Abstract
Objectives
Idiopathic inflammatory myopathies (IIM) are a class of autoimmune diseases with high heterogeneity that can be divided into different subtypes based on clinical manifestations and myositis‐specific autoantibodies (MSAs). However, even in each IIM subtype, the clinical symptoms and prognoses of patients are very different. Thus, the identification of more potential biomarkers associated with IIM classification, clinical symptoms, and prognosis is urgently needed.
Methods
Plasma and urine samples from 79 newly diagnosed IIM patients (mean disease duration 4 months) and 52 normal control (NC) samples were analysed by high‐performance liquid chromatography of quadrupole time‐of‐flight mass spectrometry (HPLC‐Q‐TOF‐MS)/MS‐based untargeted metabolomics. Orthogonal partial least‐squares discriminate analysis (OPLS‐DA) were performed to measure the significance of metabolites. Pathway enrichment analysis was conducted based on the KEGG human metabolic pathways. Ten machine learning (ML) algorithms [linear support vector machine (SVM), radial basis function SVM, random forest, nearest neighbour, Gaussian processes, decision trees, neural networks, adaptive boosting (AdaBoost), Gaussian naive Bayes and quadratic discriminant analysis] were used to classify each IIM subtype and select the most important metabolites as potential biomarkers.
Results
OPLS‐DA showed a clear separation between NC and IIM subtypes in plasma and urine metabolic profiles. KEGG pathway enrichment analysis revealed multiple unique and shared disturbed metabolic pathways in IIM main [dermatomyositis (DM), anti‐synthetase syndrome (ASS), and immune‐mediated necrotizing myopathy (IMNM)] and MSA‐defined subtypes (anti‐Mi2+, anti‐MDA5+, anti‐TIF1γ+, anti‐Jo1+, anti‐PL7+, anti‐PL12+, anti‐EJ+, and anti‐SRP+), such that fatty acid biosynthesis was significantly altered in both plasma and urine in all main IIM subtypes (enrichment ratio > 1). Random forest and AdaBoost performed best in classifying each IIM subtype among the 10 ML models. Using the feature selection methods in ML models, we identified 9 plasma and 10 urine metabolites that contributed most to separate IIM main subtypes and MSA‐defined subtypes, such as plasma creatine (fold change = 3.344, P = 0.024) in IMNM subtype and urine tiglylcarnitine (fold change = 0.351, P = 0.037) in anti‐EJ+ ASS subtype. Sixteen common metabolites were found in both the plasma and urine samples of IIM subtypes. Among them, some were correlated with clinical features, such as plasma hypogeic acid (r = −0.416, P = 0.005) and urine malonyl carnitine (r = −0.374, P = 0.042), which were negatively correlated with the prevalence of interstitial lung disease.
Conclusions
In both plasma and urine samples, IIM main and MSA‐defined subtypes have specific metabolic signatures and pathways. This study provides useful clues for understanding the molecular mechanisms, searching potential diagnosis biomarkers and therapeutic targets for IIM.
Keywords: Idiopathic inflammatory myopathies, Machine learning algorithm, Metabolomics, Biomarkers
Introduction
Idiopathic inflammatory myopathies (IIM) comprise a class of systemic autoimmune diseases characterised by chronic muscle weakness, inflammation, and extramuscular manifestations. 1 IIM are rare disorders with the prevalence ranging from approximately 2.3 to 20 per 100 000 persons around the world, but severely affect patients' quality of life. 2 These disorders can be mainly classified into six subtypes: dermatomyositis (DM), anti‐synthetase syndrome (ASS), polymyositis (PM), inclusion body myositis (IBM), immune‐mediated necrotizing myopathy (IMNM), and overlap myositis. However, certain clinical or histopathological manifestations overlap among these subtypes. Therefore, myositis‐specific autoantibodies (MSAs) are used to diagnose and classify IIM. For example, anti‐MDA5 + DM is correlated with amyopathic myositis and interstitial lung disease (ILD); anti‐TIF1γ + DM has a higher risk of cancer, and anti‐Mi2 + DM presents with severe muscle manifestations. 3 Due to the high heterogeneity of IIM, identifying valuable biomarkers associated with disease classification, clinical symptoms, and prognosis is urgently needed.
Recently, with the development of immunometabolism, it has been reported that disturbed energy metabolism can result in irreversible muscle damage. 4 Proteomics and targeted lipidomic analysis have indicated that some proteins and metabolites related to glycolysis and fatty acid metabolism are disturbed in the muscle and serum of DM and PM patients. 5 , 6 Our previous study also found that multiple glycolysis processes are dysregulated in the muscle tissues of DM/PM. 6
Metabolomics is an increasingly used approach in the post‐genomics era and performs qualitative and quantitative analysis of small‐molecule metabolites that explain the final response to genomic, transcriptomic, proteomic, or environmental changes. It can describe the metabolic status in physiological or pathological conditions and provide valuable insights into the early‐stage pathogenesis of disease. 7 Metabolomics also presents promising prospects for discovering new biomarkers, guiding individualised therapies, and predicting therapeutic effects. The application of metabolomics in autoimmune diseases such as gout and rheumatoid arthritis has attracted much attention. 8 , 9 In a previous study, NMR‐based metabolomics was performed to identify the metabolic changes in the sera and muscle tissues of IIM patients. It revealed that serum and muscle tissue metabolites had the potential to distinguish IIM from NC and active IIM from inactive IIM. 10 However, disease‐specific metabolic profiles have not been studied in IIM subtypes.
In this study, we performed an integrated analysis of plasma and urine metabolomes in 79 newly diagnosed IIM patients and identified a set of differentially expressed (DE) metabolites among IIM subtypes. Based on these DE metabolites, we used 10 machine learning (ML) algorithms to identify specific metabolic biomarkers to classify main IIM subtypes (DM, ASS, IMNM) and the MSA‐defined subtypes (anti‐Mi2+, anti‐MDA5+, anti‐TIF1γ+, anti‐Jo1+, anti‐PL7+, anti‐PL12+, anti‐EJ+, and anti‐SRP+). We also found common metabolites and functional pathways in both plasma and urine in each IIM subtype and explored the correlations between metabolites and clinical parameters. This study applied ML algorithms to systematically analyse the plasma and urine metabolome and identified potential biomarkers in IIM subtypes.
Materials and methods
Patients
In this study, a total of 79 newly diagnosed IIM patients, namely, 45 DM [anti‐Mi2+ (n = 8), anti‐TIF1γ+ (n = 11), and anti‐MDA5+ (n = 26)], 27 ASS [anti‐Jo1+ (n = 10), anti‐PL7+ (n = 3), anti‐PL12+ (n = 7), anti‐EJ+ (n = 7)], and 7 IMNM [anti‐SRP+ (n = 5), anti‐HMGCR+ (n = 2)] patients, and 52 normal control (NC) samples were enrolled at the Department of Rheumatology and Immunology of Xiangya Hospital from September 2018 to February 2021. The inclusion and exclusion criteria of patients were described in the Supporting Information. The demographic and clinical information of the samples was listed in Table S1 .
Sample preparation
Morning fasting blood and first morning midstream urine samples were collected. Detailed procedures were described in Data S1 .
High‐performance liquid chromatography of quadrupole time‐of‐flight mass spectrometry‐based untargeted metabolomics analysis
Detailed procedures were described in Data S1 .
Metabolomics date analysis
Data cleaning was performed using MetaboScape 3.0 software, including noise reduction, peak detection, peak extraction, and alignment. Detailed procedures were described in Data S1 .
Statistical analysis
The study design flow chart for metabolite‐based model was shown in Figure 1. The metabolic variables were normalized using log transformation and Pareto scaling before analysis. To identify the different metabolic profiles between the IIM subtype patients and NC samples, we performed fold change (FC) analysis and multivariate analyses using SIMCA 14.0 software (Umetrics, Umeå, Sweden) and MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/). In multivariate analysis, supervised analysis orthogonal partial least‐squares discriminate analysis (OPLS‐DA) was used to identify important metabolites by the variable importance in the projection (VIP) method. The metabolites with variable importance were filtered out according to the FC higher than 1.2 or less than 0.83 and VIP scores >1.0 in OPLS‐DA. Then, these metabolites were mapped to metabolic pathway analysis. Pathway enrichment analysis was conducted using MetaboAnalyst 5.0 based on the KEGG human metabolic pathways. The correlation between metabolites and clinical parameters was conducted by Pearson's or Spearman's correlation using the R package rstatix V.0.6.0 and correlation V.0.6.1. Bubble plots were generated with the R package ggplot2 V.3.3.3.
Figure 1.
