calculated from the total number of colonies regardless the isolated microorganism and the use of an indicator microorganism of epidemiological importance on HCAI context for that specific hospital site [19,20].
Despite the existing literature on the subject, no such data exist for Mozambican hospitals. This study aims to evaluate the environmental hygiene of a paediatric intensive care unit in Maputo Central Hospital, using as assessment tools the total ACC of indoor air and selected high touch surfaces and the resident microorganisms and antimicrobial susceptibility.
Methods
Study design
A cross-sectional study using convenience sampling methods, was carried out in the paediatric intensive care unit (PICU) of Maputo Central Hospital and in the microbiology laboratory of the University of Eduardo Mondlane Faculty of Medicine from April to May 2018 for four weeks. The PICU had 20 beds divided into five rooms. Environment sampling was conducted for the indoor air and for eight patient zone high touch surfaces (referred to in the remaining manuscript as surfaces) comprising the headboard bed rail, footboard bed rail, lateral bed rail, over bed table, intravenous pump, bedside table, hand washing sink, and infusion stand, as the sample collection points. Sample collection was done over a four-week period. Once a week an occupied room was visited twice, before and after room cleaning, to assess cleaning efficacy. The ward cleaning routine was twice per day between 04.00 to 06.00 and at 08.00. For ward cleaning, commercial bleach solutions (commonly with a sodium hypochlorite concentration of 3.5%) were used on the floor according to manufacturer's specification for dilution and alcohol at 70% for surfaces. In each visit, samples were taken from the indoor air and from four selected study surfaces.
Environment sampling
Study methods for sample collection were adapted from Tagoe et. al. [21] and Galvin et al. [22]. Indoor air samples were collected using a passive method, involving opening two sterile culture plates, one with blood agar medium and another with Sabouraud medium, in the room for 8 hours. At the time of the culture plates being collected before the first round of cleaning, three sterile swabs were used to collect samples from each one of the selected surfaces and then inoculated in three autoclaved 10ml tubes, one with meat broth, one with selenite broth and the last one with saline solution. The second environmental sampling took place, at least 30 minutes after the ward cleaning applying the same methods for the surfaces. The cultures plaques were left in the room and collected eight hours after.
In the laboratory, plates and swabs in broth were incubated at 35°C for 24 hours and theswabs in saline solution were immediately inoculated onto nutrient agar plates and then incubated in the same conditions. After incubation, the meat broth swabs were inoculated on blood agar, chocolate agar and Sabouraud plates and the selenite broth was inoculated on xylose lysine deoxycholate (XLD) and MacConkey plates and incubated at 35°C for 24 hours. After incubation, nutrient agar plates (from swab surfaces), blood agar and Sabouraud (indoor air samples) plates were assessed for the direct colony count according to the (1), (2) for the air indoor and for surfaces, respectively, to calculate the total ACC.
| (1) |
| (2) |
where N = colony forming unit per cubic meter of air (cfu/m3) or surface (cfu/cm2), a = number of colonies per Petri dish, b = surface area of Petri dish in cm2, t = time exposure (minutes) and cfu= colony forming unit.
Microbial isolation and identification
Microorganisms were recovered from plates following culture [23,24]. Single colonies from cultured plates, were assessed for catalase, oxidase, and Gram staining. Species were identified using the following biochemical tests API 20E (for identification of Enterobacteriaceae species), API® 20NE (for identification of Non-Enterobacteriaceae Gram negative) API® NH (for identification of Neisseria sp., Haemophilus sp. and Moraxella catarrhalis) and API® Staph (for identification of Staphylococcus sp. and Micrococcus sp.) (bioMérieux, Marcy-l’Etoile, France) used according to the manufacturer's instructions.
Antibiotic susceptibility assays
Antibiotic susceptibility testing was done using the Kirby Bauer method and WHO guidelines for antimicrobial resistance surveillance testing [25]. Twelve antibiotic discs, 9 Oxoid (Oxoid Ltd.) and 3 Mastdiscs® (Mast Group Ltd.) (Ceftriaxone, Ceftazidime and Nalidixic Acid) were used. Both Gram positive and Gram negative organisms were tested against ceftazidime (CAZ) 30 μg; ciprofloxacin (CIP) 5 μg; chloramphenicol (C) 30μg; gentamicin (CN) 10 μg; trimethoprim-sulfamethoxazole (SXT) 25 μg; tetracycline (TE) 30 μg.