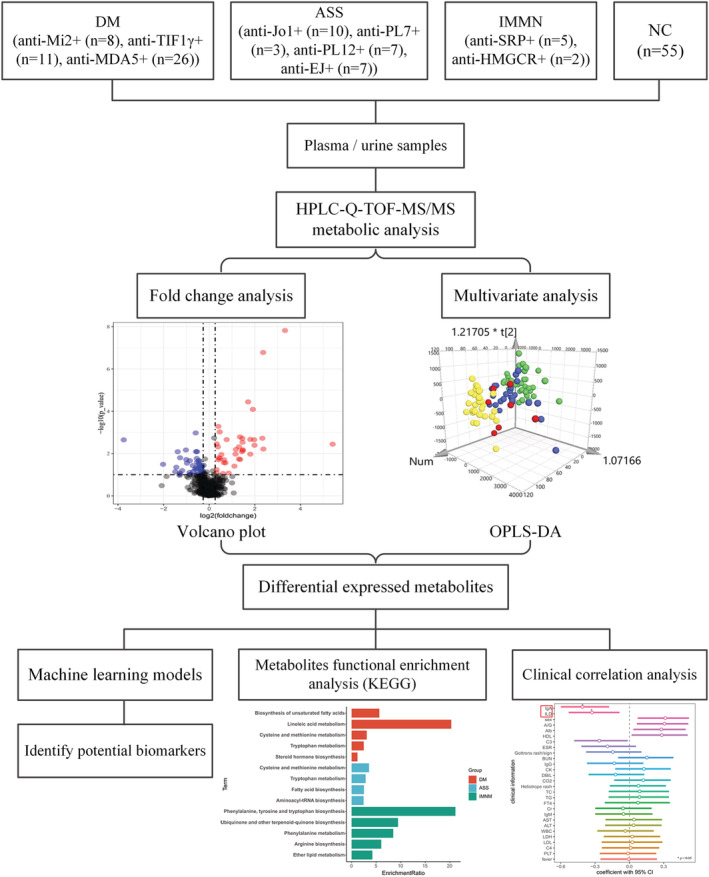
Study design flow chart for metabolite‐based model. An untargeted metabolomics analysis was carried out in the plasma and urine cohort of 79 newly diagnosed idiopathic inflammatory myopathies (IIM) patients and 52 normal control (NC) samples. Orthogonal partial least‐squares discriminate analysis (OPLS‐DA) and fold change analysis were performed to measure the significance of metabolites. Then, the differentially expressed metabolites were included in 10 machine learning models to identify potential biomarkers in each IIM subtype. Pathway enrichment analysis was conducted based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) human metabolic pathways. The correlation between the shared metabolites in the plasma and urine samples of the IIM patients and clinical parameters was conducted by Pearson's or Spearman's correlation.
ML model construction and specific metabolite selection
ML algorithms were performed using Python. DE metabolites (VIP scores >1.0, FC > 1.2 or<0.83) in each IIM subtype compared with NC were included in ML model construction. To construct the best classifiers for discriminating IIM subtypes, we used 10 ML classifiers [linear support vector machines (SVMs), radial basis function SVMs (RBF SVMs), random forests, nearest neighbours, Gaussian processes, decision trees, neural networks, adaptive boosting (AdaBoost), Gaussian naive Bayes and quadratic discriminant analysis (QDA)]. Stratified threefold cross‐validation was performed to estimate the accuracy of each model. In detail, the samples were stratified and divided into three parts in each IIM subtype, two parts of patients were used as training dataset (containing 67% of the samples), and the other part of patients were used as validation dataset (containing 33% of the samples). This process (training and validation) was repeated 1000 times with random allocation of patients in each IIM subtype. Then, we compared the 10 different ML methods according to the model accuracy scores (mean and 95% confidential interval) and chose the optimal performance classifiers. In the end, we used feature selection method in the ML models to select the most important metabolites. Linear discriminant analysis (LDA) plots were used to show the distribution of IIM subtypes based on the ML selected metabolites. All steps were performed with Python V.3.9.1, scikit‐learn V.0.24.2, NumPy V.1.19.5, Pandas V.1.2.1, and R package MASS V7.3‐53.
Results
Clinical characteristics of enrolled patients
Plasma and urine specimens were available from 52 NC samples and 79 newly diagnosed IIM patients, namely, 45 with DM [anti‐Mi2+ (n = 8), anti‐TIF1γ+ (n = 11), and anti‐MDA5+ (n = 26)], 27 with ASS [anti‐Jo1+ (n = 10), anti‐PL7+ (n = 3), anti‐PL12+ (n = 7), anti‐EJ+ (n = 7)], and 7 with IMNM [anti‐SRP+ (n = 5), anti‐HMGCR+ (n = 2)]. All the participants were age‐matched and sex‐matched. The individual demographic and clinical characteristics were provided in Table S1 .
Plasma metabolomic profiles in IIM main subtypes
LC‐MS and gas chromatography (GC)‐MS‐based untargeted metabolomics were used to study the plasma metabolomic profiles. After filtering metabolites that were absent in more than 20% of the samples, 149 and 440 metabolites were annotated in negative and positive modes, respectively. Then, OPLS‐DA was applied to further identify the metabolomic profile alterations in IIM subtypes, which was widely used to identify important metabolites by the variable importance in the VIP method. OPLS‐DA showed a clear separation of the NC from the other three IIM subtypes in both positive and negative modes (Figure 2A and 2B ). Next, we performed FC analysis on each IIM subtype compared with the NC group. Given VIP > 1.0 and FC > 1.2 or <0.83, a total of 117 metabolites were identified in each IIM subtype (DM vs. NC, ASS vs. NC, IMNM vs. NC). The UpSet plot and Tables S2 and S3 showed the unique and common DE metabolites in IIM subtype (Figure 2C ).
Figure 2.
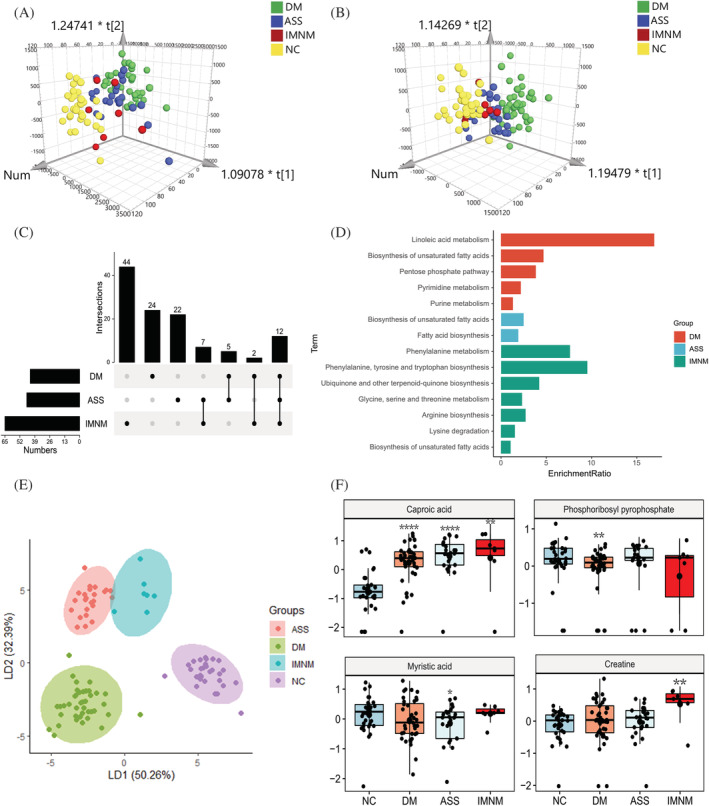
Plasma metabolomic profiles in main IIM subtypes. (A and B) The orthogonal partial least‐squares discriminate analysis (OPLS‐DA) score plots of plasma metabolomics data compared dermatomyositis (DM), anti‐synthetase syndrome (ASS), and immune‐mediated necrotizing myopathy (IMNM) to normal control (NC) samples in positive (A) and negative (B) ion mode, respectively. (C) The UpSet plot analysis based on the selected important metabolites in plasma [variable importance in the projection (VIP) > 1, fold change (FC) > 1.2 or <0.83]. (D) KEGG pathway analysis of exclusively important metabolites in each main IIM subtype. (E) The linear discriminant analysis (LDA) plot based on the Top 30 metabolites in random forest machine model. (F) The most specific metabolite identified by the random forest machine model to classify plasma samples from NC, DM, ASS, and IMNM, respectively (P value compared with NC, *, <0.05; **, <0.01; ***, <0.001, ****, <0.0001).
Furthermore, KEGG pathway enrichment analysis was used to reveal the function of these DE metabolites. The biosynthesis of unsaturated fatty acid (UFA) pathway was significantly altered in all the IIM subtypes; linoleic acid metabolism, pentose phosphate pathway, pyrimidine metabolism, and purine metabolism were enriched in DM, fatty acid biosynthesis was obviously altered in ASS, the amino acid metabolism (phenylalanine metabolism, phenylalanine, tyrosine and tryptophan biosynthesis, glycine, serine and threonine metabolism, arginine biosynthesis, and lysine degradation), and ubiquinone and other terpenoid–quinone biosynthesis were predominantly changed in IMNM (Figure 2D).
To select the most specific metabolites in each IIM subtype, the 117 DE metabolites were included in the 10 ML algorithms. From these ML classifiers, the random forest model performed the best based on accuracy scores (mean and 95% confidential interval) (Table S4 ). Then, we used random forest feature selection method and identified the most important metabolites to classify IIM main subtypes. Using the Top 30 metabolites, LDA plot showed a clear separation among DM, ASS, IMNM, and NC (Figure 2E). The Top 10 metabolites were listed in Table S5 . Among them, the levels of caproic acid, phosphoribosyl pyrophosphate, and myristic acid were lowest in NC, DM and ASS, respectively, and the creatine has the highest level in IMNM (Figure 2F).
Plasma metabolomic profiles in the MSA‐defined IIM subtypes
The NC group and MSA‐defined IIM subtypes showed a clear separation in OPLS‐DA model (Figure 3A and 3B). A total of 196 metabolites (VIP > 1.0 and FC > 1.2 or <0.83) were selected for further analysis. The exclusive and common DE metabolites in each MSA‐defined IIM subtype compared with the NC were shown in the UpSet plot and Table S6 (Figure 3C), followed by a KEGG pathway enrichment analysis with the exclusive DE metabolites. Diverse pathways were significantly dysregulated in MSA‐defined IIM subtypes, such as cysteine and methionine metabolism in the anti‐EJ + DM; arginine biosynthesis in the anti‐MDA5 + DM; phenylalanine, tyrosine, and tryptophan biosynthesis in the anti‐Mi2 + DM; and taurine and hypotaurine metabolism in the anti‐TIF1γ + DM. (Figure 3D). Based on the 196 DE metabolites, the random forest model achieved the best discrimination of MSA‐defined IIM subtypes among the 10 ML models (Table S7). The LDA plot showed a clear separation of each group using the Top 12 most important metabolites (Figure 3E). The Top 10 metabolites were shown in Table S8 . Among them, high levels of caproic acid in all MSA‐defined IIM subtypes, increased anandamide in anti‐EJ + ASS, low levels of orciprenaline‐3‐O‐sulfate in anti‐Jo1 + ASS, decreased lysoPC_14:0_0:0_in anti‐MDA5 + DM, low levels of oleic acid in anti‐Mi2 + DM, down‐regulated uridine 5‐diphosphate in anti‐TIF1γ + DM, up‐regulated prostaglandin D3 in anti‐PL12 + ASS, and the highest creatine levels in anti‐SRP + IMNM were found (Figure 3F).
Figure 3.
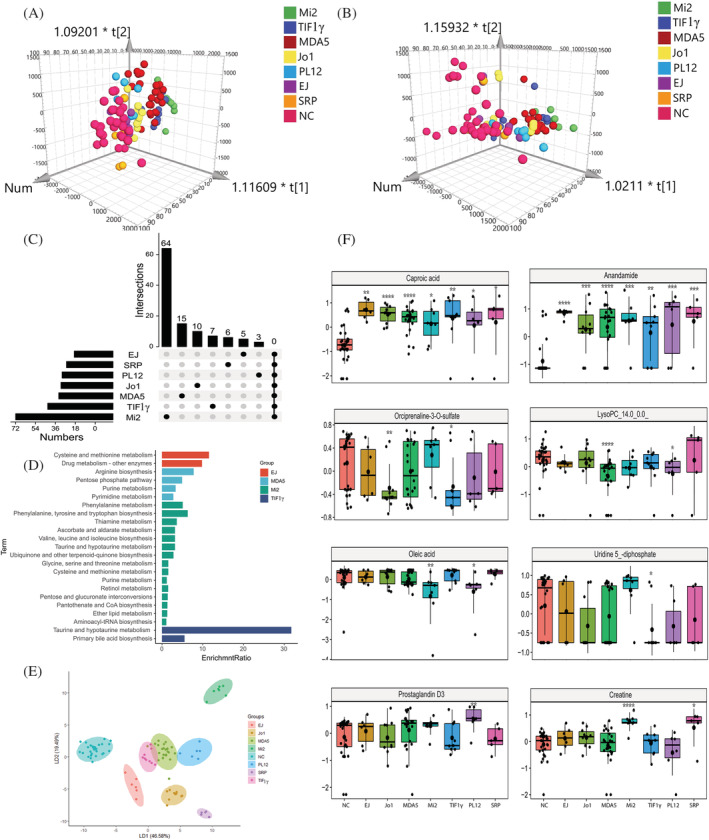
Plasma metabolomic profiles in the MSA‐defined IIM subtypes. (A and B) The Orthogonal partial least‐squares discriminate analysis (OPLS‐DA) score plots of plasma metabolomics data compared anti‐EJ+, anti‐Jo1+, anti‐MDA5+, anti‐Mi2+, anti‐TIF1γ+, anti‐PL12+, and anti‐SRP + IIM patients to normal control (NC) samples in positive (A) and negative (B) ion mode, respectively. (C) The UpSet plot analysis based on the selected important metabolites in plasma (VIP > 1, FC > 1.2 or <0.83). (D) KEGG pathway analysis of exclusively important metabolites in each IIM subtype. (E) The linear discriminant analysis (LDA) plot based on the Top 12 metabolites in random forest machine model. (F) The most specific metabolite identified by the random forest machine model to classify plasma samples from NC, anti‐EJ+, anti‐Jo1+, anti‐MDA5+, anti‐Mi2+, anti‐TIF1γ+, anti‐PL12+, and anti‐SRP + IIM patients, respectively (P value compared with NC, *, <0.05; **, <0.01; ***, <0.001, ****, <0.0001).
Urine metabolomic profiles in IIM main subtypes
Urine metabolomics has been a promising approach for the discovery of non‐invasive biomarkers in response to specific diseases or therapeutic intervention. 11 Untargeted metabolomics analysis of LC‐MS and GC‐MS was also performed in the matched IIM urine samples. After data pre‐pressing, 335 and 588 metabolites were annotated in negative and positive modes, respectively. OPLS‐DA showed a clear separation between IIM main subtypes and NC (Figure 4A and 4B). In total, 169 metabolites were identified as DE metabolites in IIM main subtypes compared with the NC group (VIP > 1.0 and FC > 1.2 or<0.83). Figure 4C and Tables S9 and S10 revealed the distinctive and intersecting DE metabolites. KEGG pathway enrichment analysis found distinguished pathways in each IIM main subtype, such as sulfur metabolism in DM, lysine degradation in ASS, and phosphonate and phosphinate metabolism in IMNM (Figure 4D). To better identify the most specific metabolites in each IIM main subtype, 169 metabolites were included in 10 ML models, and the AdaBoost model had the highest degree of accuracy (Table S11 ). Then, we identified the most specific metabolites in each IIM main subtype by the feature selection method. Using the Top 30 metabolites, the LDA plot showed a clear division among each group (Figure 4E). The Top 10 metabolites were listed in Table S12 . Among them, the levels of dodecanoic acid, isovalerylglucuronide, aminoadipic acid, and beta‐alanine were highest in the NC, DM, ASS, and IMNM group, respectively (Figure 4F).
Figure 4.
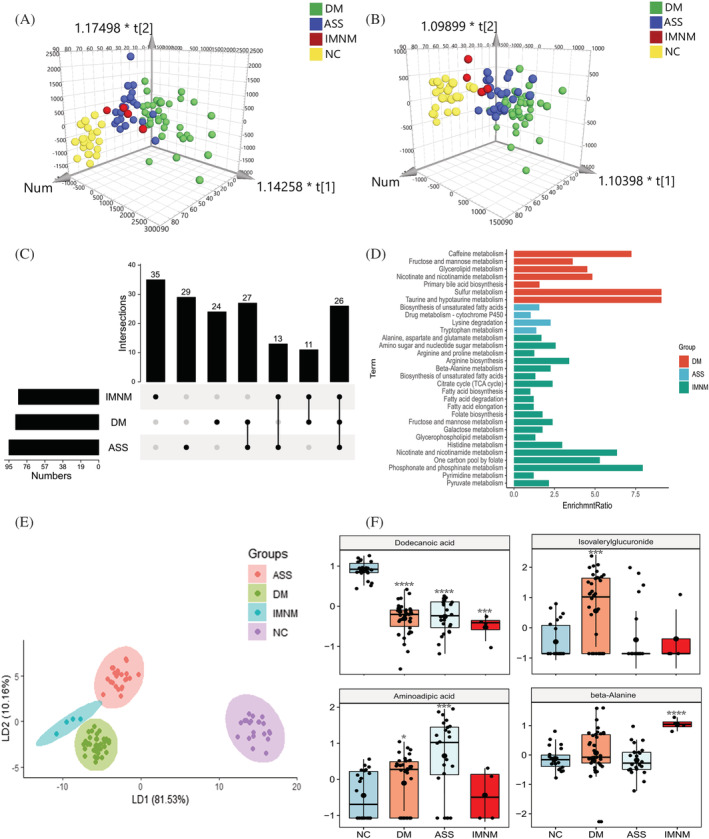
Urine metabolomic profiles in main IIM subtypes. (A and B) The orthogonal partial least‐squares discriminate analysis (OPLS‐DA) score plots of urine metabolomics data compared dermatomyositis (DM), anti‐synthetase syndrome (ASS), and immune‐mediated necrotizing myopathy (IMNM) to normal control (NC) samples in positive (A) and negative (B) ion mode, respectively. (C) The UpSet plot analysis based on the selected plasma important metabolites (VIP > 1, FC > 1.2 or <0.83). (D) KEGG pathway analysis of exclusively important metabolites in each IIM subtype. (E) The linear discriminant analysis (LDA) plot based on the Top 30 metabolites in the AdaBoost machine model. (F) The most specific metabolites used by the AdaBoost machine model to classify urine samples from NC, DM, ASS, and IMNM, respectively (P value compared with NC, *, <0.05; **, <0.01; ***, <0.001, ****, <0.0001).