Gram negative organisms were tested against ampicillin (AMP) 10 μg; ceftriaxone (CRO) 30 μg; nalidixic acid (NA) 30 μg.
Gram positive organisms were tested against penicillin (P) 10 units; oxacillin (OX) 5 μg; erythromycin (E) 15 μg. Interpretation of susceptibility assays was done using the Clinical and Laboratory Standards Institute: CLSI 2018 guidelines [26].
Isolates intermediate or resistant to three or more antimicrobial categories were classified as multi-drug resistant organisms (MDR) in accordance with Magiorakis A et al. [27].
Statistical analysis
The data was analysed generating graphics for the sampled locations and frequency tables for isolated microorganisms and susceptibility results. Indoor air and surfaces median was paired by time of collection (before or after room cleaning) using the Wilcoxon paired test, with a significance of 0.05, on the SPSS® version 20 statistical package.
Results
Sample description
A total of 96 swabs and 16 plates of air indoor were collected in the PICU. One of the five rooms in PICU was empty during sampling period therefore no samples were collected in this room. The antibiotic susceptibility test assay of Bacillus sp. was done using the same antibiotics tested for CoNS but due to the lack of breakpoint guidelines for disc diffusion tests for those species, the results could not be interpreted. From the collected samples, nine inoculated swabs did not show any growth. The fungi isolated from PICU were not tested for antifungal susceptibility due to lack of resources.
Surfaces and air indoor total ACC levels
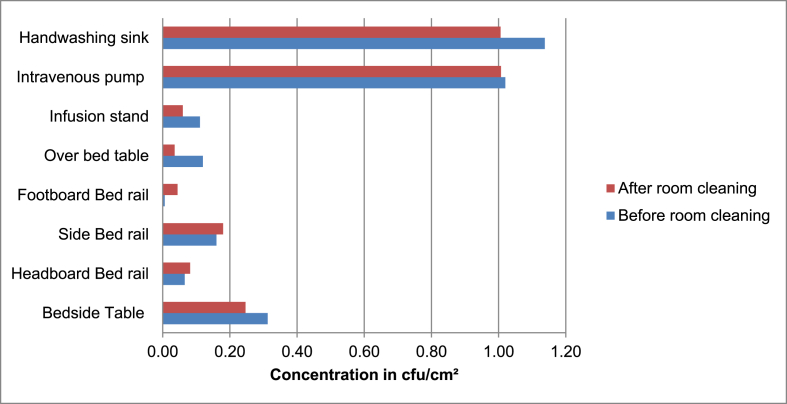
We observed in the indoor air a median concentration 287 cfu/m3 before cleaning and 195 cfu/m3 after cleaning without significative differences (p=0.144), with values ranging from 176 cfu/m3 to 287.35 cfu/m3. The surfaces presented a median concentration of 0.38 cfu/cm2 before cleaning and 0.33 cfu/cm2 after cleaning, with no significant differences (p=0.183), intravenous pump and hand washing sink presented the largest total ACC with a median of 1.02 cfu/cm2 and 1.13 cfu/cm2 respectively (Figure 1).
Figure 1.
Surfaces median total ACC, before and after room cleaning.
Microbial identification
From the total of 144 microorganisms isolated on the PICU ward, 118 (81.9%) were bacteria. There was a predominance of Gram-positive bacteria with a total of 75 (63.6%) versus 43 (36.4%) Gram negative. The frequency of isolation by surfaces showed the presence of CoNS in all sampled locations with a frequency of 51 (35.4%) isolates, followed by Klebsiella pneumoniae and Enterobacter cloacae both with five (3.5%) isolates, which are distributed as presented in Table I.
Table I.