Urine metabolomic profiles in the MSA‐defined IIM subtypes
In the urine metabolic profiles, OPLS‐DA also revealed a clear separation among MSA‐defined IIM subtypes (anti‐EJ+, anti‐Jo1+, anti‐MDA5+, anti‐Mi2+, anti‐TIF1γ+, anti‐PL7+, anti‐PL12+, and anti‐SRP+) in negative and positive modes (Figure 5A and 5B). Given VIP > 1.0 and FC > 1.2 or <0.83, 269 DE metabolites in total were identified in MSA‐defined IIM subtypes, and the number of unique and shared DE metabolites in each group was plotted in Figure 5C and Tables S13 and S14 . KEGG pathway enrichment analysis also showed specific pathways in each subtype, such as cysteine and methionine metabolism in anti‐Jo1 + ASS; arginine biosynthesis in anti‐MDA5 + DM; valine, leucine, and isoleucine biosynthesis in anti‐Mi2 + DM; ubiquinone and other terpenoid–quinone biosynthesis in anti‐PL12 + ASS; D‐arginine and D‐ornithine metabolism in anti‐PL7 + ASS; and one carbon pool by folate in anti‐SRP + IMNM (Figure 5D). Based on the 269 DE metabolites, 10 ML models were constructed, and the AdaBoost model performed the best with accuracies >80% to classify MSA‐defined IIM subtypes (Table S15 ). Using the AdaBoost feature selection method, the most specific metabolites in each group were identified. Based on the Top 12 metabolites, the LDA plot showed clear clusters among these MSA‐defined IIM subtypes (Figure 5E). The Top 10 variables were shown in Table S16 , high levels of dodecanoic acid in NC, decreased levels of tiglylcarnitine in anti‐EJ + ASS, low levels of 2‐phenylethanol glucuronide in anti‐Jo1 + ASS, down‐regulated acetamide levels in anti‐MDA5 + DM, increased beta‐alanine levels in anti‐Mi2 + DM, the lowest levels of tyramine glucuronide in anti‐TIF1γ + DM, up‐regulated levels of 5,6‐dihydrouridine in anti‐PL12 + ASS, the lowest levels of D‐ornithine in anti‐PL7 + ASS, and the highest levels of N‐acetyl‐L‐aspartic acid in anti‐SRP + IMNM were found (Figure 5F).
Figure 5.
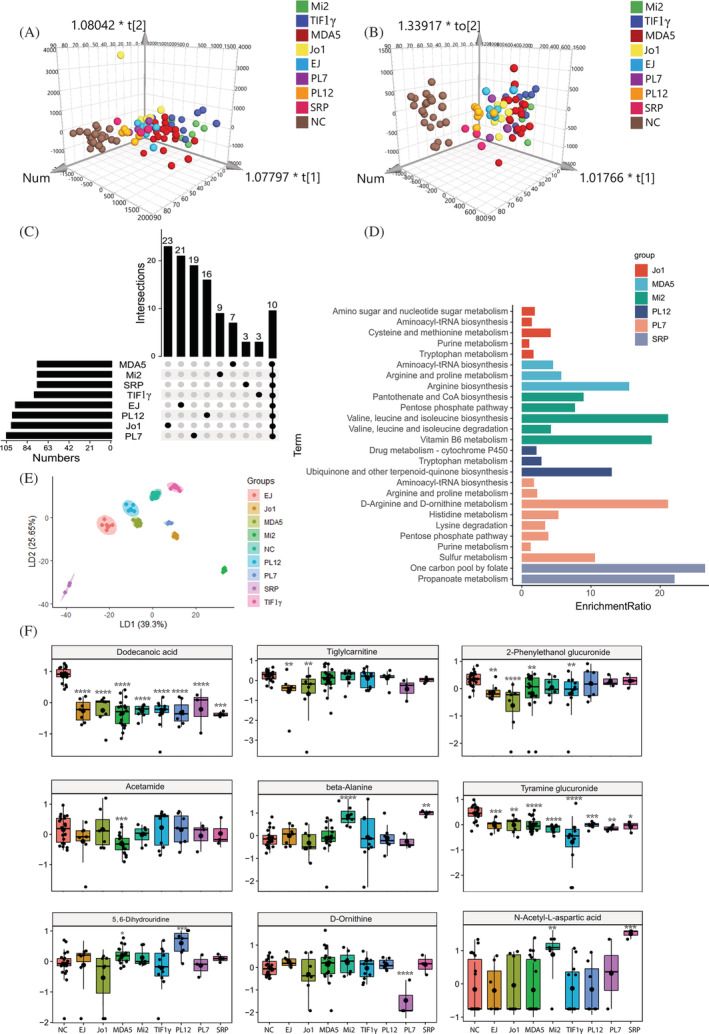
Urine metabolomic profiles in the MSA‐defined IIM subtypes. (A and B) The orthogonal partial least‐squares discriminate analysis (OPLS‐DA) score plots of urine metabolomics data compared anti‐EJ+, anti‐Jo1+, anti‐MDA5+, anti‐Mi2+, anti‐TIF1γ+, anti‐PL12+, anti‐PL7+, and anti‐SRP + IIM patients to normal control (NC) samples in positive (A) and negative (B) ion mode, respectively. (C) The UpSet plot analysis based on the selected urine important metabolites (VIP > 1, FC > 1.2 or <0.83). (D) KEGG pathway analysis of exclusively important metabolites in each IIM subtype. (E) The linear discriminant analysis (LDA) plot based on the top 12 metabolites in the AdaBoost machine model. (F) The most specific metabolites identified by the AdaBoost machine model to classify urine samples from NC, anti‐EJ+, anti‐Jo1+, anti‐MDA5+, anti‐Mi2+, anti‐TIF1γ+, anti‐PL12+, anti‐PL7+, and anti‐SRP+ myositis patients, respectively (P value compared with NC, *, <0.05; **, <0.01; ***, <0.001, ****, <0.0001).
Common pathways and metabolites in the plasma and urine of IIM main subtypes
We performed KEGG pathway analysis based on the important metabolites (VIP > 1) by comparing the global metabolomic profiles of the DM, ASS, and IMNM patients with the NC. Common perturbed pathways in the plasma and urine samples of IIM main subtypes were ranked by enrichment ratio. From these pathways, all IIM main subtypes (DM, ASS, and IMNM) exhibited disturbed retinol metabolism, pyrimidine metabolism, fatty acid biosynthesis, and biosynthesis of unsaturated fatty acids. In DM, purine metabolism, fatty acid elongation, and fatty acid degradation were exclusively altered. In IMNM, amino acid metabolism (tyrosine metabolism, lysine degradation, glycine, serine and threonine metabolism, arginine biosynthesis, arginine and proline metabolism), glycerophospholipid metabolism (phosphonate and phosphinate metabolism, glycerophospholipid metabolism), and aminoacyl‐tRNA biosynthesis were predominantly changed (Figure 6A). Moreover, we also found common DE metabolites in both the plasma and urine samples of IIM main subtypes, such as up‐regulated thromboxane B2 (TXB2) in DM, increased TXB2 and decreased myristic acid levels in ASS and up‐regulated TXB2, creatine, phosphorylcholine, proline betaine and down‐regulated (S)‐3,4‐dihydroxybutyric acid levels in IMNM. Among them, the TXB2 levels were increased in all the IIM main subtypes in both plasma and urine (Figure 6B).
Figure 6.
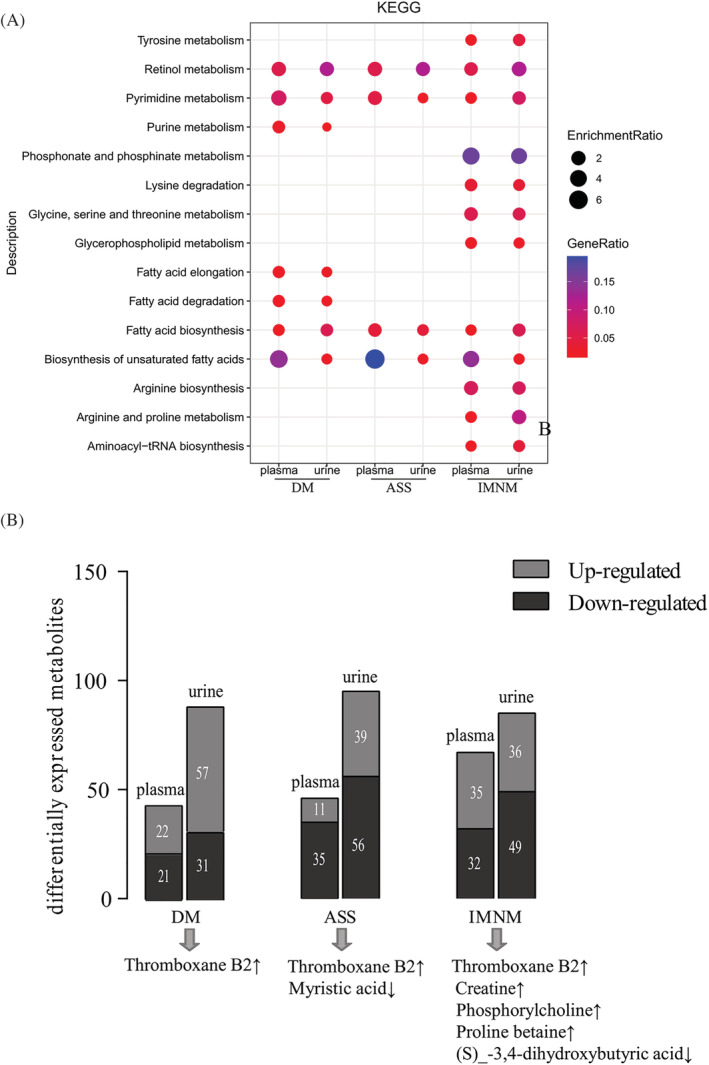
Common pathways and metabolites in the plasma and urine of main IIM subtypes. (A) KEGG pathway analysis of the dysregulated pathways in both the plasma and urine of dermatomyositis (DM), anti‐synthetase syndrome (ASS), and immune‐mediated necrotizing myopathy (IMNM) patients compared with normal control (NC) samples based on important metabolites (VIP > 1). (B) The important metabolites altered in both plasma and urine of DM, ASS, and IMNM patients, respectively (VIP > 1, FC > 1.2 or <0.83).
Common pathways and metabolites in the plasma and urine of MSA‐defined IIM subtypes
From the enrichment metabolic pathway analysis (VIP > 1) of MSA‐defined IIM subtypes, pyrimidine metabolism and fatty acid biosynthesis were disturbed in all seven MSA‐defined IIM subtypes (anti‐EJ+, anti‐Jo1+, anti‐MDA5+, anti‐Mi2+, anti‐TIF1γ+, anti‐PL12+, and anti‐SRP+). In anti‐EJ + ASS, purine metabolism, glycerolipid metabolism, fructose and mannose metabolism, cysteine and methionine metabolism, and biosynthesis of unsaturated fatty acids were significantly enriched. Obvious changes in sulfur metabolism, retinol metabolism, purine metabolism, glycerolipid metabolism, phosphonate and phosphinate metabolism, and fructose and mannose metabolism were found in anti‐Jo1 + ASS. Purine metabolism, the pentose phosphate pathway, and arginine biosynthesis appeared to be involved in the biological processes of the MDA5 + DM. In anti‐Mi2 + DM, multiple amino acid metabolism pathways, vitamin B6 metabolism, primary bile acid biosynthesis, pantothenate and CoA biosynthesis, glutathione metabolism, galactose metabolism and aminoacyl‐tRNA biosynthesis were profoundly affected. Sulfur metabolism, retinol metabolism, glycerolipid metabolism, fructose and mannose metabolism, and fatty acid metabolism were enriched in the anti‐PL12 + ASS. In SRP + IMNM, sulfur metabolism, retinol metabolism, pyrimidine and purine metabolism, phosphonate and phosphinate metabolism, glyoxylate and dicarboxylate metabolism, glycolysis/gluconeogenesis, glycerophospholipid metabolism, the citrate cycle [tricarboxylic acid (TCA) cycle], and amino acid metabolism were significantly changed. Dysregulation of sulfur metabolism, retinol metabolism, pyrimidine and purine metabolism, primary bile acid biosynthesis, glyoxylate and dicarboxylate metabolism, fructose and mannose metabolism, and fatty acid and amino acid metabolism were implicated in anti‐TIF1γ + DM (Figure 7A). Moreover, we also found overlapped metabolites (VIP > 1, FC > 1.2 or<0.83) in the plasma and urine samples of the MSA‐defined IIM subtypes, such as glyceraldehyde and myristic acid in anti‐EJ + ASS, TXB2, phosphorylcholine, glyceraldehyde, trans‐aconitic acid and myristic acid in anti‐Jo1 + ASS, TXB2 and myristic acid in anti‐MDA5 + DM, N‐acetyl‐L‐aspartic acid, creatine, pyridoxal 5′‐phosphate, malonyl‐carnitin, and galactitol in anti‐Mi2 + DM, TXB2, palmitic acid, and myristic acid in anti‐PL12 + ASS, phosphorylcholine and pyruvic acid in anti‐SRP + IMNM, and TXB2 and hypogeic acid in anti‐TIF1γ + DM (Figure 7B).
Figure 7.
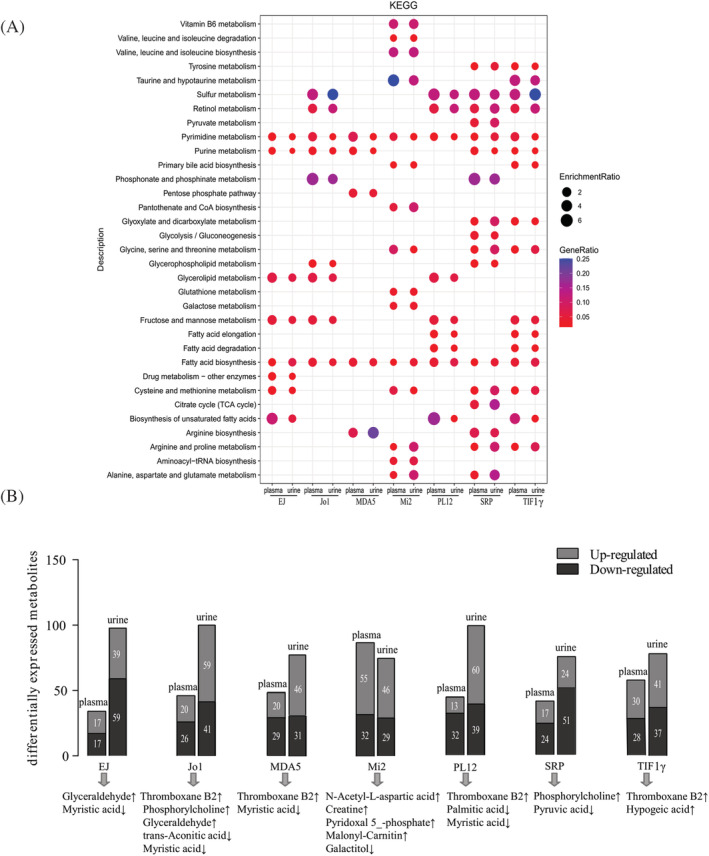
Common pathways and metabolites in the plasma and urine of the MSA‐defined IIM subtypes. (A) KEGG pathway analysis of the dysregulated pathways in both the plasma and urine of the seven pairs (anti‐EJ+ vs. NC, anti‐Jo1+ vs. NC, anti‐MDA5+ vs. NC, anti‐Mi2+ vs. NC, anti‐PL12+ vs. NC, anti‐SRP+ vs. NC, and anti‐TIF1γ+ vs. NC) (VIP > 1). (B) The important metabolites altered in both plasma and urine of anti‐EJ+, anti‐Jo1+, anti‐MDA5+, anti‐Mi2+, anti‐PL12+, anti‐SRP+, and anti‐TIF1γ + IIM patients, respectively (VIP > 1, FC > 1.2 or <0.83).
Correlations between metabolites and clinical parameters
To evaluate the relationships between metabolites alterations and clinical parameters, correlation analysis was performed on the common metabolites in the plasma and urine samples of the IIM patients. Creatine was found to be positively associated with plasma creatine kinase (CK), CO2 combining power (CO2CP), glutamic–pyruvic transaminase (ALT), total cholesterol (TC), and high‐density lipoprotein (HDL). The level of plasma hypogeic acid was negatively correlated with the prevalence of interstitial lung disease (ILD). Plasma trans‐aconitic acid showed a strongly positive association with the prevalence of Gottron's rash/sign and a negative association with the plasma CO2CP (Figure 8A–C). Moreover, plasma N‐acetyl‐L‐aspartic acid showed a positive association with plasma CK levels and CO2CP (Figure S1A ). In the urine samples of the IIM patients, our results revealed that (S)‐3,4‐dihydroxybutyric acid was negatively correlated with plasma direct bilirubin (DBIL) and positively correlated with IgA and platelets (PLTs) (Figure 8D). Urine palmitic acid and malonylcarnitine showed a strongly negative association with complement3 (C3) and the prevalence of ILD, respectively (Figure 8E and 8F). Urine glyceraldehyde was negatively related to urine pondus hydrogenii (PH) (Figure S1B ).
Figure 8.
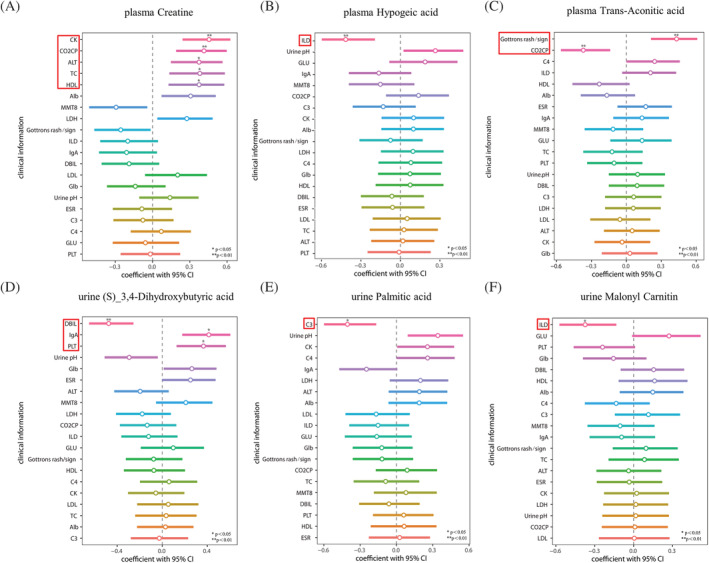
Correlations between metabolites and clinical parameters.(A and B) Spearman's or Pearson's correlation analysis was carried out to assess the associations between clinical parameters and the shared metabolites in the plasma (A–C) and urine (D–F) samples of dermatomyositis (DM), anti‐synthetase syndrome (ASS), immune‐mediated necrotizing myopathy (IMNM), and myositis‐specific autoantibody (MSA)‐defined IIM subtypes (VIP > 1, FC > 1.2 or <0.83).
Discussion
Metabolomics is emerging as a useful method for diagnosing and identifying therapeutic targets for a variety of diseases. 12 Our study first systematically analysed the metabolome profiles in IIM main and MSA‐defined subtypes in both plasma and urine samples and applied ML algorithms to identify the most specific metabolites in each subtype. We found unique metabolite signatures in the patients with IIM main subtypes (DM, ASS, IMNM) and MSA‐defined subtypes (anti‐EJ+, anti‐Jo1+, anti‐MDA5+, anti‐Mi2+, anti‐TIF1γ+, anti‐PL12+, anti‐PL7+, and anti‐SRP+).