Isolated microorganisms and distribution among the air indoor and surfaces
| Sampling site | Isolated microorganismsa |
|---|---|
| Indoor air | Mucor sp. (6); Aspergillus flavus (4); Aspergillus niger (4); Fusarium verticillioides (4); Paecilomyces variotii (3); Rhizopus sp. (3); CoNS (23) Serratia ficaria (2); Serratia odorifera (1); Burkholderia cepacia (1) |
| Bedside Table | Bacillus sp. (5), CoNS (3), Serratia plymuthica (2), Citrobacter koseri (2); Citrobacter braakii (1); Klebsiella pneumoniae (1), Acinetobacter baumannii (1) |
| Headboard rail | CoNS (2); Serratia odorifera (2); Pantoea sp. (2); Cronobacter (1); Mucor sp (1) |
| Side Bed rail | CoNS (5); Bacillus sp. (3); Raoultella ornithinolytica (1); Shigella sp. (1); Enterobacter cloacae (1); Pantoea sp. (1); Serratia odorifera (1); Haemophilus influenzae (1) |
| Footboard rail | CoNS (7); Bacillus sp. (2); Enterobacter amnigenus (1); Pseudomonas luteola (1); Moellerella wisconsensis (1) |
| Over bed table | CoNS (2); Klebsiella pneumoniae (2); Enterobacter cloacae (2); Bacillus sp. (1); Haemophilus parainfluenzae (1); Shigella sp. (1) |
| Infusion stand | Bacillus sp. (2); Serratia odorifera (2); Serratia rubidaea (1); Klebsiella oxytoca (1); Moraxella lacunata (1); Enterobacter cloacae (1). |
| Intravenous pump | CoNS (3); Klebsiella pneumoniae (1); Bacillus sp. (1) |
| Handwashing sink | CoNSb(6); Bacillus sp. (4); Klebsiella pneumoniae (1); Enterobacter cloacae (1); Acinetobacter baumanii (1) |
The numbers in brackets correspond to number of species isolates on the sampling site.
CoNS- coagulase negative staphylococci.
Antibiotic susceptibility assay
The isolated bacteria genera were grouped to show the resistance frequency. Ampicillin and trimethoprim-sulfamethoxazole had a resistance frequency of above 50% among all the groups (except for Enterobacter spp.). All Klebsiella spp. isolates were resistant to ceftriaxone and nalidixic acid as presented in Table II.
Table II.
Frequency of resistance to antibiotics by genus of epidemiological importance
| Antibiotic |
Serratia spp. (n=12) |
Klebsiella spp. (n=6)∖ |
CoNS (n=52) |
Enterobacter spp. (n=6) |
Citrobacter spp. (n=3) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | N | % | |
| Ampicillin | 8 | 67 | 5 | 83 | - | - | 6 | 100 | 3 | 100 |
| Ceftriaxone | 4 | 33 | 6 | 100 | - | - | 2 | 33 | - | - |
| Ciprofloxacin | 1 | 8 | 4 | 67 | 5 | 10 | - | - | - | - |
| Chloramphenicol | 6 | 50 | 3 | 50 | 17 | 33 | - | - | - | - |
| Trimethoprim-sulfamethoxazole | 6 | 50 | 5 | 83 | 27 | 52 | 1 | 17 | 2 | 67 |
| Erythromycin | - | - | - | - | 33 | 63 | - | - | - | - |
| Gentamicin | 4 | 33 | 5 | 83 | 18 | 35 | - | - | - | - |
| Penicillin | - | - | - | - | 42 | 81 | - | - | - | - |
| Tetracycline | 1 | 8 | 1 | 17 | 26 | 50 | 1 | 17 | 2 | 67 |
| Nalidixic Acid | 3 | 25 | 6 | 100 | - | - | - | - | - | - |
| Oxacillin | - | - | - | - | 34 | 65 | - | - | - | - |
| Ceftazidime | 6 | 50 | 2 | 33 | - | - | - | - | - | - |
Frequency of multi-drug resistant (MDR) bacteria was 63 (53.3%) and among them, 38% were CoNS with 12 different antibiotic resistance patterns. One isolate of A. baumannii was MDR with three antibiotic resistance patterns as presented on Table III. At least one of the K. pneumoniae and S. odorifera isolates, showed resistance to seven out of nine antibiotics evaluated on the Gram-negative bacteria.
Table III.
Pattern of antibiotic resistance of MDR bacteria
| Bacteria | Antibiotic resistance patterna | N of MDR (n=63) | N of non-susceptible antibiotic categories |
|---|---|---|---|
| CoNSb | C; STX; ER; CN; PE; TE; Ox | 12 | 6 |
| CIP; STX; ER; CN; PE; TE; Ox | 1 | 6 | |
| STX; ER; CN; PE; TE; Ox | 2 | 5 | |
| CIP; STX; CN; PE; TE; Ox | 1 | 5 | |
| CIP; STX; ER; CN; PE; Ox | 1 | 5 | |
| CIP; ER; CN; PE; Ox | 1 | 4 | |
| STX; CN; PE; TE; Ox | 1 | 4 | |
| STX; PE; TE; Ox | 2 | 3 | |
| STX; ER; PE; TE | 2 | 4 | |
| ER; PE; TE; Ox | 1 | 3 | |
| CIP; STX; ER; PE | 1 | 4 | |
| ER; PE; TE | 1 | 3 | |
| Serratia odorifera | AMP; CRO; CIP; C; STX; CN; TE; NA | 1 | 7 |
| AMP; CRO; CIP; C; STX; CN; NA | 1 | 5 | |
| AMP; CRO; C; STX; NA | 1 | 5 | |
| AMP; CZN; STX | 1 | 3 | |
| Serratia ficaria | AMP; CRO; C; STX; CN; CAZ | 2 | 5 |
| Klebsiella pneumoniae | AMP; CRO; CIP; C; STX; CN; TE; NA; CAZ | 1 | 7 |
| AMP; CRO; CIP; C; STX; CN; NA | 3 | 5 | |
| AMP; CRO; CIP; NA; CAZ | 1 | 3 | |
| Klebsiella oxytoca | AMP; NA; CN; STX; C; CIP; CRO | 1 | 6 |
| Enterobacter cloacae | AMP; CRO; STX; TE | 1 | 4 |
| AMP; CRO; NA | 2 | 3 | |
| Citrobacter koseri | AMP; STX; TE | 2 | 3 |
| Acinetobacter baumannii | AMP; STX; TE | 1 | 3 |
| Pantoea sp. | AMP; C; STX; NA | 1 | 4 |
Antibiotic: AMP- Ampicillin; CRO- Ceftriaxone; CIP- Ciprofloxacin; C- Chloramphenicol; SXT- Trimethoprim-sulfamethoxazole; E- Erythromycin; CN- Gentamicin; P- Penicillin; TE-Tetracycline; NA- Nalidicic Acid; OX- Oxacillin; CAZ- Ceftazidime.
CoNS- Coagulase-negative staphylococci.
Discussion
This study assessed the environmental contamination of a hospital ward, using two standard indicators of cleanliness, the total aerobic colony count (ACC) and indicator microorganisms [19,20].
For the ACC from all sampled surfaces, the study found a median concentration below 1.2 cfu/cm2 which can be considered low if compared the standard values of 2.5 cfu/cm2 [28] and 5 cfu/cm2 [19]. These results suggest that at the given time point the PICU ward surfaces may not have represented a risk for patients. On the other hand, the median indoor air total ACC was above 150 cfu/m3, the standard concentration [20] suggesting that the indoor air was potentially a riskier contamination site for patients.
Although low, the hand washing sink and the intravenous pump surfaces, presented the higher total ACC levels among the surfaces suggesting that those may be the main reservoirs of microorganisms in the PICU. Interestingly, a study in University hospital in North Carolina (USA) classified the hand washing sink as a low touch surface, and only selected bed rails, bed surface, supply cart, over-bed table, and intravenous pump as high touch [29]. This might be related to the study only observed the number of contacts that healthcare workers had with the surfaces and did not take in count the potential contribution of patient visitors. In the present study PICU, all children were accompanied by a relative who helped with the daily care of the patient. Therefore, the hand washing sink would be highly touched. Another explanation for the hand washing sink contamination values may be the possibility of contamination of the water supply system as observed in a study in Cameroon Hospital [30] where water samples had high colony counts and species such as Burkholderia cepacia and CoNS were isolated from water. In the present study no water samples were collected.
The intravenous pump is equipment solely used by healthcare professionals and may also be a main point of cross contamination. One interesting finding was that bed rails presented an increase on total ACC after cleaning suggesting that somehow, the method used on those locations is contaminating rather than cleaning. In addition, no significant difference was found between the median before and after cleaning in surfaces and indoor air, showing that cleaning method are not efficient in reducing the contamination levels in the ward, and raising concerns about the efficacy of the current disinfection methods to prevent or control HCAI and outbreaks.
Overall this finding somewhat surprising given that previous studies in hospital wards outside [18] and inside [30,31] Africa, that found surface ACC levels above 2.5 cfu/cm2, and indoor air ACC levels above 480 cfu/m3 [24,31], more than twice as high than found in the present study. The difference might be explained by several reasons such as differences in the indoor air collection method such as the 1-1-1 (1 hour open, 1m from the floor, 1m from the walls) for the passive indoor air sampling used on the studies [24,31]. Another explanation may be that a neutraliser was not used for residual chemical of cleaning on swabs as done in [18]. It is possible that the low bed occupancy compared with the other studies [31] may have impacted on ward surfaces and indoor air contamination load.