Potential metabolic biomarkers in IIM main subtypes
In plasma, phosphoribosyl pyrophosphate was specifically decreased in DM, myristic acid was specifically decreased in ASS, and creatine was significantly increased in IMNM. Phosphoribosyl pyrophosphate, also known as PRPP, is a rate‐limiting substrate for de novo and salvage purine synthesis. It plays an important role in the cell cycle regulation of purine synthesis. 13 Reduced phosphoribosyl pyrophosphate may affect myocyte development in DM. Myristic acid is an saturated fatty acid (SFA), and moderate intake of myristic acid has beneficial lipidic effects. 14 In a mouse model of congenital Type 2 diabetes, chronic administration of myristic acid improved hyperglycaemia and reduced body weight. Moreover, myristic acid could increase glucose uptake in C2C12 myotube cells. 15 , 16 Moderate supplementation with myristic acid may also have a positive effect on muscle inflammation and performance by regulating energy metabolism in ASS patients. Creatine was up‐regulated in both the plasma and urine samples of the IMNM patients. The plasma creatine levels were strongly correlated with CO2CP and the serum levels of CK, ALT, TC, and HDL, whereas the urine creatine levels did not correlate with clinical indices, suggesting that the creatine level in the plasma of IMNM patients may be a better biomarker. This phenomenon may arise from the fact that the increased circulating creatine most likely results from damaged muscle cell leakage, whereas circulating creatine exceeds the renal threshold, resulting in creatinuria. 17
In urine, isovalerylglucuronide, aminoadipic acid, and beta‐alanine were specifically increased in the DM, ASS, and IMNM patients, respectively. Isovalerylglucuronide has only been reported in the urine of patients with isovaleric acidemia when the amount of urinary 3‐hydroxyisovaleric acid excretion is high. 18 Aminoadipic acid is closely associated with oxidative stress and reactive oxygen species production. 19 Similarly, beta‐alanine can act as a mitochondrial toxin that reduces cellular respiration and oxidative phosphorylation. 20 Up‐regulated aminoadipic acid and beta‐alanine may lead to deficits in energy‐generating metabolic pathways in skeletal muscle by damaging mitochondria, finally contributing to muscle inflammation, weakness, and fatigue. 21
In both the plasma and urine samples, TXB2, myristic acid, creatine, phosphorylcholine, proline betaine, and (S)‐3,4‐dihydroxybutyric acid were changed. Of those six metabolites, TXB2 was increased in all types of myositis. TXB2 is a stable metabolite of thromboxane A2 (TXA2), which is derived from cyclooxygenase (COX) and PLTs. TXA2 has been linked to cardiovascular diseases, Type 1 and 2 diabetes, chronic inflammatory diseases, and tumour metastasis. 22 , 23 Consistent with our study, increased expression of enzymes in the COX pathway was discovered in muscles from DM/PM patients. 24 These prostanoid signalling molecules play an important role in muscle inflammation and atrophy. Elevated (S)‐3,4‐dihydroxybutyric acid has previously been identified in the serum of patients with dementia and may serve as a predictive biomarker. 25 We found decreased (S)‐3,4‐dihydroxybutyric acid in both the plasma and urine of the IMNM patients. Moreover, the expression level of urine (S)‐3,4‐dihydroxybutyric acid was negatively correlated with the plasma DBIL level and positively correlated with the IgA and PLT levels, which may participate in the early pathogenesis of IMNM.
Potential metabolic biomarkers in MSA‐defined IIM subtypes
We also identified the most specific metabolites in the plasma of the MSA‐defined IIM subtypes. Plasma oleic acid was exclusively down‐regulated in the anti‐Mi2 + DM patients. Oleic acid, a monounsaturated fatty acid, has an anti‐inflammatory effect on collagen‐induced arthritis and can also increase Type 1 fibre levels in C2C12 myotubes. 26 , 27 Although the role of anti‐Mi2 autoantibodies in anti‐Mi2 + DM is still unclear, the autoantigen Mi2 is associated with muscle cell differentiation. 3 We hypothesize that decreased oleic acid expression may affect this process. Prostaglandin D3, a kind of prostaglandin that mediates muscle inflammation and atrophy, was only up‐regulated in the plasma of anti‐PL12 + ASS patients. 28
Tiglylcarnitine, which is the most abundant group of carnitines in tissues and biofluids, was expressed at the lowest levels in the urine of anti‐EJ + ASS patients. 29 It is also decreased in the blood of patients with familial Mediterranean fever, metabolic syndrome, Type 2 diabetes, and cardiovascular diseases. 30 , 31 Supplementation with some short‐chain carnitines has been studied as a treatment for a variety of diseases, and tiglylcarnitine may also have a beneficial effect on anti‐EJ + ASS patients. Uridine 5‐diphosphate, a component of nucleotide metabolism, has been identified to be elevated in multiple malignancies, suggesting rapid tRNA degradation due to metabolic interactions between the tumour and host. 32 Significant up‐regulation of 5,6‐dihydrouridine was evident in the anti‐PL12 + ASS, but no studies have reported the relationship between the level of 5,6‐dihydrouridine and the risk of cancer in anti‐PL12 + ASS.
We proposed 13 overlapping metabolite changes in the plasma and urine of MSA‐defined IIM subtypes, including three fatty acids, namely, myristic acid, palmitic acid, and hypogeic acid; two amino acids, namely, N‐acetyl‐L‐aspartic acid and creatine; glyceraldehyde; TXB2; phosphorylcholine; trans‐aconitic acid; pyridoxal 5‐phosphate; malonyl‐carnitin; galactitol; and pyruvic acid. In the anti‐TIF1γ + DM patients, the long‐chain fatty acid hypogeic acid (cis‐7‐hexadecenoic acid, 16:1n‐9) was significantly increased in the plasma and urine samples. Interestingly, plasma hypogeic acid was negatively correlated with the percentage of ILD in IIM patients. Hypogeic acid is a possible biomarker for foamy cell formation during atherosclerosis and exerts an anti‐inflammatory effect in immune cells. 33 Adult DM patients are at risk of metabolic syndrome, and whether anti‐TIF1γ + DM patients have a higher risk of developing the disease warrants future studies. 34 Pyruvic acid was obviously decreased in the plasma and urine of anti‐SRP + IMNM patients. Pyruvic acid is the end product of glycolysis and is the central metabolite in the TCA cycle. 35 The anomalous expression of pyruvic acid in anti‐SRP + IMNM patients may reflect dysfunction of the mitochondrial respiratory chain. Given the high heterogeneity of IIM, a combined ML algorithm and metabolomics analysis offers the prospect of identifying metabolically defined IIM subtypes. The specific metabolites in each group have a high accuracy of disease prediction and are useful in IIM subsetting.
Potential mechanism exploration
From the metabolic pathway enrichment analysis, specific pathways were also revealed in both the plasma and urine samples in the IIM subtypes. In all the main IIM subtypes, we found general disturbed metabolic pathways, such as retinol metabolism, pyrimidine metabolism, and fatty acid biosynthesis pathways. In the DM patients, fatty acid metabolism‐related pathways were enriched the most. Consistent with our previous results, down‐regulated HDL and up‐regulated triglyceride levels were common in the serum of the DM patients. 36 Many amino acid metabolic pathways were disturbed in the plasma and urine profiles of the IMNM patients, including tyrosine metabolism, lysine degradation, glycine, serine and threonine metabolism, arginine biosynthesis, and arginine and proline metabolism. The synthesis of arginine and tyrosine metabolism has been associated with inflammatory reactions. 37 The lysine degradation pathways are confined to the mitochondria, ultimately yielding two acetyl‐CoAs via the mitochondrial saccharopine pathway. 38 Glycine can be converted to serine and threonine, and serine can flux to the TCA cycle, finally inhibiting adenosine triphosphate production. 39 Taken together, the significantly disturbed amino acid metabolic pathways in the IMNM patients may correlate with abnormal muscle mitochondrial function and inflammation.
In both the plasma and urine profiles of the MSA‐defined IIM subtypes, diverse pathways were dysregulated. Among them, pyrimidine metabolism and fatty acid biosynthesis were involved in most of the MSA‐defined IIM subtypes. In anti‐Mi2 + DM, amino acid biosynthesis and metabolism were enriched the most, participating in the synthesis of proteins, nucleic acids, and lipids and energy generation and contributing to inflammation, immunity, insulin resistance, and obesity. 37 , 39 , 40 Many lipid metabolism pathways, including glycerolipid metabolism, fatty acid elongation, fatty acid degradation, fatty acid biosynthesis, and biosynthesis of unsaturated fatty acids, were significantly perturbed in anti‐PL12 + ASS. Interestingly, the glycerolipid/free fatty acid (GL/FFA) cycle is integrated by lipolysis. Lipids are not only an essential part of energy metabolism but also participate in multiple biological processes. Disturbed lipid metabolism is strongly associated with metabolic syndrome, inflammation, and the pathogenesis of senescence and cancers. 41 Our results suggested that the disturbances in lipid metabolism were likely related to the processes of anti‐PL12 + ASS.
Limitations
There are some limitations in our study. First, to discard exogenous metabolites related to diets, drugs and treatments that guaranteed robust metabolite markers, metabolites absent in 20% or more samples were removed from the analysis. This filtering criterion was a common method for dealing with missing data in untargeted metabolomics analysis 42 , 43 but that may also eliminate some IIM subtype‐specific metabolite markers. Further studies are needed to focus on the validation of potential biomarkers in the metabolites absent in 20% or more samples. Second, our study focused on the metabolomic profiles in the plasma and urine of newly diagnosis IIM patients and applied ML algorithms to classify IIM subtypes to search potential biomarkers. However, we did not analysis the metabolites alterations in the IIM follow‐up cohort, which might response to disease activities and drug treatment. In addition, as muscle tissue is the most important organ involved in IIM, combined analysis of the muscle tissue metabolomic profiles might provide more valuable clues for IIM pathologic mechanism research.