Regarding the assessment of cleaning by isolation of indicator microorganisms, we isolated bacteria and fungi of epidemiological importance in the context of HCAI in the PICU [2,3,14] and also isolated in the environment in previous studies [9,17,18]. The Gram-negative bacteria were the major isolated group on the sampled sites, with bacteria such as Klebsiella spp., and Enterobacter spp., known as important pathogens associated with HCAI including neonatal sepsis [32,33]. CoNS were the most isolated pathogens as seen in studies also in referral hospital environment in Ethiopia [34] and Cameroon [30]. All isolated fungi, were found in the indoor air, and included Aspergillus spp., a pathogen of high risk for immunocompromised patients [14,19].
We observed low susceptibility to AMP, SXT, P and Ox for the isolated pathogens, as reported in previous studies [33,35,36].
There were high susceptibility rates for most of the isolated species to CIP and CRO, but MDR Klebsiella sp. with a resistance rate of 67% and 100% respectively is a concern as these antibiotics are included in WHO guidelines as treatment for nosocomial meningitis and diarrhoea [37].
Together with Klebsiella spp. most of the isolated Enterobacteriaceae and CoNS was found to be MDR, increasing the potential burden of HCAI on the unit. The possibility of infection by one of these pathogens has raised concerns about the empirical antimicrobial treatment in hospitals in low resource countries, with the lack of availability of a rapid and accurate pathogen identification. This may lead to a waste of already scarce resources, especially when the MDR species isolated in this study were resistant to more than five antibiotics.
The results from this study cannot be extrapolated to any other unit or hospital. There is a need to set a vigorous surveillance program for the national hospital to reduce environmental contamination and HCAI cases, to reduce the hospital antibiotic resistance rates and to generate information that will help inform required measures for the prevention and control of HCAI outbreaks.
Conclusions
This study found robust evidence of indoor air contamination and the presence of fungi and MDR bacteria of epidemiological importance for HCAI in the PICU. The current cleaning method showed no effect on the contamination load on the surfaces in PICU, which might be a risk for HCAI including outbreaks. Further studies on the PICU should measure the prevalence of HCAI and the main microorganisms related to HCAI. Additionally, molecular typing is needed to help determine if there is a link between the patient and the environmental isolates. As immediate measures, we suggest, the use of filters and air conditioner systems for the reduction of indoor air contamination on PICU, standardisation of cleaning methods and the implementation of total ACC and indicator microorganisms as routine cleanliness evaluation tools in all the hospital wards.
Acknowledgements
We would like to thank all staff members of the Paediatric Intensive Care Unit of the Hospital Central de Maputo for the support on the study conduction.
CRediT author statement
Vânia Maphossa: Conceptualization, Methodology, Investigation, Writing - Original Draft. José Carlos Langa: Conceptualization and Methodology, Samuel Simbine: Investigation. Fabião Manusse: Investigation. Darlene Kenga: Investigation, Ventura Relvas: Investigation. Valéria Chicamba: Investigation, Alice Manjate: Validation Jahit Sacarlal- Resources, Writing - Review & Editing, Validation.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interest statement
Authors declare they do not have any conflict of interest.
References
- 1.Storr J., Twyman A., Zingg W., Damani N., Kilpatrick C., Reilly J., et al. Core components for effective infection prevention and control programmes: New WHO evidence-based recommendations. Antimicrob Resist Infect Control. 2017;6 doi: 10.1186/s13756-016-0149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nejad S.B., Allegranzi B., Syed S.B., Ellisc B., Pittetd D. Health-care-associated infection in Africa: a systematic review. Bull World Health Organ. 2011;89:757–765. doi: 10.2471/BLT.11.088179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allegranzi B., Nejad S.B., Combescure C., Graafmans W., Attar H., Donaldson L., et al. Burden of endemic health-care-associated infection in developing countries: Systematic review and meta-analysis. Lancet. 2011;377:228–241. doi: 10.1016/S0140-6736(10)61458-4. [DOI] [PubMed] [Google Scholar]
- 4.(WHO) World Health Organization . vol. 34. 2011. https://apps.who.int/iris/bitstream/handle/10665/80135/9789241501507_eng.pdf;jsessionid=B020F4E894405420137DF61D82FF78E6?sequence=1 (Report on the burden of endemic health care-associated infection worldwide: clean care is safer care). accessed. [Google Scholar]
- 5.Monteiro L.G.S., Chaúque A., Barros M.P., Irá T.R. Determinants of antibiotic prescription in paediatric patients: The case of two hospitals in Maputo, Mozambique. SAJCH South African J Child Heal. 2017;11:109–111. doi: 10.7196/SAJCH.2017.v11i3.1224. [DOI] [Google Scholar]
- 6.Nhatave C., Lucas G., Wate I., Sáu Z., Patel S., Noormahomed E.V., et al. Profile of bacterial infectious disease and antimicrobial choices in an urban hospital at Maputo, Mozambique: a prospective observational study. Lancet Global Health. 2019;7:S14. doi: 10.1016/s2214-109x(19)30099-3. [DOI] [Google Scholar]
- 7.Torres N.F., Solomon V.P., Middleton L.E. Patterns of self-medication with antibiotics in Maputo City: A qualitative study. Antimicrob Resist Infect Control. 2019;8:1–12. doi: 10.1186/s13756-019-0618-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mate I., Come C.E., Gonçalves M.P., Cliff J., Gudo E.S. Knowledge, attitudes and practices regarding antibiotic use in Maputo City, Mozambique. PLoS One. 2019;14:1–15. doi: 10.1371/journal.pone.0221452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russotto V., Cortegiani A., Fasciana T., Iozzo P., Raineri S.M., Gregoretti C., et al. What Healthcare Workers Should Know about Environmental Bacterial Contamination in the Intensive Care Unit. BioMed Res Int. 2017;2017 doi: 10.1155/2017/6905450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dancer S.J. The role of environmental cleaning in the control of hospital-acquired infection. J Hosp Infect. 2009;73:378–385. doi: 10.1016/j.jhin.2009.03.030. [DOI] [PubMed] [Google Scholar]
- 11.Harris A.D. How Important Is the Environment in the Emergence of Nosocomial Antimicrobial-Resistant Bacteria. Clin Infect Dis. 2008;46:686–688. doi: 10.1086/527395. [DOI] [PubMed] [Google Scholar]
- 12.Carling P.C., Parry M.F., Bruno-Murtha L.A., Dick B. Improving environmental hygiene in 27 intensive care units to decrease multidrug-resistant bacterial transmission. Crit Care Med. 2010;38:1054–1059. doi: 10.1097/CCM.0b013e3181cdf705. [DOI] [PubMed] [Google Scholar]
- 13.Paz V., Paniagua M., Santillan A., Alaniz M., D’Agostino L., Orellana R., et al. Hospital environment hygiene nurse: a key player to reduce healthcare associated infections by multi-resistant organisms. Infect Prev Pract. 2020;2:2019–2021. doi: 10.1016/j.infpip.2019.100030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegel J.D., Rhinehart E., Jackson M., Chiarello L. Guideline for Isolation Precautions: Preventing Transmission of Infectious Agents in Health Care Settings. Am J Infect Control. 2007;35:S65–S164. doi: 10.1016/j.ajic.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(WHO) World Health Organization . 2009. WHO guidelines on hand hygiene in health care; p. 262.https://www.who.int/publications/i/item/9789241597906 (accessed March 16, 2021) [Google Scholar]
- 16.Cobrado L., Silva-Dias A., Azevedo M.M., Rodrigues A.G. High-touch surfaces: microbial neighbours at hand. Eur J Clin Microbiol Infect Dis. 2017;36:2053–2062. doi: 10.1007/s10096-017-3042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nwankwo E. Isolation of pathogenic bacteria from fomites in the operating rooms of a specialist hospital in Kano, North-western Nigeria. Pan Afr Med J. 2012;12:90. [PMC free article] [PubMed] [Google Scholar]