Conclusion
Taken together, by applying ML algorithms to the metabolome data, we identified specific plasma and urine metabolites that can classify IIM main subtypes and MSA‐defined IIM subtypes. We also discovered the overlapped key metabolites and perturbed metabolic pathways in the plasma and urine of each subtype. Our study reveals the plasma and urine metabolic signatures in IIM, which may provide useful clues in understanding the molecular mechanisms and search potential biomarkers for the diagnosis and prognosis of IIM subtypes. These newly detected metabolic biomarkers and pathways may also serve as promising therapeutic targets in IIM, such as dietary supplementation with taurine to reduce oxidant stress catabolism and dietary supplementation or pharmacological intervention to regulate the ratio imbalance of SFAs.
Supporting information
Figure S1. Correlations between plasma N‐Acetyl‐L‐aspartic acid, urine glyceraldehyde and clinical parameters.
(A‐B) Spearman or pearson correlation analysis was carried out to assess the associations between clinical parameters and the shared metabolites in the plasma N‐Acetyl‐L‐aspartic acid (A) and urine glyceraldehyde (B) samples of dermatomyositis (DM), anti‐synthetase syndrome (ASS), immune mediated necrotizing myopathy (IMNM) and myositis‐specific autoantibody (MSA)‐defined IIM subtypes (VIP>1, FC > 1.2 or<0.83).
Table S1. Comparison of clinical manifestations and laboratory data among DM, ASS, IMNM and NC
Table S2. The fold change of plasma commonly differentially expressed metabolites in DM, ASS, IMNM compared with NC
Table S3. The plasma exclusively differentially expressed metabolites in DM, ASS, IMNM compared with NC
Table S4. Comparison of machine learning models to classify plasma specimens of DM, ASS and IMNM based on metabolic profiles
Table S5. The top 10 most useful metabolites to differentiate plasma specimens of DM, ASS and IMNM using the random forest model
Table S6. The plasma exclusively differentially expressed metabolites in MSA‐defined IIM subtypes compared with NC
Table S7. Comparison of machine learning models to classify plasma specimens of different MSA‐defined IIM subtypes based on metabolic profiles
Table S8. The top 10 most useful metabolites to differentiate plasma specimens of different MSA‐defined IIM subtypes using the Random Forest model
Table S9. The fold change of urine commonly differentially expressed metabolites in DM, ASS, IMNM compared with NC
Table S10. The urine exclusively differentially expressed metabolites in DM, ASS, IMNM compared with NC
Table S11. Comparison of machine learning models to classify urine specimens of DM, ASS and IMNM based on metabolic profiles
Table S12. The top 10 most useful metabolites to differentiate urine specimens of DM, ASS and IMNM using the AdaBoost model
Table S13. The fold change of urine commonly differentially expressed metabolites in MSA‐defined IIM subtypes compared with NC
Table S14. The urine exclusively differentially expressed metabolites in MSA‐defined IIM subtypes compared with NC
Table S15. Comparison of machine learning models to classify urine specimens of different MSA‐defined IIM subtypes based on metabolic profiles
Table S16. The top 10 most useful metabolites to differentiate urine specimens of different MSA‐defined IIM subtypes using the AdaBoost model
Data S1. Supporting Information
Acknowledgements
This research was supported by grants from the National Natural Science Foundation of China (82101880) and Natural Science Foundation of Hunan Province (2020JJ8056).
Conflict of interest
The authors have declared no conflicts of interest.
Liu D., Zhao L., Jiang Y., Li L., Guo M., Mu Y., and Zhu H. (2022) Integrated analysis of plasma and urine reveals unique metabolomic profiles in idiopathic inflammatory myopathies subtypes, Journal of Cachexia, Sarcopenia and Muscle, 13, 2456–2472, 10.1002/jcsm.13045
References
- 1. Selva‐O'Callaghan A, Pinal‐Fernandez I, Trallero‐Araguas E, Milisenda JC, Grau‐Junyent JM, Mammen AL. Classification and management of adult inflammatory myopathies. Lancet Neurol 2018;17:816–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wilfong EM, Aggarwal R. Role of antifibrotics in the management of idiopathic inflammatory myopathy associated interstitial lung disease. Ther Adv Musculoskelet Dis 2021;13:1759720X211060907. 10.1177/1759720X211060907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stuhlmuller B, Schneider U, Gonzalez‐Gonzalez JB, Feist E. Disease specific autoantibodies in idiopathic inflammatory myopathies. Front Neurol 2019;10:438. 10.3389/fneur.2019.00438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu D, Zuo X, Luo H, Zhu H. The altered metabolism profile in pathogenesis of idiopathic inflammatory myopathies. Semin Arthritis Rheum 2020;50:627–635. [DOI] [PubMed] [Google Scholar]
- 5. Raouf J, Idborg H, Englund P, Alexanderson H, Dastmalchi M, Jakobsson PJ, Lundberg IE, Korotkova M. Targeted lipidomics analysis identified altered serum lipid profiles in patients with polymyositis and dermatomyositis. Arthritis Res Ther 2018;20:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu D, Xiao Y, Zhou B, Gao S, Li L, Zhao L, Chen W, Dai B, Li Q, Duan H, Zuo X, Luo H, Zhu H. PKM2‐dependent glycolysis promotes skeletal muscle cell pyroptosis by activating the NLRP3 inflammasome in dermatomyositis/polymyositis. Rheumatology (Oxford) 2021;60:2177–2189. [DOI] [PubMed] [Google Scholar]
- 7. Wishart DS. Metabolomics for investigating physiological and pathophysiological processes. Physiol Rev 2019;99:1819–1875. [DOI] [PubMed] [Google Scholar]
- 8. Shen X, Wang C, Liang N, Liu Z, Li X, Zhu ZJ, Merriman TR, Dalbeth N, Terkeltaub R, Li C, Yin H. Serum metabolomics identifies dysregulated pathways and potential metabolic biomarkers for hyperuricemia and gout. Arthritis Rheumatol 2021;73:1738–1748. [DOI] [PubMed] [Google Scholar]
- 9. You S, Koh JH, Leng L, Kim WU, Bucala R. The tumor‐like phenotype of rheumatoid synovium: Molecular profiling and prospects for precision medicine. Arthritis Rheumatol 2018;70:637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anupam G, Umesh K, Dinesh K, Naveen R, Anamika KA, Mantabya KS, et al. NMR‐based serum and muscle metabolomics for diagnosis and activity assessment in idiopathic inflammatory myopathies. Anal Sci Adv 2021;6:515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khamis MM, Adamko DJ, El‐Aneed A. Mass spectrometric based approaches in urine metabolomics and biomarker discovery. Mass Spectrom Rev 2017;36:115–134. [DOI] [PubMed] [Google Scholar]
- 12. Perakakis N, Stefanakis K, Mantzoros CS. The role of omics in the pathophysiology, diagnosis and treatment of non‐alcoholic fatty liver disease. Metabolism 2020;111S:154320. 10.1016/j.metabol.2020.154320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fridman A, Saha A, Chan A, Casteel DE, Pilz RB, Boss GR. Cell cycle regulation of purine synthesis by phosphoribosyl pyrophosphate and inorganic phosphate. Biochem J 2013;454:91–99. [DOI] [PubMed] [Google Scholar]
- 14. Dabadie H, Peuchant E, Bernard M, LeRuyet P, Mendy F. Moderate intake of myristic acid in sn‐2 position has beneficial lipidic effects and enhances DHA of cholesteryl esters in an interventional study. J Nutr Biochem 2005;16:375–382. [DOI] [PubMed] [Google Scholar]
- 15. Takato T, Iwata K, Murakami C, Wada Y, Sakane F. Chronic administration of myristic acid improves hyperglycaemia in the Nagoya‐Shibata‐Yasuda mouse model of congenital type 2 diabetes. Diabetologia 2017;60:2076–2083. [DOI] [PubMed] [Google Scholar]
- 16. Iwata K, Sakai H, Takahashi D, Sakane F. Myristic acid specifically stabilizes diacylglycerol kinase delta protein in C2C12 skeletal muscle cells. Biochim Biophys Acta Mol Cell Biol Lipids 2019;1864:1031–1038. [DOI] [PubMed] [Google Scholar]
- 17. Chung YL, Wassif WS, Bell JD, Hurley M, Scott DL. Urinary levels of creatine and other metabolites in the assessment of polymyositis and dermatomyositis. Rheumatology (Oxford) 2003;42:298–303. [DOI] [PubMed] [Google Scholar]
- 18. Dorland L, Duran M, Wadman SK, Niederwieser A, Bruinvis L, Ketting D. Isovalerylglucuronide, a new urinary metabolite in isovaleric acidemia. Identification problems due to rearrangement reactions. Clin Chim Acta 1983;134:77–83. [DOI] [PubMed] [Google Scholar]
- 19. Estaras M, Ameur FZ, Estevez M, Diaz‐Velasco S, Gonzalez A. The lysine derivative aminoadipic acid, a biomarker of protein oxidation and diabetes‐risk, induces production of reactive oxygen species and impairs trypsin secretion in mouse pancreatic acinar cells. Food Chem Toxicol 2020;145:111594. 10.1016/j.fct.2020.111594 [DOI] [PubMed] [Google Scholar]