- 18.Claro T., Reilly M.O., Daniels S., Humphreys H. American Journal of Infection Control Surface microbial contamination in hospitals: A pilot study on methods of sampling and the use of proposed microbiologic standards. Am J Infect Control. 2015;43:1000–1002. doi: 10.1016/j.ajic.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Dancer S.J. How do we assess hospital cleaning? A proposal for microbiological standards for surface hygiene in hospitals. J Hosp Infect. 2004;56:10–15. doi: 10.1016/j.jhin.2003.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabino R. In: Environ. Mycol. Public heal. Fungi mycotoxins risk assess. Manag. Viegas C., Pinheiro A.C., Sabino R., Viegas S., Brandão J., Veríssimo C., editors. Elesevier; London: 2016. Hospital Environment; pp. 193–210. [DOI] [Google Scholar]
- 21.Tagoe DN a, Baidoo S., Dadzie I., Tengey D., Agede C. Potential sources of transmission of healthcare-associated infections in the volta regional hospital in ghana. Ghana Med J. 2011;45:22–26. doi: 10.4314/gmj.v45i1.68918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galvin S., Dolan A., Cahill O., Daniels S., Humphreys H. Microbial monitoring of the hospital environment : why and how. J Hosp Infect. 2012;82:143–151. doi: 10.1016/j.jhin.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 23.Cheesbrough M. Second Edi. Part 2. Cambridge University Press; New York: 2006. (District laboratory practice in tropical countries). [DOI] [Google Scholar]
- 24.Gizaw Z., Gebrehiwot M., Yenew C. High bacterial load of indoor air in hospital wards: The case of University of Gondar teaching hospital, Northwest Ethiopia. Multidiscip Respir Med. 2016;11 doi: 10.1186/s40248-016-0061-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.World Health Organization Surveillance standards for antimicrobial resistance world health organization. Surveillance. 2002;16 WHO/CDS/CSR/DRS/20015. [Google Scholar]
- 26.Clinical and Laboratory Standards Institute (CLSI) 30th Edition. 2020. M100Ed30 | performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 27.Magiorakos A., Srinivasan A., Carey R., Carmeli Y., Falagas M., Giske C., et al. Bacteria : an International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 28.Cooper R.A., Griffith C.J., Malik R.E., Obee P., Looker N. Monitoring the effectiveness of cleaning in four British hospitals. Am J Infect Control. 2007;35:338–341. doi: 10.1016/j.ajic.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 29.Huslage K., Rutala W.A., Sickbert-Bennett E., Weber D.J. A Quantitative Approach to Defining “High-Touch” Surfaces in Hospitals. Infect Control Hosp Epidemiol. 2010;31:850–853. doi: 10.1086/655016. [DOI] [PubMed] [Google Scholar]
- 30.Gonsu K.H., Guenou E., Toukam M., Ndze V.N., Mbakop C.D., Tankeu D.N., et al. Bacteriological assessment of the hospital environment in two referral hospitals in Yaoundé-Cameroon. Pan Afr Med J. 2015;20:1–7. doi: 10.11604/pamj.2015.20.224.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fekadu S., Getachewu B. Microbiological Assessment of Indoor Air of Teaching Hospital Wards: A case of Jimma University Specialized Hospital. Ethiop J Health Sci. 2015;25:117–122. doi: 10.4314/ejhs.v25i2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaidi A.K.M., Thaver D., Ali S.A. Pathogens Associated With Sepsis in Newborns and Young Infants in Developing Countries. Pediatr Infect Dis J. 2009;28:10–18. doi: 10.1097/INF.0b013e3181958769. [DOI] [PubMed] [Google Scholar]
- 33.Pokherl B., Koirala T., Shah G., Joshi S., Baral P. Bacteriological Profile of Neonatal Sepsis in Neonatal Intensive Care Unit (Nicu) in a Tertiary Care Hospital : Prevalent Bugs and. BMC Pediatr. 2018;18 doi: 10.1186/s12887-018-1176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solomon F.B., Wadilo F.W., Arota A.A., Abraham Y.L. Antibiotic resistant airborne bacteria and their multidrug resistance pattern at University teaching referral Hospital in South Ethiopia. Ann Clin Microbiol Antimicrob. 2017;16:1–7. doi: 10.1186/s12941-017-0204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torkar K.G., Ivić S. Surveillance of bacterial colonisation on contact surfaces in different medical wards. Arh Hig Rada Toksikol. 2017;68:116–126. doi: 10.1515/aiht-2017-68-2892. [DOI] [PubMed] [Google Scholar]
- 36.Sabharwal E.R., Sharma R. Estimation of microbial air contamination by settle plate method : are we within acceptable limit. Scholars Acad J Biosci. 2015;3:703–707. [Google Scholar]
- 37.World Health Organization . 20th ed. vol.1006. 2017. (The selection and use of essential medicines : report of the WHO expert committee). Geneva. [Google Scholar]