- 20. Shetewy A, Shimada‐Takaura K, Warner D, Jong CJ, Mehdi AB, Alexeyev M, Takahashi K, Schaffer SW. Mitochondrial defects associated with beta‐alanine toxicity: relevance to hyper‐beta‐alaninemia. Mol Cell Biochem 2016;416:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller FW, Lamb JA, Schmidt J, Nagaraju K. Risk factors and disease mechanisms in myositis. Nat Rev Rheumatol 2018;14:255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simeone P, Boccatonda A, Liani R, Santilli F. Significance of urinary 11‐dehydro‐thromboxane B2 in age‐related diseases: Focus on atherothrombosis. Ageing Res Rev 2018;48:51–78. [DOI] [PubMed] [Google Scholar]
- 23. Kiely M, Milne GL, Minas TZ, Dorsey TH, Tang W, Smith CJ, Baker F, Loffredo CA, Yates C, Cook MB, Ambs S. Urinary thromboxane B2 and lethal prostate cancer in African American men. J Natl Cancer Inst 2021;114:123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Korotkova M, Helmers SB, Loell I, Alexanderson H, Grundtman C, Dorph C, Lundberg IE, Jakobsson PJ. Effects of immunosuppressive treatment on microsomal prostaglandin E synthase 1 and cyclooxygenases expression in muscle tissue of patients with polymyositis or dermatomyositis. Ann Rheum Dis 2008;67:1596–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mousavi M, Jonsson P, Antti H, Adolfsson R, Nordin A, Bergdahl J, Eriksson K, Moritz T, Nilsson LG, Nyberg L. Serum metabolomic biomarkers of dementia. Dement Geriatr Cogn Dis Extra 2014;4:252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Perez‐Martinez PI, Rojas‐Espinosa O, Hernandez‐Chavez VG, Arce‐Paredes P, Estrada‐Parra S. Anti‐inflammatory effect of omega unsaturated fatty acids and dialysable leucocyte extracts on collagen‐induced arthritis in DBA/1 mice. Int J Exp Pathol 2020;101:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Watanabe N, Komiya Y, Sato Y, Watanabe Y, Suzuki T, Arihara K. Oleic acid up‐regulates myosin heavy chain (MyHC) 1 expression and increases mitochondrial mass and maximum respiration in C2C12 myoblasts. Biochem Biophys Res Commun 2020;525:406–411. [DOI] [PubMed] [Google Scholar]
- 28. Korotkova M, Lundberg IE. The skeletal muscle arachidonic acid cascade in health and inflammatory disease. Nat Rev Rheumatol 2014;10:295–303. [DOI] [PubMed] [Google Scholar]
- 29. Makarova E, Makrecka‐Kuka M, Vilks K, Volska K, Sevostjanovs E, Grinberga S, Zarkova‐Malkova O, Dambrova M, Liepinsh E. Decreases in circulating concentrations of long‐chain acylcarnitines and free fatty acids during the glucose tolerance test represent tissue‐specific insulin sensitivity. Front Endocrinol (Lausanne) 2019;10:870. 10.3389/fendo.2019.00870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kiykim E, Aktuglu Zeybek AC, Barut K, Zubarioglu T, Cansever MS, Alsancak S, Kıykım E. Screening of free carnitine and acylcarnitine status in children with familial Mediterranean fever. Arch Rheumatol 2016;31:133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu ZR, Ning Y, Yu H, Tang NJ. A HPLC‐Q‐TOF‐MS‐based urinary metabolomic approach to identification of potential biomarkers of metabolic syndrome. J Huazhong Univ Sci Technolog Med Sci 2014;34:276–283. [DOI] [PubMed] [Google Scholar]
- 32. Seidel A, Brunner S, Seidel P, Fritz GI, Herbarth O. Modified nucleosides: an accurate tumour marker for clinical diagnosis of cancer, early detection and therapy control. Br J Cancer 2006;94:1726–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Astudillo AM, Meana C, Bermudez MA, Perez‐Encabo A, Balboa MA, Balsinde J. Release of anti‐inflammatory palmitoleic acid and its positional isomers by mouse peritoneal macrophages. Biomedicine 2020;8. 10.3390/biomedicines8110480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Moraes MT, de Souza FH, de Barros TB, Shinjo SK. Analysis of metabolic syndrome in adult dermatomyositis with a focus on cardiovascular disease. Arthritis Care Res (Hoboken) 2013;65:793–799. [DOI] [PubMed] [Google Scholar]
- 35. Zhang S, Wakai S, Sasakura N, Tsutsumi H, Hata Y, Ogino C, Kondo A. Pyruvate metabolism redirection for biological production of commodity chemicals in aerobic fungus Aspergillus oryzae . Metab Eng 2020;61:225–237. [DOI] [PubMed] [Google Scholar]
- 36. Wang H, Tang J, Chen X, Li F, Luo J. Lipid profiles in untreated patients with dermatomyositis. J Eur Acad Dermatol Venereol 2013;27:175–179. [DOI] [PubMed] [Google Scholar]
- 37. Capuron L, Schroecksnadel S, Feart C, Aubert A, Higueret D, Barberger‐Gateau P, Féart C, Layé S, Fuchs D. Chronic low‐grade inflammation in elderly persons is associated with altered tryptophan and tyrosine metabolism: role in neuropsychiatric symptoms. Biol Psychiatry 2011;70:175–182. [DOI] [PubMed] [Google Scholar]
- 38. Leandro J, Houten SM. The lysine degradation pathway: Subcellular compartmentalization and enzyme deficiencies. Mol Genet Metab 2020;131:14–22. [DOI] [PubMed] [Google Scholar]
- 39. Cheng ZX, Guo C, Chen ZG, Yang TC, Zhang JY, Wang J, Zhu JX, Li D, Zhang TT, Li H, Peng B, Peng XX. Glycine, serine and threonine metabolism confounds efficacy of complement‐mediated killing. Nat Commun 2019;10:3325. 10.1038/s41467-019-11129-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yu D, Richardson NE, Green CL, Spicer AB, Murphy ME, Flores V, Jang C, Kasza I, Nikodemova M, Wakai MH, Tomasiewicz JL, Yang SE, Miller BR, Pak HH, Brinkman JA, Rojas JM, Quinn WJ III, Cheng EP, Konon EN, Haider LR, Finke M, Sonsalla M, Alexander CM, Rabinowitz JD, Baur JA, Malecki KC, Lamming DW. The adverse metabolic effects of branched‐chain amino acids are mediated by isoleucine and valine. Cell Metab 2021;33:905, e6–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prentki M, Madiraju SR. Glycerolipid metabolism and signaling in health and disease. Endocr Rev 2008;29:647–676. [DOI] [PubMed] [Google Scholar]
- 42. Bijlsma S, Bobeldijk I, Verheij ER, Ramaker R, Kochhar S, Macdonald IA, van Ommen B, Smilde AK. Large‐scale human metabolomics studies: a strategy for data (pre‐) processing and validation. Anal Chem 2006;78:567–574. [DOI] [PubMed] [Google Scholar]
- 43. Fan Y, Li Y, Chen Y, Zhao YJ, Liu LW, Li J, Wang SL, Alolga RN, Yin Y, Wang XM, Zhao DS, Shen JH, Meng FQ, Zhou X, Xu H, He GP, Lai MD, Li P, Zhu W, Qi LW. Comprehensive metabolomic characterization of coronary artery diseases. J Am Coll Cardiol 2016;68:1281–1293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Correlations between plasma N‐Acetyl‐L‐aspartic acid, urine glyceraldehyde and clinical parameters.
(A‐B) Spearman or pearson correlation analysis was carried out to assess the associations between clinical parameters and the shared metabolites in the plasma N‐Acetyl‐L‐aspartic acid (A) and urine glyceraldehyde (B) samples of dermatomyositis (DM), anti‐synthetase syndrome (ASS), immune mediated necrotizing myopathy (IMNM) and myositis‐specific autoantibody (MSA)‐defined IIM subtypes (VIP>1, FC > 1.2 or<0.83).
Table S1. Comparison of clinical manifestations and laboratory data among DM, ASS, IMNM and NC
Table S2. The fold change of plasma commonly differentially expressed metabolites in DM, ASS, IMNM compared with NC
Table S3. The plasma exclusively differentially expressed metabolites in DM, ASS, IMNM compared with NC
Table S4. Comparison of machine learning models to classify plasma specimens of DM, ASS and IMNM based on metabolic profiles
Table S5. The top 10 most useful metabolites to differentiate plasma specimens of DM, ASS and IMNM using the random forest model
Table S6. The plasma exclusively differentially expressed metabolites in MSA‐defined IIM subtypes compared with NC
Table S7. Comparison of machine learning models to classify plasma specimens of different MSA‐defined IIM subtypes based on metabolic profiles
Table S8. The top 10 most useful metabolites to differentiate plasma specimens of different MSA‐defined IIM subtypes using the Random Forest model
Table S9. The fold change of urine commonly differentially expressed metabolites in DM, ASS, IMNM compared with NC
Table S10. The urine exclusively differentially expressed metabolites in DM, ASS, IMNM compared with NC
Table S11. Comparison of machine learning models to classify urine specimens of DM, ASS and IMNM based on metabolic profiles
Table S12. The top 10 most useful metabolites to differentiate urine specimens of DM, ASS and IMNM using the AdaBoost model
Table S13. The fold change of urine commonly differentially expressed metabolites in MSA‐defined IIM subtypes compared with NC
Table S14. The urine exclusively differentially expressed metabolites in MSA‐defined IIM subtypes compared with NC
Table S15. Comparison of machine learning models to classify urine specimens of different MSA‐defined IIM subtypes based on metabolic profiles
Table S16. The top 10 most useful metabolites to differentiate urine specimens of different MSA‐defined IIM subtypes using the AdaBoost model
Data S1. Supporting Information


